Supplemental Digital Content is available in the text
Abstract
Adjuvant oxaliplatin-based chemotherapy is widely used for stage III colorectal cancer (CRC) after curative surgery. CRC is a molecularly heterogeneous disease, and our current knowledge of therapeutic response-related genetic factors remains limited. N-acetylgalactosaminyltransferase 14 (GALNT14)-rs9679162 genotype is a prognostic predictor for chemotherapy response in advanced hepatocellular carcinoma. Here, we investigated whether this genotype was related to the therapeutic outcome of stage III CRC.
A cohort of 300 stage III CRC patients receiving curative resection followed by oxaliplatin-based chemotherapy was retrospectively recruited. GALNT14 genotypes and the clinicopathological factors were correlated with posttherapeutic prognosis.
Of these patients, 18% patients had GALNT14-rs9679162 “TT” and 82% had the “GT” + “GG” genotypes. The analysis showed that the “TT” genotype was associated with unfavorable overall survival (OS, P = 0.009) but not with recurrence-free survival (RFS, P = 0.700). The subgroup analysis showed that the “TT” genotype was associated with unfavorable OS in the following subgroups: age ≤65 years, men, left side CRC, N2 stage, carcinoembryonic antigen >5 ng/mL, and mucinous histology (P = 0.012, 0.011, 0.009, 0.025, 0.013, and 0.007, respectively). Within the latter 2 subgroups, the “TT” genotype was the only independent predictor for OS. Finally, the “TT” genotype was associated with the T4 tumor stage (P = 0.017) and in patients with T4 tumors, the “TT” genotype was the only independent predictor for unfavorable RFS (P = 0.007).
GALNT14 “TT” genotype was associated with unfavorable OS in stage III CRC patients receiving curative surgery and adjuvant oxaliplatin-based chemotherapy.
INTRODUCTION
Colorectal cancer (CRC) is the third most common cause of cancer-related deaths worldwide. Among all newly diagnosed CRCs, stage III CRC constitutes the greatest proportion.1 Oxaliplatin-based systemic adjuvant chemotherapy has been shown to be beneficial for stage III CRC patients in a number of trials and therefore has been suggested as a standard treatment for patients with stage III diseases.2–4 The benefit of adjuvant therapy includes an ∼30% reduction in risk of disease recurrence and a 22% to 32% reduction of mortality. It has been found that patients with stage III CRC are not a single aggregate with homogeneous risk but rather consisted of variable subsets of patients with different outcomes. The 5-year survival rate varies from 73% in stage IIIA (T1–2N1) to only 28% in stage IIIC CRC (N2).5,6 Moreover, CRC is a molecularly heterogeneous disease, and genetic analysis of the cancerous tissues revealed candidate prognostic markers such as 18q loss of heterozygosity, microsatellite instability, and large deletions in heat shock protein 110.7–10 Despite the success of these elegant studies, clinical applications of these markers are greatly limited by the accessibility to well-established molecular laboratories and the availability of standardized clinical assays. Therefore, there remain unmet needs for easily accessible genetic makers that can predict prognosis of CRC.
By the use of genome-wide association method followed by prospective validation, it was found that the genotype of polypeptide N-acetylgalactosaminyltransferase 14 (GALNT14) could be used as a prognostic predictor for chemotherapy for hepatocellular carcinoma.11 A leading single nucleotide polymorphism (SNP) marker, rs9679162, was capable of predicting the time-to-tumor progression, overall survival (OS), and responses to chemotherapy. In patients with advanced hepatocellular carcinoma receiving palliative chemotherapy, the “TT” genotype predicted a favorable treatment outcome.12,13 The product of GALNT14 gene was a catalytic enzyme that catalyzed O-glycosylation of many proteins including the death receptor (DR)-4 and -5. Because the O-glycosylation of DR 4 of 5 increased their sensitivity to apoptotic signals, this genotype association of chemotherapy sensitivities might not be restricted to hepatocellular carcinoma. In CRC cells, it has been demonstrated that the increase of DR-5 enhances cytotoxic effects of 5-fluorouracil and oxaliplatin.14 Here, we hypothesized that the GALNT14 genotype might also affect the outcome of stage III CRC patients and examined the clinicopathological parameters in relationship to GALNT14 genotype and the prognosis predictive values of these factors.
METHODS
Patients
This study was conducted under the approval of the institutional review board of Chang Gung Memorial Hospital, Taiwan. Surgical samples of 300 stage III CRC patients resected between years 2003 and 2013 were retrieved from the hospital's tissue bank without other particular selection criteria. The stage was based on tumor-node-metastasis classification. CRCs with regional lymph nodes involvement (N1, metastasis in 1 to 3 regional lymph nodes; N2, metastasis in ≥4 regional lymph nodes), and tumor invasion (T1, submucosa; T2, muscularis propria; T3, pericolorectal tissues; T4, direct invasion or adherent to other organ or structures) but without distant metastasis (M0) were classified as stage III. All the patients received oxaliplatin-based adjuvant chemotherapies after curative resection. The clinicopathological data were collected including age, sex, carcinoembryonic antigen (CEA), tumor location, tumor size, tumor surface area, tumor-free margin, tumor invasion, regional lymph node involvement, tumor differentiation, histology type, and chemotherapy regimen used.
Chemotherapy Regimens
Two oxaliplatin-based regimens, modified Folinic acid, Fluorouracil, Oxaliplatin 6 (mFOLFOX6) and capecitabine plus oxaliplatin (XELOX), were given to patients after curative resection. The mFOLFOX6 was given as the following: oxaliplatin (dose 85 mg/m2) and leucovorin (400 mg/m2) were administered continuously over 2 hours via intravenous route on day 1. The 5-fluorouracil was given intravenous bolus at the dose of 400 mg/m2 on day 1, followed by 1200 mg/m2/day for 2 days (total 2400 mg/m2 over 46–48 hours) with continuous intravenous infusion. The mFOLFOX6 regimen was repeated every 2 weeks for 12 cycles. The XELOX protocol was given as the following: oxaliplatin (130 mg/m2) was continuously administered via intravenous route for 2 hours on day 1. Capecitabine (1700–2000 mg/m2) was given via oral route on day 1 to day 14. The XELOX regimen was repeated every 3 weeks for 8 cycles.
GALNT14 Genotyping
Genotyping of GALNT14 was performed as described in our previous publications.12,13 Briefly, tissue DNA in the paraffin blocks of CRC was extracted and purified. Two primers, 5’-TCACGAGGCCAACATTCTAG-3’ and 5’-TTAGATTCTGCATGGCTCAC-3’, were synthesized, which flanked a 172-bp intronic region of GALNT14 gene containing rs9679162. The SNP genotype was determined by direct sequencing after polymerase chain reaction amplification.
Statistical Analysis
The details of statistical methods performed in this study have been described in our previous publication.12
RESULTS
Basic Clinical Data of the Stage III CRC Patients Included
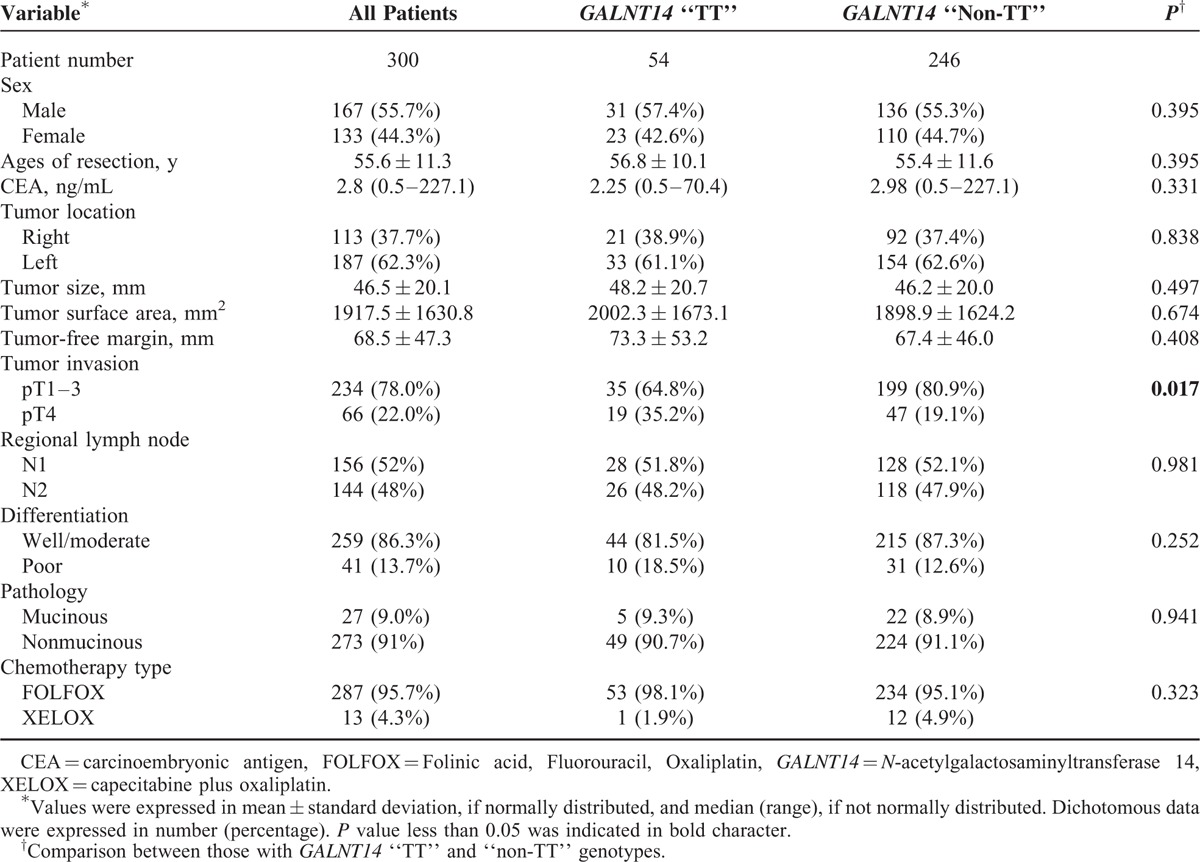
A total of 300 stage III CRC patients, who received radical resection and oxaliplatin-based adjuvant chemotherapy, were included. The basic clinical data were listed in Table 1. The GALNT14 genotype was determined based on the sequence of DNA obtained from tissue blocks. Of them, 54 (18%) patients were “TT” type and 246 (82%) were “non-TT” type (“GG” + “GT”). The proportion of “TT” type was significantly lower than those in the HapMap Chinese Han Beijing (30.15%) and Metropolitan Denver (28.44%) cohorts (χ2 test P = 0.0044 and 0.0214) respectively.15 Most clinicopathological factors showed no significant association to GALNT14 genotype (Table 1). Only 1 exception, the T4 tumor stage, which was significantly more prevalent in patients with the “TT” genotype compared with those with “non-TT” genotype (35.2% vs 19.1%, P = 0.017, Table 1).
TABLE 1.
Basic Clinicopathological Factors of Patients Included
GALNT14 Genotype in Association With OS
To study whether GALNT14 genotype was associated with prognosis, we retrieved the longitudinal data of recurrence-free survival (RFS) and OS. The median posttherapeutic follow-up period was 47 (5–133) months. Cox proportional hazard method and Kaplan–Meier plot with log-rank analysis showed there was no significance between the “TT” genotype and RFS (Cox P = 0.701, log-rank P = 0.700). However, the patients with “TT” genotype had a significantly shorter OS compared with patients with “non-TT” genotype (Figures 1 and 2A, Cox P = 0.019, log-rank P = 0.009).
FIGURE 1.
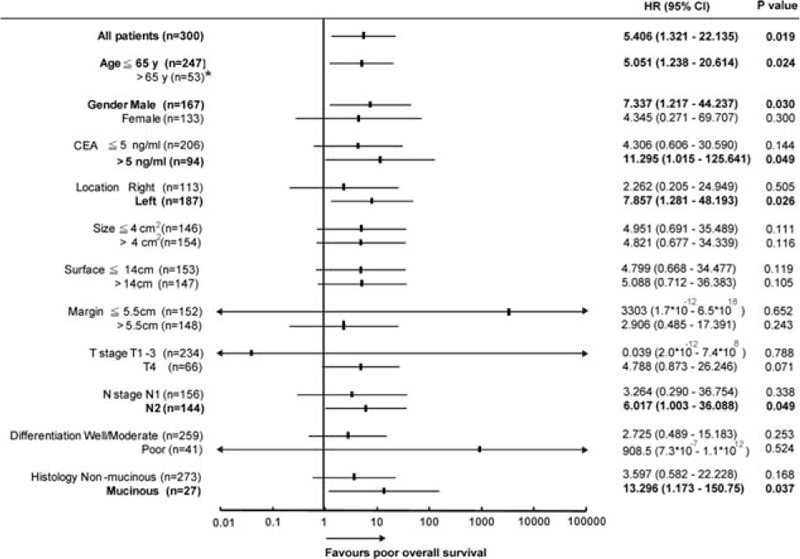
Forest plot of HRs for the impact of GALNT14 “TT” genotype in terms of OS by clinicopathological parameters. The subgroup-specific odds ratios (95% CI) were denoted by black boxes (black lines). Bold text indicate a statistically significant difference with P < 0.05. ∗No event was observed in patients with ages >65 years. CI = confidence interval, GALNT14 = N-acetylgalactosaminyltransferase 14, HR = hazard ratio, OS = overall survival.
FIGURE 2.
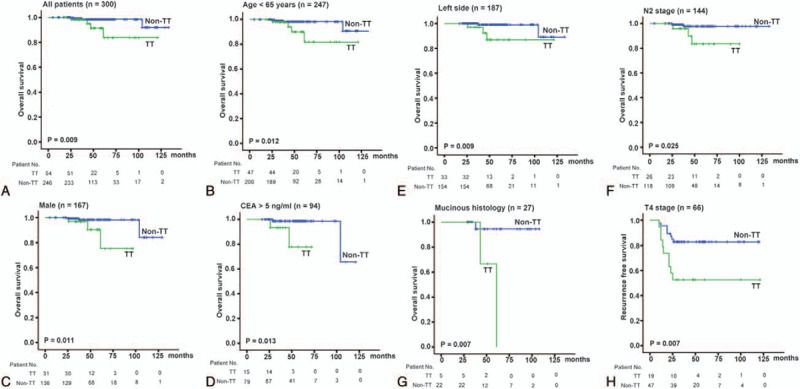
Kaplan–Meier survival curves with log-rank test stratified by GALNT14 “TT” and “non-TT” genotypes. (A) The OS of stage III CRC patients (n = 300, P = 0.009). (B) The OS of patients with ages ≤65 years (n = 247, P = 0.012). (C) The OS of male patients (n = 167, P = 0.011). (D) The OS of patients with CEA >5 ng/mL (n = 94, P = 0.013). (E) The OS of patients with left-side CRC (n = 187, P = 0.009). (F) The OS of patients with N2 stage (n = 144, P = 0.025). (G) The OS of patients with mucinous CRC (n = 27, P = 0.007). (H) The RFS of patients with T4 stage (n = 66, P = 0.007). CEA = carcinoembryonic antigen, CRC = colorectal cancer, GALNT14 = N-acetylgalactosaminyltransferase 14, OS = overall survival, RFS = recurrence-free survival.
Subsequently, we examined the predictive role of GALNT14 “TT” genotype in various clinical subgroups using Cox proportional hazard method (Figure 1). These data were further examined using the Kaplan–Meier plot with log-rank analysis. The “TT” and “non-TT” patients had distinguishable survival curves not only in all stage III CRC patients included but also in the following subgroups: age ≤65 years (Figure 2B, P = 0.012), men (Figure 2C, P = 0.011), CEA >5 ng/mL (Figure 2D, P = 0.013), left CRC (Figure 2E, P = 0.009), N2 stage (Figure 2F, P = 0.025), and mucinous histology (Figure 2G, P = 0.007).
Other Clinicopathological Predictors for RFS and OS
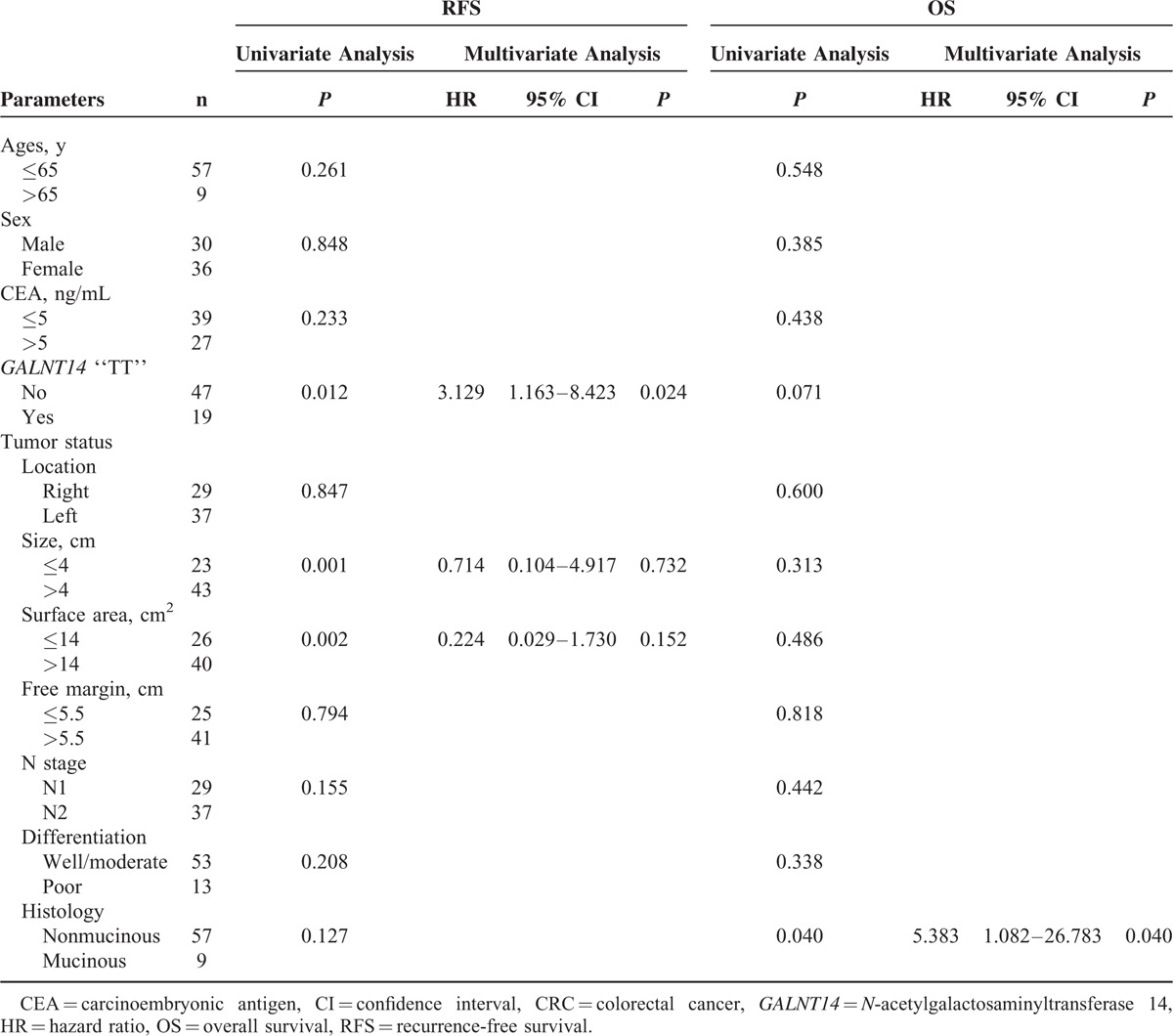
To identify other clinicopathological predictors for RFS and OS, univariate followed by multivariate Cox proportional hazard analysis was performed (Table 2). The CEA level (P = 0.030), tumor size (P = 0.012), surface area (P = 0.022), and N stage (P = 0.014) were associated with RFS by univariate analysis. After adjusting for the confounding factors, multivariate analysis showed that CEA level (P = 0.027) and N stage (P = 0.004) were independent predictors for RFS.
TABLE 2.
Analysis of Factors That Influenced RFS and OS Using Data of All Stage III CRC Patients (n = 300)
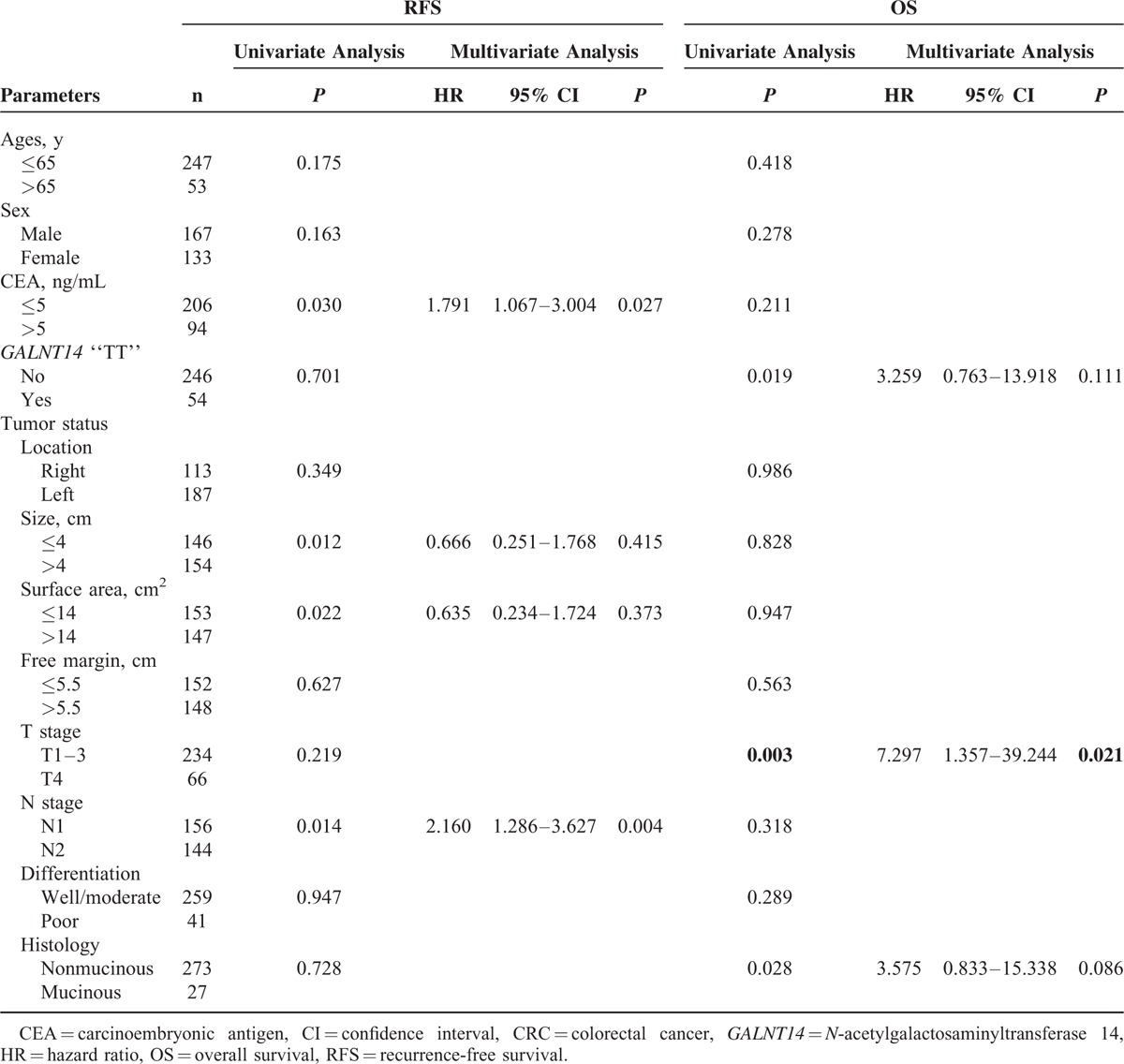
For OS, the GALNT14 “TT” genotype (P = 0.019), the T4 stage (P = 0.003), and the mucinous histology (P = 0.028) were associated with poor prognosis by univariate Cox proportional models. Multivariate analysis revealed that only T4 stage (P = 0.021) was the independent predictor for OS.
GALNT14 “TT” Genotype as an Independent Outcome Predictor in Clinical Subgroups
Because GALNT14 genotypes were associated with both T4 stage (Table 1) and OS (Figures 1 and 2), we subsequently examined the predictive value of GALNT14 “TT” genotype in the T4 stage patient subgroup (n = 66, Table 3). The result showed that “TT” genotype was the only independent predictor for RFS in this subgroup (Figure 2H, Cox P = 0.024, log-rank P = 0.007). For OS, the mucinous histology was the only unfavorable predictor (Cox P = 0.040, log-rank P = 0.021).
TABLE 3.
Analysis of Factors That Influenced RFS and OS Using Data of Patients With T4 Stage (n = 66)
In addition to T4 stage subgroup, we also analyzed the prognosis predictive value of GALNT14 genotype in other clinical subgroups associated with poor OS (Figure 1). It was found that in patients with CEA >5 ng/mL (n = 94, supplementary Table 1), “TT” genotype was the only unfavorable factor for OS (Cox P = 0.049). In patients with mucinous histology (n = 27, supplementary Table 2), the “TT” genotype was the only unfavorable predictor for OS (Cox P = 0.037). Other analysis for the predictive value of GALNT14 genotype in clinical subgroups was given in supplementary Tables 3–6.
DISCUSSION
At this time, routine adjuvant chemotherapeutic treatment is recommended for patients with high-risk stage II and stage III CRC. It has been documented that oxaliplatin-based adjuvant chemotherapy confers survival advantage in stage III CRC patients. However, no pretreatment prognostic molecular biomarkers for stage III CRC receiving oxaliplatin-based adjuvant chemotherapy regimen has been established until now. The present study demonstrated that the GALNT14 “TT” genotype was positively associated with T4 stage, a well-known poor prognostic predictor. Further analysis showed that the “TT” genotype was associated with poor OS in all included patients and several clinical subgroups, including age ≤65 years, men, CEA >5 ng/mL, left-side CRC, N2 stage, and mucinous histology.
In addition to GALNT14 “TT” genotype, several poor prognostic subgroups were identified including CEA >5 ng/mL, N2 stage, T4 stage, and mucinous histology. This is consistent with previous studies from other groups.1,16,17 To identify useful prognostic predictors within these subgroups, we further analyzed the RFS and OS with respect to the clinicopathological parameters and GALNT14 genotype. Intriguingly, GALNT14 was again identified as a valuable prognostic predictor in these subgroups. For patients in T4 stage, the “TT” genotype was significantly associated with poor RFS. For patients with CEA >5 ng/mL or mucinous histology, the “TT” genotype was the only predictor associated with poor OS.
Like many other cancers, development and progression of CRC are affected by multiple factors. Most research efforts have been focused on the underlying molecular mechanisms of CRC carcinogenesis, and a variety of candidate genetic markers with predictive value of outcome have been discovered by studying the cancerous tissues (e.g., KRAS expression, levels of DNA mismatch repair, 18q deletion, and p53 expression).7–9,18,19 However, personal genetic background might contribute greatly to this disease. Our study showed that patients with GALNT14 “TT” genotype had a higher rate to develop tumor invasion (T4 stage) and thus have an unfavorable OS. Furthermore, the “TT” genotype was associated with poor RFS in T4 stage patients. Thus, GALNT14 “TT” genotype has 2 major clinicopathological implications. First, tumors with GALNT14 “TT” genotype have more aggressive cancer invasion property during oncogenesis. Second, tumors with “TT” genotype may have decreased drug sensitivity to oxaliplatin, which is reflected from its association with poor RFS in T4 stage patients. Therefore, other more potent cytotoxic agents should be considered in patients with GALNT14 “TT” genotype.
Oxaliplatin is a platin analogue that has been widely used in treating a variety of solid tumors, especially in CRCs. It induces apoptosis and cell death through the inhibition of DNA replication by forming intrastrand cross-link DNA adducts.20 Several factors may affect the therapeutic outcome of oxaliplatin-based chemotherapy including genetic polymorphism. It has been demonstrated that metastatic CRC with the SNP located within excision repair cross-complementing rodent repair deficiency complementation group1 (ERCC1) codon 118C/T or T/T genotype was associated with worse survival,21–23 whereas other groups showed that metastatic CRC patients with the same genotype had better response or survival compared with C/C patients.24,25 Although the results on the predictive value of ERCC1 codon 118 polymorphism have been inconsistent, these studies raise the possibility that genetic SNP might play a role in therapeutic response. Our current study of GALNT14 genotype supported this view. Moreover, the gene product of GALNT14 is an enzyme that catalyzes O-glycosylation of many proteins including the DR-4 and -5. It has been reported that the cytotoxic effects of oxaliplatin in CRC can be enhanced by tenovin-6 through upregulating DR-5,14 suggesting that modulation of DR-mediated signaling may affect the effects of oxaliplatin-based chemotherapy. In this study, 1 convenient explanation for the GALNT14 genotypes–associated therapeutic outcomes is the differential intensities of DR-mediated apoptosis signaling between the “TT” and “non-TT” genotypes. However, because O-glycosylation is a body-wise process, it is unclear whether there are other unrecognized important glycoproteins involved. Moreover, several genetic alternations in colorectal cancerous tissues, such as 18q loss of heterozygosity, microsatellite instability, and large deletions in heat shock protein 110, have been proposed as candidate prognostic markers, but the association between GALNT14 genotypes and these genetic makers is unclear. Further studies to clarify these issues are needed.
This study had several limitations, including its retrospective nature, limited case number, and restriction of the sample to a Chinese population from a single medical center. Our eligibility criteria confined the treatment to oxaliplatin-based chemotherapies (mFOLFOX6 and XELOX) for interpretative clarity. Therefore, it may not pertain to other regimens not containing oxaliplatin. The decision of which oxaliplatin-based regimens to be used was not only dependent on clinical criteria but also on nonclinical considerations such as doctors’ decision and patients’ willingness. The age of patients in this cohort study was relatively young compared with other CRC cohort studies. This was because elderly CRC patients were more often willing to have chemotherapy via oral route than intravenous route. Despite all these drawbacks, our study results are considered useful in routine clinical practice.
In conclusion, GALNT14 “TT” genotype was correlated with T4 stage and associated with poor OS in stage III CRC patients receiving curative resection and adjuvant oxaliplatin-based chemotherapy. It was further associated with poor treatment outcome in subgroups of T4 stage, CEA >5 ng/mL, or mucinous histology. Taken together, GALNT14 genotype is a valuable prognostic predictor in advanced CRC.
Supplementary Material
Acknowledgments
The authors thank the staff members (Yen-Ling Chuang, Tzu-Chi Yu, Chien Chih Wang, and Hsiu Min Tseng) of the Liver Research Center and Tissue Bank in Linkou Chang Gung Memorial Hospital for their technical assistance. The authors also thank the volunteer technical work from Miss Cassandra Yeh.
Footnotes
Abbreviations: CEA = carcinoembryonic antigen, CRC = colorectal cancer, DR = death receptor, ERCC1 = excision repair cross-complementing rodent repair deficiency complementation group1, GALNT14 = N-acetylgalactosaminyltransferase 14, OS = overall survival, RFS = recurrence-free survival, SNP = single nucleotide polymorphism
W-RL and C-TY were responsible for study concept and design; W-RL, J-MC, S-NL, M-WL, T-YH, and C-KH acquired data; W-RL drafted the article; J-MC provided material support; K-HL and Y-KT analyzed and interpreted data; K-HL was responsible for statistical analysis; C-TY did a critical revision of the article for important intellectual content.
W-RL and J-MC contributed equally to this study.
This study was funded by grants from the Ministry of Science and Technology (ministry of science and technology 103-2314-B-182–021) and Chang Gung Medical Research Program, Taiwan (CMRPG3B0380, CIRPG3B0032). The sponsors were not related to the study design in the collection, analysis, and interpretation of data.
The authors have no conflict of interests to declare.
REFERENCES
- 1.Benson AB., 3rd Adjuvant chemotherapy of stage III colon cancer. Semin Oncol 2005; 32 6 suppl 9:S74–S77. [DOI] [PubMed] [Google Scholar]
- 2.Andre T, Boni C, Mounedji-Boudiaf L, et al. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N Engl J Med 2004; 350:2343–2351. [DOI] [PubMed] [Google Scholar]
- 3.Kuebler JP, Wieand HS, O’Connell MJ, et al. Oxaliplatin combined with weekly bolus fluorouracil and leucovorin as surgical adjuvant chemotherapy for stage II and III colon cancer: results from NSABC-07. J Clin Oncol 2007; 25:2198–2204. [DOI] [PubMed] [Google Scholar]
- 4.Sanoff HK, Carpenter WR, Martin CF, et al. Comparative effectiveness of oxaliplatin vs non-oxaliplatin-containing adjuvant chemotherapy for stage III colon cancer. J Natl Cancer Inst 2012; 104:211–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gunderson LL, Jessup JM, Sargent DJ, et al. Revised TN categorization for colon cancer based on national survival outcomes data. J Clin Oncol 2010; 28:264–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol 2010; 17:1471–1474. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe T, Wu TT, Catalano PJ, et al. Molecular predictors of survival after adjuvant chemotherapy for colon cancer. N Engl J Med 2001; 344:1196–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med 2003; 349:247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carethers JM, Smith EJ, Behling CA, et al. Use of 5-fluorouracil and survival in patients with microsatellite-unstable colorectal cancer. Gastroenterology 2004; 126:394–401. [DOI] [PubMed] [Google Scholar]
- 10.Collura A, Lagrange A, Svrcek M, et al. Patients with colorectal tumors with microsatellite instability and large deletions in HSP110 T17 have improved response to 5-fluorouracil-based chemotherapy. Gastroenterology 2014; 146:401–411. [DOI] [PubMed] [Google Scholar]
- 11.Liang KH, Lin CC, Yeh CT. GALNT14 SNP as a potential predictor of response to combination chemotherapy using 5-FU, mitoxantrone and cisplatin in advanced HCC. Pharmacogenomics 2011; 12:1061–1073. [DOI] [PubMed] [Google Scholar]
- 12.Lin WR, Hsu CW, Chen YC, et al. GALNT14 genotype, alpha-fetoprotein and therapeutic side effects predict post-chemotherapy survival in patients with advanced hepatocellular carcinoma. Mol Clin Oncol 2014; 2:630–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeh CT, Liang KH, Lin CC, et al. A single nucleotide polymorphism on the GALNT14 gene as an effective predictor of response to chemotherapy in advanced hepatocellular carcinoma. Int J Cancer 2013; 134:1214–1224. [DOI] [PubMed] [Google Scholar]
- 14.Ueno T, Endo S, Saito R, et al. The sirtuin inhibitor tenovin-6 upregulates death receptor 5 and enhances cytotoxic effects of 5-fluorouracil and oxaliplatin in colon cancer cells. Oncol Res 2013; 21:155–164. [DOI] [PubMed] [Google Scholar]
- 15.Liang KH, Yang PC, Yeh CT. Genotyping the gene by joint analysis of two linked single nucleotide polymorphisms using liver tissues for clinical and geographical comparisons. Oncol Lett 2014; 8:2215–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Divitiis C, Nasti G, Montano M, et al. Prognostic and predictive response factors in colorectal cancer patients: between hope and reality. World J Gastroenterol 2014; 20:15049–15059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Connor ES, Greenblatt DY, LoConte NK, et al. Adjuvant chemotherapy for stage II colon cancer with poor prognostic features. J Clin Oncol 2011; 29:3381–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gangadhar T, Schilsky RL. Molecular markers to individualize adjuvant therapy for colon cancer. Nat Rev Clin Oncol 2010; 7:318–325. [DOI] [PubMed] [Google Scholar]
- 19.Tejpar S, Bertagnolli M, Bosman F, et al. Prognostic and predictive biomarkers in resected colon cancer: current status and future perspectives for integrating genomics into biomarker discovery. Oncologist 2010; 15:390–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Funke S, Brenner H, Chang-Claude J. Pharmacogenetics in colorectal cancer: a systematic review. Pharmacogenomics 2008; 9:1079–1099. [DOI] [PubMed] [Google Scholar]
- 21.Chua W, Goldstein D, Lee CK, et al. Molecular markers of response and toxicity to FOLFOX chemotherapy in metastatic colorectal cancer. Br J Cancer 2009; 101:998–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruzzo A, Graziano F, Loupakis F, et al. Pharmacogenetic profiling in patients with advanced colorectal cancer treated with first-line FOLFOX-4 chemotherapy. J Clin Oncol 2007; 25:1247–1254. [DOI] [PubMed] [Google Scholar]
- 23.Stoehlmacher J, Park DJ, Zhang W, et al. A multivariate analysis of genomic polymorphisms: prediction of clinical outcome to 5-FU/oxaliplatin combination chemotherapy in refractory colorectal cancer. Br J Cancer 2004; 91:344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pare L, Marcuello E, Altes A, et al. Pharmacogenetic prediction of clinical outcome in advanced colorectal cancer patients receiving oxaliplatin/5-fluorouracil as first-line chemotherapy. Br J Cancer 2008; 99:1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viguier J, Boige V, Miquel C, et al. ERCC1 codon 118 polymorphism is a predictive factor for the tumor response to oxaliplatin/5-fluorouracil combination chemotherapy in patients with advanced colorectal cancer. Clin Cancer Res 2005; 11:6212–6217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


