Abstract
MicroRNAs (miRNAs) play an important role as regulators of tumor suppressors and oncogenes in cancer-related processes. Single nucleotide polymorphisms (SNPs) in miRNAs have been shown to be relevant to various different cancers, including breast cancer (BC). The aim of this study was to estimate the associations between miRNA-related gene polymorphisms (miR-196a2, miR-499, and miR-608) and the risk of BC in a Chinese population.
Gene polymorphisms were analyzed in 1143 subjects (controls = 583; BC = 560). The 3 SNPs were genotyped using the Sequenom Mass-ARRAY platform. The associations between the SNP frequencies and BC were assessed by computing odds ratios (ORs) and 95% confidence intervals (95% CIs), as well as by applying Chi-square tests.
The miR-196a2 (rs11614913) T allele was associated with a decreased risk of BC based on results from dominant (OR = 0.67, 95% CI = 0.52–0.86), recessive (OR = 0.65, 95% CI = 0.48–0.86), and allele models (OR = 0.73, 95% CI = 0.62–0.86). In contrast, the miR-499 (rs3746444) AG/GG genotypes were associated with an increased risk of BC (OR = 1.45, 95% CI = 1.10–1.91), and miR-608 (rs4919510) was not significantly associated with BC risk.
Our study suggested that the polymorphisms of rs11614913 and rs3746444 may be associated with BC risk in Chinese individuals.
INTRODUCTION
MicroRNAs (miRNAs) are a class of newly identified nonprotein-coding transcripts that are present in the cells of many species, ranging from worms to humans.1–3 miRNAs are approximately 22 nucleotide sequences long and participate in the regulation of several biological functions, including cell differentiation, cell cycle progression, and apoptosis.1,4,5 It is well known that miRNAs regulate the expression of approximately 50% of human genes.6 Interestingly, more than 52.5% of miRNA genes are located within carcinoma-related genetic regions or in fragile sites, which implies that miRNAs may play a pivotal role in the pathogenesis of a limited range of human diseases, especially including many cancers.7,8 However, the study of correlations between miRNAs and human cancers is in its infancy.9
Recently, 1872 precursors and 2578 mature miRNAs have been identified and described in humans.10 Numerous studies have indicated that aberrant expression of miRNAs may influence diverse carcinogenic processes through mRNA targets that encode oncogenes or tumor suppressor genes for several types of cancer, including breast cancer (BC).11,12 Some miRNAs, such as miR-183, miR-494, and miR-21, were upregulated in metastatic BC tissues which was associated with a poor prognosis. These data indicated that these miRNAs represent new risk biomarkers of metastatic BC and may be useful for future-targeted studies.13
The role of single nucleotide polymorphisms (SNPs) in miRNAs has been regarded as being potentially linked with many different cancers. As the most frequent cancer among women, BC is an important topic for studies of the links between miRNAs and human cancers. While previous studies have investigated the relationships between common SNPs in miRNA genes and BC (such as for rs11614913 in miR-196a2, rs3746444 in miR-499, and rs4919510 in miR-608), only some of these studies have found significant associations with increased susceptibility to BC. Recently, 1 case-control study indicated that polymorphisms of rs11614913 and rs3746444 were associated with susceptibility to BC in southern Chinese women.14 However, another previous study suggested that SNPs rs11614913 and rs3746444 were not associated with BC risk or age at BC onset.15 To date, only one case-control study has focused on the link between variant genotypes of rs4919510 and BC risk, and negative results were observed in the overall population.16 Thus, we decided to genotype 3 different SNPs (rs11614913, rs3746444, and rs4919510) and assess their associations with BC risk in independent case-control sets of Chinese women.
MATERIALS AND METHODS
Patients and Controls
The cases were recruited from the Department of Oncology of the Second Affiliated Hospital of Xi’an Jiaotong University, Xi’an City, China. The eligible patients were women who had been newly diagnosed with BC that was treated at the hospital. These women were consecutively recruited between January 2012 and October 2014 with an overall response rate of 92.6% (560/605). All subjects had nonfamilial cases of BC and were Han Chinese individuals from Xi’an City and surrounding regions in Shaanxi Province. Women were excluded from the study if they had a self-reported history of prior cancer or a self-reported history of prior radiotherapy and/or chemotherapy for unknown conditions. The patient cohort consisted of 560 subjects (mean age 49.09 ± 11.02 years) with histologically confirmed BC, whereas the control cohort included a total of 583 cancer-free female volunteers who were recruited from the same hospital and had a similar age distribution (mean age 48.80 ± 8.28 years). All study subjects were genetically unrelated Chinese individuals.
The study was approved by the Institutional Review Board of Xi’an Jiaotong University (Xi’an, China). The study methods were carried out in accordance with approved guidelines.17 All of the participants were surveyed using a self-administered questionnaire after written informed consent had been obtained. After the survey, about 2 mL of venous blood was collected from each subject.
DNA Extraction and Genotyping
The blood samples were collected in tubes containing ethylenediaminetetraacetic acid. Subsequently, the samples were centrifuged at 8000g for 180 s at room temperature. After centrifugation, the specimens were stored at −80°C. Genomic DNA from each sample was isolated from the leucocytes of the peripheral blood using the Qiagen DNA Blood Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. We selected 3 tag SNPs for genotyping in this study: rs11614913, rs3746444, and rs4919510. These SNPs, which captured the majority of known common variations of miRNAs, were listed in the public database of the Chinese population from HapMap (http://www.hapmap.org). SNP genotyping was performed using the Sequenom MassARRAY RS1000 (Sequenom, San Diego, CA) according to the standard protocol recommended by the manufacturer.18
The following, corresponding primers were used for each SNP: for rs11614913, forward primer 5′-ACGTTGGATGTCGACGAAAACCGACTGATG-3′ and reverse primer 5′-ACGTTGGATGCTGATCTGTGGCTTAGGTAG-3′; for rs3746444, forward primer 5′-ACGTTGGATGACGGGAAGCAGCACAGACTT-3′ and reverse primer 5′-ACGTTGGATGGGCTGTTAAGACTTGCAGTG-3′; and for rs4919510, forward primer 5′-ACGTTGGATGATTCCCAAGATCCACTGGGC-3′ and reverse primer 5′-ACGTTGGATGATGGAAGCTCTTGGAGATGC-3′. Sequenom MassARRAY Assay Design 3.0 software was used for the data analyses,18,19 and the data were managed using Sequenom Typer 4.0 software.
Statistical Analysis
Differences in demographic variables and risk factors between BC cases and controls were compared using Student t test for continuous variables and the Chi-square test for categorical variables. The Hardy-Weinberg equilibrium (HWE) of the control subjects was evaluated for each of the SNPs using a goodness-of-fit Chi-square test before the analysis. The associations between miRNA SNPs, BC risk, and the patients’ clinical characteristics were determined by computing the odds ratios (ORs) and 95% confidence intervals (CIs) from both univariate and multivariate logistic regression analyses. Stratified analyses was used to assess BC risk in subgroups based on tumor size, axillary lymph node metastasis, progesterone receptor expression, estrogen receptor expression, and human epidermal growth factor receptor 2 (HER2) expression. The analysis was carried out using co-dominant, dominant, recessive, and allele models.
All of the statistical analyses were performed using SPSS version 18.0 for Windows (PASW Statistics, SPSS Inc, Chicago, IL). The statistical power was calculated to evaluate the significant findings. Only the significant result with a statistical power value >0.8 was considered a noteworthy finding. The statistical power of the case-control study was calculated using QUANTO software 1.2.4 (University of Southern California, Los Angeles, CA; http://biostats.usc.edu/Quanto.html). All statistical tests were 2-sided, P-values <0.05 were considered statistically significant.
RESULTS
General Characteristics of the Subjects
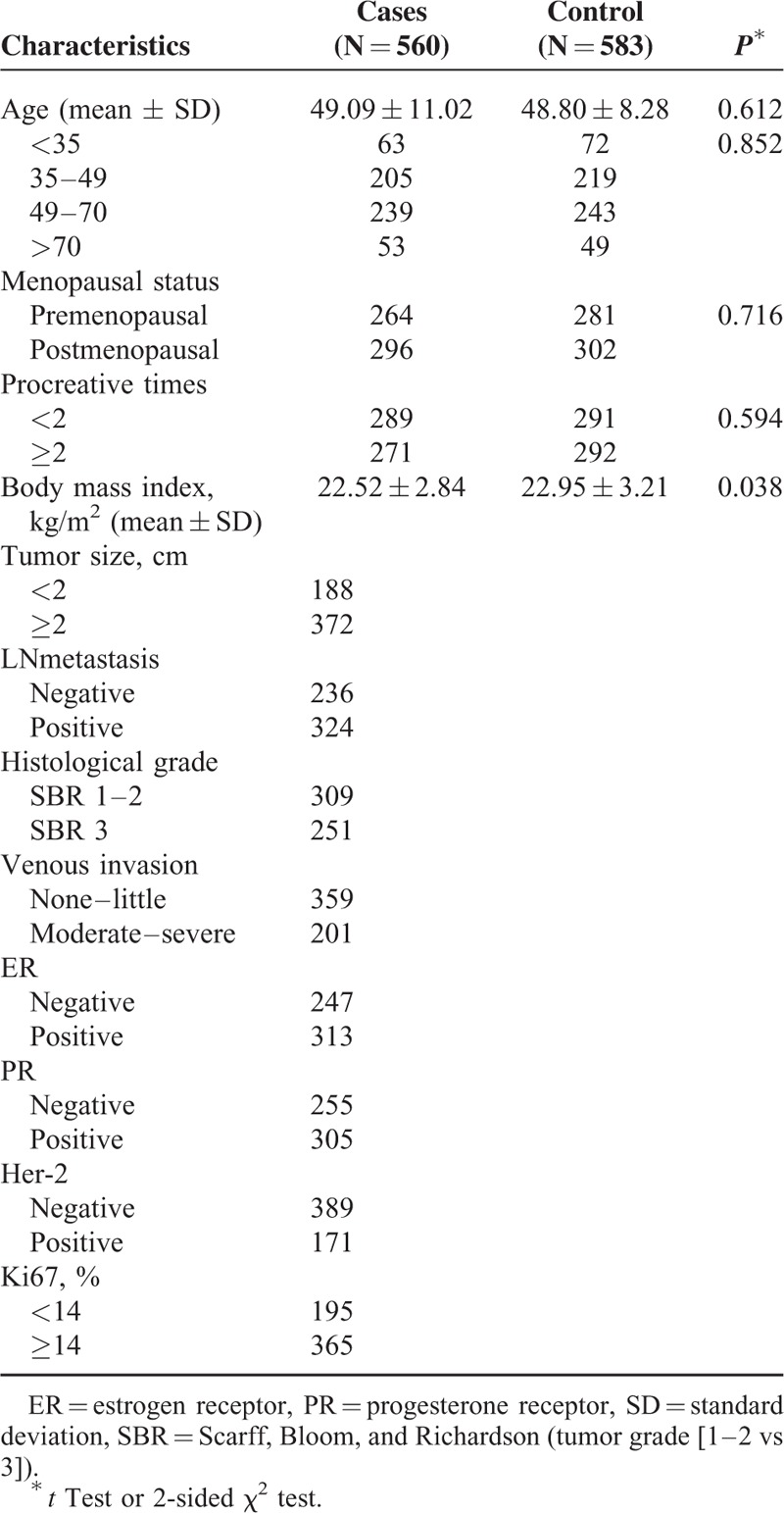
The characteristics of both the cases and controls are presented in Table 1. The study group involved 560 BC patients with an average age of 49.09 ± 11.02 years and 583 healthy subjects with an average age of 48.80 ± 8.28 years (P = 0.26). There was no significant difference in the distributions of menopausal status or procreative times between the case and control groups (P > 0.05). However, body mass index was significantly higher in the control group than in the case group (22.95 ± 3.21 vs 22.52 ± 2.84; P = 0.038), which might have resulted from weight loss after the onset of breast cancer. In light of this difference, our statistical analysis of the case-control comparisons was adjusted for body mass index.
TABLE 1.
Frequency Distribution of Selected Variables in Breast Cancer Cases and Cancer-free Controls
Associations of miRNA SNPs and Risk of BC
The results indicated that all 3 of the SNPs were in HWE among the controls (rs11614913, P = 0.54; rs3746444, P = 0.13; and rs4919510, P = 0.98), which excludes the possibility of experimental artefacts. The frequencies of genotypes and alleles of the 3 miRNA SNPs in BC patients and healthy controls are shown in Table 2. For rs11614913, the minor allele T was associated with a decreased risk of BC based on results from dominant (OR = 0.67, 95% CI = 0.52–0.86, P = 0.002), recessive (OR = 0.65, 95% CI = 0.48–0.86, P = 0.003), and allele models (OR = 0.73, 95% CI = 0.62–0.86, P = 0.0002). In the analysis of rs3746444, the individuals who carried the AG or GG genotype had an increased risk of BC as compared with the individuals who had wild type AA (OR = 1.45, 95% CI = 1.10–1.91, P = 0.008). For rs4919510, no significant associations were observed in any of the genetic comparison models of controls and BC patients.
TABLE 2.
Genotype Frequencies of miR-196a2, miR-499, miR-608 SNPs in Controls and Breast Cancer Patients
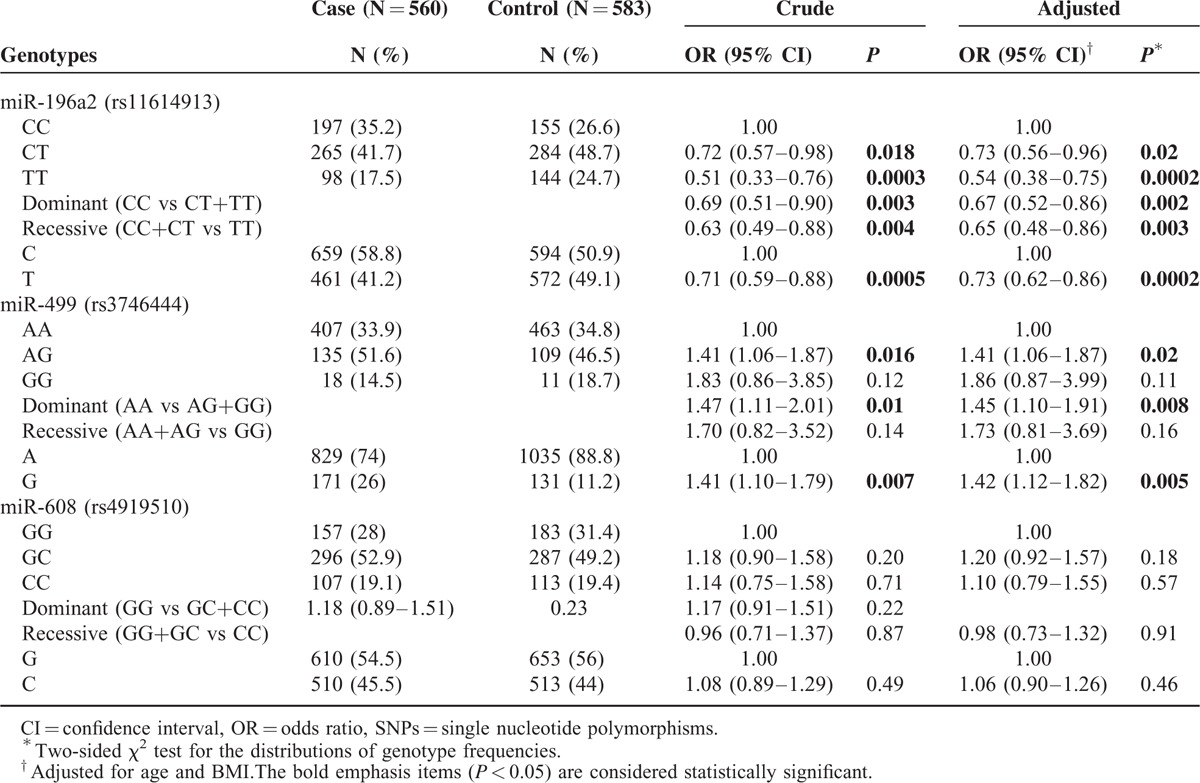
We also obtained the statistical power of 0.876 and 0.824 for the 2 significant polymorphisms identified, rs11614913 and rs3746444, respectively. This showed that our sample size of 1143 was adequate and the study was sufficiently able to detect the true association of these 2 polymorphisms with BC.
Associations Between rs3746444 Polymorphism and Clinical Parameters of BC Patients
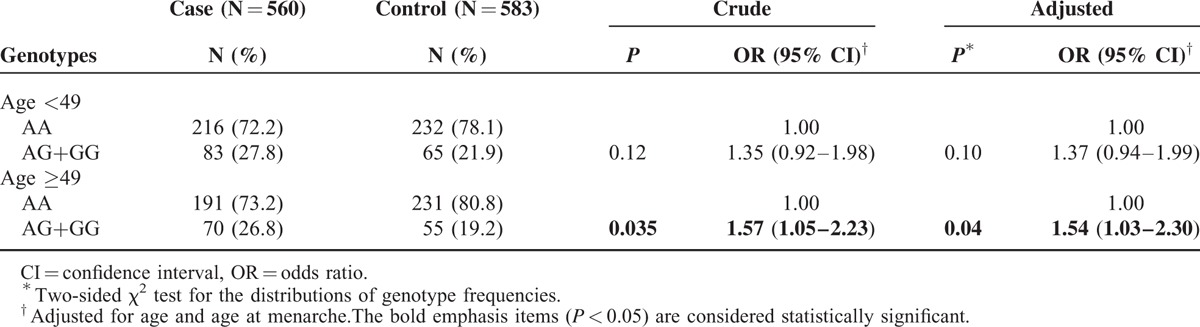
As shown in Table 3, we performed an age-stratified analysis of the associations between rs3746444 polymorphisms in miR-499 and BC. The results indicated that the rs3746444 variant genotypes (AG/GG) were significantly increased in older subjects (OR = 1.54, 95% CI = 1.03–2.30, P = 0.04). The same analyses were also conducted for the polymorphisms of rs11614913 and rs4919510; however, no statistically significant result was observed (data not shown).
TABLE 3.
Stratification Analyses on Age between miR-499 Polymorphism and Breast Cancer Risk
Stratified Analysis of the 3 Polymorphisms and BC Risk
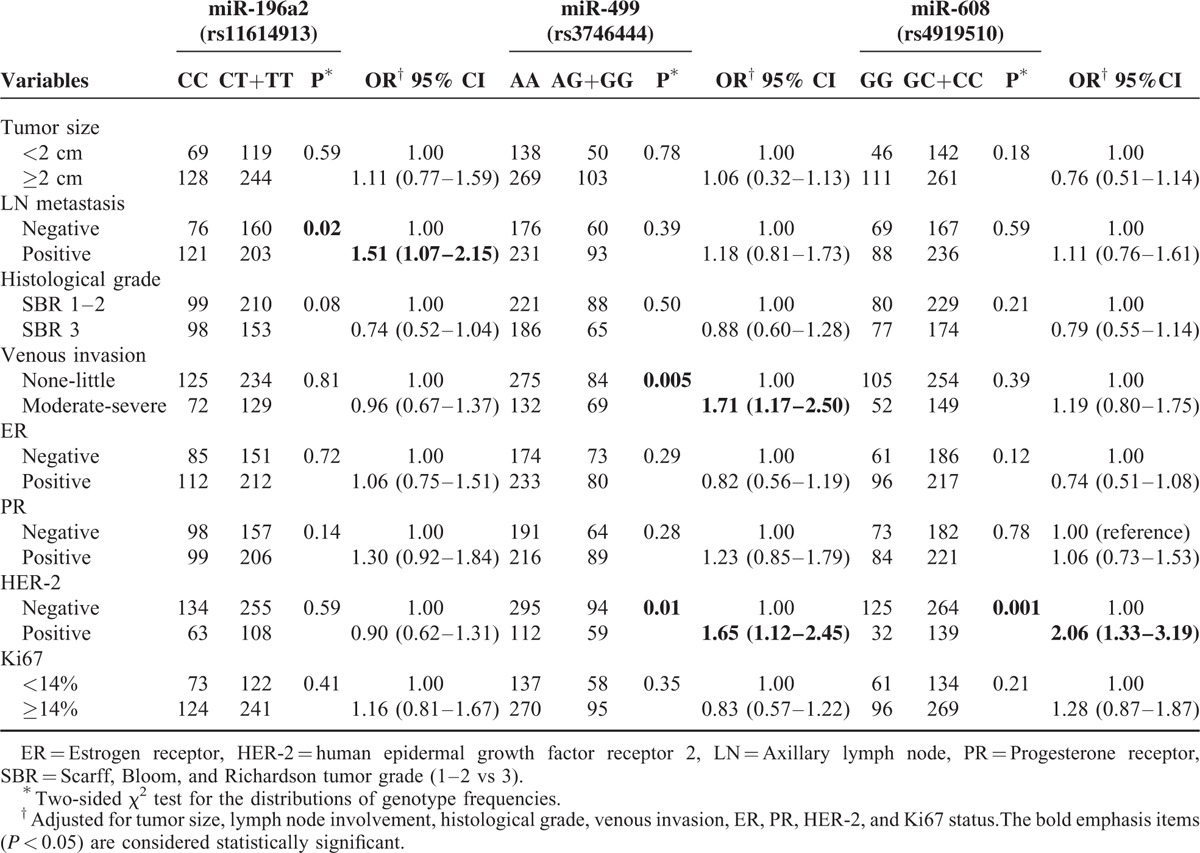
We analyzed the associations between the 3 SNPs and a series of clinicopathologic features, including tumor size, lymph node metastasis, histological grade, venous invasion, estrogen receptor status, progesterone receptor status, HER-2 status, and Ki67 index (Table 4). We found that the increased risk associated with the rs3746444 variant genotypes (AG/GG) was more pronounced in patients who were positive for HER-2 (OR = 1.65, 95% CI = 1.12–2.45, P = 0.01) and in those with moderate-severe vascular invasion (OR = 1.71, 95% CI = 1.17–2.50, P = 0.005). The risk associated with rs4919510 GC/CC variant genotypes was also significantly higher in patients who were positive for HER-2 (OR = 2.06, 95% CI = 1.33–3.19, P = 0.001). In addition, a significant association was observed between rs11614913 polymorphisms and patients who had lymph node metastasis (OR = 1.51, 95% CI = 1.07–2.15, P = 0.02).
TABLE 4.
Stratified Analyses on Association between miR-196a2, miR-499, and miR-608 Polymorphism and Clinical Characteristics of Breast Cancer Patients
DISCUSSION
It has recently been shown that, as pathway regulators, miRNAs are involved in several human biological processes and that abnormal expression of miRNAs is associated with many human malignancies.20,21 Further, SNPs or mutations in the genes encoding miRNAs may influence the translation of a target mRNA and contribute to its aberrant expression, thereby potentially affecting the risk of developing cancer.22 Located in the mature sequence of miR-196a-3P, rs11614913 variation could lead to less efficient processing of the miRNA precursor to its mature form and diminished capacity to regulate target genes.23 Hsa-mir-499 rs3746444 A > G polymorphism-resulted mismatch may affect target mRNA expression. Epidemiological studies found that various effects of rs3746444 polymorphism on different genes may result in different associations with diseases such as cancer at phenotype level.24
We conducted a case-control study to explore polymorphisms in miR-196a2 (rs11614913), miR-499 (rs3746444), and miR-608 (rs4919510) and their associations with BC susceptibility. We included 583 controls and 560 BC patients, all of whom were of Chinese descent. The results showed that the rs11614913 variation (CT/TT) was significantly associated with a decreased risk of BC. Further, the rs3746444 variant genotypes (AG/GG) were associated with a significantly increased risk of BC, as compared with the TT genotype. However, rs4919510 polymorphism had no significant association with BC susceptibility. The results suggest that common SNPs in miRNAs, as candidate biomarkers, could contribute to BC susceptibility in Chinese women.
Currently, gene polymorphism of rs11614913 is one of the most investigated SNPs in case-control studies of several types of cancer. Genotype-phenotype correlation analyses found that the CC homozygote in miR-196a2 was associated with significantly increased miR-196a expression.25,26 miR-196a has been identified as partially directing the cleavage of the mRNA of the HOX gene clusters as an upstream regulator.27 Recent data have shown that HOX genes were expressed aberrantly and that HOXD10 initiated tumor invasion and metastasis in BC.28 Using a whole-genome expression microarray, Hoffman et al23 showed significantly more enhanced gene expression in the C allele of rs11614913 than in the T allele, indicating that the miR-196a2 variant might have a potential oncogene role in breast tumorigenesis. Recent findings by Hu et al14 have indicated that CC/CT genotypes were associated with significantly increased risks of BC, as compared with the TT genotype. However, the link between rs11614913 and BC risk was not observed in other studies by Catucci et al15 and Jedlinski et al.29 Furthermore, the results of several meta-analyses showed that the CC genotype of rs11614913 polymorphism had significant associations with an increased risk of BC,30–34 and our results supported these findings.
Considering the existing evidence regarding miR-499 (rs3746444) in cancer development, we evaluated related associations in our study of patients with BC. The rs3746444 polymorphism, which is located at the 3p mature miRNA regions of miR-499a, involves an A to G nucleotide substitution and influences the binding of target mRNAs to 3p mature miRNAs. miR-499 can target regulation of the expression of FOXO4, PDCD4, and SOX6 genes, which play important roles in the etiology of cancers.35,36 Recent analyses of rs3746444 polymorphisms in BC patients have shown mixed results. Some studies reported that rs3746444 was associated with increased BC risk for multiple ethnicities.37–39 In contrast, negative results were observed in Caucasian15 and in Chinese individuals.40 Although some meta-analytic results showed that the association was more notable in Asian populations than in Caucasian populations,32,41 other meta-analyses found no significant association between rs3746444 and BC risk.34,42 In our study, the results suggested that individuals who carried rs3746444 AG/GG genotypes had an increased BC risk. Moreover, the rs3746444 AG/GG genotypes were revealed to be associated with an increased risk of moderate-severe vascular invasion and positive HER-2 in BC. Interestingly, our subgroup analysis showed that the increased risk associated with the rs3746444 AG/GG genotypes was more prominent in individuals older than 49 years. It is commonly known that individuals become more susceptible to many types of cancer with increasing age. However, the finding regarding rs3746444 should be interpreted with caution because of the finite number of subjects in this study.
The rs4919510 polymorphism in miR-608 has been poorly investigated in BC, as compared with other types of cancer. A recent meta-analysis did not find significant associations with rs4919510 polymorphism, in terms of either the overall risk of cancer or the risks of specific types of cancer.43 A case-control study that included 1138 patients with sporadic BC and 1434 community-based controls found no significant association with BC risk, but variant genotypes (GC/GG) were significantly associated with an increased risk of HER2-positive BC. Our study supports the negative findings of this previous study in that no significant association was detected between the rs4919510 polymorphism and the risk of BC. Furthermore, when analyzing the associations between the rs4919510 variant and clinicopathologic features of BC, we also found that GC/GG genotypes were specifically associated with an increased risk of HER2-positive BC. The association between rs4919510 and prognosis has also been investigated in BC patients, and the results showed that rs4919510 polymorphisms were significantly correlated with recurrence and survival.44
The present study had several limitations that should be acknowledged. First, the numbers of enrolled women with and without BC were not large enough for this genotyping study to be conclusive. Therefore, the associations that were observed for the 3 SNPs should be investigated in further, larger-scale studies. Second, since most of the women in the control group came to our hospital for breast examinations, the enrolled control group might have had an elevated prevalence of benign breast disease. More generally, the choice of the control group could have biased the analysis because it was composed of hospital-based patients without previous cancer histories. It is important to note that all subjects in the case and control groups were genotyped using the same genotyping platform and comparable technical procedures, in order to control for errors associated with the detection method.
In summary, our study found significant associations between miR-196a2 (rs11614913) and miR-499 (rs3746444) and the risk of BC in a Chinese population. These results have provided us with further motivation to investigate and clarify the function of miRNA SNPs in normal and malignant human cells. However, the present study had limitations that should be acknowledged when considering its conclusions. Data were often unavailable on factors that are commonly associated with gene–environment interactions, such as alcohol consumption and smoking, which prevented us from performing stratified analyses for these factors. Further, sample size is an important parameter in any study of gene polymorphisms. The BC and control groups were not large enough for this genotyping study to be conclusive; therefore, studies with larger patient cohorts are needed to confirm the roles of miR-196a2, miR-499, and miR-608 SNPs in BC risk.
Acknowledgments
The authors would like to thank Editage for language editing.
Footnotes
Abbreviations: BC = breast cancer, BMI = body mass index, CI = confidence interval, ER = estrogen receptor, HER-2 = human epidermal growth factor receptor-2, HWE = Hardy-Weinberg equilibrium, OR = odds ratio, PR = progesterone receptor, SBR = Scarff, Bloom and Richardson, SNP = single-nucleotide polymorphism.
Author contributions: DZJ, DZM, and KHF designed the research. DZM, LHB, ZSQ, MXB, LS, and LK performed the experiments throughout this research. LHB, ZWG, WM, and LXH contributed to the reagents and participated in its design and coordination. DZM and FYJ analyzed the data; DZM, LHB, and XP contributed to the writing of the article. Co-first authors: DZM, KHF and ZWG. All authors have read and approved the final article.
Z-MD, H-FK, and W-GZ contributed equally to this work and share joint first authorship.
This study was supported by National Natural Science Foundation, China (No. 81471670; 81274136); China Postdoctoral Science Foundation (No. 2014M560791; 2015T81037); the Fundamental Research Funds for the Central Universities, China (No. 2014qngz-04); and specialized Research Fund of the Second Affiliated Hospital of Xi’an Jiaotong University, China [RC (GG) 201203].
The authors have no conflict of interest to disclose.
REFERENCES
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75:843–854. [DOI] [PubMed] [Google Scholar]
- 2.Pasquinelli AE. Demystifying small RNA pathways. Dev Cell 2006; 10:419–424. [DOI] [PubMed] [Google Scholar]
- 3.Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000; 403:901–906. [DOI] [PubMed] [Google Scholar]
- 4.Liu JD. Control of protein synthesis and mRNA degradation by microRNAs. Curr Opin Cell Biol 2008; 20:214–221. [DOI] [PubMed] [Google Scholar]
- 5.Chen T, Li ZG, Yan J, et al. MicroRNA expression profiles distinguish the carcinogenic effects of riddelliine in rat liver. Mutagenesis 2012; 27:59–66. [DOI] [PubMed] [Google Scholar]
- 6.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010; 11:597–610. [DOI] [PubMed] [Google Scholar]
- 7.Zhang BH, Pan XP, Cobb GP, et al. microRNAs as oncogenes and tumor suppressors. Dev Biol 2007; 302:1–12. [DOI] [PubMed] [Google Scholar]
- 8.Nugent M, Miller N, Kerin MJ. MicroRNAs in colorectal cancer: function, dysregulation and potential as novel biomarkers. Eur J Surg Oncol 2011; 37:649–654. [DOI] [PubMed] [Google Scholar]
- 9.Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res 2005; 65:3509–3512. [DOI] [PubMed] [Google Scholar]
- 10.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 2011; 39:D152–D157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 2006; 6:259–269. [DOI] [PubMed] [Google Scholar]
- 12.Lagos-Quintana M, Rauhut R, Meyer J, et al. New microRNAs from mouse and human. RNA 2003; 9:175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marino AL, Evangelista AF, Vieira RA, et al. MicroRNA expression as risk biomarker of breast cancer metastasis: a pilot retrospective case-cohort study. BMC Cancer 2014; 14:739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Z, Liang J, Wang Z, et al. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum Mutat 2009; 30:79–84. [DOI] [PubMed] [Google Scholar]
- 15.Catucci I, Yang R, Verderio P, et al. Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as low-penetrance alleles in German and Italian familial breast cancer cases. Hum Mutat 2010; 31:E1052–E1057. [DOI] [PubMed] [Google Scholar]
- 16.Huang AJ, Yu KD, Li J, et al. Polymorphism rs4919510:C > G in Mature Sequence of Human MicroRNA-608 Contributes to the Risk of HER2-Positive Breast Cancer but Not Other Subtypes. Plos One 2012; 7:e35252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao J, Kang HF, Ma XB, et al. Functional promoter -765 G > C variant in COX-2 gene is associated with the susceptibility of breast cancer in Chinese Han women. Cancer Cell Int 2014; 14:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet 2009; 12: Chapter 2,Unit 2. [DOI] [PubMed] [Google Scholar]
- 19.Thomas RK, Baker AC, Debiasi RM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet 2007; 39:347–351. [DOI] [PubMed] [Google Scholar]
- 20.Baltimore D, Boldin MP, O’Connell RM, et al. MicroRNAs: new regulators of immune cell development and function. Nat Immunol 2008; 9:839–845. [DOI] [PubMed] [Google Scholar]
- 21.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer 2010; 10:389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Negrini M, Nicoloso MS, Calin GA. MicroRNAs and cancer—new paradigms in molecular oncology. Curr Opin Cell Biol 2009; 21:470–479. [DOI] [PubMed] [Google Scholar]
- 23.Hoffman AE, Zheng T, Yi C, et al. microRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res 2009; 69:5970–5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005; 433:769–773. [DOI] [PubMed] [Google Scholar]
- 25.Xu J, Hu Z, Xu Z, et al. Functional variant in microRNA-196a2 contributes to the susceptibility of congenital heart disease in a Chinese population. Hum Mutat 2009; 30:1231–1236. [DOI] [PubMed] [Google Scholar]
- 26.Hu Z, Chen J, Tian T, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest 2008; 118:2600–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mansfield JH, Harfe BD, Nissen R, et al. MicroRNA-responsive 'sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet 2004; 36:1079–1083. [DOI] [PubMed] [Google Scholar]
- 28.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science 2004; 304:594–596. [DOI] [PubMed] [Google Scholar]
- 29.Jedlinski DJ, Gabrovska PN, Weinstein SR, et al. Single nucleotide polymorphism in hsa-mir-196a-2 and breast cancer risk: a case control study. Twin Res Hum Genet 2011; 14:417–421. [DOI] [PubMed] [Google Scholar]
- 30.Gao LB, Bai P, Pan XM, et al. The association between two polymorphisms in pre-miRNAs and breast cancer risk: a meta-analysis. Breast Cancer Res Treat 2011; 125:571–574. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H, Su YL, Yu H, et al. Meta-Analysis of the association between Mir-196a-2 polymorphism and cancer susceptibility. Cancer Biol Med 2012; 9:63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen QH, Wang QB, Zhang B. Ethnicity modifies the association between functional microRNA polymorphisms and breast cancer risk: a HuGE meta-analysis. Tumour Biol 2014; 35:529–543. [DOI] [PubMed] [Google Scholar]
- 33.Dai ZJ, Shao YP, Wang XJ, et al. Five common functional polymorphisms in microRNAs (rs2910164, rs2292832, rs11614913, rs3746444, rs895819) and the susceptibility to breast cancer: evidence from 8361 cancer cases and 8504 controls. Curr Pharm Des 2015; 21:1455–1463. [DOI] [PubMed] [Google Scholar]
- 34.Wang PY, Gao ZH, Jiang ZH, et al. The associations of single nucleotide polymorphisms in miR-146a, miR-196a and miR-499 with breast cancer susceptibility. PLoS One 2013; 8:e70656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, Wang J, Jia Z, et al. MiR-499 regulates cell proliferation and apoptosis during late-stage cardiac differentiation via Sox6 and cyclin D1. PLoS One 2013; 8:e74504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007; 129:1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alshatwi AA, Shafi G, Hasan TN, et al. Differential expression profile and genetic variants of microRNAs sequences in breast cancer patients. PLoS One 2012; 7:e30049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Omrani M, Hashemi M, Eskandari-Nasab E, et al. hsa-mir-499 rs3746444 gene polymorphism is associated with susceptibility to breast cancer in an Iranian population. Biomark Med 2014; 8:259–267. [DOI] [PubMed] [Google Scholar]
- 39.He B, Pan Y, Xu Y, et al. Associations of polymorphisms in microRNAs with female breast cancer risk in Chinese population. Tumour Biol 2015; 36:4575–4582. [DOI] [PubMed] [Google Scholar]
- 40.Qi P, Wang L, Zhou B, et al. Associations of miRNA polymorphisms and expression levels with breast cancer risk in the Chinese population. Genet Mol Res 2015; 14:6289–6296. [DOI] [PubMed] [Google Scholar]
- 41.Chen C, Yang S, Chaugai S, et al. Meta-analysis of Hsa-mir-499 polymorphism (rs3746444) for cancer risk: evidence from 31 case-control studies. BMC Med Genet 2014; 15:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang SG, Chen L, Tang JH, et al. Lack of association between Hsa-Mir-499 rs3746444 polymorphism and cancer risk: meta-analysis findings. Asian Pac J Cancer Prev 2015; 16:339–344. [DOI] [PubMed] [Google Scholar]
- 43.Hu Y, Yu CY, Wang JL, et al. MicroRNA sequence polymorphisms and the risk of different types of cancer. Sci Rep 2014; 4:3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiao L, Zhang J, Dong Y, et al. Association between miR-125a rs12976445 and survival in breast cancer patients. Am J Transl Res 2014; 6:869–875. [PMC free article] [PubMed] [Google Scholar]


