Abstract
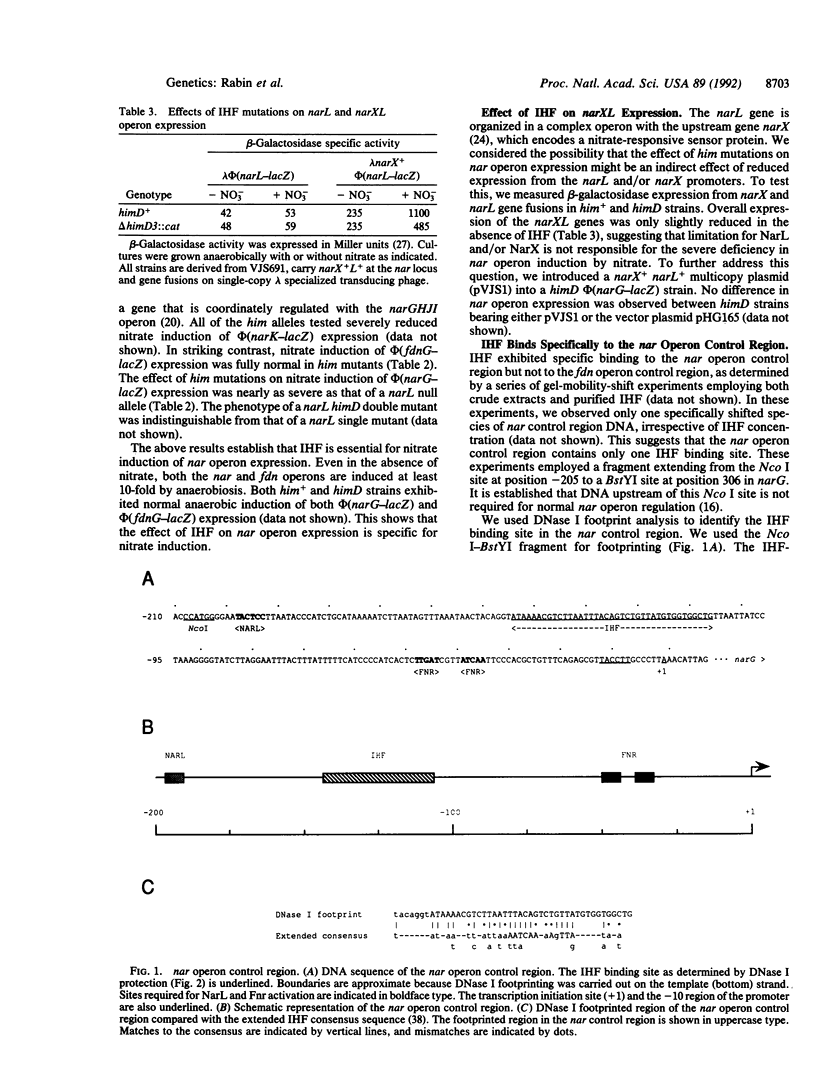
The nitrate reductase operon (narGHJI) of Escherichia coli encodes an anaerobic respiratory enzyme. Previous work has identified two cis-acting sites in the nar operon control region: a proximal site required for anaerobic induction mediated by the activator Fnr and a remote upstream site required for nitrate induction mediated by the activator NarL [Li, S. & DeMoss, J. A. (1988) J. Biol. Chem. 263, 13700-13705]. Our search for nar regulatory mutants yielded one strain with a mutation in himD, the structural gene for one of the subunits of integration host factor (IHF). Strains carrying null alleles of the IHF structural genes, himD and himA, had severe defects in nitrate induction of the nar operon but were normal for nitrate induction of the coordinately regulated fdn operon. Anaerobic expression of both operons was normal in him mutants. Gel-mobility-shift and DNase I protection experiments revealed a single IHF binding site in the nar operon control region, located midway between the upstream activation site and the promoter. We conclude that an IHF-mediated DNA bend is essential for efficient nitrate induction of the sigma 70-dependent nar operon promoter. This requirement of IHF for transcriptional activation had been noted for several sigma 54-dependent promoters.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg B. L., Li J., Heider J., Stewart V. Nitrate-inducible formate dehydrogenase in Escherichia coli K-12. I. Nucleotide sequence of the fdnGHI operon and evidence that opal (UGA) encodes selenocysteine. J Biol Chem. 1991 Nov 25;266(33):22380–22385. [PubMed] [Google Scholar]
- Berg B. L., Stewart V. Structural genes for nitrate-inducible formate dehydrogenase in Escherichia coli K-12. Genetics. 1990 Aug;125(4):691–702. doi: 10.1093/genetics/125.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biek D. P., Cohen S. N. Involvement of integration host factor (IHF) in maintenance of plasmid pSC101 in Escherichia coli: characterization of pSC101 mutants that replicate in the absence of IHF. J Bacteriol. 1989 Apr;171(4):2056–2065. doi: 10.1128/jb.171.4.2056-2065.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkmann A., Sawers R. G., Böck A. Involvement of the ntrA gene product in the anaerobic metabolism of Escherichia coli. Mol Gen Genet. 1987 Dec;210(3):535–542. doi: 10.1007/BF00327209. [DOI] [PubMed] [Google Scholar]
- Castilho B. A., Olfson P., Casadaban M. J. Plasmid insertion mutagenesis and lac gene fusion with mini-mu bacteriophage transposons. J Bacteriol. 1984 May;158(2):488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie-Martin F., Magasanik B. Role of integration host factor in the regulation of the glnHp2 promoter of Escherichia coli. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1631–1635. doi: 10.1073/pnas.88.5.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado-Vides J., Magasanik B., Gralla J. D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991 Sep;55(3):371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X. R., Li S. F., DeMoss J. A. Upstream sequence elements required for NarL-mediated activation of transcription from the narGHJI promoter of Escherichia coli. J Biol Chem. 1992 Jul 15;267(20):14122–14128. [PubMed] [Google Scholar]
- Egan S. M., Stewart V. Mutational analysis of nitrate regulatory gene narL in Escherichia coli K-12. J Bacteriol. 1991 Jul;173(14):4424–4432. doi: 10.1128/jb.173.14.4424-4432.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan S. M., Stewart V. Nitrate regulation of anaerobic respiratory gene expression in narX deletion mutants of Escherichia coli K-12. J Bacteriol. 1990 Sep;172(9):5020–5029. doi: 10.1128/jb.172.9.5020-5029.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamm E. L., Weisberg R. A. Primary structure of the hip gene of Escherichia coli and of its product, the beta subunit of integration host factor. J Mol Biol. 1985 May 25;183(2):117–128. doi: 10.1016/0022-2836(85)90206-2. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. I. Integration host factor: a protein for all reasons. Cell. 1988 Nov 18;55(4):545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giladi H., Gottesman M., Oppenheim A. B. Integration host factor stimulates the phage lambda pL promoter. J Mol Biol. 1990 May 5;213(1):109–121. doi: 10.1016/S0022-2836(05)80124-X. [DOI] [PubMed] [Google Scholar]
- Goodrich J. A., Schwartz M. L., McClure W. R. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF). Nucleic Acids Res. 1990 Sep 11;18(17):4993–5000. doi: 10.1093/nar/18.17.4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins N. P., Collier D. A., Kilpatrick M. W., Krause H. M. Supercoiling and integration host factor change the DNA conformation and alter the flow of convergent transcription in phage Mu. J Biol Chem. 1989 Feb 15;264(5):3035–3042. [PubMed] [Google Scholar]
- Hoover T. R., Santero E., Porter S., Kustu S. The integration host factor stimulates interaction of RNA polymerase with NIFA, the transcriptional activator for nitrogen fixation operons. Cell. 1990 Oct 5;63(1):11–22. doi: 10.1016/0092-8674(90)90284-l. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kustu S., North A. K., Weiss D. S. Prokaryotic transcriptional enhancers and enhancer-binding proteins. Trends Biochem Sci. 1991 Nov;16(11):397–402. doi: 10.1016/0968-0004(91)90163-p. [DOI] [PubMed] [Google Scholar]
- Kustu S., Santero E., Keener J., Popham D., Weiss D. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989 Sep;53(3):367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. C., Hales L. M., Gumport R. I., Gardner J. F. The isolation and characterization of mutants of the integration host factor (IHF) of Escherichia coli with altered, expanded DNA-binding specificities. EMBO J. 1992 Jan;11(1):305–313. doi: 10.1002/j.1460-2075.1992.tb05053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. C., MacWilliams M. P., Gumport R. I., Gardner J. F. Genetic analysis of Escherichia coli integration host factor interactions with its bacteriophage lambda H' recognition site. J Bacteriol. 1991 Jan;173(2):609–617. doi: 10.1128/jb.173.2.609-617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Stewart V. Localization of upstream sequence elements required for nitrate and anaerobic induction of fdn (formate dehydrogenase-N) operon expression in Escherichia coli K-12. J Bacteriol. 1992 Aug;174(15):4935–4942. doi: 10.1128/jb.174.15.4935-4942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. F., DeMoss J. A. Location of sequences in the nar promoter of Escherichia coli required for regulation by Fnr and NarL. J Biol Chem. 1988 Sep 25;263(27):13700–13705. [PubMed] [Google Scholar]
- Li S., Rabi T., DeMoss J. A. Delineation of two distinct regulatory domains in the 5' region of the nar operon of Escherichia coli. J Bacteriol. 1985 Oct;164(1):25–32. doi: 10.1128/jb.164.1.25-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Noji S., Nohno T., Saito T., Taniguchi S. The narK gene product participates in nitrate transport induced in Escherichia coli nitrate-respiring cells. FEBS Lett. 1989 Jul 31;252(1-2):139–143. doi: 10.1016/0014-5793(89)80906-8. [DOI] [PubMed] [Google Scholar]
- Pagel J. M., Hatfield G. W. Integration host factor-mediated expression of the ilvGMEDA operon of Escherichia coli. J Biol Chem. 1991 Jan 25;266(3):1985–1996. [PubMed] [Google Scholar]
- Reitzer L. J., Magasanik B. Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter. Cell. 1986 Jun 20;45(6):785–792. doi: 10.1016/0092-8674(86)90553-2. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53(1):85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Singer M., Baker T. A., Schnitzler G., Deischel S. M., Goel M., Dove W., Jaacks K. J., Grossman A. D., Erickson J. W., Gross C. A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989 Mar;53(1):1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G. S., Lubinsky-Mink S., Jackson C. G., Cassel A., Kuhn J. pHG165: a pBR322 copy number derivative of pUC8 for cloning and expression. Plasmid. 1986 May;15(3):172–181. doi: 10.1016/0147-619x(86)90035-1. [DOI] [PubMed] [Google Scholar]
- Stewart V., MacGregor C. H. Nitrate reductase in Escherichia coli K-12: involvement of chlC, chlE, and chlG loci. J Bacteriol. 1982 Aug;151(2):788–799. doi: 10.1128/jb.151.2.788-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V. Nitrate respiration in relation to facultative metabolism in enterobacteria. Microbiol Rev. 1988 Jun;52(2):190–232. doi: 10.1128/mr.52.2.190-232.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V., Parales J., Jr Identification and expression of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli K-12. J Bacteriol. 1988 Apr;170(4):1589–1597. doi: 10.1128/jb.170.4.1589-1597.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V. Requirement of Fnr and NarL functions for nitrate reductase expression in Escherichia coli K-12. J Bacteriol. 1982 Sep;151(3):1320–1325. doi: 10.1128/jb.151.3.1320-1325.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W., Porter S., Kustu S., Echols H. DNA-looping and enhancer activity: association between DNA-bound NtrC activator and RNA polymerase at the bacterial glnA promoter. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5504–5508. doi: 10.1073/pnas.87.14.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint B., Bosc C., Richaud P., Colbeau A., Vignais P. M. A mutation in a Rhodobacter capsulatus gene encoding an integration host factor-like protein impairs in vivo hydrogenase expression. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10749–10753. doi: 10.1073/pnas.88.23.10749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M. S., DeMoss J. A. Role of alternative promoter elements in transcription from the nar promoter of Escherichia coli. J Bacteriol. 1992 Feb;174(4):1119–1123. doi: 10.1128/jb.174.4.1119-1123.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedel A., Weiss D. S., Popham D., Dröge P., Kustu S. A bacterial enhancer functions to tether a transcriptional activator near a promoter. Science. 1990 Apr 27;248(4954):486–490. doi: 10.1126/science.1970441. [DOI] [PubMed] [Google Scholar]





