Abstract
Colorectal cancer (CRC) has become the third most common cancer worldwide and leads to a high mortality rate. Although colorectal cancer has been studied widely, the underlying molecular mechanism remains unclear. Increasing evidence shows that the abnormal expression of microRNAs (miRNAs) is involved in tumorigenesis. Previous studies have reported that miRNA-103 (miR-103) is dysregulated in CRC; however, the expression, function and mechanism of miR-103 in CRC are not well known. The present study showed that miR-103 was overexpressed in the primary tumor tissues of patients with CRC and was significantly associated with a more aggressive phenotype of CRC in patients. Survival rate analysis demonstrated that CRC patients with high miR-103 expression had a poorer overall survival compared with CRC patients with low miR-103 expression. In CRC cell lines, miR-103 inhibition significantly decreased the proliferation, invasion and migration of the cells in vitro. Furthermore, miR-103 repressed large tumor suppressor kinase 2 (LATS2) expression by directly binding to the LATS2-3′-untranslated region, and an inverse correlation was identified between the expression of miR-103 and LATS2 messenger RNA in primary CRC tissues. In addition, the restoration of LATS2 led to suppressed proliferation, invasion and migration of CRC cells. In vivo, miR-103 promotes tumor growth in nude mice. In summary, miR-103 performs a critical role in the promotion of the invasive and metastatic capacities of CRC, possibly by directly targeting LATS2. This miRNA may be involved in the development and progression of CRC.
Keywords: microRNA-103, LATS2, colorectal cancer, proliferation, invasion, migration
Introduction
Colorectal cancer (CRC) has become the third most common cancer worldwide and leads to a high mortality, with frequent tumor migration and invasion (1,2). CRC is a biologically heterogeneous disease that evolves in the background of various genetic and epigenetic alterations (3). The development of CRC from normal epithelial cells to malignant carcinoma is hypothesized to be a multistage process involving genetic changes that lead to the activation of oncogenes and inactivation of tumor suppressor genes. A large number of oncogenes and tumor suppressor genes have been reported to be responsible for the development of CRC, but the molecular mechanisms underlying the migration and invasion of advanced CRC remain unclear.
MicroRNAs (miRNAs) are non-coding RNAs that are 18–24 nucleotides long and inhibit translation or induce mRNA decay through binding to the 3′-untranslated region (3′-UTR) of their target RNAs (4). miRNAs are highly conserved between species and play important roles in various physiological and pathological processes, including cancers (5). Numerous miRNAs are highly tissue-specific and important for cell development and differentiation. Therefore, the aberrant expression of miRNAs may lead to cellular dedifferentiation, oncogenesis, cancer metastasis and tumor invasion (6). An increasing number of miRNAs have been revealed to be involved in the growth and metastasis of CRC, including miRNA-27a (7,8), miRNA-145 (9,10), miRNA-29 (11), miRNA-221 (12), miRNA-96 (13) and miRNA-375 (14). miRNA-103 (miR-103), which belongs to the miR-103/107 family, is capable of inducing epithelial-to-mesenchymal transition (EMT) of mammary epithelial cells. miR-103 has been found to be elevated in CRC, and high expression of miR-103 has been associated with the metastatic potential of colorectal cancer cell lines and ensuing poor prognosis (15–17). Despite an increasing number of studies on the biogenesis and mechanisms of miR-103 in the pathogenesis of CRC, the mechanisms of miR-103 dysregulation remains unclear.
In the present study, it was confirmed that miR-103 expression was increased in CRC cells and inhibition of miR-103 significantly inhibited the proliferation, invasion and migration of CRC cells. The current study found that miR-103 directly targeted large tumor suppressor kinase 2 (LATS2), a tumor suppressor (18), and LATS2 overexpression partially attenuated the effect of miR-103 in CRC. Furthermore, the present data showed that miR-103 directly downregulated LATS2 expression by binding to the 3′-UTR of LATS2. In addition, the expression of LATS2 was negatively associated with miR-103 in CRC tissues.
Materials and methods
CRC tissues, cell lines and transfection
In total, 38 CRC tissues and matched normal tissues were surgically collected at the Department of Gastrointestinal Surgery of Renmin Hospital of Wuhan University (Wuhan, Hubei, China), and informed consent was obtained from all patients. The present study was approved by the Ethics Committee of Renmin Hospital of Wuhan University. Collected tissues were immediately frozen in liquid nitrogen and stored at −80°C prior to RNA isolation. The CRC SW480, HT-29, HCT-116, SW620 and LoVo cell lines were acquired from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and grown in Invitrogen Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% HyClone fetal bovine serum (FBS; GE Healthcare Life Sciences, Logan, UT, USA). The normal colon epithelial FHC and NCM460 cell lines were grown in Gibco DMEM:F12 (Thermo Fisher Scientific, Inc.) supplemented with 10% FBS. Transfection was performed using Invitrogen Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
TRIzol reagent (Invitrogen) was used to isolate total RNAs from frozen tissues and CRC cells according to the manufacturer's protocol. RT-qPCR assays for LATS2 and miR-103 were performed were performed using SYBR Green Reagents (Takara, Tokyo, Japan) on Applied Biosystems Prism 7700 system (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The primers used were as follows: LATS2 forward, 5′-AAGAGCTACTCGCCATACGCCTTT-3′ and reverse, 5′-AGCTTTGGCCATTTCTTGCTCCAG-3′; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward, 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse, 5′-GAAGATGGTGATGGGATTTC-3′. Primers for U6 and miR-103 were purchased from GeneCopoeia (Rockville, MD, USA; Guangzhou RiboBio Co., Ltd., Guangzhou, China). The LATS2 level was normalized against GAPDH and the miR-103 level was normalized against U6. The relative expression was quantified using the 2−ΔΔCq method (19).
Cell proliferation assay
SW620 and LoVo cells were seeded at a density of 103 cells per well in 96-well plates. The cells were transfected with anti-miR-NC, anti-miR-103, negative control or cotransfected with miR-103 and LATS2 vector. Cell viability was assessed for 4 consecutive days by absorbance at 570 nm using a microplate reader (iMark™; Bio-Rad Laboratories, Inc., Hercules, CA, USA). All experiments were repeated three times and the mean values were calculated.
Cell migration and invasion assays
For the invasion and migration assays, 1×105 SW620 and LoVo cells/ml were prepared subsequent to transfection with anti-miR-NC or anti-miR-103 for 24 h, respectively, and 1×105 SW620 and LoVo cells/ml were prepared subsequent to transfection with miR-NC or miR-103, or cotransfection with miR-103 and LATS2 vector. Cell migration and invasion assays were determined using Transwell insert chambers (Costar; Corning Incorporated, Corning, NY, USA). Cells that migrated or invaded through the membrane were fixed with 4% polyoxymethylene (Merck Millipore, Darmstadt, Germany), stained with 0.2% crystal violet (Merck Millipore), and visualized and counted under an inverted microscope (CXK41; Olympus, Tokyo, Japan).
Vector construct
For the 3′-UTR of LATS2, the DNA oligonucleotides containing the 53-nt wild-type 3′-UTR of LATS2 or 52-nt mutant 3′-UTR of LATS2 were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) with flanking SpeI and HindIII restriction enzyme digestion sites, respectively. The DNAs and pMIR-REPORT Luciferase vectors (Promega Corporation, Madison, WI, USA) were used to build the luciferase report vectors. The mutant 3′-UTR of LATS2 acted as a control (where TGCTGCT was changed to GCGTAC). For the LATS2 vector, the Homo sapiens full open reading frame cDNA clone for LATS2 was transcribed, and the product was amplified using primers with flanking SpeI and HindIII restriction enzyme digestion sites, and the DNAs were then inserted into the pcDNA3.1 vector.
Luciferase assay
The HEK293T cells (Cell Bank of the Chinese Academy of Sciences) were transfected with wild-type pMIR-miR-103-3′-UTR or mutant pMIR-miR-103-3′-UTR with Renilla luciferase control vectors (pRL-TK; Promega Corporation) using Lipofectamine 2000. The cells were then transfected with miR-NC or miR-103. Luciferase activity was examined 48 h subsequent to transfection using the Dual-Luciferase Reporter Assay system (Promega Corporation) with an Centro XS3 LB 960 Luminometer (Molecular Devices, LLC, Sunnyvale, CA, USA). Renilla luciferase activity was normalized against firefly luciferase activity.
Western blotting
Western blotting was performed using 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels. Subsequent to transferring the bands to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA), 5% BSA was used for blocking the membranes. LATS2 protein was detected using a mouse anti-LATS2 monoclonal antibodies (Abs) (dilution, 1:500; catalogue number MAB0009; Abnova, Taipei, Taiwan) and GAPDH protein, which acted as an internal reference, was detected using anti-GAPDH rabbit monoclonal Abs (Abcam, Cambridge, UK). This was followed by incubation with horseradish peroxidase-conjugated secondary Abs (Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Signals were detected using an enhanced chemiluminescence system (Gel Doc XR; Bio-Rad Laboratories, Inc.).
Tumor growth in nude mice in vivo
A total of 16 female 5-week-old BALB/c nude mice (Beijing HFK Bioscience Co., Ltd., Beijing, China) were used under conditions approved by the Institutional Animal Care and Use Committee of Wuhan University. LV-miR-103-SW480 and LV-miR-control-SW480 cells were generated by amplification of the miR-103 and miR-control precursor sequences from human genomic DNA, followed by cloning into the lentiviral vector pLVX-shRNA1 (Clontech Laboratories, Inc., Mountainview, CA, USA). Virus packaging was performed in HEK293T cells by co-transfecting pLV-miR-103 or pLV-miR-control and Lenti-X HTX Packaging System (Clontech Laboratories, Inc.) using the Xfect transfection reagent (Clontech Laboratories, Inc.). The SW480 cells were transduced with pLV-miR-103 or pLV-miR-control, and the cell line that stably expressed miRNA-103 was named LV-miR-103-SW480, while the control vector cell line was named LV-miR-control-SW480. To determine the proliferation capacity of LV-miR-103-SW480 and LV-miR-control-SW480 cells in vivo, a total of 1×106 cells were injected subcutaneously into nude mice (n=8 mice/group). The tumor volume in mm3 was calculated as follows: Volume = length × width2 × 0.5. Mouse tumors were harvested and weighed 42 days subsequent to inoculation and the curve of tumor growth was produced.
Statistical analysis
Data are expressed as the mean ± standard deviation unless otherwise noted. Two-tailed Student's t-test was used to analyze results using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). Pearson's product-moment correlation coefficient was used to analyze the association between the expression of miR-103 and LATS2 mRNA. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-103 was increased in CRC tissues and correlated with the metastatic capacity of CRC cell lines and tissues, and miR-103 is correlated with poor survival in CRC
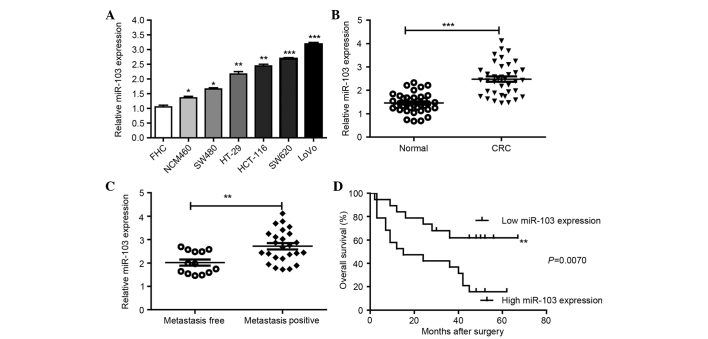
SYBR Green qPCR was performed to detect miR-103 levels in CRC cell lines and tissues. The expression levels of miR-103 were tested in 7 human colonic cell lines. The expression of miR-103 was elevated in the 5 CRC cell lines, consisting of the SW480, HT-29, HCT-116, SW620 and LoVo cell lines, compared with the 2 human normal colon epithelial cell lines, consisting of the FHC and NCM460 cell lines (Fig. 1A). Furthermore, the expression of miR-103 in the 38 CRC tissues and the paired adjacent normal tissues was also detected. The results showed that miR-103 expression was significantly increased in CRC tissues compared with the paired adjacent normal tissues (Fig. 1B). In addition, the expression of miR-103 was significantly increased in the CRC tissues of patients with metastasis compared with the expression in CRC tissues obtained from patients without metastasis (Fig. 1C). Using the Kaplan-Meier method and log-rank test, the overall survival time of CRC patients with high miR-103 expression was significantly decreased compared with the survival time of patients with low miR-103 expression (P=0.0070; Fig. 1D).
Figure 1.
miR-103 levels are correlated with the metastatic capacity in CRC cell lines and CRC tissues. (A) The expression of miR-103 in the 7 colonic FHC, NCM460, SW480, HT-29, HCT-116, SW620 and LoVo cell lines was detected by RT-qPCR. (B) Expression of miR-103 in 38 CRC tissues and matched normal tissues was detected by RT-qPCR. (C) qPCR data of miR-103 levels in primary CRC lymph nodes (metastasis positive or metastasis free). (D) Overall survival curves for the two groups defined by low and high expression of miR-103 in patients with CRC. Expression of miR-103 was normalized against U6. All assays were performed in duplicate. *P<0.05, **P<0.01, ***P<0.001 vs. control. CRC, colorectal cancer, RT-qPCR, reverse transcription-quantitative polymerase chain reaction; miR-103, microRNA-103.
Inhibition of miR-103 suppressed the proliferation, invasion and migration of CRC cells in vitro
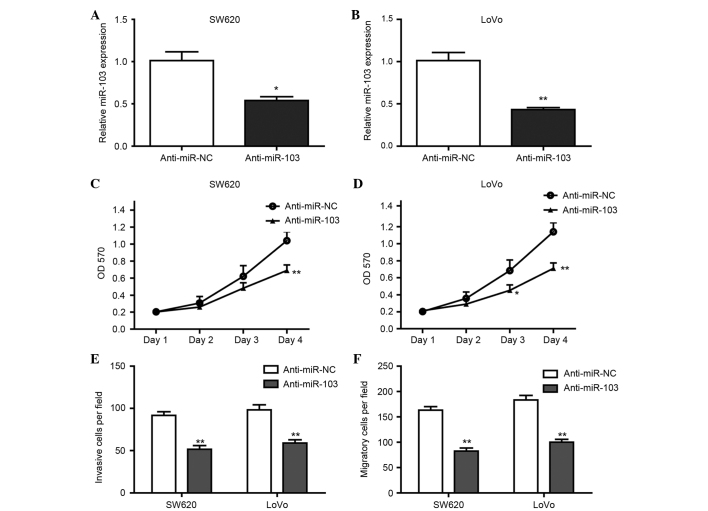
To determine whether miR-103 promotes the proliferation, migration and invasion of CRC cells, the SW620 and LoVo cells were transfected with anti-miR-103 or anti-miR-NC for 24 h, and the proliferation, invasion and migration of those cells were then analyzed. The results of RT-qPCR confirmed that the expression of miR-103 in SW620 and LoVo cells transfected with anti-miR-103 was significantly decreased (Fig. 2A and B), and anti-miR-103 significantly suppressed the proliferation (Fig. 2C and D), invasion (Fig. 2E) and migration (Fig. 2F) of SW620 and LoVo cells. These data suggest that inhibition of miR-103 suppressed the proliferation and motility of CRC cells.
Figure 2.
Inhibition of miR-103 expression suppressed the proliferation, invasion and migration of colorectal cancer cells. SW620 and LoVo cells were transfected with anti-miR-NC or anti-miR-103 for 24 h, respectively. (A and B) Expression of miR-103 in (A) SW620 and (B) LoVo cells transfected with anti-miR-NC or anti-miR-103 was detected by reverse transcription-quantitative polymerase chain reaction. Subsequent to transfection, the proliferation of the (C) SW620 and (D) LoVo cells was assessed, in addition to the (E) invasion and (F) migration of the cells. *P<0.05, **P<0.01 vs. control. All assays were repeated in duplicate. miR, microRNA; NC, negative control; OD, optical density.
LATS2 is a direct target of miR-103
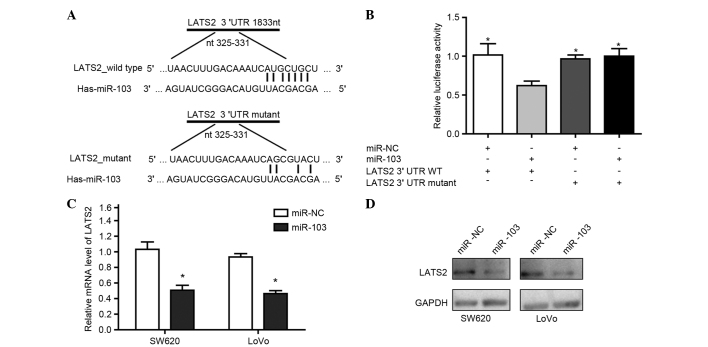
Bioinformatics analysis using TargetScan 6.2 (20) showed that LATS2 contains potential binding sites of miR-103. To identify whether LATS2 is a target of miR-103, vectors containing the wild-type 3′-UTR or mutant 3′-UTR of LATS2 mRNA were constructed, with the 3′-UTR being fused directly downstream of the firefly luciferase gene (Fig. 3A). For the luciferase assays, the wild-type or mutant vector was cotransfected into HEK293T cells with miR-NC or miR-103. As shown in Fig. 3B, miR-103 significantly reduced the relative luciferase activity of the wild-type 3′-UTR of LATS2 (P<0.05), while the luciferase activity of the mutant 3′-UTR was not significantly changed. It was also confirmed that miR-103 significantly decreased the mRNA and protein expression of LATS2 (Fig. 3C and D). Overall, miR-103 downregulated LATS2 expression through direct binding to the 3′-UTR of LATS2.
Figure 3.
LATS2 was a direct target of miR-103. (A) The putative binding sequences of miR-103 in the 3′-UTR of LATS2. (B) Luciferase activity assays to determine the activity of luciferase vectors containing wild-type or mutant LATS2-3′-UTR were performed subsequent to transfection with miR-NC or miR-103. The luciferase activity was normalized to Renilla luciferase activity. Western blot analysis and RT-qPCR identified the expression of (C) LATS2 messenger RNA and (D) LATS2 protein in SW620 and LoVo cells subsequent to transfection with miR-NC or miR-103. For western blot analysis, GAPDH acted as an internal control. For RT-qPCR assays repeated in duplicate, GAPDH acted as an internal control for LATS2 and RNU6B acted as an internal control for miR-103. *P<0.05 compared with the control. miR, microRNA; NC, negative control; LATS2, large tumor suppressor kinase 2; UTR, untranslated region; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; WT, wild-type; RNU6, U6 small nuclear RNA.
miR-103 promoted CRC cell growth and motility by targeting LATS2
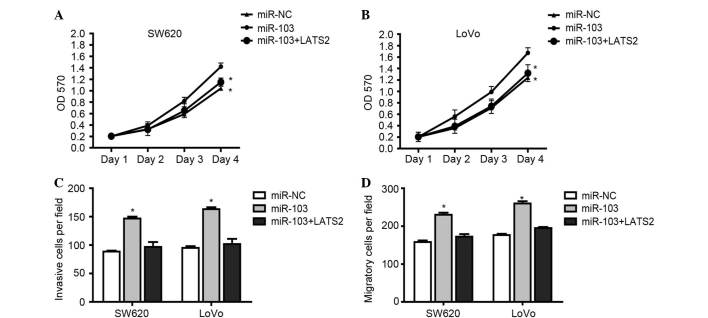
To investigate whether the effect of miR-103 on promoting the proliferation, invasion and migration of CRC cells occurred through targeting LATS2, the CRC cells were transfected with miR-NC, miR-103 or cotransfected with miR-103 and LATS2 vector. Function investigation revealed that the proliferation, invasion and migration of CRC cells was significantly enhanced subsequent to transfection with miR-103, whereas the restoration of LATS2 markedly led to suppression of the proliferation (Fig. 4A and B), invasion (Fig. 4C) and migration (Fig. 4D) of CRC cells. Overall, miR-103-induced loss of LATS2 expression promotes the metastasis, invasion and proliferation of CRC cells by targeting LATS2.
Figure 4.
miR-103 promoted the proliferation, invasion and migration of colorectal cancer cells by targeting LATS2. (A) SW620 and (B) LoVo cells were cotransfected with miR-103 and pcDNA3.1-LATS2 or a blank vector. A cell counting kit-8 assay was used to measure proliferation. (C) In vitro invasion assay of SW620 and LoVo cells cotransfected with miR-103 and pcDNA3.1-LATS2 or the blank vector. (D) In vitro migration assay of SW620 and LoVo cells cotransfected with miR-103 and pcDNA3.1-LATS2 or the blank vector. Data were drawn from three independent experiments. *P<0.05 vs. miR-103 group. miR, microRNA; NC, negative control; OD, optical density; LATS2, large tumor suppressor kinase 2.
miR-103 was inversely correlated with LATS2 expression in CRC tissues
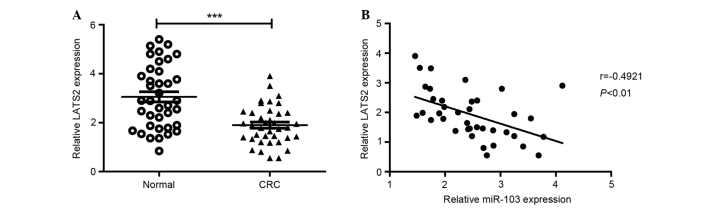
To confirm the association between miR-103 and LATS2 expression, miR-103 and LATS2 mRNA expression levels were investigated in 38 CRC and paired adjacent normal tissues. The results showed that the average level of LATS2 mRNA was significantly decreased in CRC tissues compared with the corresponding normal tissues (Fig. 5A). In addition, the LATS2 mRNA level was inversely correlated with the miR-103 expression level (Fig. 5B).
Figure 5.
miR-103 expression was inversely correlated with LATS2 expression in CRC tissues. (A) Expression of LATS2 in 40 CRC tissues and corresponding nontumor tissues was detected by reverse transcription-quantitative polymerase chain reaction. (B) Correlation analysis between miR-103 and LATS2 messenger RNA level in CRC tissues (Spearman's rank correlation analysis; r=−0.4921; P<0.01). miR-103, microRNA-103; LATS2, large tumor suppressor kinase 2; CRC, colorectal cancer. ***P<0.001 vs. control.
miR-103 promotes tumor growth in nude mice in vivo
To investigate the role of miR-103 in vivo, a lentiviral vector was constructed to mediate the expression of miR-103, and 2 stable cell lines were established, termed LV-miR103-SW480 and LV-miR-control-SW480. These cells were injected into the flanks of nude mice, and tumor progression was observed over time. To evaluate tumor growth, the length and width of orthotopic tumors was measured every 7 days post-inoculation. Progressive solid tumors were observed in all mice. The volumes of the tumors resulting from the LV-miR-control-SW480 injection were significantly smaller compared with the tumors resulting from the LV-miR-103-sw480 injection (Fig. 6A). The mice were sacrificed 42 days post-inoculation. In agreement with the tumor volumes, the weight of tumors from the LV-miR-control-SW480 group was significantly decreased compared with the tumors in the LV-miR-103-SE480 group (Fig. 6B). The data suggest that miR-103 promotes the growth of SW480-engrafted tumors in vivo.
Figure 6.
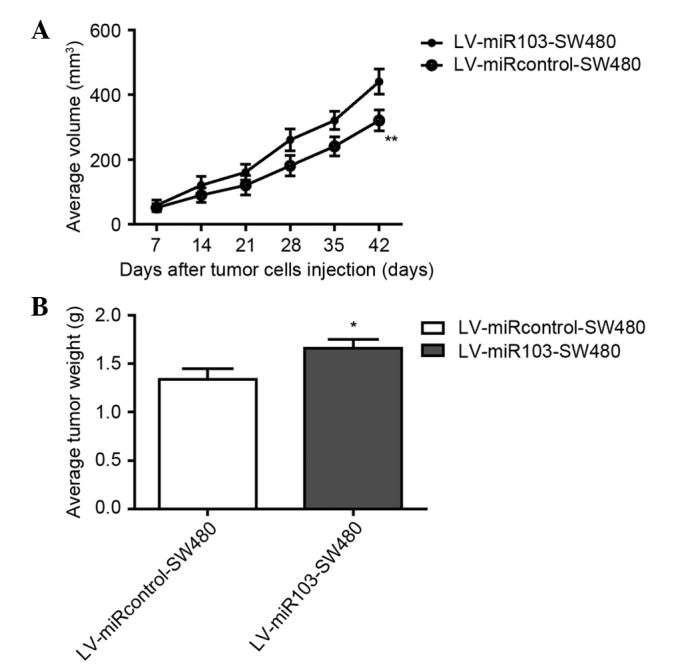
miR-103 promotes tumor growth in nude mice in vivo. (A) Overexpression of miR-103 promotes colorectal cancer growth in vivo, (n=8; **P=0.0072). (B) Tumor weights 42 days post-inoculation (n=8; *P=0.0314). Data are presented as mean ± standard deviation. miR-103, microRNA-103.
Discussion
In the present study, the RT-qPCR results showed that miR-103 was significantly increased in the CRC SW480, HT-29, HCT-116, SW620 and LoVo cell lines compared with the normal colon epithelium FHC and NCM460 cell lines. miR-103 expression was significantly increased in CRC tissues compared with the paired adjacent normal tissues. In addition, the expression of miR-103 was significantly increased in lymph node and liver metastases compared with metastases-free CRC tumors. Statistical analyses revealed that miR-103 overexpression was significantly correlated with tumor progression in CRC patients, and miR-103 expression was correlated with poor survival of patients with CRC. Furthermore, inhibition of miR-103 suppressed cell proliferation, migration and invasion in SW620 and LoVo cells. Using a luciferase reporter assay, miR-103 was shown to promote cell migration and invasion and appeared to be associated with the silencing of LAST2. Furthermore, the current observation of an inverse correlation between miR-103 expression and LATS2 expression in CRC tissues reduces the lack of experimental evidence for the function of miR-103 and LATS2 in CRC pathogenesis in the literature. In vivo, miR-103 promotes tumor growth in nude mice. In summary, these data suggested that miR-103 may be used to design novel strategies against CRC growth and metastasis in the future.
Previous studies have shown that miR-103 is involved in various biological and pathological processes, including glucose homeostasis, insulin sensitivity (21) and heart failure (22). miR-103 has also been reported to associate with several human cancers. For example, miR-103 downregulates the expression of the tumor suppressor gene tissue inhibitor of metalloproteinase 3 (TIMP-3) and promotes the growth and invasion of endometrial cancer cells (23). Although studies have reported that miR-103 is expressed in CRC as an oncogenic miRNA by targeting death-associated protein kinase, Kruppel-like factor 4, period circadian clock 3, Dicer and phosphatase and tensin homolog (15–17), the detailed mechanism of the effects of miR-103 in CRC growth and metastasis remains largely unknown. The present data confirmed that miR-103 was overexpressed in the primary tumor tissues of CRC patients compared with the matched non-tumor tissues. These data indicated that miR-103 overexpression is involved in CRC carcinogenesis. In addition, the present study showed that miR-103 inhibition repressed the proliferation, invasion and migration of CRC cells in vitro, suggesting that miR-103 may partially control the metastasis, invasion and proliferation potential of CRC. The present data further explored the oncogenic role of miR-103 in CRC by targeting LATS2.
The Hippo signaling pathway plays a critical role in oncogenesis by regulating cell proliferation, epithelial-mesenchymal transition and apoptosis (24). LATS2 is a key component of the Hippo signaling pathway. In canonical Hippo signaling, LATS2 phosphorylates Yes-associated protein (YAP) and promotes YAP cytoplasmic retention and degradation, resulting in the inhibition of cell proliferation and oncogenesis. LATS2 has been reported to be decreased and act as a tumor suppressor in various cancers, including breast cancer (25), lung cancer (26) and hepatocellular carcinoma (27). Li et al reported that LATS2 expression was significantly lower in CRC tissues. In addition, nocodazole, an antimicrotubule drug, potently induced LATS2 to suppress CRC growth in vivo by targeting β-catenin/B-cell lymphoma 9 (28). The present data were consistent with these previous findings, as it also indicated that miR-103 induced the loss of LATS2, which enhanced the proliferation, invasion and metastasis of CRC cells, and restoration of LATS2 led to suppression of the proliferation, invasion and migration.
In conclusion, the present study highlighted the regulatory mechanism that miR-103 induced the loss of LATS2, which promotes the metastasis, invasion and proliferation of CRC cells through direct binding to the 3′-UTR of LATS2. The present study also indicated that miR-103 may be a biomarker for the prognosis of CRC patients.
Acknowledgements
This study was supported by a grant from the National Natural Science Foundation of China (grant no. 81372553).
References
- 1.Talbot R, Kirkham S. Colorectal cancer. Lancet. 2010;376:330. doi: 10.1016/S0140-6736(10)61182-8. [DOI] [PubMed] [Google Scholar]
- 2.Fujita T. Colorectal cancer. Lancet. 2010;376:331–332. doi: 10.1016/S0140-6736(10)61183-X. [DOI] [PubMed] [Google Scholar]
- 3.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 4.Visone R, Croce CM. MiRNAs and cancer. Am J Pathol. 2009;174:1131–1138. doi: 10.2353/ajpath.2009.080794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Croce CM. miRNAs in the spotlight: Understanding cancer gene dependency. Nat Med. 2011;17:935–936. doi: 10.1038/nm0811-935. [DOI] [PubMed] [Google Scholar]
- 7.Bao Y, Chen Z, Guo Y, Feng Y, Li Z, Han W, Wang J, Zhao W, Jiao Y, Li K, et al. Tumor suppressor microRNA-27a in colorectal carcinogenesis and progression by targeting SGPP1 and Smad2. PLoS One. 2014;9:e105991. doi: 10.1371/journal.pone.0105991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Sun X, Wang Y, Liu X, Xuan Y, Hu S. Association between miR-27a genetic variants and susceptibility to colorectal cancer. Diagn Pathol. 2014;9:146. doi: 10.1186/1746-1596-9-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan W, Sui C, Liu Q, Tang W, An H, Ma J. Up-regulation of microRNA-145 associates with lymph node metastasis in colorectal cancer. PLoS One. 2014;9:e102017. doi: 10.1371/journal.pone.0102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng Y, Zhu J, Ou C, Deng Z, Chen M, Huang W, Li L. MicroRNA-145 inhibits tumour growth and metastasis in colorectal cancer by targeting fascin-1. Br J Cancer. 2014;110:2300–2309. doi: 10.1038/bjc.2014.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding Q, Chang CJ, Xie X, Xia W, Yang JY, Wang SC, Wang Y, Xia J, Chen L, Cai C, et al. APOBEC3G promotes liver metastasis in an orthotopic mouse model of colorectal cancer and predicts human hepatic metastasis. J Clin Invest. 2011;121:4526–4536. doi: 10.1172/JCI45008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yau TO, Wu CW, Dong Y, Tang CM, Ng SS, Chan FK, Sung JJ, Yu J. microRNA-221 and microRNA-18a identification in stool as potential biomarkers for the non-invasive diagnosis of colorectal carcinoma. Br J Cancer. 2014;111:1765–1771. doi: 10.1038/bjc.2014.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ress AL, Stiegelbauer V, Winter E, Schwarzenbacher D, Kiesslich T, Lax S, Jahn S, Deutsch A, Bauernhofer T, Ling H, et al. MiR-96-5p influences cellular growth and is associated with poor survival in colorectal cancer patients. Mol Carcinog. 2015;54:1442–1450. doi: 10.1002/mc.22218. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Tang Q, Li M, Jiang S, Wang X. MicroRNA-375 inhibits colorectal cancer growth by targeting PIK3CA. Biochem Biophys Res Commun. 2014;444:199–204. doi: 10.1016/j.bbrc.2014.01.028. [DOI] [PubMed] [Google Scholar]
- 15.Geng L, Sun B, Gao B, Wang Z, Quan C, Wei F, Fang XD. MicroRNA-103 promotes colorectal cancer by targeting tumor suppressor DICER and PTEN. Int J Mol Sci. 2014;15:8458–8472. doi: 10.3390/ijms15058458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen HY, Lin YM, Chung HC, Lang YD, Lin CJ, Huang J, Wang WC, Lin FM, Chen Z, Huang HD, et al. miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Cancer Res. 2012;72:3631–3641. doi: 10.1158/0008-5472.CAN-12-0667. [DOI] [PubMed] [Google Scholar]
- 17.Hong Z, Feng Z, Sai Z, Tao S. PER3, a novel target of miR-103, plays a suppressive role in colorectal cancer in vitro. BMB Rep. 2014;47:500–505. doi: 10.5483/BMBRep.2014.47.9.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao B, Lei QY, Guan KL. The Hippo-YAP pathway: New connections between regulation of organ size and cancer. Curr Opin Cell Biol. 2008;20:638–646. doi: 10.1016/j.ceb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 21.Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 22.Ellis KL, Cameron VA, Troughton RW, Frampton CM, Ellmers LJ, Richards AM. Circulating microRNAs as candidate markers to distinguish heart failure in breathless patients. Eur J Heart Fail. 2013;15:1138–1147. doi: 10.1093/eurjhf/hft078. [DOI] [PubMed] [Google Scholar]
- 23.Yu D, Zhou H, Xun Q, Xu X, Ling J, Hu Y. microRNA-103 regulates the growth and invasion of endometrial cancer cells through the downregulation of tissue inhibitor of metalloproteinase 3. Oncol Lett. 2012;3:1221–1226. doi: 10.3892/ol.2012.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi Y, Miyoshi Y, Morimoto K, Taguchi T, Tamaki Y, Noguchi S. Low LATS2 mRNA level can predict favorable response to epirubicin plus cyclophosphamide, but not to docetaxel, in breast cancers. J Cancer Res Clin Oncol. 2007;133:501–509. doi: 10.1007/s00432-007-0194-0. [DOI] [PubMed] [Google Scholar]
- 26.Strazisar M, Mlakar V, Glavac D. LATS2 tumour specific mutations and down-regulation of the gene in non-small cell carcinoma. Lung Cancer. 2009;64:257–262. doi: 10.1016/j.lungcan.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 27.Yang X, Yu J, Yin J, Xiang Q, Tang H, Lei X. MiR-195 regulates cell apoptosis of human hepatocellular carcinoma cells by targeting LATS2. Pharmazie. 2012;67:645–651. [PubMed] [Google Scholar]
- 28.Li J, Chen X, Ding X, Cheng Y, Zhao B, Al Hezaimik, Hakem R, Guan KL, Wang CY, Lai ZC. LATS2 suppresses oncogenic Wnt signaling by disrupting beta-catenin/BCL9 interaction. Cell Rep. 2013;5:1650–1663. doi: 10.1016/j.celrep.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]