Abstract
Osteoarthritis (OA) is a complex disorder characterized by degenerative articular cartilage and is largely attributed to genetic risk factors. Single nucleotide polymorphisms (SNPs) are common DNA variants that have shown promising and efficiency, compared with positional cloning, to map candidate genes of complex diseases, including OA.
In this study, we aim to provide an overview of multiple SNPs from a number of genes that have recently been linked to OA susceptibility. We also performed a comprehensive meta-analysis to evaluate the association of SNP rs7639618 of double von Willebrand factor A domains (DVWA) gene with OA susceptibility.
A systematic search of studies on the association of SNPs with susceptibility to OA was conducted in PubMed and Google scholar. Studies subjected to meta-analysis include human and case-control studies that met the Hardy–Weinberg equilibrium model and provide sufficient data to calculate an odds ratio (OR). A total of 9500 OA cases and 9365 controls in 7 case-control studies relating to SNP rs7639618 were included in this study and the ORs with 95% confidence intervals (CIs) were calculated.
Over 50 SNPs from different genes have been shown to be associated with either hip (23), or knee (20), or both (13) OA. The ORs of these SNPs for OA and the subtypes are not consistent. As to SNP rs7639618 of DVWA, increased knee OA risk was observed in all genetic models analyzed. Specifically, people from Asian with G-allele showed significantly increased risk of knee OA (A versus G: OR = 1.28, 95% CI 1.13–1.46; AA versus GG: OR = 1.60, 95% CI 1.25–2.05; GA versus GG: OR = 1.31, 95% CI 1.18–1.44; AA versus GA+GG: OR = 1.34, 95% CI 1.12–1.61; AA+GA versus GG: OR = 1.40, 95% CI 1.19–1.64), but not in Caucasians or with hip OA.
Our results suggest that multiple SNPs play different roles in the pathogenesis of OA and its subtypes; SNP rs7639618 of DVWA gene is associated with a significantly increased risk of knee OA in Asians. Given the limited sample size, further studies are needed to evaluate this observation.
INTRODUCTION
Osteoarthritis (OA) is the most common joint disease worldwide, affecting approximately 10% of men and 18% of women over 60 years of age.1,2 Multiple factors, including advanced age, excess body weight, repeated trauma or surgery to the joint structures, abnormal joints at birth, gout, diabetes, and hormone-related disorders, have been demonstrated to contribute to increased risk of OA.3,4 Previous epidemiological studies from twin-pair and family-based segregation analyses have provided clear evidence of a heritable component in susceptibility of OA. However, the specific genetic factors that lead to OA are currently largely unknown. It therefore remains a challenge to identify candidate genes or risk alleles that contribute to OA pathogenesis.
Identification of candidate genes responsible for numerous monogenic disorders has been successful over the past decades with the technology of positional cloning.5,6 Similar strategy has been used to target many complex diseases, including asthma, heart disease, cancer, and OA.5,7,8 However, due to lack of suitable genetic markers, not many candidate genes have been identified that show a clear OA etiology. Single nucleotide polymorphisms (SNPs) are common genomic DNA variations within a population. SNPs, in combination with genome-wide association studies (GWAS), have significantly accelerated complex disease gene localization.5,8,9 SNPs of genes COL11A1, VEGF, GDF5, and IL-8, etc., have been associated with OA. Given the heterogeneity and complexity of OA, it is not surprising that these gene polymorphisms showed different levels of association with increased risk of OA. Some polymorphisms may be specific to OA subtypes (hip-, knee-, or hand-OA) and ethnic groups, but the results are with a wide range of discrepancy.
Recently, multiple studies have indicated an association of double von Willebrand factor A domains (DVWA) gene with susceptibility to OA. DVWA gene variants encode 2 protein isoforms, the long (L-DVWA, 385 amino acids) and short (S-DVWA, 276 amino acids) proteins. DVWA protein is predicted to have 2 domains homologous to the VWA domain, which typically is involved in cell adhesion and protein–protein interactions.10,11 Interestingly, L-DVWA and S-DVWA were mainly expressed in articular cartilage, suggesting a potential function of these isoforms in OA.12 Indeed, DVWA has been associated with susceptibility to knee OA and linked to OA etiology, possibly by interacting with β-tubulin. Data from later replication study and meta-analyses of one of the DVWA polymorphisms further confirmed its association with OA in European and Asian populations.13 However, no significant association between DVWA and OA was found in UK patient samples and neither independent association with OA was observed in Europeans.14 In addition, there are multiple SNPs in DVWA gene with allelic difference. The association between DVWA and OA has been inconsistent due to allelic effect and effect size of case-controlled studies analyzed.14
In this study, we summarized and analyzed the SNPs that have recently been associated with OA and calculated their ORs with OA susceptibility. We also provided an updated and comprehensive meta-analysis to evaluate the association of SNP rs7639618 of DVWA with OA susceptibility with consideration of publication bias and the source of heterogeneity.
MATERIALS AND METHODS
Literature Search
A systematic search of studies on the association of SNPs with susceptibility to OA was conducted in PubMed and Google scholar. “Single Nucleotide Polymorphism”, “SNP”, and “Osteoarthritis” were used as key words for the searching. The odds ratios (ORs) with 95% confidence intervals were calculated on the basis of data provided in the literature. For meta-analysis, the PubMed was searched for eligible articles up to the end of June 2015. Following keywords were used for the searching “polymorphism”, “SNP”, “rs7639618”, “von Willebrand factor A domains”, “DVWA”, “Osteoarthritis”, “OA.” Google academic searching was also performed to obtain additional information.15
Study Selection
Studies subjected to meta-analysis were selected according to the following inclusion criteria: human studies; studies investigating the association between the SNP rs7639618 and osteoarthritis (OA); case-control studies providing sufficient data on genotypes to calculate an OR; the genotype distribution of the control population met the Hardy–Weinberg equilibrium (HWE) model. Two reviewers were assigned to independently assess the studies using the inclusion criteria and disagreement was subjected to discussion with a third reviewer for a consensus agreement.
Data Extraction
The following data were collected from each eligible study: name of first author, years of publication, ethnicity, OA sites, HWE, number of cases and controls, genotype frequency in cases and controls. Different ethnicity descents were classified as Caucasian and Asian. When HWE in the controls was not reported, an online program (https://ihg.gsf.de/cgi-bin/hw/hwa1.pl) was used to test the HWE by χ2 test for goodness of fit.16
Evaluation of the Study Quality
The quality of the studies was evaluated by 2 reviewers according to the predefined scale for quality assessment, which was modified from previous meta-analysis.16,17 In this scale, 5 items, including the representativeness of cases, source of controls, sample size, quality control of genotyping methods, HWE, were carefully checked. Total scores were recorded with a range from 0 to 10 and a higher score indicated a better quality of the study. Any disagreement of the score was modified based on discussion between the 2 reviewers.
Statistical Analysis
Crude ORs together with their corresponding 95% CIs were used to assess the strength of association between the rs7639618 SNP and the risk of OA. The pooled ORs were calculated for allelic comparison (A versus G), heterozygote model (GA versus GG), homozygote model (AA versus GG), dominant model (AA+GA versus GG), recessive model (AA versus GA+GG), respectively. Heterogeneity among the trials was analyzed in this study using the Q statistic (significance level of P value <0.10) and the I2 test (greater than 50% as evidence of significant inconsistency). When significant heterogeneity (P < 0.10 or I2>50%) was achieved, the random effect model was used to combine the effect sizes of the included studies. If no significant heterogeneity was found, fixed effect was selected to pool the data. Meta-regression analysis was performed to detect the source of heterogeneity. The variance (τ2) between studies was used to quantify the degree of heterogeneity and the percentage of τ2 was used to describe the extent of heterogeneity.17,18 Sensitivity analyses were performed to identify individual study effect on pooled results and test the reliability of results.17 In addition, subgroup analyses were stratified by ethnicity. Potential publication bias was estimated using the Begg test and a forest plot was used to analyze and display the results. All calculations were measured and analyzed using the STATA (version 11.0).
Ethical Statement
No ethical approval was needed for this manuscript, as the data used in this review and the meta-analyses have all been published.
RESULTS
Multiple SNPs From a Number of Genes are Associated with OA
Based on the quality of the studies and the selection criteria as described above, we summarized most of the SNPs that have recently been linked to OA susceptibility. As illustrated in Figure 1, there are 56 SNPs from 50 genes or gene loci that have been associated with OA or OA subtypes. Specifically, 23 SNPs from 21 genes, including COL11A1 (rs1241164, rs4907986, rs2615977),19,20VEGF (rs833058), and IGF1 (rs2195239) etc., are associated with hip OA; 20 SNPs from 17 genes, including GDF5 (rs143383), ADAMTS14 (rs4747096), and DVWA (rs7639618, rs11713836, and rs3773472), etc., are frequently seen in knee OA; while individuals containing 13 SNPs of 12 genes, including IL8 (rs4073, rs2227306), TGFβ1 (2227306), and SMAD3 (rs12901499), etc., tend to develop both hip and knee OA. These results demonstrate involvement of multiple SNPs/genes in susceptibility to OA and its subtypes.
FIGURE 1.
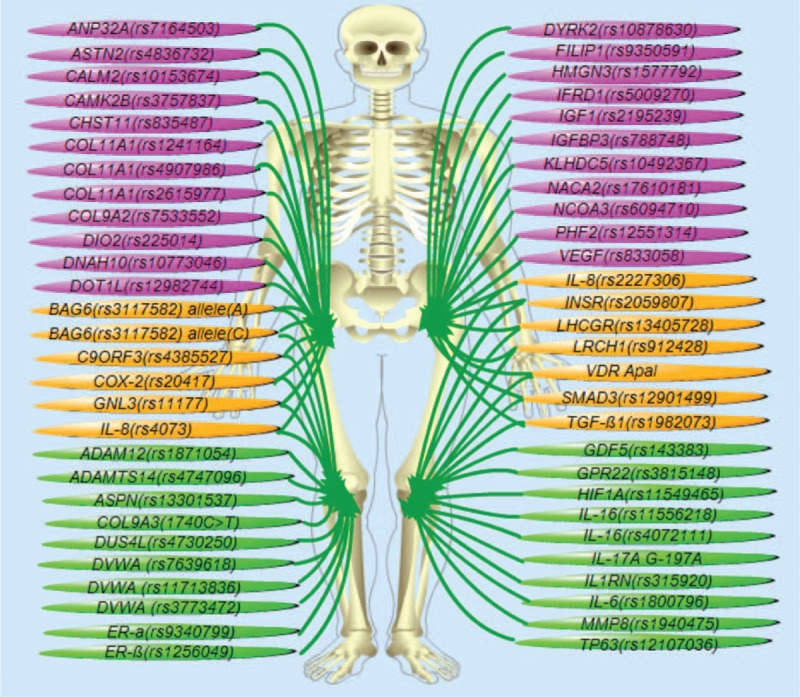
Multiple polymorphisms are associated with the risk of OA. Illustrated are 56 SNPs from 50 genes that have been linked to OA risk. Among these, 23 SNPs from 21 genes have been associated with hip OA (pink ones); 20 SNPs from 17 genes are frequently altered in knee OA (green ones); while other 13 SNPs of 12 genes are reportedly associated with both knee- and hip-OA (yellow ones).
Odds Ratios of Candidate SNPs Associated With OA
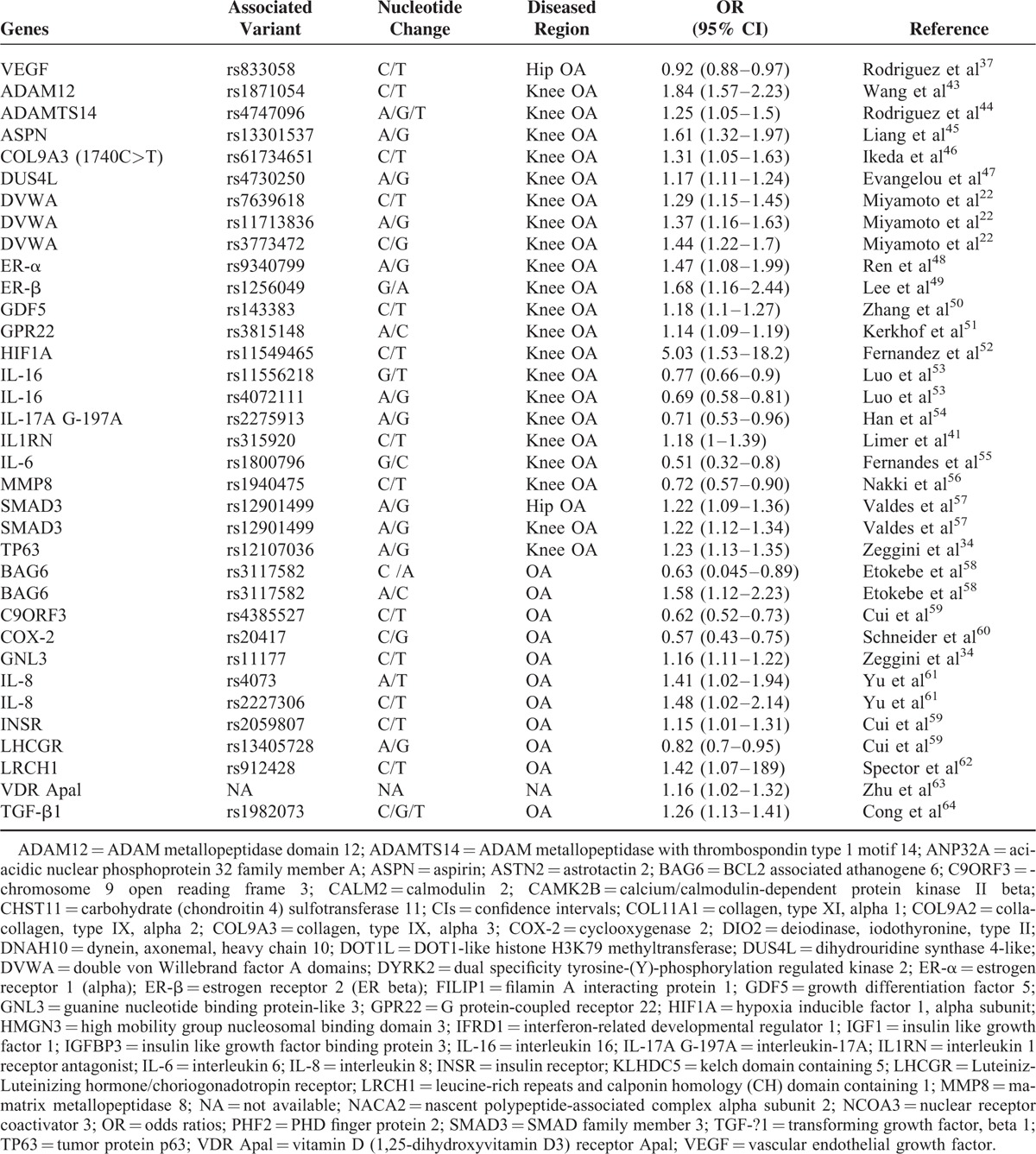
To evaluate the strength of association with OA, we have summarized the crude ORs of each of the candidate SNPs with 95% confidence intervals (CIs). Listed in Table 1 are detailed information about the names of associated genes, the SNP numbers, locations of OA (knee- or hip-OA), OR values, and literature source. Most of the SNPs associated with hip OA showed a value of odds ratio greater than 1.0, with SNP rs225014 of DIO2 showing the highest value of 1.79 (1.37–2.34, Figure 2A, Table 1 ). The odds ratios of SNPs that are associated with knee OA were also calculated and most are above the value of 1.2 with SNP rs4747096 of ADAM12 showing the highest value of 1.84 (1.57–2.23, Figure 2B, Table 1 ). Similar results were observed for SNPs of genes that are associated with both knee and hip OA (Figure 2C, Table 1 ), although some SNPs showed an OR value less than 1.0 as seen in groups of hip- or knee-OA (Figure 2, Table 1 ). Together, these results suggest that multiple SNPs from a variety of genes showed different levels of association with knee and/or hip OA.
TABLE 1.
List of Genes and SNPs Associated With OA
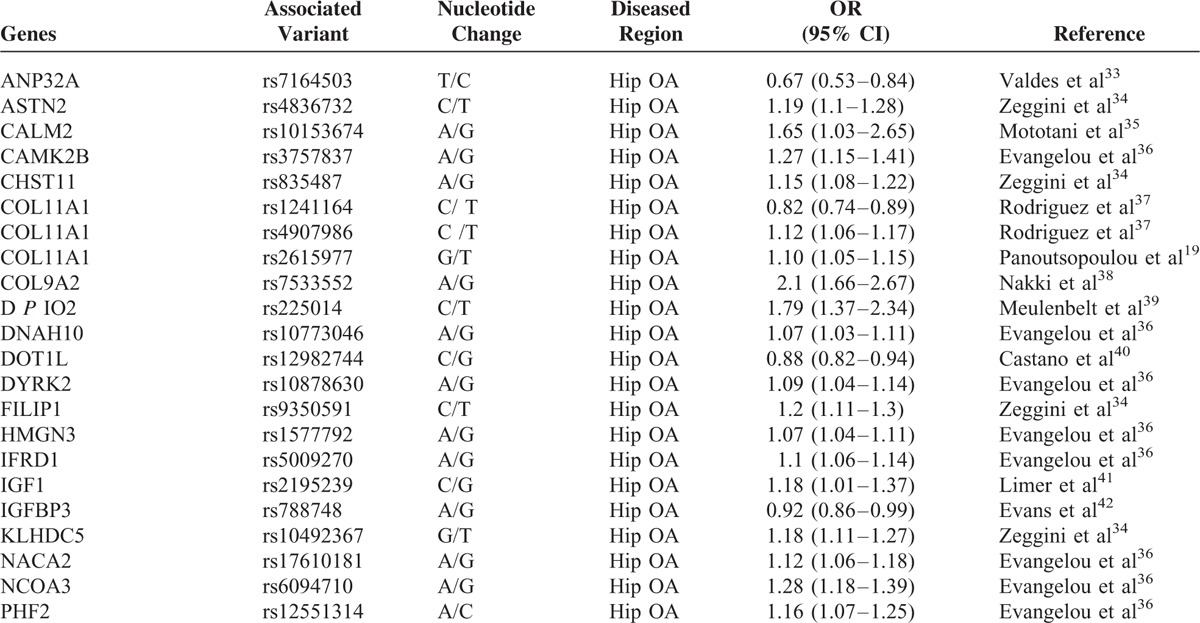
FIGURE 2.
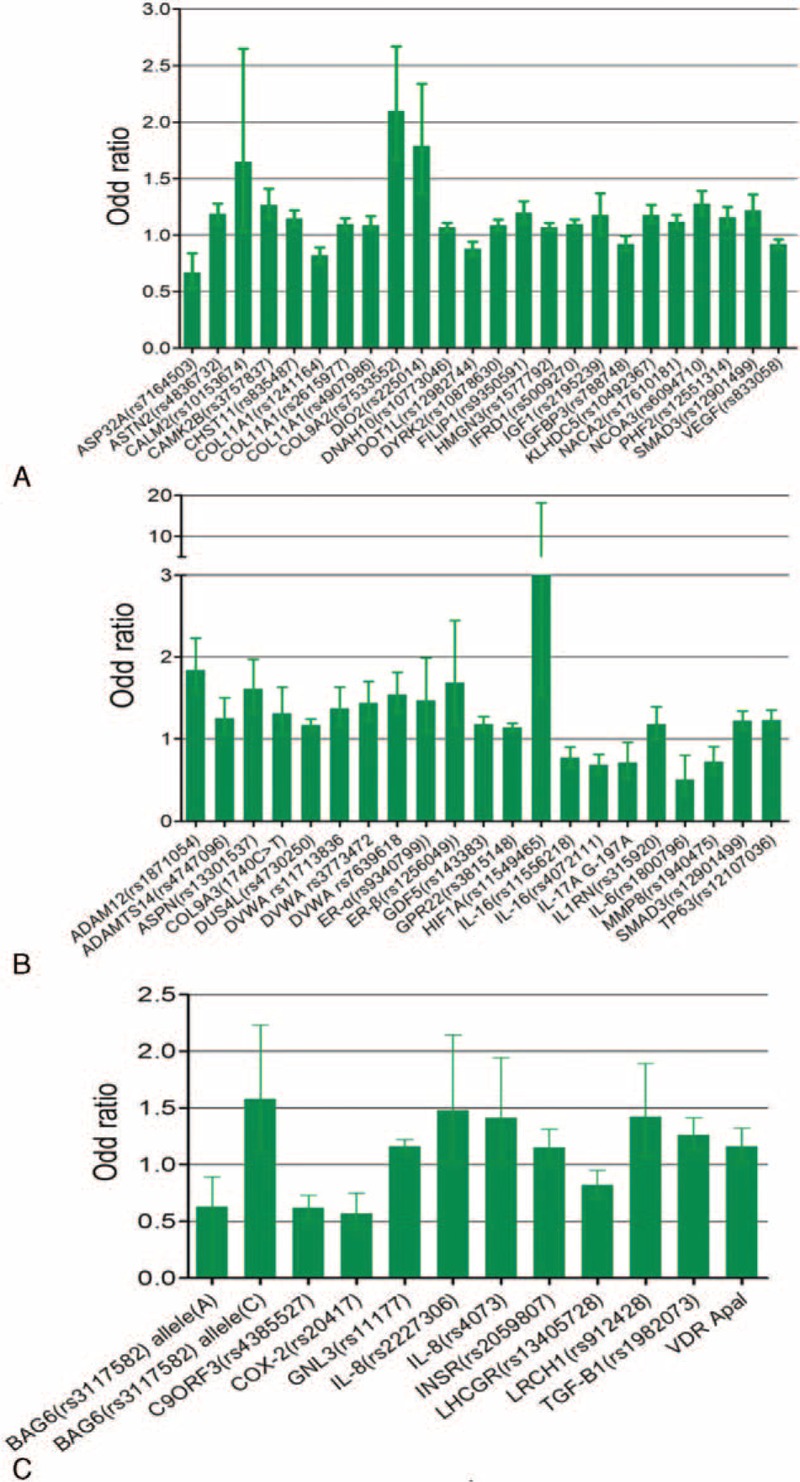
The odd ratios (OR) of the SNPs associated with OA or OA subtypes. A, The ORs of most of the SNPs associated with hip OA showed a value greater than 1.0. SNP rs225014 of DIO2 showed the highest value of 1.79 (1.37–2.34). B, The ORs of SNPs associated with knee OA were mostly above the value of 1.2. SNP rs4747096 of ADAM12 showed the highest value of 1.84 (1.57–2.23). C, SNPs of genes associated with both knee and hip OA showed similar OR values. The OR value of SMAD3 is only available for individual knee- or hip-OA.
Eligible Studies Selected for Meta-Analysis of DVWA and OA
By searching the PubMed and Google scholar database using above key words (SNP, rs7639618, DVWA, and OA), we have identified approximately 50 relevant papers, including 38 studies that potentially show an association between DVWA and OA. After exclusion of duplicated studies and improper studies determined by reading the title and abstracts, 9 studies meet the selection criteria. Due to lack of genotype frequency in 2 of the studies, 7 studies were eventually subjected to meta-analysis of the association between SNP rs7639618 of DVWA and OA. The detailed process of literature screening is outlined in Figure 3. In these studies, 9500 OA cases and 9365 controls were identified according to the inclusion and exclusion criteria. Six knee-OA-related studies contained information from 6807 knee OA cases and 7785 controls,13,14,21–24 while 3 studies involving hip OA include 2693 hip OA cases and 1580 controls.13,14,25 The HWE of genotype distribution in the controls was tested in all studies and they were all in consistent with HWE. Detailed information about the OA types, the year and countries the studies conducted, allelic difference, power of HWE, and the quality score of the studies is provided in Table 2.
FIGURE 3.
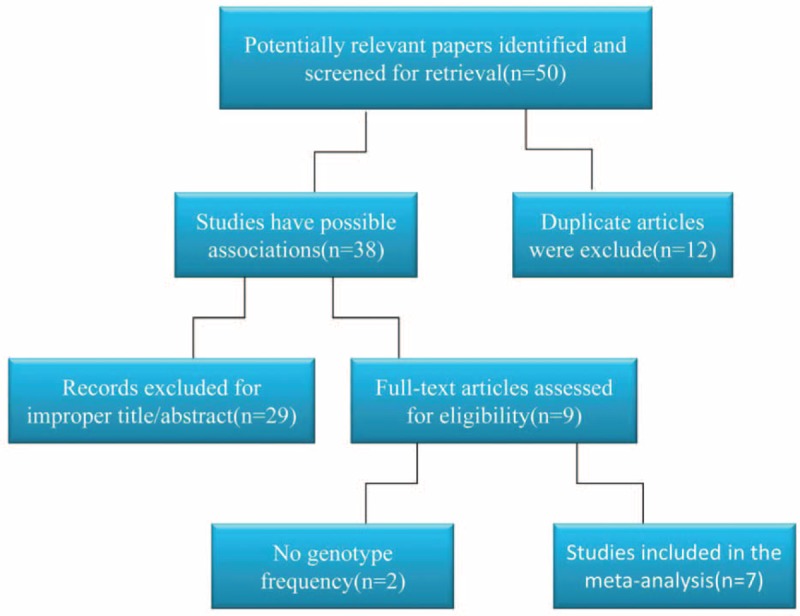
Flow chart of study selection. Studies selected for current meta-analysis are as illustrated. Thirty-eight studies from 50 relevant papers show a potential association between DVWA and OA. Only 9 studies meet the selection criteria. Seven studies were eventually subjected to meta-analysis as 2 of the studies lack genotype frequency.
TABLE 1 (Continued).
List of Genes and SNPs Associated With OA
SNP rs7639618 is Significantly Associated with Knee OA
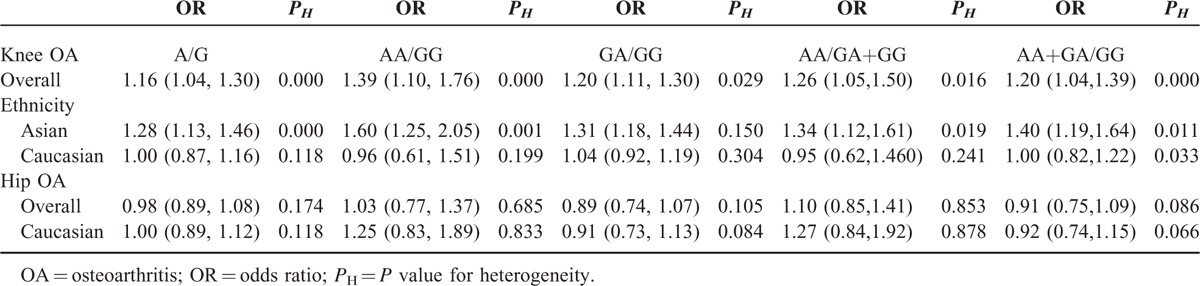
Through meta-analysis of SNP rs7639618 of DVWA, we observed a significantly increased risk of knee OA susceptibility in allelic comparison in Asians (A versus G: OR = 1.16, 95% CI 1.04–1.30 Figure 4). The OR values for each of the allelic models are listed in Table 2 and illustrated in Figure 5A (homozygote model, AA versus GG: OR = 1.39, 95% CI 1.10–1.76), Figure 5B (heterozygote model, GA versus GG: OR = 1.20, 95% CI 1.11–1.30), Figure 6A (recessive model, AA versus GA+GG: OR = 1.26, 95% CI 1.05–1.50), and Figure 6B (dominant model, AA+GA versus GG: OR = 1.20, 95% CI 1.04–1.39). As shown in Table 3, there was significant heterogeneity between studies, ranging from 0 to 0.029. We therefore performed subgroup analysis according to ethnicity. No heterogeneity was shown in heterozygote model and thus, a fixed model was applied for its pooled OR. For other allelic models that show significant heterogeneity, a randomized effect model was used. The results showed that there was a statistically increased knee OA risk in all allelic models (A versus G: OR = 1.28, 95% CI 1.13–1.46, Figure 4; AA versus GG: OR = 1.60, 95% CI 1.25–2.05, Figure 5A; GA versus GG: OR = 1.31, 95% CI 1.18–1.44, Figure 5B; AA versus GA+GG: OR = 1.34, 95% CI 1.12–1.61 Figure 6A; AA+GA versus GG: OR = 1.40, 95% CI 1.19–1.64, Figure 6B and Table 3). The results in Asians were similar to that of overall comparisons of pooled eligible researches (Table 3), while in Caucasians, even with increased sample size, there is no significant association in any allelic models compared as illustrated in Figure 4 (A versus G: OR = 1.00, 95% CI 0.87–1.16), Figure 5A (AA versus GG: OR = 0.96, 95% CI 0.61–1.51), Figure 5B (GA versus GG: OR = 1.04, 95% CI 0.92–1.19), Figure 6A (AA versus GA+GG: OR = 0.95, 95% CI 0.62–1.46), Figure 6B (AA+GA versus GG: OR = 1.00, 95% CI 0.82–1.22), and Table 3. Together, these results support that SNP rs7639618 of DVWA was only associated with an increased risk of knee OA in Asians.
FIGURE 4.
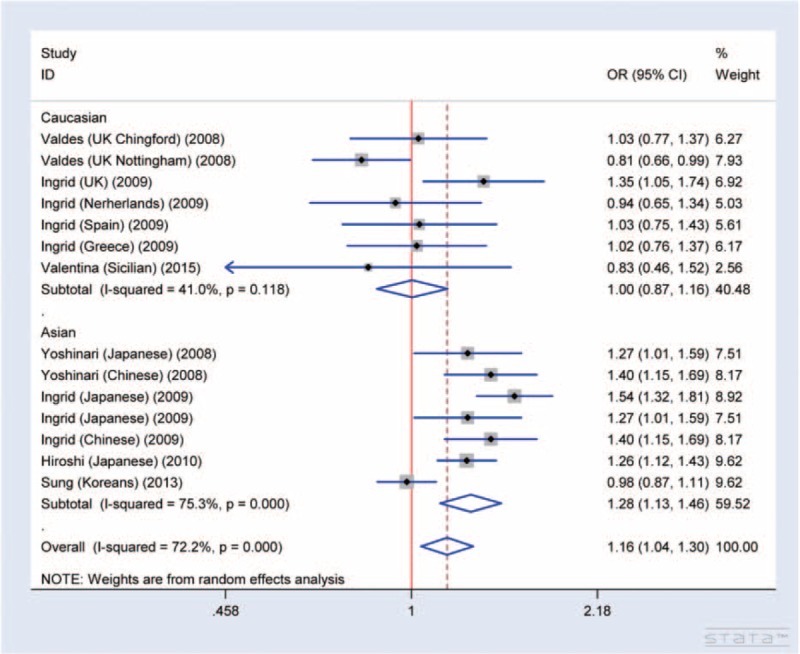
Forest plot of allele comparison of DVWA rs7639618 for overall comparison (A versus G, association of rs7639618 and knee OA). SNP rs7639618 is associated with a significantly increased risk of knee OA in allelic comparison in Asian (A versus G: OR: 1.16, 95% CI 1.04–1.30), and in all allelic models (A versus G: OR: 1.28, 95% CI 1.13–1.46), but not in Caucasians (A versus G: OR: 1.00, 95% CI 0.87–1.16).
FIGURE 5.
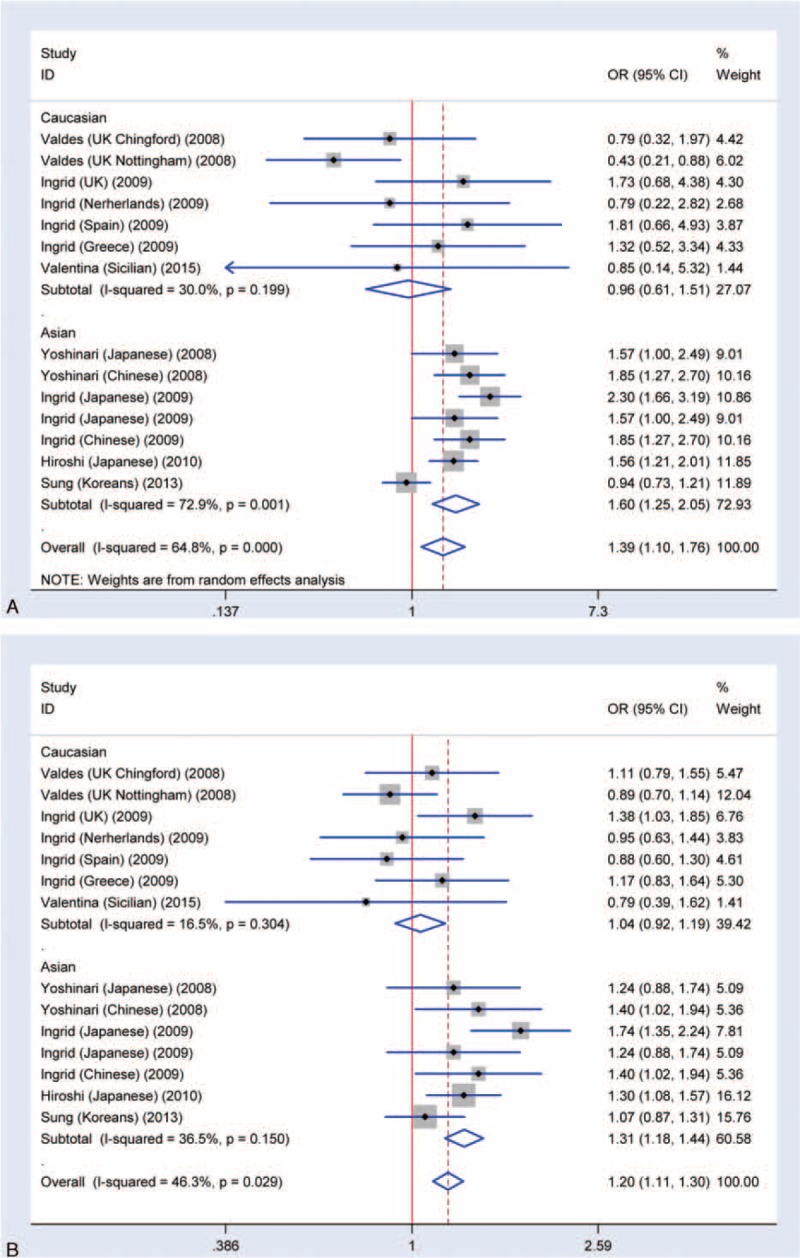
Meta-analysis of the association rs7639618 and knee OA. A, Forest plot of homozygote model for overall comparison (AA versus GG: OR: 1.39, 95% CI 1.10–1.76). B, Forest plot of heterozygote model for overall comparison (GA versus GG: OR: 1.20, 95% CI 1.11–1.30).
FIGURE 6.
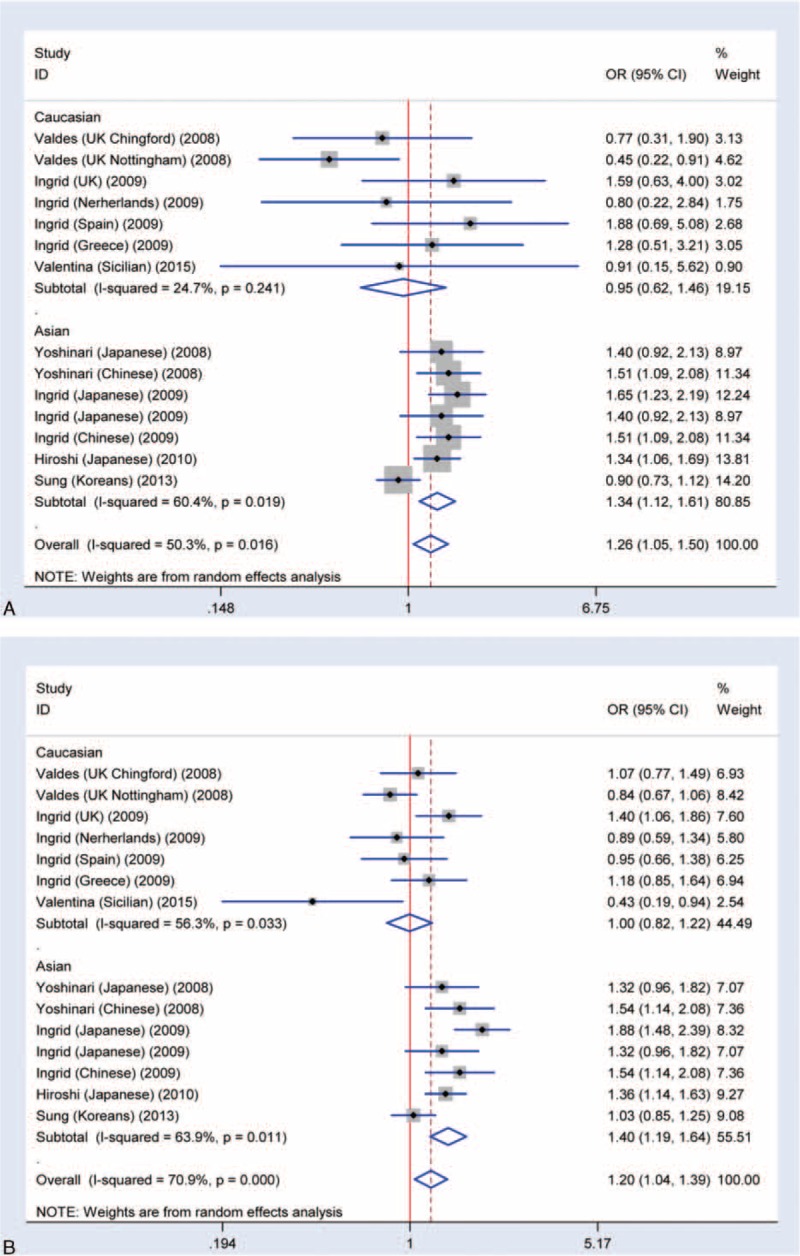
Meta-analysis of the association rs7639618 and knee OA. A, Forest plot of recessive model for overall comparison (AA versus GA+GG: OR: 1.26, 95% CI 1.05–1.50). B, Forest plot of dominant model for overall comparison (AA+GA versus GG: OR: 1.20, 95% CI 1.04–1.39).
TABLE 2.
Characteristics of Eligible Studies
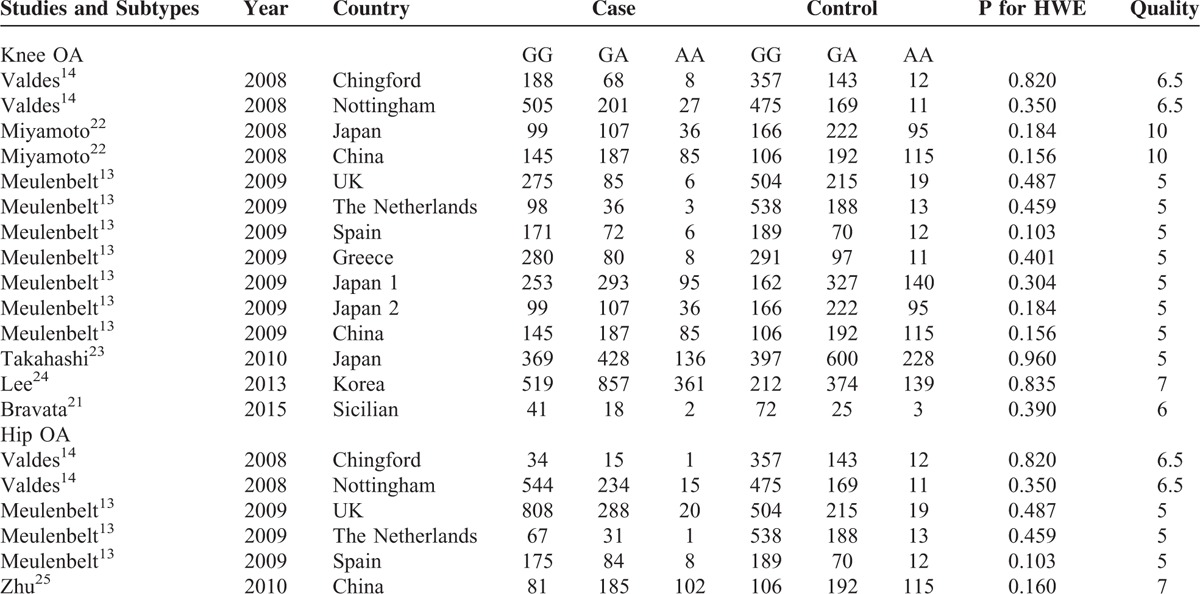
TABLE 3.
Results of Meta-Analysis
No Significant Association Between SNP rs7639618 and Hip OA
The association between SNP rs7639618 of DVWA and the risk of hip OA was analyzed in 3 independent studies. Random-effects model was used in the dominant model and heterozygote model due to the presence of heterogeneity, while fixed-effects model was used in other models without significant heterogeneity. Overall, no significant association was identified in any of the allelic models analyzed (A versus G: OR = 0.98, 95% CI 0.89–1.08, AA versus GG: OR = 1.03, 95% CI 0.77–1.37, GA versus GG: OR = 0.89, 95% CI 0.74–1.07, AA versus GA+GG: OR = 1.10, 95% CI 0.85–1.41, and AA+GA versus GG: OR = 0.91, 95% CI 0.75–1.09, Figure 7). Five studies in 2 papers were carried out in Caucasian population. Subgroup analysis of Caucasian population was also assessed, and there is no significant association between SNP rs7639618 and hip OA in any allelic models analyzed (A versus G: OR = 1.00, 95% CI 0.89–1.12, AA versus GG: OR = 1.25, 95% CI 0.83–1.89, GA versus GG: OR = 0.91, 95% CI 0.73–1.13, AA versus GA+GG: OR = 1.27, 95% CI 0.84–1.92, and AA+GA versus GG: OR = 0.92, 95% CI 0.74–1.15, Figure 7).
FIGURE 7.
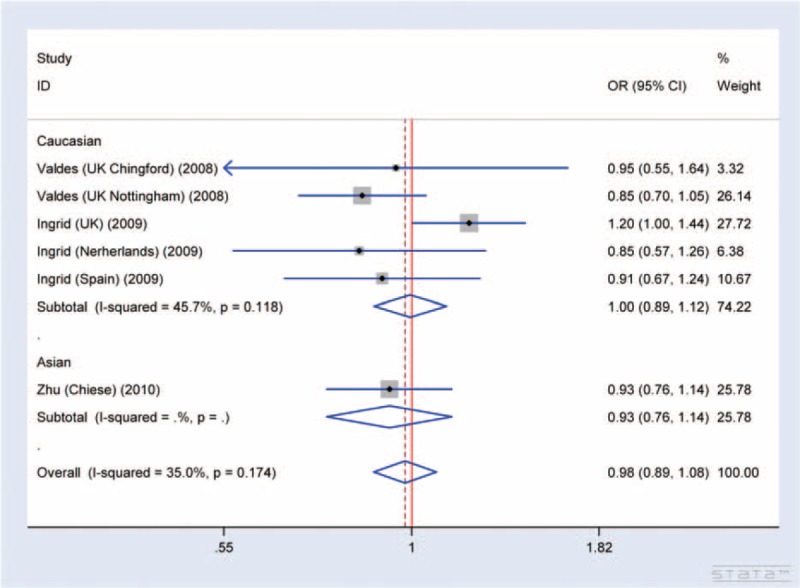
Forest plot of allele comparison of DVWA rs7639618 for overall comparison (A versus G, association of rs7639618 and hip OA). No significant association between SNP rs7639618 and hip OA in any allelic models was detected (A versus G: OR: 1.00, 95% CI 0.89–1.12, AA versus GG: OR: 1.25, 95% CI 0.83–1.89, GA versus GG: OR: 0.91, 95% CI 0.73–1.13, AA versus GA+GG: OR: 1.27, 95% CI 0.84–1.92, and AA+GA versus GG: OR: 0.92, 95% CI 0.74–1.15).
Test of Heterogeneity and Sensitivity
As significant heterogeneity was shown between studies in each comparison, we therefore investigated the source of heterogeneity by ethnicity, year of report, and sample size using meta-regression analysis and subjected to allelic comparison (A versus G). Studies containing more than 1000 participants were categorized as “large,” while studies with less than 1000 participants were categorized as “small.” Meanwhile, group of cases with an average age greater than 65 were categorized as “high-risk,” while cases younger than 65 were assigned to “low-risk” group. Group of “mixed” cases indicate that no data of age are available. The results suggested that ethnicity (P = 0.002), group average age (P = 0.024), and the year the study conducted (P = 0.038), but not the sample size (P = 0.438), contributed to the source of heterogeneity. In addition, ethnicity, the average age, and year of study could explain 37% of the variance (τ2). We also performed sensitivity analysis to evaluate whether the present meta-analysis is stable and the results showed that no individual study affected the pooled OR (Figure 8A).
FIGURE 8.
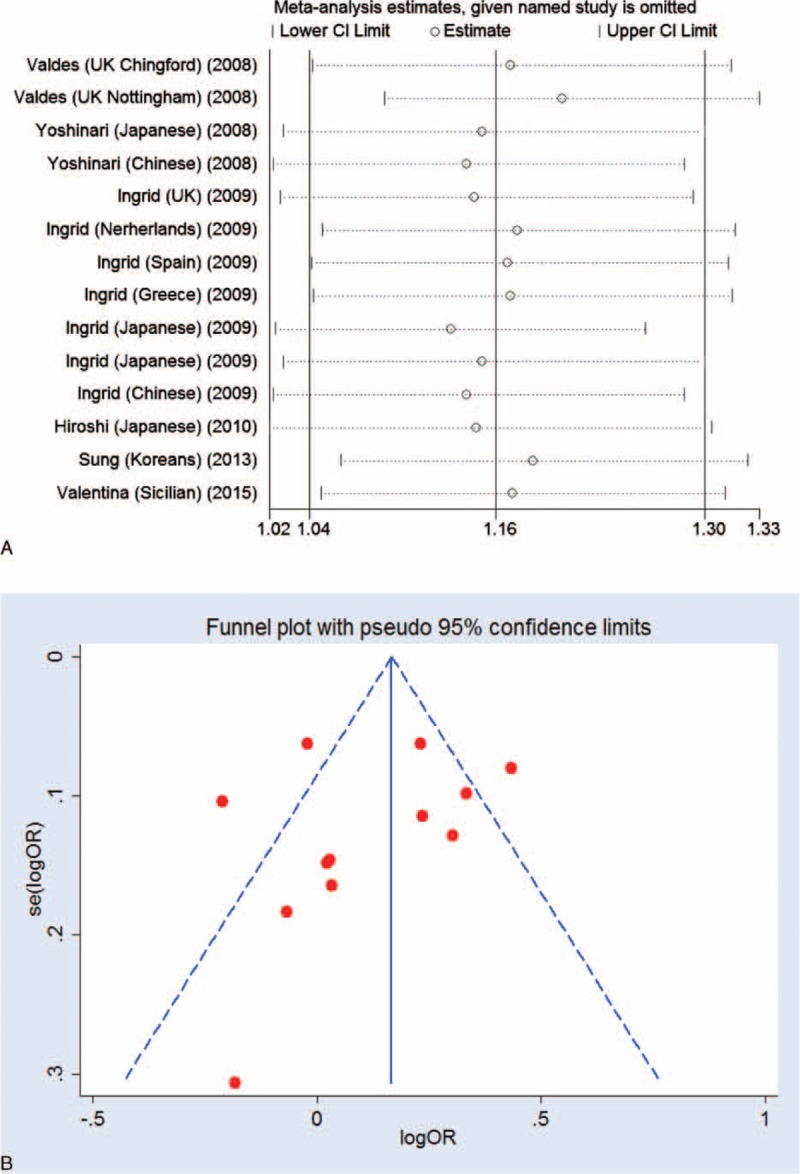
The result of sensitivity and Funnel plot analysis. A, Sensitivity analysis showed no individual study affected the pooled OR. B, Funnel plot analysis showed no potential publication bias.
Publication Bias
A Begg funnel plot and a Begg test were used to assess for publication bias for all allelic models. The result showed no obvious asymmetry, indicating no publication bias (Figure 8B).
DISCUSSION
OA is characterized by progressive cartilage matrix degradation, subchondral bone sclerosis, and osteophyte formation.26,27 These are nonreversible processes due to the limited repair capacity of cartilage. With the disease advances, pain becomes the most prominent symptom which is almost unbearable and eventually leads to joint replacement.27 Thanks to the better life and medical conditions, people live longer nowadays, but it also raises the incidence and the population suffering from OA. Hence, OA is a major source of pain, disability, and a tremendous socioeconomic burden worldwide.1 Unfortunately, given the heterogeneity of multiple subtypes and the complexity of OA pathogenesis, it remains a challenge to find unanimous biomarkers that help with the early diagnosis and targeted therapy for all OA patients. Recently, personalized medicine becomes a hot topic in health care and may be achieved due to existence of SNPs, the most common genetic variations, within a population.28 There are many SNPs within different genes that have shown various associations with OA. Identification of these gene SNPs and their correlation with OA would further our understanding of the molecular mechanisms involved in the pathogenesis of OA, so as to develop better diagnostics and more targeted therapy at early stages of OA.
In this effort, we provided an overview of multiple SNPs in association with OA susceptibility. We identified more than 50 SNPs from a number of genes that have been linked to either hip (COL11A1, VEGF, etc.), or knee (COL9A3, ASPN, GDF5, etc.), or both (IL-8, TGF-β1, etc.) OA. Among these genes, GDF5, which encodes the growth differentiation factor 5, is a member of the transforming growth factor-β superfamily. GDF5 has previously been shown to play a role in development and maintenance of bone and cartilage.14 Notably, SNP rs143383 of GDF5 is known to be associated with high risk of OA.14 The difference of GDF5 SNPs in relationship with OA subtypes and ethnic groups was also observed.29 In a recent GWAS associated meta-analysis of OA candidate genes, COL11A1 and VEGF were significantly associated with OA. Interestingly, SNPs rs4907986 and rs1241164 of COL11A1 and SNP rs833058 of VEGF all showed association with hip OA.14 It was previously shown that mutation in Col11a1 may cause deposition of degraded type II collagen in articular cartilage and eventually lead to OA.30 While it is relatively established for a role of GDF5, COL11A1, and VEGF in OA pathogenesis, the association between SNPs of these genes with OA susceptibility is not always consistent. SNP rs2615977 of COL11A1 was also shown to be associated with OA. However, another COL11A1 SNP rs1676486, which has been linked to lumbar disc herniation (LDH, also a degenerative musculoskeletal disease), was found not associated with OA.20 In our analysis, we tried to summarize most of the gene SNPs that have shown some correlation with OA or its subtypes. The OR of many of these SNPs/genes are not always consistent, further meta-analysis of multiple studies may overcome the limited sample size and inadequate statistical power of single case-control studies to provide more reliable results.31
DVWA gene contains multiple SNPs, including rs11718863, rs7639618, rs7651842, rs7639807, rs17040821, etc. These SNPs may cause protein functional changes or diseases, as described in studies associated with OA.21 However, as indicated above, the results of their association with OA are not consistent. In this study, we specifically analyzed the association of SNP rs7639618 of DVWA with OA through comprehensive meta-analysis. We have analyzed all available eligible studies that include 9500 OA cases and 9365 controls. Our results confirmed that SNP rs7639618 is associated with a significantly increased risk of knee OA, especially in Asian populations. Similar results were found in subgroup analysis by ethnicity. No evidence was found for the association between rs7639618 SNP and hip OA susceptibility in any genetic allelic models. We have performed heterogeneity analysis and the results showed that ethnicity, average age of case group, and the year of the study were the source of heterogeneity. When we restricted the ethnicity to Asian, there was also heterogeneity, suggesting that ethnicity was not the main source of heterogeneity. We have shown that ethnicity, age, and year of study account for 37% of the variance (τ2) by meta-regression analysis, while the sensitivity analysis demonstrated that our meta-analysis is stable. In addition, no limitation was made in the search, and the selection bias was well controlled as demonstrated by Begg funnel plot analysis, which showed no publication bias.
In summary, it should be noted that due to limited number of studies on SNP rs7639618 and OA, the relatively small sample size may affect the power and statistics of the meta-analysis.32 The heterogeneity in some of the genetic/allelic models also needs to be treated with caution when interpreting the results. The insufficient sample size in single pioneer or replication studies of multiple SNPs, including DVWA, did result in a wide range of values of the ORs. However, we have provided a comprehensive overview of most of the relevant SNPs in OA or its subtypes.33–64 We have also updated all available studies on DVWA SNPs, and our results further support an association of SNP rs7639618 with OA as recently indicated.65,66 In addition, we have performed multiple analyses including subgroup, heterogeneity, sensitivity, and meta-regression assessment. The results showed that our meta-analysis is stable and we have analyzed the sources of the heterogeneity as indicated above. Together, our results support that rs7639618 SNP is significantly associated with increased risk of knee OA in Asians. There was insufficient data to support an association between SNP rs7639618 and the risk of hip OA or OA in Caucasians, although further studies are required to validate this genetic epidemiology and to functionally characterize this DVWA variant with OA pathophysiology.
Acknowledgments
We thank the families for participating in this research project. This work is supported by clinical medicine science and technology projects of Jiangsu province (BL2013019), Jiangsu provincial health department scientific research project (Q201412), Suzhou science and technology support program (SS201429), the innovation program of Jiangsu province (QZ), and the NSFC grants, China (Nos. 31271399 and 81472047, QZ).
Footnotes
Abbreviations: CIs = confidence intervals, DVWA = double von Willebrand factor A domains, HWE = Hardy–Weinberg equilibrium, ORs = odds ratios, SNPs = single nucleotide polymorphisms.
TW and YL contributed equally to this work.
The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Glyn-Jones S, Palmer AJ, Agricola R, et al. Osteoarthritis. Lancet (London) 2015; 386:376–387. [DOI] [PubMed] [Google Scholar]
- 2.Woolf AD, Pfleger B. Burden of major musculoskeletal conditions. Bull World Health Organization 2003; 81:646–656. [PMC free article] [PubMed] [Google Scholar]
- 3.Yin YW, Sun QQ, Hu AM, et al. Association of rs9340799 polymorphism in estrogen receptor alpha gene with the risk of osteoarthritis: evidence based on 8,792 subjects. Mol Genet Genomics 2015; 290:513–520. [DOI] [PubMed] [Google Scholar]
- 4.Brandt KD, Dieppe P, Radin E. Etiopathogenesis of osteoarthritis. Med Clin North Am 2009; 93:1–24. [DOI] [PubMed] [Google Scholar]
- 5.Riley JH, Allan CJ, Lai E, et al. The use of single nucleotide polymorphisms in the isolation of common disease genes. Pharmacogenomics 2000; 1:39–47. [DOI] [PubMed] [Google Scholar]
- 6.Puliti A, Caridi G, Ravazzolo R, et al. Teaching molecular genetics: chapter 4—positional cloning of genetic disorders. Pediatr Nephrol 2007; 22:2023–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abel K, Reneland R, Kammerer S, et al. Genome-wide SNP association: identification of susceptibility alleles for osteoarthritis. Autoimmunity Rev 2006; 5:258–263. [DOI] [PubMed] [Google Scholar]
- 8.Srinivasan S, Clements JA, Batra J. Single nucleotide polymorphisms in clinics: fantasy or reality for cancer? Crit Rev Clin Lab Sci 2016; 53:29–39. [DOI] [PubMed] [Google Scholar]
- 9.Huang Q. Genetic study of complex diseases in the post-GWAS era. J Genet Genomics 2015; 42:87–98. [DOI] [PubMed] [Google Scholar]
- 10.Whittaker CA, Hynes RO. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol Biol Cell 2002; 13:3369–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becker AK, Mikolajek H, Paulsson M, et al. A structure of a collagen VI VWA domain displays N and C termini at opposite sides of the protein. Structure 2014; 22:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakajima M, Miyamoto Y, Ikegawa S. Cloning and characterization of the osteoarthritis-associated gene DVWA. J Bone Miner Metab 2011; 29:300–308. [DOI] [PubMed] [Google Scholar]
- 13.Meulenbelt I, Chapman K, Dieguez-Gonzalez R, et al. Large replication study and meta-analyses of DVWA as an osteoarthritis susceptibility locus in European and Asian populations. Hum Mol Genet 2009; 18:1518–1523. [DOI] [PubMed] [Google Scholar]
- 14.Valdes AM, Spector TD, Doherty S, et al. Association of the DVWA and GDF5 polymorphisms with osteoarthritis in UK populations. Ann Rheum Dis 2009; 68:1916–1920. [DOI] [PubMed] [Google Scholar]
- 15.Qiao L, Liang Y, Li N, et al. Endothelin-A receptor antagonists in prostate cancer treatment: a meta-analysis. Int J Clin Exp Med 2015; 8:3465–3473. [PMC free article] [PubMed] [Google Scholar]
- 16.Wang F, Ma YL, Zhang P, et al. A genetic variant in microRNA-196a2 is associated with increased cancer risk: a meta-analysis. Mol Biol Rep 2012; 39:269–275. [DOI] [PubMed] [Google Scholar]
- 17.Qiu MT, Hu JW, Ding XX, et al. Hsa-miR-499 rs3746444 polymorphism contributes to cancer risk: a meta-analysis of 12 studies. PLoS One 2012; 7:e50887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitehead A, Whitehead J. A general parametric approach to the meta-analysis of randomized clinical trials. Stat Med 1991; 10:1665–1677. [DOI] [PubMed] [Google Scholar]
- 19.Panoutsopoulou K, Southam L, Elliott KS, et al. Insights into the genetic architecture of osteoarthritis from stage 1 of the arcOGEN study. Ann Rheum Dis 2011; 70:864–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raine EV, Dodd AW, Reynard LN, et al. Allelic expression analysis of the osteoarthritis susceptibility gene COL11A1 in human joint tissues. BMC Musculoskelet Disord 2013; 14:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bravata V, Minafra L, Forte GI, et al. DVWA gene polymorphisms and osteoarthritis. BMC Res Notes 2015; 8:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyamoto Y, Shi D, Nakajima M, et al. Common variants in DVWA on chromosome 3p24.3 are associated with susceptibility to knee osteoarthritis. Nat Genet 2008; 40:994–998. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi H, Nakajima M, Ozaki K, et al. Prediction model for knee osteoarthritis based on genetic and clinical information. Arthritis Res Ther 2010; 12:R187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SJ, Kim MJ, Kee SJ, et al. Association study of the candidate gene for knee osteoarthritis in Koreans. Rheumatol Int 2013; 33:783–786. [DOI] [PubMed] [Google Scholar]
- 25.Zhu L, Shi D, Dai J, et al. Lack of evidence for association between DVWA gene polymorphisms and developmental dysplasia of the hip in Chinese Han population. Rheumatol Int 2011; 31:883–887. [DOI] [PubMed] [Google Scholar]
- 26.Saito T, Fukai A, Mabuchi A, et al. Transcriptional regulation of endochondral ossification by HIF-2alpha during skeletal growth and osteoarthritis development. Nat Med 2010; 16:678–686. [DOI] [PubMed] [Google Scholar]
- 27.Miller RE, Tran PB, Das R, et al. CCR2 chemokine receptor signaling mediates pain in experimental osteoarthritis. Proc Natl Acad Sci U S A 2012; 109:20602–20607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawitzke AD. Personalized medicine for osteoarthritis: where are we now? Ther Adv Musculoskelet Dis 2013; 5:67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chapman K, Takahashi A, Meulenbelt I, et al. A meta-analysis of European and Asian cohorts reveals a global role of a functional SNP in the 5’ UTR of GDF5 with osteoarthritis susceptibility. Hum Mol Genet 2008; 17:1497–1504. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez RR, Seegmiller RE, Stark MR, et al. A type XI collagen mutation leads to increased degradation of type II collagen in articular cartilage. Osteoarthritis Cartilage 2004; 12:314–320. [DOI] [PubMed] [Google Scholar]
- 31.He XF, Wei W, Liu ZZ, et al. Association between the CYP1A1 T3801C polymorphism and risk of cancer: evidence from 268 case-control studies. Gene 2014; 534:324–344. [PubMed] [Google Scholar]
- 32.Qiao L, Liang Y, Mira RR, et al. Mammalian target of rapamycin (mTOR) inhibitors and combined chemotherapy in breast cancer: a meta-analysis of randomized controlled trials. Int J Clin Exp Med 2014; 7:3333–3343. [PMC free article] [PubMed] [Google Scholar]
- 33.Valdes AM, Lories RJ, van Meurs JB, et al. Variation at the ANP32A gene is associated with risk of hip osteoarthritis in women. Arthritis Rheum 2009; 60:2046–2054. [DOI] [PubMed] [Google Scholar]
- 34.Zeggini E, Panoutsopoulou K, Southam L, et al. Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet 2012; 380:815–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mototani H, Iida A, Nakamura Y, et al. Identification of sequence polymorphisms in CALM2 and analysis of association with hip osteoarthritis in a Japanese population. J Bone Miner Metab 2010; 28:547–553. [DOI] [PubMed] [Google Scholar]
- 36.Evangelou E, Kerkhof HJ, Styrkarsdottir U, et al. A meta-analysis of genome-wide association studies identifies novel variants associated with osteoarthritis of the hip. Ann Rheum Dis 2014; 73:2130–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez-Fontenla C, Calaza M, Evangelou E, et al. Assessment of osteoarthritis candidate genes in a meta-analysis of nine genome-wide association studies. Arthritis Rheumatol 2014; 66:940–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakki A, Videman T, Kujala UM, et al. Candidate gene association study of magnetic resonance imaging-based hip osteoarthritis (OA): evidence for COL9A2 gene as a common predisposing factor for hip OA and lumbar disc degeneration. J Rheumatol 2011; 38:747–752. [DOI] [PubMed] [Google Scholar]
- 39.Meulenbelt I, Min JL, Bos S, et al. Identification of DIO2 as a new susceptibility locus for symptomatic osteoarthritis. Hum Mol Genet 2008; 17:1867–1875. [DOI] [PubMed] [Google Scholar]
- 40.Castano Betancourt MC, Cailotto F, Kerkhof HJ, et al. Genome-wide association and functional studies identify the DOT1L gene to be involved in cartilage thickness and hip osteoarthritis. Proc Natl Acad Sci U S A 2012; 109:8218–8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Limer KL, Tosh K, Bujac SR, et al. Attempt to replicate published genetic associations in a large, well-defined osteoarthritis case-control population (the GOAL study). Osteoarthritis Cartilage 2009; 17:782–789. [DOI] [PubMed] [Google Scholar]
- 42.Evans DS, Cailotto F, Parimi N, et al. Genome-wide association and functional studies identify a role for IGFBP3 in hip osteoarthritis. Ann Rheum Dis 2015; 74:1861–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L, Guo L, Tian F, et al. Analysis of single nucleotide polymorphisms within ADAM12 and risk of knee osteoarthritis in a Chinese Han population. BioMed Res Int 2015; 2015:518643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez-Lopez J, Pombo-Suarez M, Loughlin J, et al. Association of a nsSNP in ADAMTS14 to some osteoarthritis phenotypes. Osteoarthritis Cartilage 2009; 17:321–327. [DOI] [PubMed] [Google Scholar]
- 45.Liang W, Gao B, Xu G, et al. Association between single nucleotide polymorphisms of asporin (ASPN) and BMP5 with the risk of knee osteoarthritis in a Chinese Han population. Cell Biochem Biophys 2014; 70:1603–1608. [DOI] [PubMed] [Google Scholar]
- 46.Ikeda T, Mabuchi A, Fukuda A, et al. Association analysis of single nucleotide polymorphisms in cartilage-specific collagen genes with knee and hip osteoarthritis in the Japanese population. J Bone Miner Res 2002; 17:1290–1296. [DOI] [PubMed] [Google Scholar]
- 47.Evangelou E, Valdes AM, Kerkhof HJ, et al. Meta-analysis of genome-wide association studies confirms a susceptibility locus for knee osteoarthritis on chromosome 7q22. Ann Rheum Dis 2011; 70:349–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren Y, Tan B, Yan P, et al. Association between polymorphisms in the estrogen receptor alpha gene and osteoarthritis susceptibility: a meta-analysis. BMC Musculoskelet Disord 2015; 16:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee SW, Song JH, Choi WS, et al. The single nucleotide polymorphism (SNP) of the estrogen receptor-beta gene, rs1256049, is associated with knee osteoarthritis in Korean population. Knee 2014; 21:242–246. [DOI] [PubMed] [Google Scholar]
- 50.Zhang R, Yao J, Xu P, et al. A comprehensive meta-analysis of association between genetic variants of GDF5 and osteoarthritis of the knee, hip and hand. Inflamm Res 2015; 64:405–414. [DOI] [PubMed] [Google Scholar]
- 51.Kerkhof HJ, Lories RJ, Meulenbelt I, et al. A genome-wide association study identifies an osteoarthritis susceptibility locus on chromosome 7q22. Arthritis Rheum 2010; 62:499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fernandez-Torres J, Hernandez-Diaz C, Espinosa-Morales R, et al. Polymorphic variation of hypoxia inducible factor-1 A (HIF1A) gene might contribute to the development of knee osteoarthritis: a pilot study. BMC Musculoskelet Disord 2015; 16:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luo SX, Li S, Zhang XH, et al. Genetic polymorphisms of interleukin-16 and risk of knee osteoarthritis. PLoS One 2015; 10:e0123442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han L, Lee HS, Yoon JH, et al. Association of IL-17A and IL-17F single nucleotide polymorphisms with susceptibility to osteoarthritis in a Korean population. Gene 2014; 533:119–122. [DOI] [PubMed] [Google Scholar]
- 55.Fernandes MT, Fernandes KB, Marquez AS, et al. Association of interleukin-6 gene polymorphism (rs1800796) with severity and functional status of osteoarthritis in elderly individuals. Cytokine 2015; 75:316–320. [DOI] [PubMed] [Google Scholar]
- 56.Nakki A, Rodriguez-Fontela C, Gonzalez A, et al. Association study of MMP8 gene in osteoarthritis. Connect Tissue Res 2015; Nov 17:1-9. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 57.Valdes AM, Spector TD, Tamm A, et al. Genetic variation in the SMAD3 gene is associated with hip and knee osteoarthritis. Arthritis Rheum 2010; 62:2347–2352. [DOI] [PubMed] [Google Scholar]
- 58.Etokebe GE, Jotanovic Z, Mihelic R, et al. Susceptibility to large-joint osteoarthritis (hip and knee) is associated with BAG6 rs3117582 SNP and the VNTR polymorphism in the second exon of the FAM46A gene on chromosome 6. J Orthopaed Res 2015; 33:56–62. [DOI] [PubMed] [Google Scholar]
- 59.Cui L, Li G, Zhong W, et al. Polycystic ovary syndrome susceptibility single nucleotide polymorphisms in women with a single PCOS clinical feature. Hum Reprod 2015; 30:732–736. [DOI] [PubMed] [Google Scholar]
- 60.Schneider EM, Du W, Fiedler J, et al. The (-765 G–>C) promoter variant of the COX-2/PTGS2 gene is associated with a lower risk for end-stage hip and knee osteoarthritis. Ann Rheum Dis 2011; 70:1458–1460. [DOI] [PubMed] [Google Scholar]
- 61.He Y, Liang X, Wu X, et al. Association between interleukin 8-251 A/T and +781 C/T polymorphisms and osteoarthritis risk. Immunol Lett 2014; 162:207–211. [DOI] [PubMed] [Google Scholar]
- 62.Spector TD, Reneland RH, Mah S, et al. Association between a variation in LRCH1 and knee osteoarthritis: a genome-wide single-nucleotide polymorphism association study using DNA pooling. Arthritis Rheum 2006; 54:524–532. [DOI] [PubMed] [Google Scholar]
- 63.Zhu ZH, Jin XZ, Zhang W, et al. Associations between vitamin D receptor gene polymorphisms and osteoarthritis: an updated meta-analysis. Rheumatology (Oxford) 2014; 53:998–1008. [DOI] [PubMed] [Google Scholar]
- 64.Cong Y, Ru JY, Bao NR, et al. A single nucleotide polymorphism in the TGF-beta1 gene (rs1982073 C>T) may contribute to increased risks of bone fracture, osteoporosis, and osteoarthritis: a meta-analysis. Clin Rheumatol 2014. [DOI] [PubMed] [Google Scholar]
- 65.Zhang R, Yao J, Xu P, et al. Association between genetic variants of DVWA and osteoarthritis of the knee and hip: a comprehensive meta-analysis. Int J Clin Exp Med 2015; 8:9430–9437. [PMC free article] [PubMed] [Google Scholar]
- 66.Wang D, Zhou K, Chen Z, et al. The association between DVWA polymorphisms and osteoarthritis susceptibility: a genetic meta-analysis. Int J Clin Exp Med 2015; 8:12566–12574. [PMC free article] [PubMed] [Google Scholar]


