Abstract
Background
Glioblastoma highly expresses the proto-oncogene MET in the setting of resistance to bevacizumab. MET engagement by hepatocyte growth factor (HGF) results in receptor dimerization and autophosphorylation mediating tumor growth, invasion, and metastasis. Evasive revascularization and the recruitment of TIE2-expressing macrophages (TEMs) are also triggered by anti-VEGF therapy.
Methods
We investigated the activity of altiratinib (a novel balanced inhibitor of MET/TIE2/VEGFR2) against human glioblastoma stem cell lines in vitro and in vivo using xenograft mouse models. The biological activity of altiratinib was assessed in vitro by testing the expression of HGF-stimulated MET phosphorylation as well as cell viability after altiratinib treatment. Tumor volume, stem cell and mesenchymal marker levels, microvessel density, and TIE2-expressing monocyte infiltration were evaluated in vivo following treatment with a control, bevacizumab alone, bevacizumab combined with altiratinib, or altiratinib alone.
Results
In vitro, HGF-stimulated MET phosphorylation was completely suppressed by altiratinib in GSC17 and GSC267, and altiratinib markedly inhibited cell viability in several glioblastoma stem cell lines. More importantly, in multiple xenograft mouse models, altiratinib combined with bevacizumab dramatically reduced tumor volume, invasiveness, mesenchymal marker expression, microvessel density, and TIE2-expressing monocyte infiltration compared with bevacizumab alone. Furthermore, in the GSC17 xenograft model, altiratinib combined with bevacizumab significantly prolonged survival compared with bevacizumab alone.
Conclusions
Together, these data suggest that altiratinib may suppress tumor growth, invasiveness, angiogenesis, and myeloid cell infiltration in glioblastoma. Thus, altiratinib administered alone or in combination with bevacizumab may overcome resistance to bevacizumab and prolong survival in patients with glioblastoma.
Keywords: altiratinib, bevacizumab, glioma, MET, TIE2–expressing monocytes
Glioblastoma is the most common malignant brain tumor and is characterized by cellular heterogeneity, vascular proliferation, and extensive tissue infiltration. The treatment for glioblastoma includes surgery, radiation, and chemotherapy. The prognosis remains poor, with a median survival time of 12–15 months in patients with this disease.1 A new, effective approach for prolonging survival and improving quality of life in patients with glioblastoma is urgently needed.
The antiangiogenic agent bevacizumab (Avastin; Roche/Genentech), a recombinant humanized monoclonal antibody directed against vascular endothelial growth factor (VEGF), has been approved by the U.S. FDA for the treatment of glioblastoma.2 Bevacizumab alone or combined with other chemotherapy inhibits tumor angiogenesis and tumor growth; however, after initially responding to bevacizumab treatment, glioblastoma develops resistance to bevacizumab, becoming increasingly invasive and growing faster.3 One mechanism of this resistance is the inhibition of the VEGF pathway, which causes vascular pruning in the tumor, induces a hypoxic environment, and subsequently triggers MET expression, which enhances tumor invasion and metastasis.4–6 A previous study by our group showed that after xenograft mouse models of glioblastoma developed resistance to bevacizumab therapy, MET was expressed at high levels in those tumors compared with the vehicle control group.7 Anti-VEGF therapy has also been shown to lead to enhanced and sustained MET phosphorylation status in gliomas by blocking VEGF-mediated downregulation of MET activation.8
MET, a transmembrane tyrosine kinase receptor that binds only to the hepatocyte growth factor (HGF) ligand, is overexpressed, activated, amplified, or mutated in several types of human cancer.9–13 HGF is secreted by cells of mesenchymal origin—including fibroblasts, macrophages, and endothelial cells.14 MET gene amplification has been detected in glioblastoma,15–17 and MET is expressed in primary glioblastoma.13,18 MET plays a wide-ranging role in tumor cell invasion,19 proliferation,20 and antiapoptotic effects21 in various cancers. High MET expression is associated with poor prognosis in patients with glioblastoma.22–24 According to gene expression profiling analyses of glioblastoma, MET is a signature gene associated with the glioblastoma mesenchymal subtype.25,26 Interestingly, MET is a functional marker of the glioblastoma stem cell subset.22,27 More importantly, MET confers resistance to radiation therapy in patients with glioblastoma.28
Tunica interna endothelial cell receptor 2 (TIE2)-expressing monocytes are a subpopulation of circulating blood monocytes that contribute to angiogenesis in human glioblastoma orthotopic xenografts.29 According to a recent report, TIE2-expressing monocytes contribute to the refractoriness of glioblastoma to bevacizumab treatment in a U87 MG xenograft mouse model.30 In that study, the TEMs were recruited to the normal tissue/tumor invasive boundary and were characterized by high levels of MMP9 expression. TEMs recruited to the normal/tumor boundary were also demonstrated from human biopsy samples of anti-VEGF–treated glioblastoma patients.30 Infiltration of these myeloid cells likely accounts for the mesenchymal signature that results following bevacizumab treatment31,32. Thus, TIE2-expressing monocytes are a novel, biologically relevant marker of angiogenesis and may be a promising anticancer target in glioblastoma and other tumors.
Altiratinib (currently under development by Deciphera Pharmaceuticals, LLC) is a novel inhibitor of MET, TIE2, VEGFR2, and tropomyosin receptor kinase family kinases.33 Because of its balanced inhibitory potencies for MET, TIE2, and VEGFR2, it was hypothesized that single agent altiratinib therapy would be efficacious in experimental glioblastoma models and further, when used in combination with bevacizumab, would prevent or delay bevacizumab-mediated resistance mechanisms.
In the current study, we evaluated the antitumor effects of altiratinib in a genetically diverse panel of human glioblastoma stem cell lines in vitro and in vivo. We found that altiratinib combined with bevacizumab significantly inhibited tumor growth, invasiveness, mesenchymal marker expression, angiogenesis, and TIE2-expressing monocyte infiltration compared with bevacizumab alone in GSC11 and GSC17 xenograft mouse models. Furthermore, altiratinib, in combination with bevacizumab, provided a significant survival benefit compared with single-agent bevacizumab. This study provides a rationale for further clinical investigation of altiratinib combined with bevacizumab in patients with glioblastoma.
Materials and Methods
Cell Lines, Reagents, and Treatment
The human glioblastoma stem cell lines GSC2, GSC262, GSC267, GSC295, GSC300, GSC6-27, GSC7-2, GSC11, GSC17, GSC231, GSC20, GSC272, GSC28, GSC8-11, GSC23, and GSC280 were derived from recurrent glioblastoma specimens as previously described.34 Glioma stem cell lines were generated under the Pathology Core of the MD Anderson Cancer Center Brain SPORE (National Institutes of Health, P50CA127001). These glioblastoma stem cells were maintained in suspension in Dulbecco's modified Eagle's medium (DMEM)/F12 containing epidermal growth factor (20 ng/mL), basic fibroblast growth factor (20 ng/mL), and B27 (2%) at 37°C in a 5% CO2 atmosphere.
To test HGF-stimulated MET phosphorylation, we prepared GSC11, GSC17, GSC20, and GSC267 cells in 6-well plates at a density of 5 × 105 cells per well. After 6 hours of incubation, cells were stimulated by 40 ng/mL HGF for 10 minutes and harvested for Western blot on phospho-MET. To investigate whether altiratinib inhibits HGF-stimulated MET phosphorylation, we prepared GSC267 and GSC17 cells in 6-well plates at a density of 5 × 105 and treated cells with different concentrations of altiratinib for 6 hours. Cells were then stimulated by 40 ng/mL HGF for 10 minutes, and cell lysates were subjected to Western blot on phospho-MET.
Cell Viability Assay
GSC6-27,GSC7-2, GSC11, GSC17, GSC231, GSC295, GSC20, GSC300, GSC28, GSC272, GSC8-11, GSC262, GSC23, and GSC280 cells were plated in 96-well black-walled plates at a density of 5 × 103 cells per well. The DMEM/F12 (2% B27, 20 ng/mL EGF, and 20 ng/mL FGF) medium contained altiratinib at concentrations of 0.001 µM, 0.01 µM, 0.1 µM, 1 µM, and 5 µM. Viable cells were quantitated 72 hours after drug exposure using the CellTiter-Glo Assay (Promega) per manufacturer's instructions. The results were generated from 3 independent experiments.
Immunoblotting Analysis
Cells were lysed in an ice-cold lysis buffer containing 50 mM Tris-Cl (pH 7.5), 100 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM PMSF, 1 μg/mL leupeptin, and 1 μg/mL pepstatin A. The protein concentration in the supernatant was determined using a Pierce bicinchoninic acid protein assay (Life Technologies). Samples were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis with an 8%–12% gradient, and the separated proteins were electrophoretically transferred to nitrocellulose membranes. The blots were incubated with the primary antibodies against phospho-MET (1:1000; Cell Signaling Technology), against MET (1:1000; Cell Signaling Technology), or against tubulin (1:3000; Sigma-Aldrich). The blots were then incubated with horseradish peroxidase–linked secondary anti-rabbit or anti-mouse antibodies (Bio-Rad Laboratories).
Mouse Experiments
For the in vivo experiments, we used 4 to 6-week-old female nude mice that were strictly inbred at The University of Texas MD Anderson Cancer Center and maintained in the MD Anderson Veterinary Facility in accordance with Laboratory Animal Resources Commission standards and were treated according to approved protocol 10-07-12132. For the in vivo experiments, GSC11 (5 × 105) or GSC17 (5 × 104) glioblastoma cells were implanted intracranially into nude mice. Beginning 4 days after tumor cell implantation, bevacizumab (10 mg/kg) was administered by i.p. injection twice per week and altiratinib (10 mg/kg) was administered by oral gavage twice daily. Control mice for these cohorts were treated with phosphate-buffered saline by i.p. injection and/or with 0.4% HPMC vehicle by oral gavage. One cohort of 10 mice per group was treated continuously and followed for survival. A separate cohort of 9 mice per group was treated continuously until the designated time point, and the tumors from these mice were extracted at 3, 4, and 5 weeks in the GSC11 xenograft mice model and at 3.5, 4.5, and 5.5 weeks in the GSC17 xenograft mice model after the start of treatment.
When the mice treated for the designated time period developed signs and symptoms of advanced tumors, they were euthanized, and their brains were removed, fixed in 4% formaldehyde for 24 hours, and embedded in paraffin. The tissues were then sectioned serially (4 µm) and stained with hematoxylin and eosin (H&E; Sigma-Aldrich). Tumor formation and phenotype were determined by histologic analysis of the H&E-stained sections. The tumor volume, greatest longitudinal diameter (length), and greatest transverse diameter (width) were measured using an external caliper and Adobe Illustrator software. The tumor volumes were calculated by the following formula: volume = ½(length × width2).35
Immunohistochemistry and Immunofluorescence
For immunohistochemical analyses, the tissue sections were deparaffinized and subjected to graded rehydration. After blocking the tissue in 5% horse and goat serum and antigen-retrieval solution (citrate buffer, pH 6.0), we incubated the tissue sections overnight at 4°C with primary antibodies against Factor VIII (1:500; A0082, Dako) to assess microvessel formation, vimentin (1:100; M0725, Dako) to assess mesenchymal marker expression, F4/80 (1:50; 123102, BioLegend) to assess monocyte infiltration, nestin (1:300; Ab6142, Abcam), HGF (1:200; LS-B4657, LSBio), and TIE2 (1:50; Santa Cruz Biotechnology) to assess TIE2-expressing monocyte infiltration. Texas red fluorescein isothiocyanate–conjugated secondary antibodies or anti-rat immunoglobulin G antibodies (Invitrogen) were used for 1 hour at room temperature.
Statistical Analyses
All statistical analyses were conducted with GraphPad 6 (InStat) software for Windows 7. Survival analysis was conducted using the Kaplan-Meier method, and differences in survival between treatment groups were assessed using the log-rank test. All other comparisons were performed using an unpaired 2-tailed Student t test. Summary statistics for continuous data are expressed as the mean ± standard error of the mean. P values <.05 were considered statistically significant. Nestin, vimentin, Factor VIII, and TIE2+/F4/80 + staining were assessed using the Image-Pro Plus system version 7.0 (Media Cybernetics) in ×10 fields of at least 3 tumor samples per group with 3–4 different sections per tumor sample.
Results
Altiratinib Inhibits MET Phosphorylation and Cell Viability in Diverse Glioblastoma Stem Cell Lines
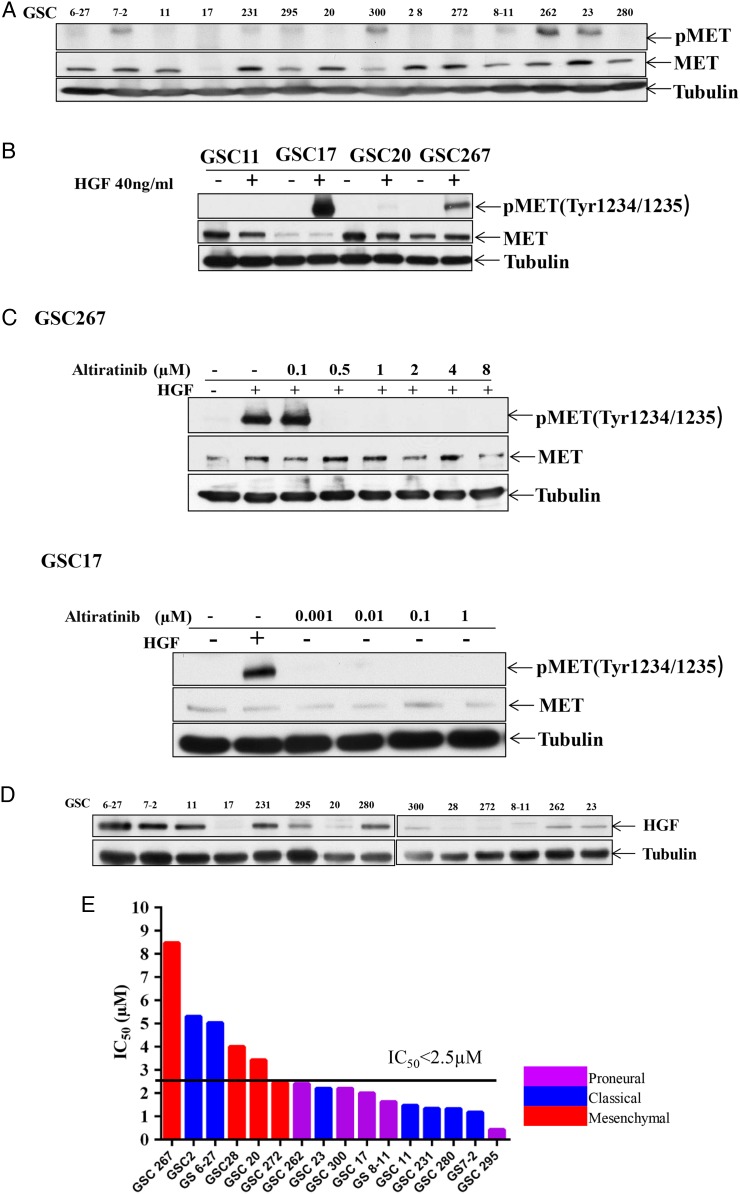
Glioblastomas are classified as proneural, neural, classical, or mesenchymal on the basis of gene expression and gene alteration;25 the glioblastoma stem cell lines we studied have diverse genetic backgrounds representing these different subtypes. First, we assessed MET and phospho-MET expression levels in these glioblastoma stem cell lines using Western blotting. MET was significantly overexpressed in GSC6-27, GSC7-2, GSC11, GSC231, GSC295, GSC20, GSC300, GSC28, GSC272, GSC8-11, GSC262, GSC23, and GSC280 and was detectable in GSC17. More importantly, MET phosphorylation was measured and was constitutively phosphorylated in the GSC7-2, GSC231, GSC295, GSC300, GSC272, GSC8-11, GSC262, and GSC23 cell lines (Fig. 1A). Second, we asked whether MET and phospho-MET are activated by HGF stimulation in glioblastoma. After the GSC11, GSC17, GSC20, and GSC267 cells were stimulated by 40 ng/mL HGF for 10 minutes, MET phosphorylation was induced in GSC17 and GSC267 but not in GSC11 or GSC20 (Fig. 1B). Third, we asked whether altiratinib inhibits HGF-stimulated MET phosphorylation in glioblastoma. Our data showed that altiratinib robustly inhibited MET phosphorylation in a dose-dependent manner in GSC17 and GSC267 (Fig. 1C). In addition, we determined HGF expression levels in the glioblastoma stem cell lines because HGF is the only ligand that binds to MET. We found HGF expression in GSC6-27, GSC7-2, GSC11, GSC231, GSC295, GSC280, GSC300, GSC262, and GSC23 (Fig. 1D).
Fig. 1.
Altiratinib inhibits MET phosphorylation and cell viability in diverse glioblastoma stem cell lines. (A) Western blotting revealed that MET and phospho-MET were expressed in multiple human glioblastoma stem cell lines. (B) Western blotting revealed that phospho-MET was expressed in GSC17 and GSC267 cells after stimulation by 40 ng/mL hepatocyte growth factor (HGF) for 10 minutes. (C) Western blotting revealed that altiratinib inhibited HGF-stimulated MET phosphorylation in GSC17 and GSC267 cells in a dose-dependent manner. (D) Western blotting revealed that HGF was expressed in multiple human glioblastoma stem cell lines. (E) The bar graph shows the IC50 values for altiratinib in several glioblastoma stem cell lines and the genetic profiles of those cell lines.
Furthermore, we performed a cell viability assay to determine the half maximal inhibitory concentration (IC50) for altiratinib in 16 glioblastoma cell lines. Altiratinib inhibited cell viability regardless of genetic background. Although GSC267, GSC2, GSC20, GSC28, and GSC6-27 were relatively resistant to altiratinib treatment (IC50 > 2.5 µM), GSC272, GSC262, GSC23, GSC300, GSC17, GSC8-11, GSC11, GSC231, GSC280, GSC7-2, and GSC295 were sensitive to altiratinib treatment (IC50 < 2.5 µM; Fig. 1E). To investigate the mechanism of altiratinib-mediated inhibition of cell viability, we performed an Annexin V assay 72 hours after treatment with 1 µM and 2 µM altiratinib in GSC17, GSC1,1 and GSC6-27 cell lines. In GSC17 cells, the percentage of Annexin V + 7-ADD + cells was 58.95% and 65.94% (vs control, 8.7%). In the GSC11, the percentage of Annexin V + 7-ADD + cells was 18.71% and 21.40% (vs control, 14.86%). And in GSC6-27, the percentage of Annexin V + 7-ADD + cells was 10.71% and 18.42% (vs control, 10.85%). Western blots demonstrated an increase in caspase 3 activation after being treated with 1 µM altiratinib in GSC11, GSC17, and GSC6-27, whereas the autophagy marker LC3B increased in only 2 lines(ie, GSC17 and GSC6-27). These data suggest that altiratinib induces apoptosis in most of the GSCs, although some of the cells also undergo autophagy (Supplementary data S1).
Altiratinib Combined With Bevacizumab Reduces Glioblastoma Tumor Volume in Xenograft Mouse Models
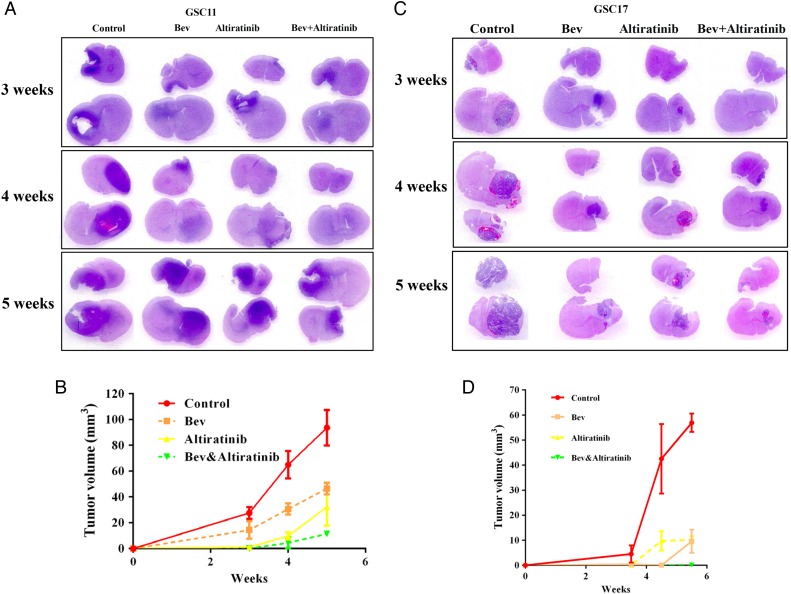
MET was previously shown to be highly expressed in glioblastomas that developed resistance to bevacizumab therapy7 (and Supplementary data S2). To investigate whether altiratinib combined with bevacizumab inhibits glioblastoma growth in vivo, we measured tumor volumes in glioblastoma stem cell xenograft mouse models (Fig. 2A). In the GSC11 model, tumor volumes at 3, 4, and 5 weeks were 27.6 ± 4.5 mm3, 64.9 ± 10.6 mm3, and 93.6 ± 13.7 mm3, respectively; in the control-treated mice and 14.3 ± 6.9 mm3, 30.7 ± 4.2 mm3, and 46.6 ± 4.5 mm3, respectively; and in the bevacizumab-treated mice (vs control: P= .049, P= .006, and P= .005, respectively). Altiratinib treatment alone significantly inhibited the tumor volume, which was 1.2 ± 0.5 mm3, 9.8 ± 2.8 mm3, and 32.3 ± 14.6 mm3 at 3, 4, and 5 weeks, respectively (vs control: P = .001, P= .001, and P= .006, respectively). Altiratinib combined with bevacizumab dramatically reduced the tumor volume to 0.1 ± 0.1 mm3, 4.5 ± 4.3 mm3, and 11.4 ± 2.0 mm3 at 3, 4, and 5 weeks, respectively (vs control: P= .004, P= .001, and P= .001, respectively; vs bevacizumab alone: P= .027, P= .002, and P= 0.001, respectively; Fig. 2B).
Fig. 2.
Altiratinib combined with bevacizumab reduced tumor volume in GSC11 and GSC17 xenograft mouse models. The line graph shows the sizes of the tumors in each treatment group for the GSC11 (A and B) and GSC17 (C and D) xenograft mouse models.
Altiratinib also reduced tumor growth in the GSC17 xenograft mouse model (Fig. 2C). At 3.5, 4.5, and 5.5 weeks, the tumor volumes were 4.5 ± 3.4 mm3, 42.5 ± 13.9 mm3, and 56.9 ± 7.4 mm3, respectively, in the control-treated mice and 0.4 ± 0.4 mm3, 0.05 ± 0.03 mm3, and 9.6 ± 4.6 mm3 in the bevacizumab-treated mice (vs control: P= .105, P= .007, and P= .001, respectively). Altiratinib treatment alone markedly suppressed tumor volume at 0.1 ± 0.2 mm3, 9.8 ± 3.9 mm3, and 10.1 ± 1.4 mm3 at 3.5, 4.5, and 5.5 weeks, respectively (vs control: P= .087, P= .017, and P= .001, respectively). Altiratinib combined with bevacizumab dramatically reduced the tumor volume to 0.081 ± 0.1 mm3, 0.04 ± 0.02 mm3, and 0.19 ± 0.11 mm3 at 3.5, 4.5, and 5.5 weeks, respectively (vs control: P= .085, P= .006, and P= .001, respectively; vs bevacizumab alone: P= .28, P= .58, and P = .02, respectively; Fig. 2D).
Altiratinib Combined With Bevacizumab Prolongs Survival in Glioblastoma Xenograft Mouse Models
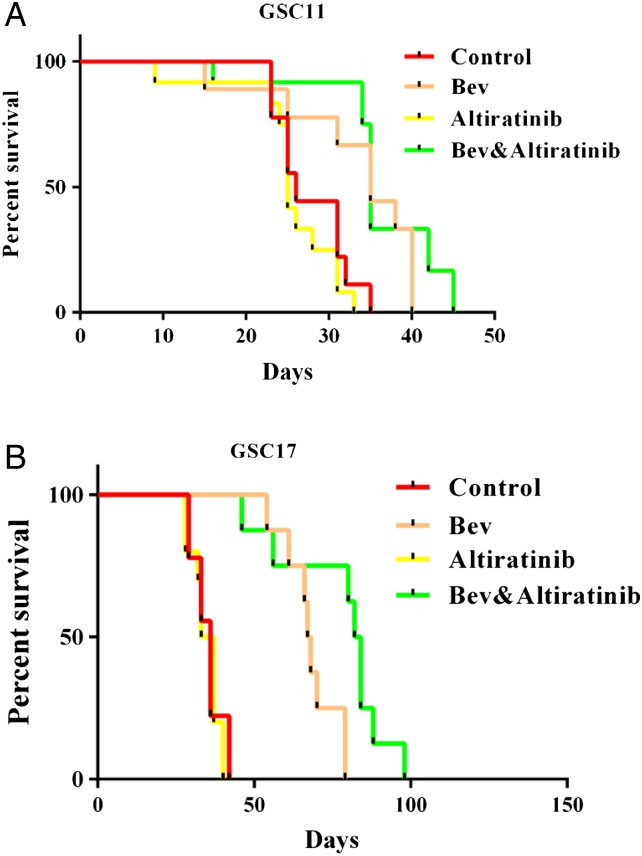
To further examine the antitumor efficiency of altiratinib in vivo, we performed a survival study in our glioblastoma stem cell xenograft mouse models. In the GSC11 model, the median survival times were 26 days in the control mice, 35 days in the bevacizumab-treated mice (vs control: P= .017), 25 days in the altiratinib-alone treated mice (vs control: P= .49), and 35 days in the mice treated with altiratinib combined with bevacizumab (vs control: P= .006, vs bevacizumab alone: P= .239; Fig. 3A). Thus, in the GSC11 mouse model, altiratinib combined with bevacizumab significantly prolonged survival compared with the control but did not significantly prolong survival compared with bevacizumab alone.
Fig. 3.
Altiratinib combined with bevacizumab prolonged survival in the GSC11 and GSC17 xenograft mouse models, as shown by the Kaplan-Meier survival curves. (A) In the GSC11 xenograft mouse model, the mice treated with altiratinib combined with bevacizumab survived longer than the control-treated mice. However, no statistically significant difference was seen between the combination treatment group and the bevacizumab-alone group. (B) In the GSC17 xenograft mouse model, the mice treated with altiratinib combined with bevacizumab survived longer than the control-treated mice and survived longer than the mice treated with bevacizumab alone.
In the GSC17 xenograft mouse model, the median survival time was 36 days in the control-treated mice and 67.5 days in the bevacizumab-treated mice (vs control: P < .001). However, the median survival time was 83 days in mice treated with altiratinib combined with bevacizumab (vs control: P < .001, vs bevacizumab alone: P= .012; Fig. 3B). Thus, altiratinib combined with bevacizumab significantly prolonged survival in the GSC17 xenograft mouse model.
To investigate whether altiratinib inhibits Met phosphorylation in vivo, we performed immunofluorescence staining for pMET in the GSC17 xenograft mouse model. After bevacizumab treatment, pMET expression was greater compared with control. However, pMET expression was lower in tumors treated with altiratinib alone and in those treated with the combination of altiratinib and bevacizumab (Supplementary data S3).
Altiratinib Combined With Bevacizumab Reduces Tumor Invasiveness in Glioblastoma Xenograft Mouse Models
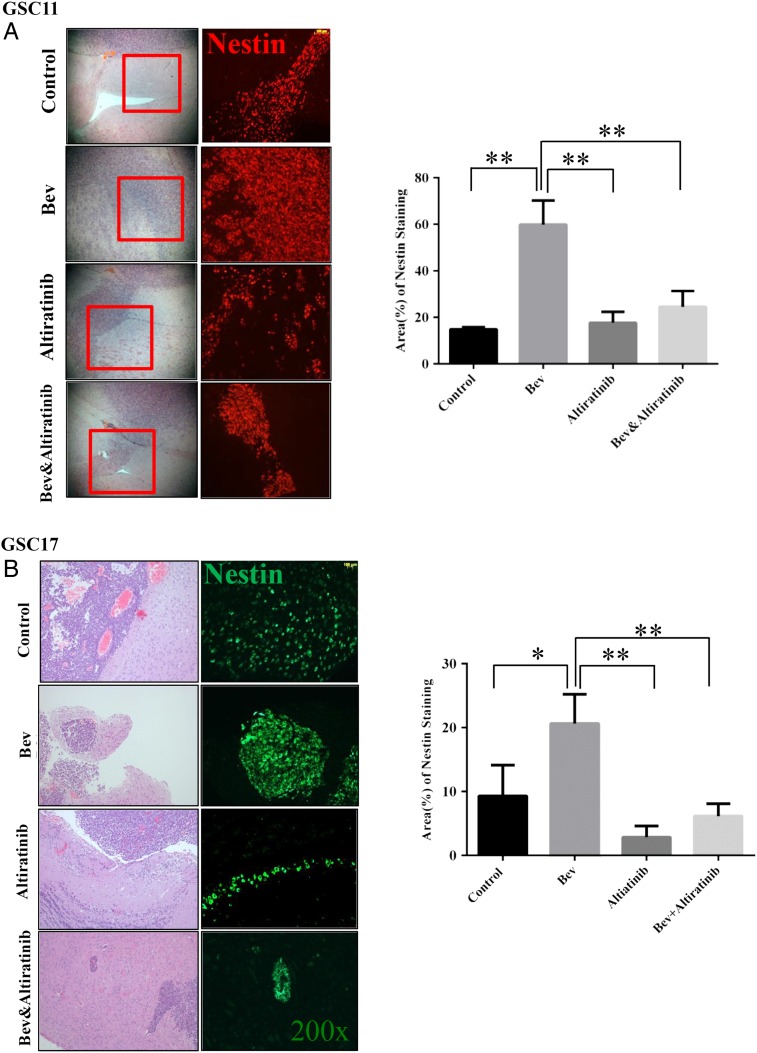
It is well known that tumor cells become more aggressive and more invasive after developing resistance to antiangiogenic therapy.31 Indeed, in our xenograft mouse models, we found that tumor cells became more invasive in bevacizumab-treated mice compared with the control-treated mice. However, the tumors treated with altiratinib alone showed very little invasiveness, and altiratinib combined with bevacizumab dramatically decreased invasiveness compared with bevacizumab alone in the xenograft mouse models, as detailed below.
To quantify invasiveness, we stained tissue sections with the glioblastoma stem cell marker nestin and quantified the nestin-positive staining area. In the GSC11 xenograft mouse model, the nestin-positive area was 14.8% ± 0.52% in the control-treated tumors and 59.8% ± 6.0% in the bevacizumab-treated tumors (vs control: P= .002). However, the nestin-positive area was only 24.5% ± 3.9% in the tumors treated with altiratinib combined with bevacizumab (vs control: P= .072; vs bevacizumab alone: P= .008) and 17.7% ± 2.7% in the tumors treated with altiratinib alone (Fig. 4A). Similarly, in the GSC17 xenograft mouse model, the nestin-positive area was 9.3% ± 2.8% in the control-treated tumors and 20.6% ± 2.7% in the bevacizumab-treated tumors (vs control: P= .042). However, the nestin-positive area was only 6.1% ± 1.1% in the tumors treated with altiratinib combined with bevacizumab (vs control: P= .354; vs bevacizumab alone: P= .007) and 2.9% ± 1.0% in the tumors treated with altiratinib alone (Fig. 4B).
Fig. 4.
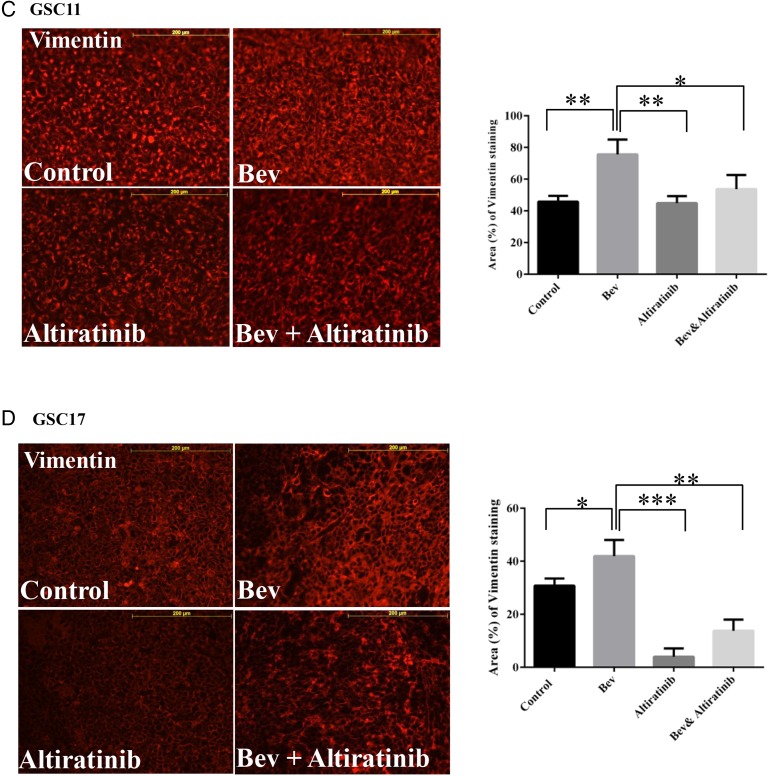
Altiratinib combined with bevacizumab reduced tumor invasiveness in the GSC11 and GSC17 xenograft mouse models. Nestin immunofluorescence staining revealed that nestin-positive staining was suppressed in tumor invasion areas in the mice treated with altiratinib alone or with altiratinib combined with bevacizumab compared with bevacizumab alone in the GSC11 xenograft model (A) and the GSC17 xenograft model (B). Representative photomicrographs are shown (magnification ×100). Vimentin immunofluorescence staining revealed that vimentin-positive staining was suppressed in mice treated with altiratinib alone or with altiratinib combined with bevacizumab compared with bevacizumab alone in the GSC11 xenograft model (C) and the GSC17 xenograft model (D). Representative photomicrographs are shown (magnification ×200).
Our previous study showed that antiangiogenic therapy promoted expression of mesenchymal markers (such as vimentin and smooth muscle actin) in glioblastoma tumors in vivo.31 We performed immunofluorescence staining for vimentin and quantified the vimentin-positive staining area at the tumor invasion sites in GSC11 and GSC17 xenograft mouse model tissue samples. In the GSC11 model, the vimentin-positive area was 45.8% ± 2.1% in the control-treated tumors and 75.6% ± 5.4% in the bevacizumab-treated tumors (vs control: P= .007). However, the vimentin-positive area was 53.7% ± 5.1% in the tumors treated with altiratinib combined with bevacizumab (vs control: P= .254; vs bevacizumab alone: P= .04) and 44.87% ± 2.6% in the tumors treated with altiratinib alone (vs bevacizumab: P= .007) (Fig. 4C). In the GSC17 xenograft mouse model, the vimentin-positive area was 41.9% ± 3.5% after treatment with bevacizumab (vs 30.78% ± 1.6% in control: P= .044) but was only 13.8% ± 2.4% in the tumors treated with altiratinib combined with bevacizumab (vs bevacizumab alone: P= .003) and 4.0% ± 1.8% in the tumors treated with altiratinib alone (vs control: P= .001; vs bevacizumab alone: P= .001; Fig. 4D).
Altiratinib Combined With Bevacizumab Decreases Blood Vessel Formation in Glioblastoma Xenograft Mouse Models
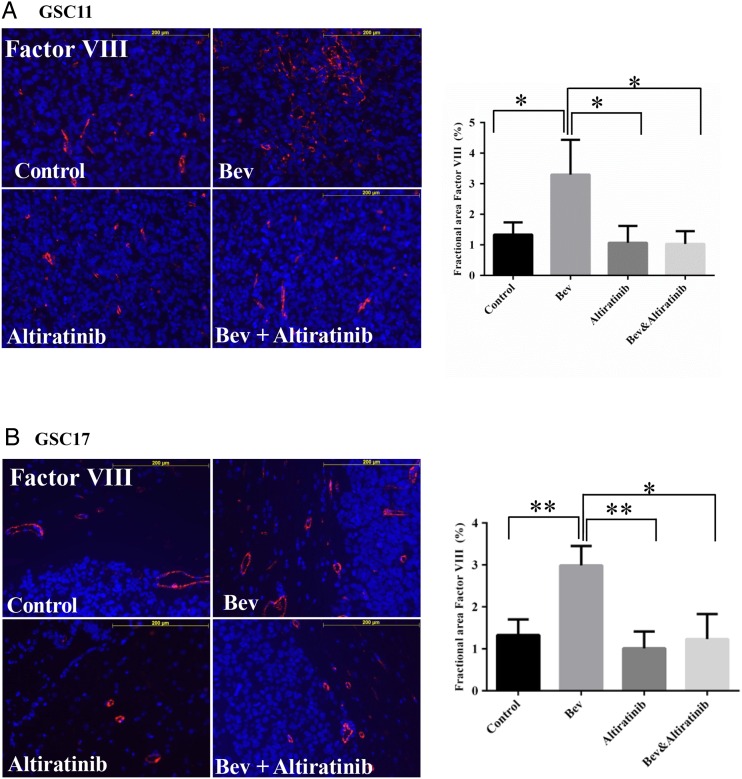
In our previous study, glioblastoma stem cells escaped anti-VEGF therapy through revascularization and became more invasive in vivo.31 We performed immunofluorescence staining for Factor VIII and quantified the Factor VIII–positive staining area at the tumor sites in GSC11 and GSC17 xenograft mouse models. In the GSC11 model, the Factor VIII–positive area was 1.3% ± 0.4% in the control-treated tumors and 3.3% ± 1.1% in the bevacizumab-treated tumors (vs control: P= .048). However, the Factor VIII–positive area was only 1.03% ± 0.4% in the tumors treated with altiratinib combined with bevacizumab (vs bevacizumab alone: P= .032) and 1.1% ± 0.5% in the tumors treated with altiratinib alone (vs bevacizumab alone: P= .038; Fig. 5A).
Fig. 5.
Altiratinib combined with bevacizumab (Bev) decreased blood vessel formation in GSC11 and GSC17 xenograft mouse models. Immunofluorescence Factor VIII staining analysis revealed that Factor VIII staining was decreased in mice treated with altiratinib or with altiratinib combined with bevacizumab compared with those treated with bevacizumab alone in the GSC11 xenograft model (A) and the GSC17 xenograft model (B). Representative photomicrographs are shown (magnification ×200).
Similarly, in the GSC17 xenograft mouse model, the Factor VIII–positive area was 3.0% ± 0.3% in the tumors treated with bevacizumab (vs control: P= .008). However, the Factor VIII–positive area was only 1.2% ± 0.4% in the tumors treated with altiratinib combined with bevacizumab (vs bevacizumab alone: P= .016) and 1.0% ± 0.2% in the tumors treated with altiratinib alone (vs bevacizumab alone: P= .005; Fig. 5B).
Altiratinib Combined With Bevacizumab Suppresses TIE2-expressing Monocyte Infiltration Into Tumors in Glioblastoma Xenograft Mouse Models
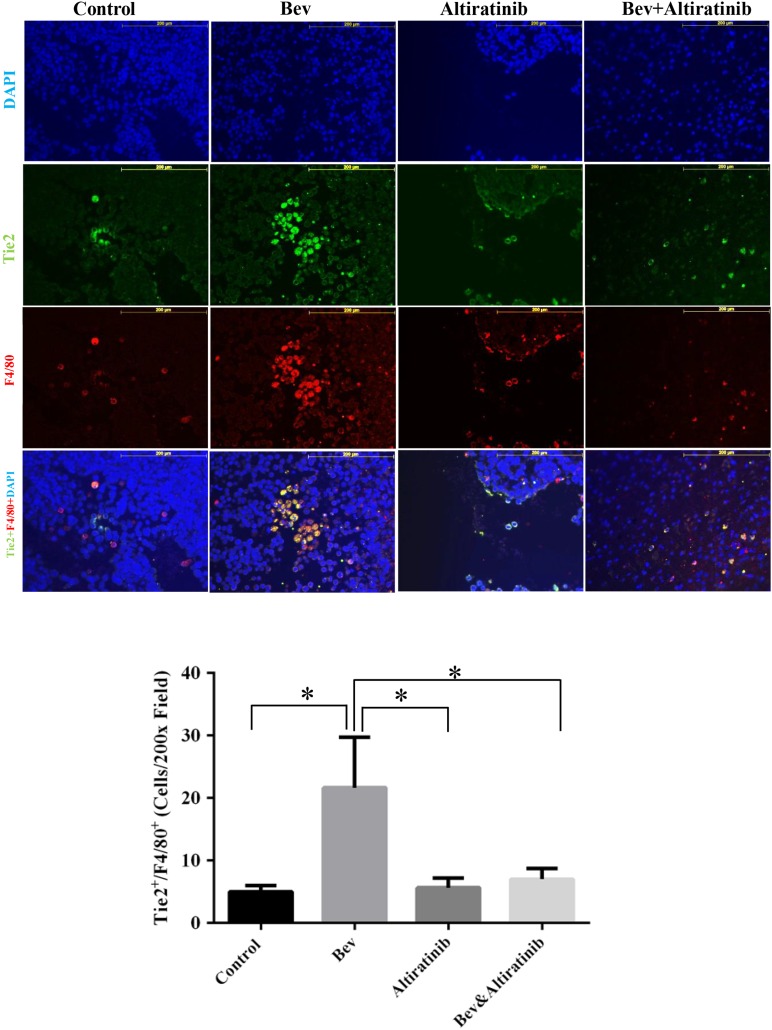
TIE2 is expressed by monocytes observed to accumulate at the invasive edges of malignant gliomas treated with VEGF inhibitors.30 We investigated whether altiratinib inhibits infiltration of these monocytes into glioblastomas by performing co-immunofluorescence staining for TIE2 and F4/80 and quantifying the TIE2-positive/F4/80-positive staining cells at the tumor sites in the GSC17 xenograft mouse model. After bevacizumab treatment, the tumor infiltration of TIE2-positive/F4/80-positive cells was greater than in controls (vs control: P= .024). However, the infiltration of TIE2-positive/F4/80-positive cells was lower in tumors treated with altiratinib alone and in those treated with altiratinib combined with bevacizumab than in tumors treated with bevacizumab alone (P= .028 and P= .038 respectively; Fig. 6). Notably, altiratinib in combination with bevacizumab completely ablated bevacizumab-induced increases in TIE2-expressing monocyte recruitment.
Fig. 6.
Altiratinib combined with bevacizumab (Bev) suppressed TIE2-expressing monocyte infiltration into tumors in GSC17 xenograft mice models. TIE2 and F4/80 co-immunofluorescence staining revealed that TIE2-positive/F4/80-positive cell infiltration into tumors was decreased in mice treated with altiratinib or with altiratinib combined with bevacizumab compared with those treated with bevacizumab alone in the GSC17 xenograft model. Representative photomicrographs are shown (magnification ×200).
Discussion
This study provides preclinical evidence that altiratinib effectively inhibits MET/TIE2/VEGFR2 activation and indicates that altiratinib, either as a single agent or combined with bevacizumab, may be a promising candidate for treating glioblastoma patients. Altiratinib was engineered to inhibit tumor MET genomic alterations and overexpression while also blocking cross talk with the tumor microenvironment by inhibition of wild-type MET, TIE2, and VEGFR2.33 All 3 of these kinases have been implicated in glioblastoma invasion and progression. Our data show that altiratinib had robust antitumor activity in vitro and in vivo—and combined with bevacizumab—can prevent the bevacizumab-induced invasiveness, epithelial-mesenchymal transition, revascularization, and TIE2-expressing monocyte infiltration.
The high activity of MET in glioblastoma tumors occurs through several different mechanisms such as gene amplification, transcriptional regulation, HGF autocrine or paracrine stimulation, ligand-independent activation, and therapy-induced MET activation.8,13 MET gene amplification, the most predictive genetic event, is associated with high sensitivity to MET inhibitors.36,37 In the present study, GSC17 and GSC262 were the only cell lines that showed MET gene amplification, and the GSC262 cell line also demonstrated constitutive phosphorylation of MET. GSC17 and GSC262 were consistently sensitive to altiratinib treatment in the cell viability assay, and altiratinib was most efficacious in the MET-amplified GSC17 in vivo xenograft model.
Another mechanism of MET upregulation that could predict sensitivity to altiratinib is HGF autocrine status. The GSC7-2, GSC231, GSC295, and GSC23 cell lines showed high HGF protein expression and high phospho-MET expression as well as sensitivity to altiratinib treatment in vitro. Altiratinib was recently shown to be sensitive in the U87 glioblastoma xenograft model, known to be dependent on HGF-mediated autocrine signaling.33 Similarly, a previous study showed that glioblastoma xenograft tumors with HGF autocrine activation were sensitive to treatment with the MET inhibitor SGX523.38 In the present study, the GSC300, GSC272, and GSC8-11 cells showed sensitivity to altiratinib treatment and expressed high phospho-MET and low HGF. Therefore, the HGF-independent phospho-MET expression level may also predict sensitivity to altiratinib treatment.
We also found that glioblastoma cells in which MET could be activated by paracrine HGF stimulation were sensitive to altiratinib treatment. MET was markedly phosphorylated by exogenous HGF stimulation in the GSC17 cells, which inherently expressed relatively low phospho-MET and HGF. Phospho-MET was more robustly induced by HGF in GSC17 than in other cell lines, perhaps at least in part due to the high level of MET amplification. Another study showed that exogenous HGF can stimulate phospho-MET expression in the glioblastoma stem cell line GSCS 1228. This short-term exogenous activation of MET by HGF enhanced the effect of the MET inhibitor crizotinib in inhibiting tumor growth in the GSCS 1228–derived xenograft model.39
Therapy-induced MET activation is a pragmatic mechanism that has been demonstrated in both preclinical and clinical practice. Bevacizumab promotes glioblastoma tumor resistance through multiple mechanisms mediated in large part by bevacizumab therapy-induced hypoxia. Such hypoxia triggers (i) upregulation of MET activity in glioma cells that mediates transition to an invasive mesenchymal phenotype, (ii) evasive revascularization mediated by ANG or HGF activation of TIE2 or MET on endothelial cells, and (iii) an increase in myeloid cell recruitment and infiltration including macrophages, granulocytes, and TIE2-expressing monocytes (TEMs). Regarding bevacizumab-mediated upregulation of glioma MET activity, it has been demonstrated that bevacizumab therapy blocks cross talk between VEGFR2 receptors and MET receptors on glioma cells.8 Under normal signaling homeostasis, VEGF activation of VEGFR2 receptors was shown to recruit PTP1B phosphatase to a complex including VEGFR2 and MET receptors. The activated VEGFR2-recruited phosphatase downregulated the phosphorylation and activity of MET. Bevacizumab sequestration of VEGF blocked this inhibitory cross talk between VEGFR2 and MET, leading to sustained MET signaling in glioma cells. Also, the angiopoietin (ANG)/TIE2 axis has been implicated in bevacizumab-mediated provocation of evasive revascularization via during anti-VEGF therapy,40,41 and stromal HGF has also been demonstrated to mediate evasive vascularization during anti-VEGF therapy.42,43 Thirdly, TIE2-expressing monocytes are a subpopulation of circulating blood monocytes that contribute to angiogenesis in human glioblastoma orthotopic xenografts.29 TEMs are expressed in the peripheral blood of cancer patients and are also recruited to human solid tumors.44 According to a recent report, TEMs contribute to the refractoriness of glioblastomas to bevacizumab treatment in a U87 MG xenograft mouse model.30 Infiltration of these myeloid cells likely accounts for the mesenchymal signature that results following bevacizumab treatment (personal communication with Roel Verhaak, MD Anderson). Thus, TIE2-expressing monocytes are a novel, biologically relevant marker of angiogenesis and invasiveness, and may be a promising anticancer target in glioblastoma and other tumors.
Altiratinib's balanced profile for inhibiting MET, TIE2, and VEGFR2 blocks these hallmark tumor microenvironment mechanisms characterizing anti-VEGF therapy resistance. Indeed, when combined with bevacizumab, altiratinib blocked tumor invasion, blocked evasive revascularization, and blocked recruitment of TIE2-expressing monocytes. Interestingly, we found that bevacizumab treatment induced HGF expression in the recruited F4/80/TIE2-positive macrophages in the GSC17 xenograft mouse model (Supplementary data S4). This finding indicated that bevacizumab could stimulate macrophages to produce HGF in the glioma tumor microenvironment, and this stromal HGF could further activate MET on glioma cells already primed by bevacizumab for increased MET expression. Since mouse HGF does not bind to human MET, our murine xenograft model could not assess the role of recruited macrophage-derived HGF on tumor progression. Nonetheless, this observation suggests that such HGF produced from recruited macrophages could play a pro-tumoral role in glioblastoma patients treated with bevacizumab.
Altiratinib significantly prolonged survival in the GSC17 xenograft mouse model when combined with bevacizumab. Thus, altiratinib may provide better inhibition of several mediators of tumor progression in the setting of potent and sustained VEGF sequestration that translates into meaningful outcomes. Taken together, these findings suggest that human glioblastomas, and perhaps other tumors, may be effectively treated by altiratinib wherein cross talk between tumors and the microenvironment leads to upregulated MET expression, evasive revascularization, and/or recruitment of pro-tumoral TEMs. However, to evaluate our findings in future clinical trials, the association between relevant biomarkers and sensitivity to altiratinib would need to be investigated.
In conclusion, the new MET/TIE2/VEGFR2 inhibitor altiratinib suppressed tumor growth and invasiveness in GSC11 and GSC17 xenograft mouse models. Altiratinib combined with bevacizumab prevents bevacizumab-induced revascularization, infiltration of TIE2-expressing monocytes, and upregulation of mesenchymal markers, and could be clinically effective for the treatment of patients with glioblastoma.
Supplementary Material
Funding
This work was supported in part by the Martha G. Williams Memorial Brain Tumor Research Fund (J.D.) and by Deciphera Pharmaceuticals, LLC.
Supplementary Material
Acknowledgments
We thank Sarah Bronson (Department of Scientific Publications, The University of Texas MD Anderson Cancer Center, Houston, TX) and Kay Hyde (Department of Neuro-Oncology, The University of Texas MD Anderson Cancer Center, Houston, TX) for editorial assistance. Portions of this study were presented at the 2014 Society of Neuro-Oncology Annual Meeting as an oral presentation.
Conflict of interest statement. J.D. is on the SAB of Deciphera Pharmaceuticals and is a paid consultant. B.S. and D.F. are employees of Deciphera Pharmaceuticals and have equity positions in the company.
References
- 1. Reardon DA, Wen PY. Glioma in 2014: unravelling tumour heterogeneity-implications for therapy. Nat Rev Clin Oncol. 2015;12 (2):69–70. [DOI] [PubMed] [Google Scholar]
- 2. Khasraw M, Ameratunga M, Grommes C. Bevacizumab for the treatment of high-grade glioma: an update after phase III trials. Expert Opin Biol Ther. 2014;14 (5):729–740. [DOI] [PubMed] [Google Scholar]
- 3. Norden AD, Young GS, Setayesh K et al. Bevacizumab for recurrent malignant gliomas: efficacy, toxicity, and patterns of recurrence. Neurology. 2008;70 (10):779–787. [DOI] [PubMed] [Google Scholar]
- 4. Pennacchietti S, Michieli P, Galluzzo M, Mazzone M, Giordano S, Comoglio PM. Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer Cell. 2003;3 (4):347–361. [DOI] [PubMed] [Google Scholar]
- 5. di Tomaso E, Snuderl M, Kamoun WS et al. Glioblastoma recurrence after cediranib therapy in patients: lack of “rebound” revascularization as mode of escape. Cancer Res. 2011;71 (1):19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Paez-Ribes M, Allen E, Hudock J et al. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell. 2009;15 (3):220–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Piao Y, Liang J, Holmes L, Henry V, Sulman E, de Groot JF. Acquired resistance to anti-VEGF therapy in glioblastoma is associated with a mesenchymal transition. Clin Cancer Res. 2013;19 (16):4392–4403. [DOI] [PubMed] [Google Scholar]
- 8. Lu KV, Chang JP, Parachoniak CA et al. VEGF inhibits tumor cell invasion and mesenchymal transition through a MET/VEGFR2 complex. Cancer Cell. 2012;22 (1):21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Edakuni G, Sasatomi E, Satoh T, Tokunaga O, Miyazaki K. Expression of the hepatocyte growth factor/c-Met pathway is increased at the cancer front in breast carcinoma. Pathol Int. 2001;51 (3):172–178. [DOI] [PubMed] [Google Scholar]
- 10. Galimi F, Torti D, Sassi F et al. Genetic and expression analysis of MET, MACC1, and HGF in metastatic colorectal cancer: response to met inhibition in patient xenografts and pathologic correlations. Clin Cancer Res. 2011;17 (10):3146–3156. [DOI] [PubMed] [Google Scholar]
- 11. Schmidt L, Duh FM, Chen F et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat Genet. 1997;16 (1):68–73. [DOI] [PubMed] [Google Scholar]
- 12. Ma PC, Jagadeeswaran R, Jagadeesh S et al. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer Res. 2005;65 (4):1479–1488. [DOI] [PubMed] [Google Scholar]
- 13. Koochekpour S, Jeffers M, Rulong S et al. Met and hepatocyte growth factor/scatter factor expression in human gliomas. Cancer Res. 1997;57 (23):5391–5398. [PubMed] [Google Scholar]
- 14. Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432 (7015):332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chi AS, Batchelor TT, Dias-Santagata D et al. Prospective, high-throughput molecular profiling of human gliomas. J Neurooncol. 2012;110 (1):89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455 (7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pierscianek D, Kim YH, Motomura K et al. MET gain in diffuse astrocytomas is associated with poorer outcome. Brain Pathol. 2013;23 (1):13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kong DS, Song SY, Kim DH et al. Prognostic significance of c-Met expression in glioblastomas. Cancer. 2009;115 (1):140–148. [DOI] [PubMed] [Google Scholar]
- 19. Boccaccio C, Comoglio PM. Invasive growth: a MET-driven genetic programme for cancer and stem cells. Nat Rev Cancer. 2006;6 (8):637–645. [DOI] [PubMed] [Google Scholar]
- 20. McCubrey JA, Steelman LS, Chappell WH et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta. 2007;1773 (8):1263–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bowers DC, Fan S, Walter KA et al. Scatter factor/hepatocyte growth factor protects against cytotoxic death in human glioblastoma via phosphatidylinositol 3-kinase- and AKT-dependent pathways. Cancer Res. 2000;60 (15):4277–4283. [PubMed] [Google Scholar]
- 22. Joo KM, Jin J, Kim E et al. MET signaling regulates glioblastoma stem cells. Cancer Res. 2012;72 (15):3828–3838. [DOI] [PubMed] [Google Scholar]
- 23. Olmez OF, Cubukcu E, Evrensel T et al. The immunohistochemical expression of c-Met is an independent predictor of survival in patients with glioblastoma multiforme. Clin Transl Oncol. 2014;16 (2):173–177. [DOI] [PubMed] [Google Scholar]
- 24. Lathia JD, Li M, Sinyuk M et al. High-throughput flow cytometry screening reveals a role for junctional adhesion molecule a as a cancer stem cell maintenance factor. Cell Rep. 2014;6 (1):117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Verhaak RG, Hoadley KA, Purdom E et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17 (1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Phillips HS, Kharbanda S, Chen R et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9 (3):157–173. [DOI] [PubMed] [Google Scholar]
- 27. De Bacco F, Casanova E, Medico E et al. The MET oncogene is a functional marker of a glioblastoma stem cell subtype. Cancer Res. 2012;72 (17):4537–4550. [DOI] [PubMed] [Google Scholar]
- 28. Lal B, Xia S, Abounader R, Laterra J. Targeting the c-Met pathway potentiates glioblastoma responses to gamma-radiation. Clin Cancer Res. 2005;11 (12):4479–4486. [DOI] [PubMed] [Google Scholar]
- 29. De Palma M, Venneri MA, Galli R et al. Tie2 identifies a hematopoietic lineage of proangiogenic monocytes required for tumor vessel formation and a mesenchymal population of pericyte progenitors. Cancer Cell. 2005;8 (3):211–226. [DOI] [PubMed] [Google Scholar]
- 30. Gabrusiewicz K, Liu D, Cortes-Santiago N et al. Anti-vascular endothelial growth factor therapy-induced glioma invasion is associated with accumulation of Tie2-expressing monocytes. Oncotarget. 2014;5 (8):2208–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Piao Y, Liang J, Holmes L et al. Glioblastoma resistance to anti-VEGF therapy is associated with myeloid cell infiltration, stem cell accumulation, and a mesenchymal phenotype. Neuro Oncol. 2012;14 (11):1379–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim H, Verhaak RG. Transcriptional mimicry by tumor-associated stroma. Nat Genet. 2015;47 (4):307–309. [DOI] [PubMed] [Google Scholar]
- 33. Smith BD, Kaufman MD, Leary CB et al. Altiratinib inhibits tumor growth, invasion, angiogenesis, and microenvironment-mediated drug resistance via balanced inhibition of MET, TIE2, and VEGFR2. Mol Cancer Ther. 2015;14 (9):2013–2034. [DOI] [PubMed] [Google Scholar]
- 34. He H, Nilsson CL, Emmett MR et al. Glycomic and transcriptomic response of GSC11 glioblastoma stem cells to STAT3 phosphorylation inhibition and serum-induced differentiation. J Proteome Res. 2010;9 (5):2098–2108. [DOI] [PubMed] [Google Scholar]
- 35. Euhus DM, Hudd C, LaRegina MC, Johnson FE. Tumor measurement in the nude mouse. J Surg Oncol. 1986;31 (4):229–234. [DOI] [PubMed] [Google Scholar]
- 36. Christensen JG, Schreck R, Burrows J et al. A selective small molecule inhibitor of c-Met kinase inhibits c-Met-dependent phenotypes in vitro and exhibits cytoreductive antitumor activity in vivo. Cancer Res. 2003;63 (21):7345–7355. [PubMed] [Google Scholar]
- 37. Smolen GA, Sordella R, Muir B et al. Amplification of MET may identify a subset of cancers with extreme sensitivity to the selective tyrosine kinase inhibitor PHA-665752. Proc Natl Acad Sci USA. 2006;103 (7):2316–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xie Q, Bradley R, Kang L et al. Hepatocyte growth factor (HGF) autocrine activation predicts sensitivity to MET inhibition in glioblastoma. Proc Natl Acad Sci USA. 2012;109 (2):570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang Y, Farenholtz KE, Yang Y et al. Hepatocyte growth factor sensitizes brain tumors to c-MET kinase inhibition. Clin Cancer Res. 2013;19 (6):1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hashizume H, Falcon BL, Kuroda T et al. Complementary actions of inhibitors of angiopoietin-2 and VEGF on tumor angiogenesis and growth. Cancer Res. 2010;70 (6):2213–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rigamonti N, Kadioglu E, Keklikoglou I, Wyser Rmili C, Leow CC, De Palma M. Role of angiopoietin-2 in adaptive tumor resistance to VEGF signaling blockade. Cell Rep. 2014;8 (3):696–706. [DOI] [PubMed] [Google Scholar]
- 42. Xin X, Yang S, Ingle G et al. Hepatocyte growth factor enhances vascular endothelial growth factor-induced angiogenesis in vitro and in vivo. Am J Pathol. 2001;158 (3):1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sennino B, Ishiguro-Oonuma T, Wei Y et al. Suppression of tumor invasion and metastasis by concurrent inhibition of c-Met and VEGF signaling in pancreatic neuroendocrine tumors. Cancer Discov. 2012;2 (3):270–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Venneri MA, De Palma M, Ponzoni M et al. Identification of proangiogenic TIE2-expressing monocytes (TEMs) in human peripheral blood and cancer. Blood. 2007;109 (12):5276–5285. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.