Abstract
Purpose
Endoscopic surveillance of Barrett’s esophagus (BE) is problematic because dysplasia/early-stage neoplasia are frequently invisible and likely to be missed due to sampling bias. Molecular abnormalities may be more diffuse than dysplasia. The aim was therefore to test whether DNA methylation; especially on imprinted and X-chromosome genes; is able to detect dysplasia/early-stage neoplasia.
Experimental design
27K methylation arrays were used to find genes best able to differentiate between 22 BE and 24 esophageal adenocarcinoma (EAC) samples. These were validated using pyrosequencing on a retrospective cohort (60 BE, 36 dysplastic and 90 EAC) and then in a prospective multicenter study (98 BE patients, including 28 dysplastic and 9 early EAC) designed to utilize biomarkers to stratify patients according to their prevalent dysplasia/EAC status.
Results
23% genes on the array, including 7% of X-linked and 69% of imprinted genes, demonstrated statistically significant changes in methylation in EAC vs. BE (Wilcoxon P<0.05). 6/7 selected candidate genes were successfully internally (Pearson’s P<0.01) and externally validated (ANOVA P<0.001). Four genes (SLC22A18, PIGR, GJA12 and RIN2) showed the greatest area under curve (0.988) to distinguish between BE and dysplasia/EAC in the retrospective cohort. This methylation panel was able to stratify patients from the prospective cohort into three risk groups based on the number of genes methylated (low risk: <2 genes, intermediate: 2 and high: >2).
Conclusion
Widespread DNA methylation changes were observed in Barrett’s carcinogenesis including ≈70% of known imprinted genes. A four-gene methylation panel stratified BE patients into three risk groups with potential clinical utility.
Keywords: Esophageal adenocarcinoma; Biomarker (from the title: Barrett’s esophagus, DNA methylation)
Introduction
Patients with Barrett’s esophagus (BE) have a substantially increased risk of progression to esophageal adenocarcinoma (EAC) compared to the general population (RR: 11.3, 95% CI: 8.8-14.4)(1). The incidence of EAC has increased 7-fold in the past 30 years (3.6 to 25.6 cases per million)(2) and the prognosis is poor with a median survival of about 11 months due to late presentation(3). Due to the improved survival in those diagnosed when the disease is confined to mucosa or sub-mucosal layers, patients with BE are recommended to undergo endoscopic surveillance for the early detection of cancer(4, 5). The cost-effectiveness and risk:benefit ratio to the patient of endoscopy has been questioned time and again since the risk of progression is relatively low(1, 6, 7); around 0.3% according to the recent estimates(8). The intermediate dysplastic stages between BE and EAC are the most reliable markers of progression; however the histological presence of dysplasia is subjective due to known sampling bias during endoscopy together with a high inter and intra-observer variability(9, 10). The wide variation in progression rates in patients categorized as having low grade dysplasia (LGD) has been highlighted by two recent studies. In a Dutch study the incidence rate of high grade dysplasia (HGD) or EAC in individuals with confirmed low grade dysplasia was high at 13.4% (95% CI 3.5 – 23.2) per patient per annum(11); whereas in a US study the progression rate of individuals with LGD was similar to that of non-dysplastic patients which is a 16-fold difference(12). In patients with HGD, data from a randomized radiofrequency ablation intervention trial suggest a rate of progression of 19% per year in the non-treatment arm(13). Hence there is a pressing need for biomarkers that can accurately detect prevalent dysplasia in flat Barrett’s mucosa and predict those patients most likely to progress to cancer.
Aberrant DNA methylation is shown to be a characteristic of cancer and these changes are known to occur early during transformation(14). It has already been shown in a number of studies that DNA methylation changes occur during progression from BE to EAC and that these alterations have the potential to be utilized as biomarkers(15–20). These studies have mostly employed a candidate approach based on known methylation targets in other cancers. However high-throughput array based platforms are now available to identify DNA methylation changes and we have employed this approach to find candidate biomarkers in Barrett’s carcinogenesis. Imprinted genes and the X-chromosome are both epigenetically controlled by DNA methylation(21), but have never been examined specifically in the context of biomarkers for EAC. Methylation changes in these genes may be ideal biomarkers since physiological inactivation of one allelic copy has already occurred due to imprinting and via X-inactivation in females.
Hence, in this study we performed DNA methylation screening of BE and EAC samples using arrays to determine candidate biomarkers. We analyzed imprinted and X-chromosome genes separately and purposefully separated males from females to allow meaningful conclusions to be drawn. We performed robust internal and external validation using pyrosequencing which is the current gold standard in DNA methylation analysis and from this determined a panel of biomarkers to discriminate between dysplastic and non-dysplastic BE. Finally we validated the biomarker panel in a prospective cohort with real-time analysis to stratify BE patients into low, intermediate and high risk groups based on their risk of having prevalent dysplasia/EAC.
Materials and Methods
Patient samples
For the retrospective studies (methylation arrays and retrospective external validation) all patient samples (H&E slides, endoscopic biopsies and surgical resection specimens), were obtained from patients who had attended Cambridge University Hospitals NHS Trust and provided individual informed consent (ethics: 04/Q2006/28, 09/H0308/118). For the prospective study patients with BE undergoing surveillance or tertiary referral for further evaluation of HGD or early EAC were recruited after obtaining informed consent from Cambridge University Hospitals NHS Trust, Queens University Hospital Nottingham and Amsterdam Medical Centre (ethics: 10/H0305/52). Pathology was verified for all cases according to the Royal College of Pathologists UK guidelines by an experienced upper GI pathologist (Dr Maria O’Donovan) and for dysplasia and EAC a minimum of two experienced pathologists reviewed the cases (referring hospital + Dr Maria O’Donovan). All BE samples were confirmed to have intestinal metaplasia and all EACs for a cellularity of ≥70%. Patient demographics are available in Supplementary Tables 1, 2 and 3.
DNA extraction and bisulphite conversion
For the methylation arrays, high molecular weight DNA was isolated from fresh frozen tissue using standard proteinase-K phenol/chloroform extraction. Samples with A260/280 of <1.8 and a fragment size of <2kb were discarded. A volume corresponding to 1µg of DNA was measured using Quant-iT™ PicoGreen® dsDNA kit (Invitrogen Ltd, UK) according to the manufacturer’s instructions. Bisulphite modification was done using EZ DNA Methylation-Gold™ Kit (Zymo Research Corporation, USA).
DNA extraction for pyrosequencing assays was also carried out using the above mentioned protocol. DNA extraction from formalin fixed paraffin embedded (FFPE) tissues was carried out using QIAamp DNA Micro Kit (Qiagen, UK) using the manufacturer’s instructions. 1µg of DNA was bisulphite modified and eluted in 30 µl of elution buffer.
Illumina Infinium assay
The Infinium assay (Illumina, UK) was run using the automated protocol at Cambridge Genomic Services. Samples were denatured prior to whole genome amplification (WGA) using 0.1N NaOH. Multi-sample amplification master mix (MSM) was then added to the DNA samples and incubated at 37°C for 20 hours. The amplified DNA was fragmented by vortexing, precipitated using isopropanol and dispensed onto the BeadChips which were incubated at 48°C for 20 hours in hybridization buffer to allow for the DNA to hybridize. Unhybridized DNA was washed off and single-base extension was carried out with extended primers and labeled nucleotides using the TECAN Freedom Evo liquid handling robot. The BeadArray Reader (Illumina) was used to read the signal and output files were generated using GenomeStudio Software (Illumina).
Array data analysis and selecting targets
Signal-to-noise ratio ranking
BE and EAC samples were separated into two groups and ranking of all genes was done using the ‘Signal2Noise’ metric (GSEA software, Broad Institute, USA). Signal2Noise uses the difference of means scaled by the standard deviation.
where μ is the mean and σ is the standard deviation. The larger the signal-to-noise ratio, the larger the difference of means (scaled by standard deviation); hence more distinct methylation is seen for each phenotype and more the gene acts as a ‘class marker’. Imprinted genes and those on the X-chromosome were also analyzed separately. The final list of genes can be obtained from Supplementary Table 4.
Wilcoxon test
As a further check to test for differential methylation, a two-sided Wilcoxon test was performed for each probe on the array. Variance of probes with low or high methylation is in general lower than variance of probes with medium methylation(22). So tests for differential methylation tend to preferentially select probes whose values are confined to the extremes of the scale. To reduce this effect we performed a Gaussian normalization prior to the Wilcoxon tests to reduce heteroscedasticity. The values' ranks, normalized between 0 and 1, were taken to be probabilities from a Gaussian distribution and transformed to variables using the distribution's quantile function. The P-values were adjusted for multiple testing using the false discovery rate method of Benjamini & Hochberg(23). We were interested in probes that had both statistically significant and large absolute differences in methylation. Therefore, for each probe we also calculated the difference between the median of the methylation values in the two phenotypes. A probe’s rank in the ordered list of Wilcoxon P-values and its rank in the ordered list of absolute difference in medians were averaged. The probes were arranged in descending order of this average.
The purpose of using two different tests to look for targets was to avoid false positives and to ensure that the selected targets not only have a statistically significant but a large absolute difference in methylation that was reproducible using pyrosequencing which is generally attributed to have an error margin of ±5%. The targets appearing high up in both these analyses were then selected for validation.
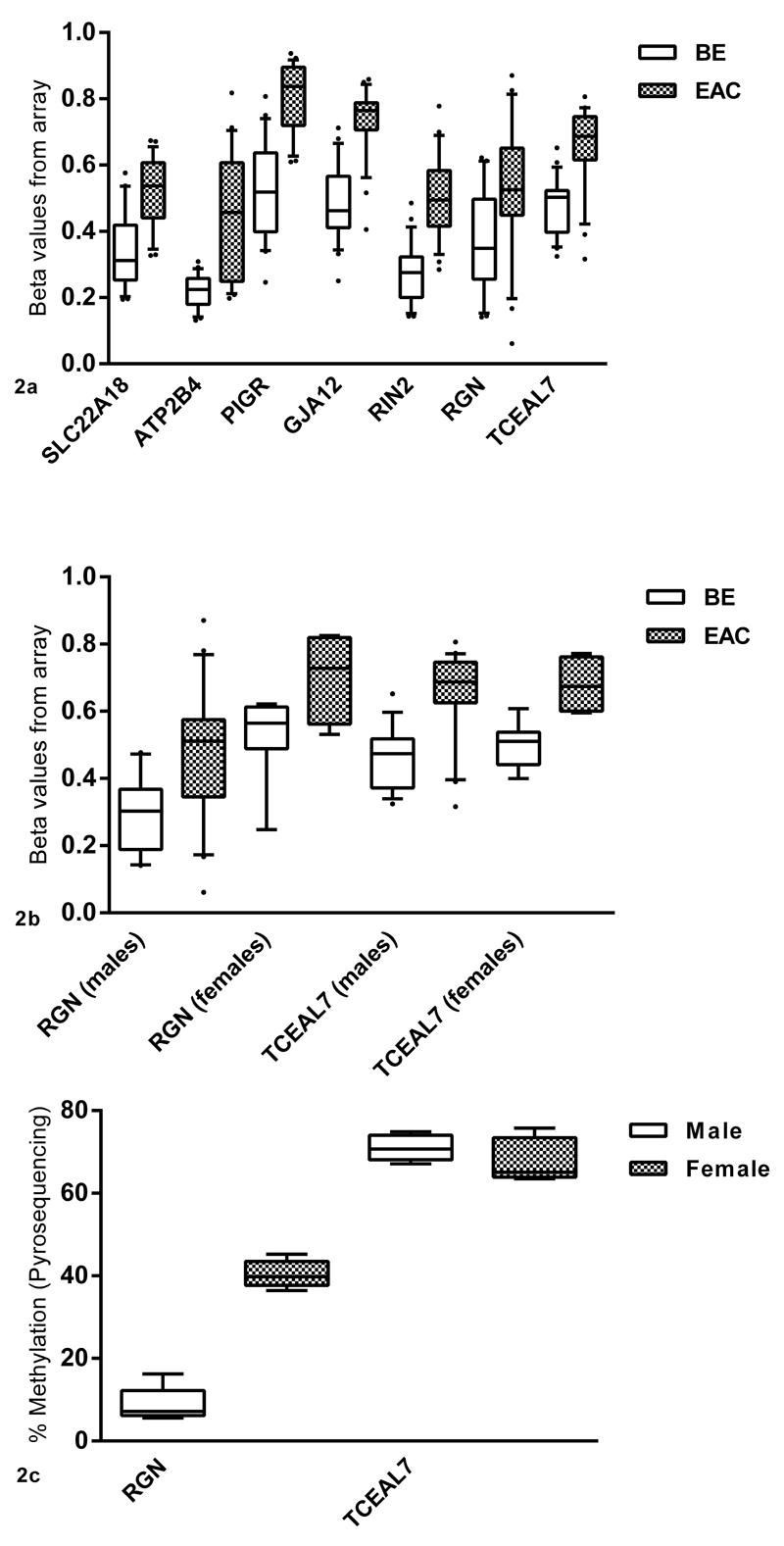
Genes were selected for validation based on the following criteria: present in both of the lists, biological importance in EAC and/or other cancers, probe’s proximity to the promoter and relatively low density of CpGs in the vicinity so that it would be possible to design robust pyrosequencing assays (Figure 2a and 2b, Supplementary Table 4).
Figure 2.
a: Genes selected from the array analysis showing the greatest difference in methylation between BE and EAC. Beta values from the array are plotted on the x-axis against the gene name and tissue type on y-axis. b: For genes on the X-chromosome, analyses were separated on the basis of gender to cater for the effects of X-inactivation in females. Since RGN lies on the region of X-chromosome that is inactivated, males and females have different levels of methylation. Females have higher methylation in both tissues (BE and EAC) compared to males. TCEAL7 does not appear to be affected by X-inactivation and males and females have similar levels of methylation in both BE and EAC. c: Methylation levels for RGN and TCEAL7 in the normal esophageal epithelium in males and females using pyrosequencing (N=5).
Pyrosequencing assays
Pyrosequencing assays were designed using PSQ Assay Design Software (version 1.0.6, Biotage, Sweden) (Supplementary Table 5). Genomic DNA sequences were obtained from NCBI map viewer (build 36). All PCR reactions were carried out in volumes of 25µl using IMMOLASE™ DNA Polymerase (Bioline, UK). 0.75μl of bisulphite converted DNA was used as a template for each reaction. 20μl of each PCR reaction was mixed with 60μl of bead mix composed of 3µl streptavidin-coated beads solution (GE Healthcare, UK), 20ul nuclease free water and 37µl PyroMark binding buffer (Qiagen) in a 96-well plate and left on a shaking platform for 10 min. The pyrosequencing reaction plate was prepared by adding 1.5μl of 10μM sequencing primer and 43.5μl of PyroMark Annealing Buffer (Qiagen) into each of the wells. The pyrosequencing vacuum machine (Biotage) was used to wash and denature the DNA bound to streptavidin-coated beads before being released into the pyrosequencing reaction plate. The plate was heated to 80°C for 3 min and then cooled down to room temperature to allow the sequencing primer to anneal onto the single-stranded DNA and the sequencing reaction was carried out according to the manufacturer’s protocol.
0%, 50% and 100% methylated controls were prepared for all the assays and used with every run. DNA synthesized by PCR was used for this. Primers were designed using the NCBI Primer Designing Tool(24) in order to amplify a region greater than but containing the sequence to be analyzed by pyrosequencing (Supplementary Table 6). Genomic DNA isolated from normal squamous esophagus was used as a template. All PCRs were performed in 50μl duplicates. One reaction was used for in-vitro methylation. Briefly 40μl of the PCR reaction was mixed with 5μl of 10×NEBuffer2, 2.5μl of 3.2mM S-adenosylmethionine (SAM), 4U (1μl) of CpG Methyltransferase (M.SssI) (NEB, UK) and incubated for 2 hours at 37°C. After 2 hours another 0.5μl of 3.2mM SAM, 2U (0.5μl) of M.SssI and 0.5μl of water were added and incubated overnight at 37°C. Both reactions (in-vitro methylated and unmethylated) were then purified using QIAquick PCR purification Kit (Qiagen). These were then bisulphite converted as mentioned before and mixed to generate a 50% methylated control along with 0% and 100% methylated controls.
Statistical analysis
To compare the difference between groups (BE, BE with dysplasia and EAC) a t-test or one way ANOVA was used for continuous variables (age, segment length and methylation) and chi-square test for categorical variables (gender). A P<0.05 was considered statistically significant. Receiver operating characteristic (ROC) curves were used to evaluate the distinguishing ability of each gene and the four gene signature and area under the curve (AUC) was reported. Methylation cut-off points for each gene were selected based on individual ROC curves to have the best accuracy. All analyses were done using GraphPad Prism and SPSS19.0.
Results
Array data analysis
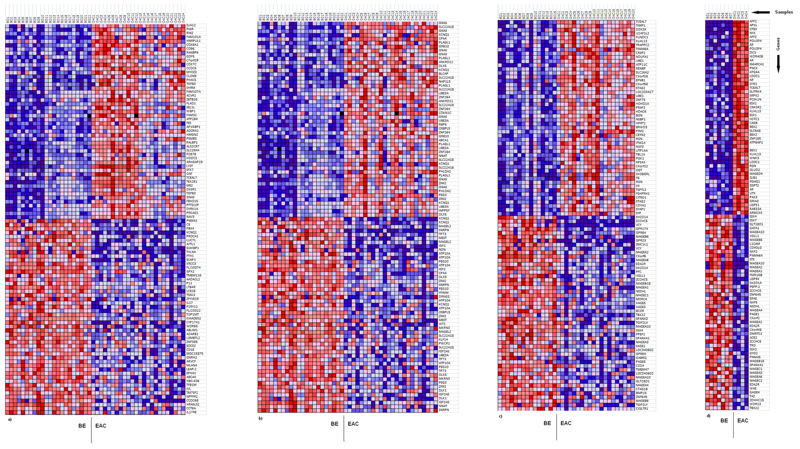
Illumina HumanMethylation27 BeadChips were used to assess and compare methylation levels of 27,578 individual CpG loci spanning 14,475 genes and 110 miRNA promoters in 22 BE and 24 EAC samples (GEO accession no: GSE32925). Signal-to-noise ratio and two-sided Wilcoxon tests were used to rank genes showing the greatest difference in methylation (both hypermethylation and hypomethylation) between the BE and EAC, and from this a ‘class marker’ gene set was identified that was able to clearly distinguish between the two phenotypes (Figure 1). 23% of all the genes present on the array showed a statistically significant difference in methylation (Wilcoxon P<0.05). On the whole hypermethylation was observed to be slightly more prevalent (1,764/14,475 – 12.18%) as compared to hypomethylation (1,590/14,475 – 10.98%) in EAC vs. BE (Wilcoxon P<0.05). Out of the 51 imprinted genes present on the array(25) 17 (33.33%) showed hypermethylation and 18 (35.29%) hypomethylation in EAC vs. BE (Wilcoxon P<0.05) (which comes to a total of 68.62% of all the imprinted genes present on the array). Separate analyses were done for males and females for genes on the X-chromosome to cater for the effects of X-inactivation in females. Genes on the X-chromosome showed similar levels of hyper and hypomethylation in EAC compared to BE (22 genes each hyper and hypomethylated out of a total 600, Wilcoxon P<0.05). Most methylation changes were confined to within known CpG islands. Detailed results can be seen in Table 1.
Figure 1.
GSEA generated heat maps for the top 50 probes showing greatest differential methylation between BE and EAC (red color = high methylation, blue color = low methylation). a – all probes (22BE vs. 24EAC), b – imprinted genes probes (22BE vs. 24 EAC), c – X-chromosome probes (15BE vs. 20EAC, males only), d – X-chromosome probes (7BE vs. 4EAC, females only).
Table 1.
DNA methylation changes observed from the Illumina Infinium 27K array analysis comparing 24 EAC and 22 BE cases (total number of probes on the array=27,578, total number of genes=14,475)
| Trends (EAC vs. BE) | Number of probes | Percentage of probes | Number of genes | Percentage of genes | Number of probes within CpG islands | Number of probes outside of CpG Islands | |
|---|---|---|---|---|---|---|---|
| All genes | Hypermethylation | 1952 | 7.1 | 1764 | 12.18 | 1389 | 563 |
| Hypomethylation | 1740 | 6.3 | 1590 | 10.98 | 1114 | 626 | |
| Total | 3692 | 13.4 | 3354 | 23.16 | 2503 | 1189 | |
| Imprinted genes | Hypermethylation | 33 | 8.5 | 17 | 33.33 | 29 | 4 |
| Hypomethylation | 27 | 6.9 | 18 | 35.29 | 24 | 3 | |
| Total | 60 | 15.4 | 35 | 68.62 | 53 | 7 | |
| X-chromosome genes (males only) | Hypermethylation | 24 | 2.2 | 22 | 3.66 | 20 | 4 |
| Hypomethylation | 24 | 2.2 | 22 | 3.66 | 12 | 12 | |
| Total | 48 | 4.4 | 44 | 7.33 | 32 | 16 | |
Identifying targets for validation
To ensure that the selected targets for validation would have a statistically significant and large absolute difference in methylation and hence be suitable as biomarkers, the results of signal-to-noise ratio ranking were compared to the results of the Wilcoxon tests. The top seven genes present in both the lists fulfilling the aforementioned selection criteria (see methods) were selected for validation (Figure 2a). For RGN which is an X-inactivated gene (p11.3-Xp11.23) it was observed that methylation levels were different in males compared to females in normal tissues (normal squamous esophageal epithelium). Therefore, separate analyses were done for both the genders for RGN in the pathological external validation samples. TCEAL7 on the other hand, also on the X-chromosome, did not appear to be affected by DNA methylation associated X-inactivation and therefore the analysis for males and females were combined in all subsequent experiments (Figure 2b and 2c).
Internal validation
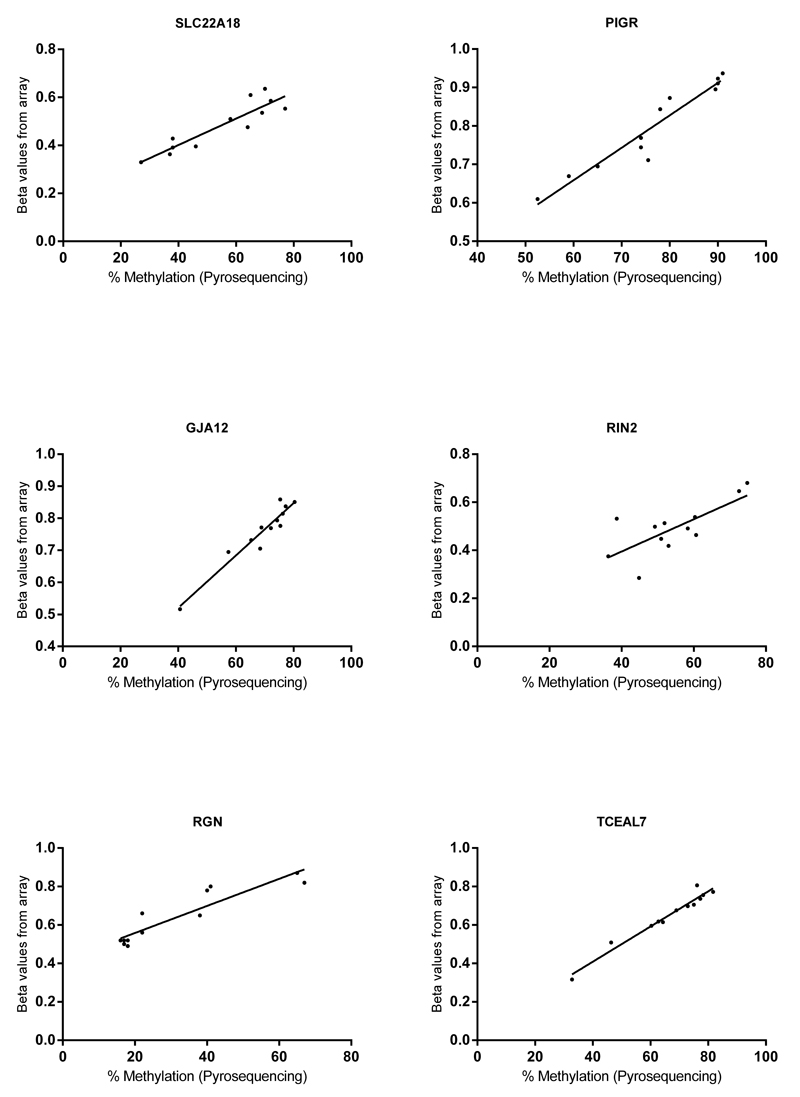
The seven genes selected were first internally validated using pyrosequencing assays on the same samples that were run on the methylation arrays. The assays were designed to analyze the same DNA sequence which was probed by the arrays. Pearson’s correlation was used to assess whether the results from pyrosequencing matched with the results from the arrays (Fig. 3). Six out of seven genes successfully validated which were SLC22A18 (tumor suppressing subtransferable candidate 5, a paternally imprinted gene) (P<0.0001, coefficient=0.9), PIGR (polymeric immunoglobulin receptor) (P<0.0001, coefficient=0.9), GJA12 (gap junction protein, gamma 2) (P<0.0001, coefficient=0.9), RIN2 (Ras and Rab interactor 2) (P<0.01, coefficient=0.7), RGN (senescence marker protein-30, X-linked gene) (P<0.0001, coefficient=0.9)and TCEAL7 (transcription elongation factor A - like 7, X-linked gene) (P<0.0001, coefficient=0.9). ATP2B4 however failed to validate (P=0.6, coefficient=0.1) as shown in Supplementary Figure 1.
Figure 3.
Internal validation. Beta values from the Illumina Infinium array (y-axis) are plotted against the % methylation from pyrosequencing (x-axis) (N=12).
Retrospective external validation
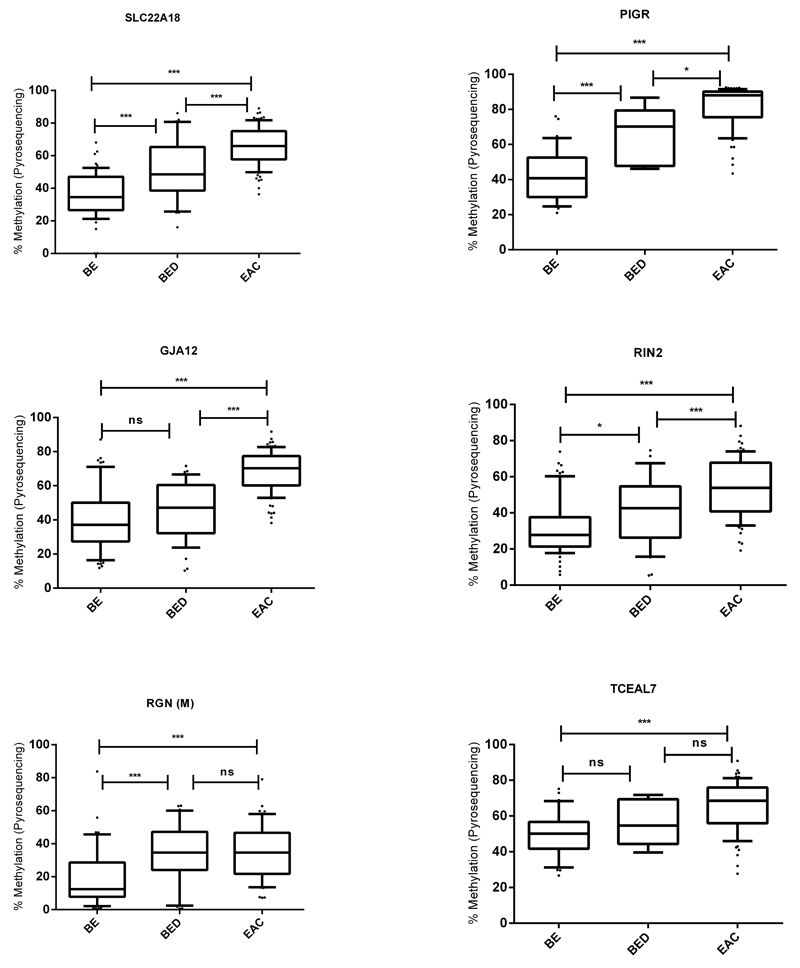
External validation by pyrosequencing was carried out on an independent set of retrospectively collected 60 BE, 36 BE with dysplasia and 90 EAC samples (Figure 4). All of these cases had the histopathological diagnosis confirmed on the actual biopsy used for analysis. This validation set also enabled an assessment to be made of when in the disease pathogenesis the methylation changes occurred. A statistically significant increase in methylation was observed for all the selected biomarker genes in EAC and/or dysplastic BE compared to non-dysplastic BE (ANOVA P<0.001). An ANOVA test was also done to confirm this trend separately for EAC samples with and without neo-adjuvant chemotherapy (P<0.001). For SLC22A18, PIGR, TCEAL7 and RIN2 genes it was a gradual increase, whereas for RGN the biggest change in methylation occurred at the onset of dysplasia and for GJA12 this occurred between dysplasia and EAC.
Figure 4.
Retrospective external validation. N(BE) = 60, N(BED) = 36, N(EAC) = 90 for SLC22A18, GJA12 and RIN2. N(BE) = 30, N(BED) = 6, N(EAC) = 70 for PIGR and TCEAL7. N(BE) = 45, N(BED) = 30, N(EAC) = 60 for RGN (Males only). Middle line = median, box = 25-75 percentile, whiskers = 10-90 percentile. *=p<0.01, **=p<0.001, ***=p<0.0001 using ANOVA (BE=Barrett’s esophagus, BED=Barrett’s esophagus with dysplasia, EAC=esophageal adenocarcinoma).
Generating a methylation cut-off
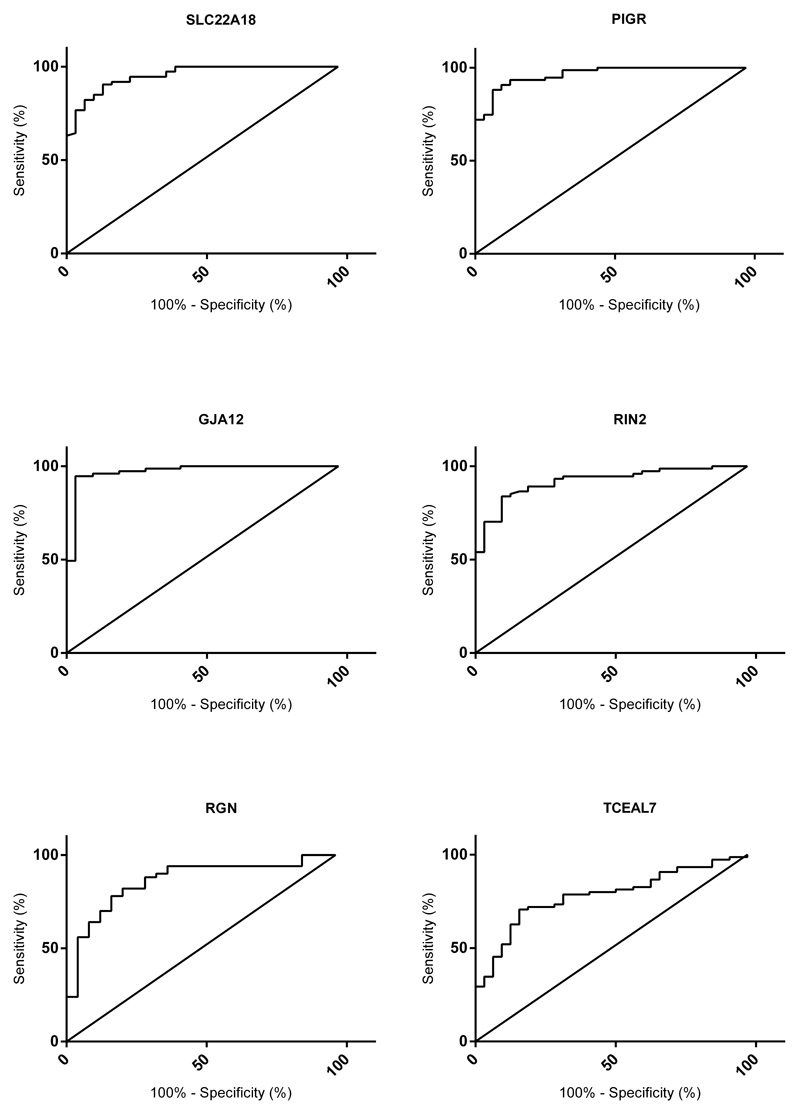
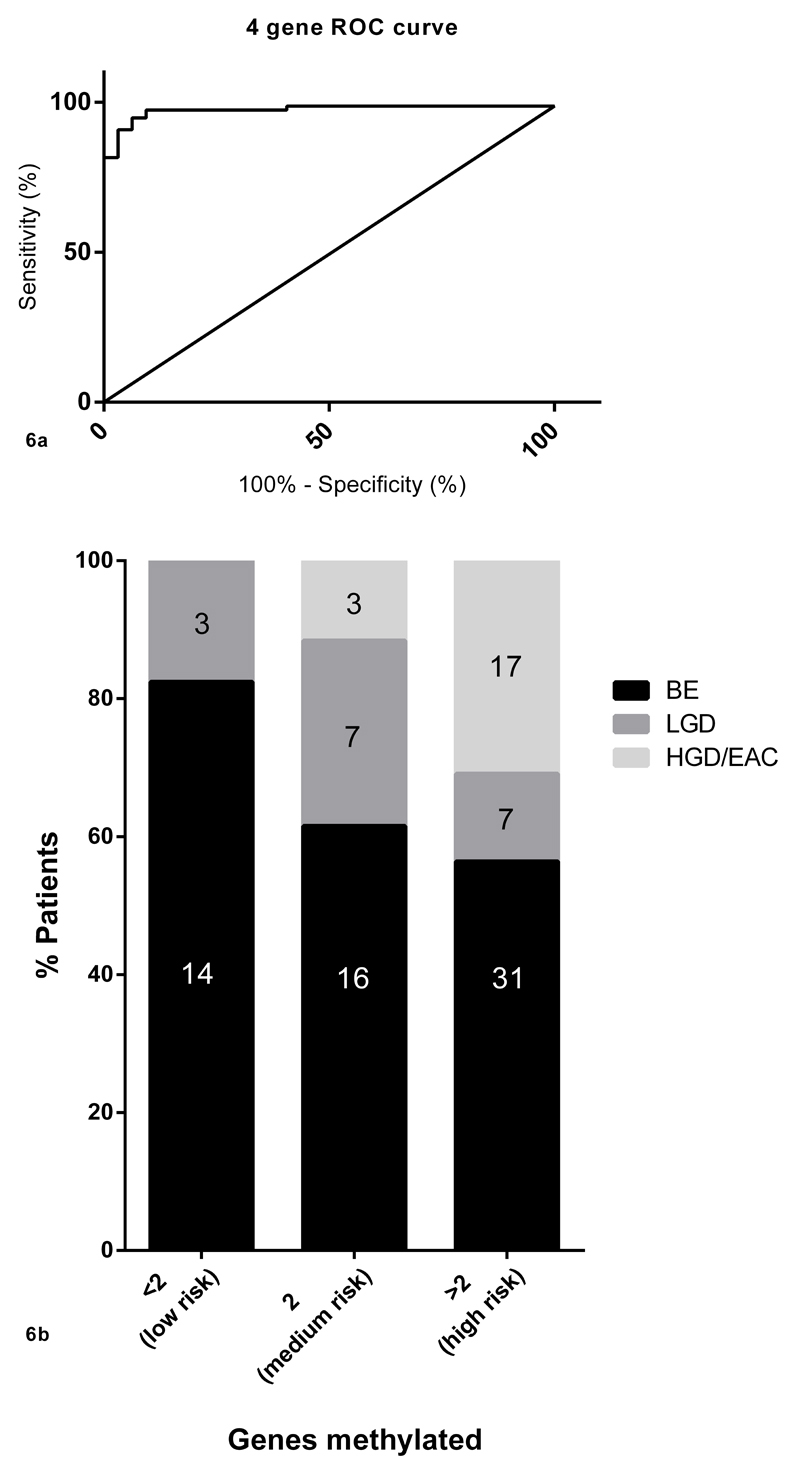
Since an increase in DNA methylation was observed in EAC and dysplastic BE compared to non-dysplastic BE, ROC curves were used to detect the power of the 6 genes individually and then in combination to differentiate between dysplastic BE/EAC and non-dysplastic BE (Figure 5, Supplementary Table 7). Individually GJA12 (AUC=0.973) was best able to distinguish between dysplasia/EAC and non-dysplastic BE followed by PIGR (AUC=0.963), SLC22A18 (AUC=0.954), RIN2 (0.922), RGN (AUC=0.865) but only in males and lastly TCEAL7 (AUC=0.788). The greatest AUC of 0.988 (P<0.01) was obtained using the four gene combination (SLC22A18 + PIGR + GJA12 + RIN2) which had a sensitivity of 94% and a specificity of 97% (Figure 6a).
Figure 5.
ROC curves for all six targets. N(BE) = 32 vs. N(BED)+ N(EAC) = 73. For RGN (Males only) N(BE) = 25 vs. N(BED)+N(EAC) = 51 (BE=Barrett’s esophagus, BED=Barrett’s esophagus with dysplasia, EAC=esophageal adenocarcinoma).
Figure 6.
a: The four gene risk score (SLC22A18 + PIGR + GJA12 + RIN2) had the best AUC of 0.988 (P<0.01).
b: Graphical representation of percentage of patients falling into each group. The probability of HDG/early EAC increases with an increase in the number of positive biomarkers (BE=Barrett’s esophagus, LGD=low grade dysplasia, HGD=high grade dysplasia, EAC=esophageal adenocarcinoma).
Prospective validation
The methylation cut-offs selected for the four genes using ROC curves (SLC22A18, PIGR, GJA12, RIN2) were then tested on a prospective cohort of 98 patients (including 17 LGD and 20 HGD/EAC) undergoing BE surveillance endoscopy in three tertiary referral centers to enrich for dysplasia and EAC. Random quadrantic biopsies every 2 cm were taken according to international guideline(26) along with 3 extra biopsies for DNA methylation taken randomly from within the BE segment. For the analysis, the biopsy with the highest methylation value per gene was selected taking advantage of the likely molecular field effect. A patient was categorized according to their highest histopathological diagnosis (LGD<HGD<EAC) on any surveillance biopsy taken at that endoscopy. The data demonstrated that the risk of both dysplasia and EAC increased with the number of genes methylated (Figure 6b). 17.6% of the cases in the <2 gene methylated group were dysplastic (low grade dysplasia only). In the group with 2 genes methylated the proportion of dysplastic cases increased to 42.3% including 11.5% high grade dysplasia/EAC. In the group with >2 genes methylated 14.5% of cases had LGD and 32.8% had HGD/EAC (combined cases of dysplasia and EAC: 47.3%). It should be noted that these data were derived from minimal sampling (3 biopsies for methylation study regardless of segment length) compared with the quadrantic biopsies taken every 2 cm to determine the histopathological diagnosis. Clinical variables such as age (t-test P=0.5) and sex (chi-square P=0.5) did not alter the risk for prevalent dysplasia and EAC. The mean segment length in non-dysplastic BE was observed to be 7.3 cm (range 2-14 cm) and 7.2 cm (range 2-16 cm) in cases with dysplasia/EAC (t-test P=0.9).
Discussion
This study has identified widespread changes in DNA methylation which distinguish between BE and EAC. Use of an array based strategy has enabled us to identify novel genes previously unknown to play a role in this disease. We hypothesized that methylation of imprinted and X-chromosome genes might provide candidate biomarkers since one copy is already inactivated. The analysis demonstrated almost 70% imprinted genes had altered methylation status in EAC and one of these, SLC22A18, was in the final stratification panel. Robust internal and external validation using pyrosequencing allowed us to select a four gene panel with an excellent receiver operating characteristic to distinguish between non-dysplastic BE and dysplastic BE/EAC samples (AUC=0.988). This panel enabled us to stratify patients into three (low, intermediate and high) risk groups based on the number of methylated genes identified from analysis of a limited number of biopsies by virtue of the field effect.
A number of previous studies have looked at DNA methylation changes in Barrett’s carcinogenesis. However none of the genes such as p16, APC(27) and MGMT(19) and a previously identified eight gene panel(28) were shown in this current study to be differentially methylated in EAC vs. BE. One reason for this might be that most biomarker studies have used a candidate, rather than an array based approach, and compared the BE associated disease states (dysplasia and EAC) to the normal squamous epithelium of the esophagus whereas we have compared dysplasia/EAC to BE in our study(17, 29, 30). Also these studies have focused on predicting the future risk of cancer whereas in our study we have focused on detecting dysplasia and early cancer in patients where it is not picked by histopathology. Metaplastic BE resembles intestinal rather than the squamous esophageal epithelium; and there is the possibility that the differences in DNA methylation observed between the normal squamous esophageal epithelium and BE/dysplasia/EAC might purely reflect differences in tissue morphology rather than playing any role in carcinogenesis. For this reason we included two duodenum samples as control in our array based methylation scan. If the methylation level of a gene was similar in both BE and duodenum; it was deemed that gene was involved in the maintenance of the columnar intestinal type epithelium rather than in the development of cancer. There were also methodological differences in the assays used; previous studies have employed methylation specific PCR (MSP) whereas here we used pyrosequencing which is a more quantitative method that has gained widespread acceptance(31). Lack of external validation has also been a problem in a number of recent studies(32, 33).
In this study more hypermethylation was seen in cancer compared to hypomethylation (Table 1), in keeping with the fact that promoter hypermethylation is a well-established phenomenon in cancer. We also observed greater methylation changes to occur within known CpG islands. However in a recent publication comparing the normal squamous mucosa with Barrett’s mucosa in 3 patients, methylation changes were reported to occur more frequently outside of CpG islands(34). It should however be noted that the majority of probes on the Illumina Infinium platform are positioned around promoter sites and 60% of human genes are associated with promoters spanning CpG islands. The recent availability of comprehensive genome wide coverage of methylation changes will enable further light to be shed on this.
For imprinted genes, as mentioned above almost 70% of genes showed statistically significant changes in methylation in EAC vs. BE (Wilcoxon P<0.05) (Table 1). Disruption of genomic imprinting is a well-established phenomenon in cancer. One imprinted gene, SLC22A18, met the criteria for validation. This gene is located in the 11p15.5 cluster which is an important tumor-suppressor gene region. Mutations, deletions and LOH of this gene have all been reported in different cancers highlighting its importance in tumorigenesis(35). Gain of imprinting of SLC22A18 has been documented in other cancers such as breast(36) and hepatocarcinomas(37) but we have shown for the first time that this can have a biomarker potential.
We looked at X-chromosome genes not only because DNA methylation plays a major role in X-inactivation in females but also because BE is more common in males who only have one copy of the X-chromosome and thus would theoretically only require one hit for the loss of the only functional allele. We were able to identify RGN, a putative tumor-suppressor gene(38, 39) that shows a successive increase in DNA methylation in the Barrett’s associated metaplasia-dysplasia-adenocarcinoma sequence in males but not in females (Supplementary Figure 2). TCEAL7 is also a candidate tumor suppressor gene shown to be epigenetically regulated in ovarian cancer and it functions by negatively regulating the NF-κB pathway(40, 41).
Of the other three genes identified by our study; RIN2 encodes a guanine nucleotide exchange factor; PIGR encodes a poly-Ig receptor downregulation of which has been shown to be associated with more frequent lymph node metastasis in gastro-esophageal junctional tumors(42); and GJA12 encodes a gap junction protein mutations in which have recently been shown to increase the risk for secondary lymphedema after treatment for breast cancer(43). It is interesting that GJA12 appears to be methylated late in carcinogenesis, at the high grade dysplasia/cancer transition compared with the other candidates described here.
The findings of our cross sectional cohort study to identify putative biomarkers have potential clinical applications. For the detection of dysplasia a four quadrant biopsy sampling technique is employed since dysplastic lesions can be focally distributed within the Barrett’s segment without any endoscopically visible lesion. Furthermore, there is substantial intra-observer disagreement among pathologists in differentiating between low and high grade dysplasia(9, 10, 44). In the prospective study we observed using our four gene methylation panel that DNA methylation is able to detect dysplasia/early-stage neoplasia in endoscopic biopsies even when the biopsy itself does not contain any visible dysplasia/early-stage neoplasia. This suggests that there is a field effect of methylation alterations in keeping with other research in the area of colon cancer(45, 46). The clonality of BE and evolving dysplastic lesions is still not clearly understood(47, 48) but there do appear to be widespread molecular changes prior to the emergence of phenotypical alterations visible by histopathology criteria(49). Our methylation panel is not intended to replace histopathology but to help reduce the chances for sampling bias and misclassification by taking advantage of the field effect and using a method that can be quantified in an objective fashion. It should be noted that there are differences between the cohorts which may explain the apparent false positives in the prospective cohort. The non-dysplastic samples from the retrospective cohort were ascertained from the index biopsy, whereas the non-dysplastic BE patients in the prospective study were BE patients under surveillance in tertiary referral centre with longer segments of BE (≥2 cm). Other studies have focused on the ability of biomarkers to predict future cancer development. Although this was not an endpoint of our multicenter trial it will be interesting to analyze the predictive power of this panel as follow-up data becomes available.
Overall, we feel that this panel has the potential as an adjunct to histopathology to flag patients who harbor prevalent high grade dysplasia and early adenocarcinoma. This needs validation in larger cohorts not skewed by referral bias in tertiary referral centers and is a promising area for further study.
Supplementary Material
Statement of translational relevance.
In Barrett’s esophagus endoscopic detection of dysplasia and its confirmation by histology suffers from sampling bias and a high inter and intra-observer variability. DNA methylation is known to play a role in cancer development and hence in this study we hypothesized that it could be used as an adjunct for detecting and diagnosing dysplasia/early cancer as molecular changes are more diffuse than dysplastic lesions which can be easily missed during endoscopy. Using Illumina Infinium arrays to select a methylation signature and pyrosequencing for both retrospective and prospective validation, we have shown that methylation changes can be used alongside histopathology to detect patients who have no visible signs of dysplasia/cancer at high risk of progression.
Acknowledgements
We would like to thank all patients who provided written consent for participation in this study; Cambridge Genomic Services where the microarray experiments were performed; Cambridge Institute of Medical Research where all the pyrosequencing assays were designed; Cambridge University Dept. Pathology where all pyrosequencing assays were run under the guidance of Dr A E K Ibrahim and all the staff over at Addenbrooke’s Hospital Tissue Bank, Addenbrooke's Centre for Clinical Investigation (ACCI) and S4 Addenbrooke’s Hospital.
Grant support:
M A Alvi was funded by Trinity College Cambridge and Cambridge Commonwealth Trust for his PhD. The laboratory work was funded by an MRC programme grant to RC Fitzgerald with additional clinical research infrastructure funding from the NHS National Institute for Health Research, Experimental Cancer Medicine Centre Network and the National Institute for Health Research Cambridge Biomedical Research Centre.
Footnotes
Conflict of interest statement:
There is no conflict of interest to declare.
References
- 1.Hvid-Jensen F, Pedersen L, Drewes AM, Sørensen HT, Funch-Jensen P. Incidence of Adenocarcinoma among Patients with Barrett's Esophagus. New England Journal of Medicine. 2011;365:1375–83. doi: 10.1056/NEJMoa1103042. [DOI] [PubMed] [Google Scholar]
- 2.Pohl H, Sirovich B, Welch HG. Esophageal Adenocarcinoma Incidence: Are We Reaching the Peak? Cancer Epidemiology Biomarkers & Prevention. 2010;19:1468–70. doi: 10.1158/1055-9965.EPI-10-0012. [DOI] [PubMed] [Google Scholar]
- 3.Crane SJ, Locke GR, Harmsen WS, Zinsmeister AR, Romero Y, Talley NJ. Survival Trends in Patients With Gastric and Esophageal Adenocarcinomas: A Population-Based Study. Mayo Clinic Proceedings. 2008;83:1087–94. doi: 10.4065/83.10.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spechler SJ, Sharma P, Souza RF, Inadomi JM, Shaheen NJ. American Gastroenterological Association Technical Review on the Management of Barrett's Esophagus. Gastroenterology. 2011;140:e18–e52. doi: 10.1053/j.gastro.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeMeester S. Evaluation and Treatment of Superficial Esophageal Cancer. Journal of Gastrointestinal Surgery. 2010;14:94–100. doi: 10.1007/s11605-009-1025-1. [DOI] [PubMed] [Google Scholar]
- 6.Yousef F, Cardwell C, Cantwell MM, Galway K, Johnston BT, Murray L. The Incidence of Esophageal Cancer and High-Grade Dysplasia in Barrett's Esophagus: A Systematic Review and Meta-Analysis. American Journal of Epidemiology. 2008;168:237–49. doi: 10.1093/aje/kwn121. [DOI] [PubMed] [Google Scholar]
- 7.Kahrilas PJ. The Problems with Surveillance of Barrett's Esophagus. New England Journal of Medicine. 2011;365:1437–8. doi: 10.1056/NEJMe1108435. [DOI] [PubMed] [Google Scholar]
- 8.Desai TK, Krishnan K, Samala N, Singh J, Cluley J, Perla S, et al. The incidence of oesophageal adenocarcinoma in non-dysplastic Barrett's oesophagus: a meta-analysis. Gut. 2011 doi: 10.1136/gutjnl-2011-300730. [DOI] [PubMed] [Google Scholar]
- 9.Goldblum JR. Controversies in the Diagnosis of Barrett Esophagus and Barrett-Related Dysplasia: One Pathologist's Perspective. Archives of Pathology & Laboratory Medicine. 2010;134:1479–84. doi: 10.5858/2010-0249-RA.1. [DOI] [PubMed] [Google Scholar]
- 10.Downs-Kelly E, Mendelin JE, Bennett AE, Castilla E, Henricks WH, Schoenfield L, et al. Poor Interobserver Agreement in the Distinction of High-Grade Dysplasia and Adenocarcinoma in Pretreatment Barrett's Esophagus Biopsies. Am J Gastroenterol. 2008;103:2333–40. doi: 10.1111/j.1572-0241.2008.02020.x. [DOI] [PubMed] [Google Scholar]
- 11.Curvers WL, ten Kate FJ, Krishnadath KK, Visser M, Elzer B, Baak LC, et al. Low-grade dysplasia in Barrett's esophagus: overdiagnosed and underestimated. Am J Gastroenterol. 2010;105:1523–30. doi: 10.1038/ajg.2010.171. [DOI] [PubMed] [Google Scholar]
- 12.Wani S, Falk GW, Post J, Yerian L, Hall M, Wang A, et al. Risk Factors for Progression of Low-Grade Dysplasia in Patients With Barrett's Esophagus. Gastroenterology. 2011;141:1179–86. e1. doi: 10.1053/j.gastro.2011.06.055. [DOI] [PubMed] [Google Scholar]
- 13.Shaheen NJ, Sharma P, Overholt BF, Wolfsen HC, Sampliner RE, Wang KK, et al. Radiofrequency Ablation in Barrett's Esophagus with Dysplasia. New England Journal of Medicine. 2009;360:2277–88. doi: 10.1056/NEJMoa0808145. [DOI] [PubMed] [Google Scholar]
- 14.Esteller M. Epigenetics in Cancer. New England Journal of Medicine. 2008;358:1148–59. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 15.Schulmann K, Sterian A, Berki A, Yin J, Sato F, Xu Y, et al. Inactivation of p16, RUNX3, and HPP1 occurs early in Barrett's-associated neoplastic progression and predicts progression risk. Oncogene. 2005;24:4138–48. doi: 10.1038/sj.onc.1208598. [DOI] [PubMed] [Google Scholar]
- 16.Clement G, Braunschweig R, Pasquier N, Bosman FT, Benhattar J. Alterations of the Wnt signaling pathway during the neoplastic progression of Barrett's esophagus. Oncogene. 2006;25:3084–92. doi: 10.1038/sj.onc.1209338. [DOI] [PubMed] [Google Scholar]
- 17.Jin Z, Cheng Y, Olaru A, Kan T, Yang J, Paun B, et al. Promoter hypermethylation of CDH13 is a common, early event in human esophageal adenocarcinogenesis and correlates with clinical risk factors. International Journal of Cancer. 2008;123:2331–6. doi: 10.1002/ijc.23804. [DOI] [PubMed] [Google Scholar]
- 18.Huang Y, Peters CJ, Fitzgerald RC, Gjerset RA. Progressive silencing of p14ARF in oesophageal adenocarcinoma. Journal of Cellular and Molecular Medicine. 2009;13:398–409. doi: 10.1111/j.1582-4934.2008.00336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuester D, El-Rifai We, Peng D, Ruemmele P, Kroeckel I, Peters B, et al. Silencing of MGMT expression by promoter hypermethylation in the metaplasia-dysplasia-carcinoma sequence of Barrett's esophagus. Cancer Letters. 2009;275:117–26. doi: 10.1016/j.canlet.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin Z, Cheng Y, Gu W, Zheng Y, Sato F, Mori Y, et al. A Multicenter, Double-Blinded Validation Study of Methylation Biomarkers for Progression Prediction in Barrett's Esophagus. Cancer Research. 2009;69:4112–5. doi: 10.1158/0008-5472.CAN-09-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taby R, Issa J-PJ. Cancer Epigenetics. CA: A Cancer Journal for Clinicians. 2010;60:376–92. doi: 10.3322/caac.20085. [DOI] [PubMed] [Google Scholar]
- 22.Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 23.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 24.Primer designing tool. [cited; Available from: http://www.ncbi.nlm.nih.gov/tools/primer-blast/
- 25.Geneimprint. [cited; Available from: www.geneimprint.com.
- 26.American Gastroenterological Association Medical Position Statement on the Management of Barrett's Esophagus. Gastroenterology. 2011;140:1084–91. doi: 10.1053/j.gastro.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 27.Wang JS, Guo M, Montgomery EA, Thompson RE, Cosby H, Hicks L, et al. DNA Promoter Hypermethylation of p16 and APC Predicts Neoplastic Progression in Barrett's Esophagus. Am J Gastroenterol. 2009;104:2153–60. doi: 10.1038/ajg.2009.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin Z, Cheng Y, Gu W, Zheng Y, Sato F, Mori Y, et al. A Multicenter, Double-Blinded Validation Study of Methylation Biomarkers for Progression Prediction in Barrett's Esophagus. Cancer Res. 2009;69:4112–5. doi: 10.1158/0008-5472.CAN-09-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin Z, Hamilton JP, Yang J, Mori Y, Olaru A, Sato F, et al. Hypermethylation of the AKAP12 Promoter is a Biomarker of Barrett's-Associated Esophageal Neoplastic Progression. Cancer Epidemiol Biomarkers Prev. 2008;17:111–7. doi: 10.1158/1055-9965.EPI-07-0407. [DOI] [PubMed] [Google Scholar]
- 30.Jin Z, Olaru A, Yang J, Sato F, Cheng Y, Kan T, et al. Hypermethylation of Tachykinin-1 Is a Potential Biomarker in Human Esophageal Cancer. Clin Cancer Res. 2007;13:6293–300. doi: 10.1158/1078-0432.CCR-07-0818. [DOI] [PubMed] [Google Scholar]
- 31.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protocols. 2007;2:2265–75. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 32.Kaz AM, Wong CJ, Luo Y, Virgin JB, Kay Washington M, Willis JE, et al. DNA methylation profiling in Barrett's esophagus and esophageal adenocarcinoma reveals unique methylation signatures and molecular subclasses. Epigenetics. 2011;6:1403–12. doi: 10.4161/epi.6.12.18199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhai R, Zhao Y, Su L, Cassidy L, Liu G, Christiani DC. Genome-wide DNA methylation profiling of cell-free serum DNA in esophageal adenocarcinoma and Barrett esophagus. Neoplasia (New York, NY) 2012;14:29–33. doi: 10.1593/neo.111626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alvarez H, Opalinska J, Zhou L, Sohal D, Fazzari MJ, Yu Y, et al. Widespread Hypomethylation Occurs Early and Synergizes with Gene Amplification during Esophageal Carcinogenesis. PLoS Genet. 2011;7:e1001356. doi: 10.1371/journal.pgen.1001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu S-H, Feng D-F, Ma Y-B, Zhang H, Zhu Z-A, Li Z-Q, et al. Promoter methylation and downregulation of SLC22A18 are associated with the development and progression of human glioma. Journal of Translational Medicine. 2011;9:156. doi: 10.1186/1479-5876-9-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gallagher E, Mc Goldrick A, Chung WY, Mc Cormack O, Harrison M, Kerin M, et al. Gain of imprinting of SLC22A18 sense and antisense transcripts in human breast cancer. Genomics. 2006;88:12–7. doi: 10.1016/j.ygeno.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Schwienbacher C, Gramantieri L, Scelfo R, Veronese A, Calin GA, Bolondi L, et al. Gain of imprinting at chromosome 11p15: A pathogenetic mechanism identified in human hepatocarcinomas. Proceedings of the National Academy of Sciences. 2000;97:5445–9. doi: 10.1073/pnas.090087497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamaguchi M, Daimon Y. Overexpression of regucalcin suppresses cell proliferation in cloned rat hepatoma H4-II-E cells: Involvement of intracellular signaling factors and cell cycle-related genes. Journal of Cellular Biochemistry. 2005;95:1169–77. doi: 10.1002/jcb.20490. [DOI] [PubMed] [Google Scholar]
- 39.Maia C, Santos C, Schmitt F, Socorro S. Regucalcin is under-expressed in human breast and prostate cancers: Effect of sex steroid hormones. Journal of Cellular Biochemistry. 2009;107:667–76. doi: 10.1002/jcb.22158. [DOI] [PubMed] [Google Scholar]
- 40.Rattan R, Narita K, Chien J, Maguire JL, Shridhar R, Giri S, et al. TCEAL7, a putative tumor suppressor gene, negatively regulates NF-[kappa]B pathway. Oncogene. 2010;29:1362–73. doi: 10.1038/onc.2009.431. [DOI] [PubMed] [Google Scholar]
- 41.Chien J, Staub J, Avula R, Zhang H, Liu W, Hartmann LC, et al. Epigenetic silencing of TCEAL7 (Bex4) in ovarian cancer. Oncogene. 2005;24:5089–100. doi: 10.1038/sj.onc.1208700. [DOI] [PubMed] [Google Scholar]
- 42.Gologan A, Acquafondata M, Dhir R, Sepulveda AR. Polymeric Immunoglobulin Receptor–Negative Tumors Represent a More Aggressive Type of Adenocarcinomas of Distal Esophagus and Gastroesophageal Junction. Archives of Pathology & Laboratory Medicine. 2008;132:1295–301. doi: 10.5858/2008-132-1295-PIRTRA. [DOI] [PubMed] [Google Scholar]
- 43.Finegold DN, Baty CJ, Knickelbein KZ, Perschke S, Noon SE, Campbell D, et al. Connexin 47 Mutations Increase Risk for Secondary Lymphedema Following Breast Cancer Treatment. Clinical Cancer Research. 2012;18:2382–90. doi: 10.1158/1078-0432.CCR-11-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kerkhof M, Van Dekken H, Steyerberg EW, Meijer GA, Mulder AH, De Bruïne A, et al. Grading of dysplasia in Barrett's oesophagus: substantial interobserver variation between general and gastrointestinal pathologists. Histopathology. 2007;50:920–7. doi: 10.1111/j.1365-2559.2007.02706.x. [DOI] [PubMed] [Google Scholar]
- 45.Belshaw NJ, Elliott GO, Foxall RJ, Dainty JR, Pal N, Coupe A, et al. Profiling CpG island field methylation in both morphologically normal and neoplastic human colonic mucosa. Br J Cancer. 2008;99:136–42. doi: 10.1038/sj.bjc.6604432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen L, Kondo Y, Rosner GL, Xiao L, Hernandez NS, Vilaythong J, et al. MGMT Promoter Methylation and Field Defect in Sporadic Colorectal Cancer. Journal of the National Cancer Institute. 2005;97:1330–8. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]
- 47.Leedham SJ, Preston SL, McDonald SAC, Elia G, Bhandari P, Poller D, et al. Individual crypt genetic heterogeneity and the origin of metaplastic glandular epithelium in human Barrett’s oesophagus. Gut. 2008;57:1041–8. doi: 10.1136/gut.2007.143339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merlo LMF, Shah NA, Li X, Blount PL, Vaughan TL, Reid BJ, et al. A Comprehensive Survey of Clonal Diversity Measures in Barrett's Esophagus as Biomarkers of Progression to Esophageal Adenocarcinoma. Cancer Prevention Research. 2010;3:1388–97. doi: 10.1158/1940-6207.CAPR-10-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reid BJ, Kostadinov R, Maley CC. New Strategies in Barrett's Esophagus: Integrating Clonal Evolutionary Theory with Clinical Management. Clinical Cancer Research. 2011;17:3512–9. doi: 10.1158/1078-0432.CCR-09-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.