Abstract
The one-carbon metabolism pathway disorder was important role in successful pregnancy. The MTHFR and TS protein were crucial factor in one-carbon metabolism. To investigate the association between recurrent implantation failure (RIF) and enzymes in the one-carbon metabolism pathway. A total of 120 women diagnosed with RIF and 125 control subjects were genotyped for MTHFR 677C>T, 1298A>C, TSER 2R/3R and TS 1494del/ins by a polymerase chain reaction-restriction fragment length polymorphism assay. According to the gene-gene combination analysis, the MTHFR 677/MTHFR 1298 (TT/AA) and MTHFR 677/TS 1494 (TT/6bp6bp) genetic combinations were associated with relatively higher risks [adjusted odds ratio (AOR), 2.764; 95% CI, 1.065–7.174; P = 0.037 and AOR, 3.186; 95% CI, 1.241–8.178; P = 0.016] in RIF patients compared to the CC/AA (MTHFR 677/MTHFR 1298) and TT/6bp6bp (MTHFR 677/TS 1494) combinations, respectively. The results suggested that the combined MTHFR 677/MTHFR 1298 genotype might be associated with increased risk of RIF. To the best of our knowledge, this study is the first to elucidate the potential association of MTHFR, TS and TSER polymorphisms with RIF risk in Korean patients.
Introduction
Recurrent implantation failure (RIF) is one of the most common reproductive disorders observed at in vitro fertilization (IVF) clinics. RIF is defined as implantation failure following three IVF cycles involving the transfer of a high-grade embryo [1]. Various factors influence successful implantation, including anatomic or endometrial factors, thrombophilia, genetics, and immunologic factors, to name a few [2,3]. However, in terms of a clinical approach, the etiology of RIF remains a complicated challenge [2,4].
Successful implantation occurs during a short period of time between days 7 to 10 of the secretory phase of the normal menstrual cycle, when the embryo develops into a blastocyst and migrates to the receptive uterus. Communication between the embryo and the uterus is critical for synchronizing embryonic development and uterine differentiation during the implantation window and is regulated by numerous pathways, including hormones and signaling factors [5]. Among these pathways, folate metabolism is reported to be an essential regulator of early development and pregnancy [6–8]. Folate is a critical molecule in the synthesis of S-adenosylmethionine (SAMe), which acts as a methyl group carrier in cellular processes including DNA synthesis, DNA methylation, and amino-acid metabolism. Additionally, folate metabolism is important for folate-homocysteine homeostasis, which is regulated by numerous enzymes in the folate-methionine cycle.
Methylenetetrahydrofolate reductase (MTHFR), which is required for the conversion of 5,10-methylenetetrahydrofolate (5,10-MTHF) to 5-methyltetrahydrofolate (5-MTHF), is one of the major regulatory enzymes in the folate-homocysteine cycle. The methyl group of 5-MTHF is transferred to homocysteine to produce methionine, which is important for the methylation of various substrates such as DNA, RNA, proteins, and lipids. Further, genetic polymorphisms in MTHFR are associated with various diseases [9]. Thymidylate synthase (TS), which catalyzes the conversion of deoxyuridine monophosphate (dUMP) to deoxythymidine monophosphate (dTMP) by the transfer of a 5,10-MTHF methyl group, is a crucial enzyme in DNA biosynthesis [10]. As well, the activity of TS is affected by polymorphisms in TS and in the TS enhancer region (TSER), which likewise results in altered metabolic reactions and the occurrence of disease [11].
In this study, we demonstrated a relationship between RIF and genetic polymorphisms in genes that encode enzymes involved in folate metabolism, including MTHFR and TS, as well as in the enhancer region of TS, in Korean patients experiencing RIF.
Materials and Methods
Participants
The study population consisted of 225 female participants recruited from the Department of Obstetrics and Gynecology of CHA Bundang Medical Center, CHA University (Seongnam, Korea) between March 2010 and December 2012 who were experiencing RIF (n = 120) or served as controls (n = 125). The clinical characteristics of the participants are summarized in Table 1. The mean age of the RIF patients and the controls was 34.23 ± 3.33 and 32.75 ± 7.47, respectively (Table 1).
Table 1. Clinical characteristics of RIF patients and control subjects.
| Characteristic | Control (n = 125) | RIF patients (n = 120) |
|---|---|---|
| Age (y, mean± SD) | 32.75 ± 7.47 | 34.23 ± 3.33 |
| BMI (kg/m2, mean± SD) | 21.72 ± 3.40 | 20.96 ± 2.59 |
| Previous implantation failure (N, mean± SD) | NA | 1.59 ± 0.50 |
| Live birth (N, mean± SD) | 1.78 ± 0.74 | NA |
| Mean gestational age (week, mean± SD) | 39.36 ± 1.66 | NA |
| Confirmed history of RPL (n) | NA | 17 (14.2) |
| tHcy (μmol/L, mean± SD) | NA | 6.56 ± 1.32 |
| Folate (ng/mL, mean± SD) | NA | 15.37 ± 11.23 |
| BUN (mg/dl, mean± SD) | NA | 10.35 ± 2.87 |
| Creatinine (mg/dl, mean± SD) | NA | 0.78 ± 0.1 |
| Uric acid (mg/dl, mean± SD) | NA | 3.93 ± 0.96 |
| Total Cholesterol (mg/dl, mean± SD) | NA | 188.93 ± 44.28 |
Note: RIF, recurrent implantation failure; SD, standard deviation; BMI, body mass index; RPL, recurrent pregnancy loss; NA, not applicable; tHcy, total plasma homocysteine; BUN, blood urea nitrogen
The study was approved by the Institutional Review Board of CHA Bundang Medical Center reviewed on 23 February 2010 (reference no. PBC09–120) and all patients provided written informed consent.
Serum human chorionic gonadotropin (hCG) concentrations were less than 5 mIU/mL 14 days after embryo transfer. All transferred embryos were examined by the embryologist before transfer and were deemed to be of good quality. We evaluated both the male and female partners in couples experiencing RIF. Subjects who were diagnosed with RIF due to anatomic, chromosomal, hormonal, infectious, autoimmune, or thrombotic causes were excluded from the study group. Anatomical abnormalities were evaluated using several imaging modalities including sonography, hysterosalpingogram, hysteroscopy, computerized tomography, and magnetic resonance imaging. Karyotyping was conducted using standard protocols. Hormonal causes including hyperprolactinemia, luteal insufficiency, and thyroid disease were excluded by measuring prolactin, thyroid-stimulating hormone, free T4, follicle-stimulating hormone, luteinizing hormone, and progesterone levels in peripheral blood. Lupus anticoagulant and anticardiolipin antibodies were examined to rule out autoimmune diseases such as lupus and antiphospholipid syndrome. Thrombotic disorders were defined as thrombophilia and were evaluated by protein C and protein S deficiencies and by the presence of anti-α2 glycoprotein antibody. Among the initial 167 patients evaluated, 47 who had intrauterine adhesion, hypothyroidism, trisomy and chromosomal translocation (patients or spouses), or antiphospholipid syndrome were excluded from the patient group, leaving 120 patients for the study. Enrollment criteria for the control group included regular menstrual cycles, normal karyotype (46XX), a history of at least one naturally-conceived pregnancy, and no history of pregnancy loss. Data were collected identically for both groups.
Genotyping
Genomic DNA was extracted from patient peripheral blood samples using a G-DEX(TM) blood extraction kit (iNtRON Biotechnology, Seongnam, South Korea). All of the genetic polymorphisms were detected by polymerase chain reaction (PCR) amplification and restriction enzyme digestion. The PCR primers for each polymorphism were as follows: MTHFR 677C>T, forward 5’-TGA AGG AGA AGG TGT CTG CGG GA-3’ and reverse 5’-AGG ACG GTG CGG TGA GAG TC-3’; MTHFR 1298A>C, forward 5'-CTT TGG GGA GCT GAA GGA CTACTA C-3' and reverse 5'-CAC TTT GTG ACC ATT CCG GTT TG-3'; TSER 2R/3R, forward 5’-CGT GGC TCC TGC GTT TCC-3’ and reverse 5’-GAG CCG GCC ACA GGC ATG-3’; and TS 1494 0bp/6bp, forward 5’-CAA ATC TGA GGG AGC TGA GT-3’ and reverse 5’-CAG ATA AGT GGC AGT ACA GA-3’.
The MTHFR 677C>T and MTHFR 1298A>C polymorphism PCR products were confirmed by restriction enzyme digestion with HinfI and Fnu4HI (New England BioLaboratories, Ipswich, MA, USA). For MTHFR 677, a 203 bp undigested PCR product indicated the CC genotype, three bands at 203, 173, and 30 bp, respectively, indicated the heterozygous CT genotype, and two bands at 170 and 30 bp, respectively, indicated the homozygous TT genotype. For MTHFR 1298, a single band at 138 bp indicated the AA genotype, and two bands at 119 and 19 bp, respectively, indicated the homozygous CC genotype. The TS 1494 0bp/6bp polymorphism fragment was 142 bp for the 0bp allele and 148 bp for the 6bp allele. The PCR products were digested with DraI (New England BioLaboratories, Ipswich, MA, USA), resulting in a band at 142 bp (0bp/0bp), three bands at 142 bp, 88 bp, and 60 bp, respectively (0bp/6bp), and two bands at 88 bp and 60 bp, respectively (6bp/6bp). A series of two (2R) or three (3R) 28 bp tandem repeats for the TSER 2R/3R polymorphism were confirmed by electrophoretic separation on 4% agarose gels. All of the digestion reactions were performed at 37°C for several hours, depending on the enzymes.
All genotypes were confirmed in triplicate to rule out genotyping errors due to a violation of the Hardy-Weinberg equilibrium (HWE). In addition, some of the PCR products were randomly chosen for DNA sequencing using an ABI 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA).
Statistical analysis
Differences in genetic frequencies of the polymorphisms between patients and control subjects were compared using Fisher’s exact test and logistic regression. The odds ratio (OR) and 95% confidence interval (CI) were used as a measure of the strength of the association between genotype frequencies and RIF. The OR and 95% CI were also used to assess the relationship between each specific polymorphism and allele combination. The polymorphisms with RIF incidence was calculated using adjusted ORs (AORs) and 95% CIs from logistic regression adjusted for age. Statistical significance was accepted at a level of P < 0.05. The false discovery rate (FDR) correction was performed to adjust for multiple comparisons. All of the polymorphisms were in HWE (P > 0.05). Statistical analyses were performed using Graphpad Prism 4.0 (Graphpad Software, Inc., San Diego, CA, USA), StatsDirect software version 2.4.4 (StatsDirect Ltd., Altrincham, UK), HaploView 4.1 (Broad Institute of MIT and Harvard, Boston, MA, USA), and HAPSTAT 3.0 (University of North Carolina, Chapel Hill, NC, USA). Gene-gene interaction analysis was performed using the open source multidimensional reduction (MDR) software package v.2.0 (www.epistasis.org). All possible combinations of the polymorphisms were studied using the MDR analysis to determine the combinations with strong synergistic effects.
Results
The clinical characteristics of all participants are summarized in Table 1. The average number of live births and length of gestation of control subjects were 1.78 ± 0.74 and 39.36 ± 1.66, respectively. RIF patients had no live births and 17 RIF patients had verified histories of recurrent pregnancy loss (Table 1).
To examine the relationship between RIF and polymorphisms in major folate metabolism enzymes and associated genetic enhancer regions, seven alleles were chosen, including MTHFR 677, MTHFR 1298, TSER, and TS 1494. The genotype and frequency of the genetic polymorphisms are summarized in Table 2, S1 Table [patients without recurrent pregnancy loss (RPL)] and S2 Table [analysis according to number of previous implantation failure (IF)]. Four of the polymorphisms analyzed were in HWE. The MTHFR 677C>T, MTHFR 1298A>C, TSER 2R/3R, and TS 1494 0bp/6bp polymorphisms were not related to the prevalence of RIF.
Table 2. Genotype frequencies of one-carbon metabolism-related gene polymorphisms between controls and RIF patients.
| Genotype | Controls | RIF patients | Reference allele | Models | AOR (95% CI) | P | FDR-P |
|---|---|---|---|---|---|---|---|
| MTHFR 677C>T | n = 125 | n = 120 | |||||
| CC | 46 (36.8) | 35 (29.2) | 677C | Additive | 1.394 (0.957―2.030) | 0.083 | 0.332 |
| CT | 64 (51.2) | 60 (50.0) | 677C | Dominant | 1.384 (0.807―2.375) | 0.238 | 0.476 |
| TT | 15 (12.0) | 25 (20.8) | 677C | Recessive | 1.834 (0.908―3.705) | 0.091 | 0.364 |
| HWE P | 0.308 | 0.939 | |||||
| MTHFR 1298A>C | |||||||
| AA | 79 (63.2) | 78 (65.0) | 1298A | Additive | 1.005 (0.631―1.600) | 0.984 | 0.984 |
| AC | 43 (34.4) | 38 (31.7) | 1298A | Dominant | 0.977 (0.576―1.657) | 0.931 | 0.931 |
| CC | 3 (2.4) | 4 (3.3) | 1298A | Recessive | 1.273 (0.277―5.851) | 0.756 | 0.907 |
| HWE P | 0.306 | 0.810 | |||||
| TSER 2R/3R | |||||||
| 3R3R | 82 (65.6) | 81 (67.5) | 3R | Additive | 0.958 (0.615―1.493) | 0.850 | 0.984 |
| 2R3R | 37 (29.6) | 34 (28.3) | 3R | Dominant | 0.953 (0.558―1.628) | 0.860 | 0.931 |
| 2R2R | 6 (4.8) | 5 (4.2) | 3R | Recessive | 0.930 (0.275―3.143) | 0.907 | 0.907 |
| HWE P | 0.497 | 0.811 | |||||
| TS 1494 0bp/6bp | |||||||
| 0bp0bp | 70 (56.0) | 59 (49.2) | 14940bp | Additive | 1.242 (0.835―1.848) | 0.285 | 0.570 |
| 0bp6bp | 45 (36.0) | 51 (42.5) | 14940bp | Dominant | 1.391 (0.836―2.316) | 0.204 | 0.476 |
| 6bp6bp | 10 (8.0) | 10 (8.3) | 14940bp | Recessive | 1.091 (0.435―2.739) | 0.852 | 0.907 |
| HWE P | 0.471 | 0.826 |
RIF, recurrent implantation failure; AOR, adjusted odds ratio; FDR, false discovery rate. Adjusted by age of female participants.
Combined genotype analysis was performed in MTHFR 677/MTHFR 1298, MTHFR 677/TSER 238, MTHFR 677/TS 1494, MTHFR 1298/TSER 238, MTHFR 1298/TS 1494, and TSER 238/TS 1494 (Table 3). The combined genotype analysis results that indicated an association with RIF risk included MTHFR 677TT/MTHFR 1298AA [adjusted odds ratio (AOR), 2.764; 95% CI, 1.0665–7.174; P = 0.037], MTHFR 677TT/TS 1494 0bp6bp+6bp6bp (AOR, 3.185; 95% CI, 1.241–8.178; P = 0.016), and MTHFR 1298AA/TS 1494 0bp6bp+6bp6bp (AOR, 1.945; 95% CI, 1.024–3.694; P = 0.042). In addition, the combined genotype analyses, ranked according to the number of implantation failure, are presented in S3 Table. However, the MTHFR 1298AA/TS 1494 0bp6bp+6bp6bp was not significant in patients without RPL (S4 Table).
Table 3. The combination model of one-carbon metabolism-related gene polymorphisms between controls and RIF patients.
| 1st SNP | 2nd SNP | Controls | RIF patients | AOR (95% CI) | P | FDR-P |
|---|---|---|---|---|---|---|
| MTHFR 677C>T | MTHFR 1298A>C | n = 125 | n = 120 | |||
| CC | AA | 21 (16.8) | 13 (10.8) | 1.000 (reference) | ||
| CC | AC | 22 (17.6) | 18 (15.0) | 1.358 (0.530―3.480) | 0.523 | 0.523 |
| CC | CC | 3 (2.4) | 4 (3.3) | 1.994 (0.375―10.612) | 0.418 | 0.523 |
| CT | AA | 43 (34.4) | 40 (33.3) | 1.486 (0.657―3.363) | 0.342 | 0.523 |
| CT | AC | 21 (16.8) | 20 (16.7) | 1.755 (0.662―4.652) | 0.258 | 0.523 |
| TT | AA | 15 (12.0) | 25 (20.8) | 2.764 (1.065―7.174) | 0.037 | 0.185 |
| MTHFR 677C>T | TSER 2R/3R | |||||
| CC+CT | 3R3R | 70 (56.0) | 65 (54.2) | 1.000 (reference) | ||
| CC+CT | 2R3R+2R2R | 40 (32.0) | 30 (25.0) | 0.843 (0.468―1.520) | 0.570 | 0.570 |
| TT | 3R3R | 12 (9.6) | 16 (13.3) | 1.388 (0.607―3.172) | 0.437 | 0.570 |
| TT | 2R3R+2R2R | 3 (2.4) | 9 (7.5) | 3.195 (0.823―12.407) | 0.093 | 0.279 |
| MTHFR 677C>T | TS 1494 0bp/6bp | |||||
| CC+CT | 0bp0bp | 62 (49.6) | 53 (44.2) | 1.000 (reference) | ||
| CC+CT | 0bp6bp+6bp6bp | 48 (38.4) | 42 (35.0) | 1.094 (0.623―1.920) | 0.754 | 0.858 |
| TT | 0bp0bp | 8 (6.4) | 6 (5.0) | 0.902 (0.291―2.793) | 0.858 | 0.858 |
| TT | 0bp6bp+6bp6bp | 7 (5.6) | 19 (15.8) | 3.186 (1.241―8.178) | 0.016 | 0.048 |
| MTHFR 1298A>C | TSER 2R/3R | |||||
| AA | 3R3R | 53 (42.4) | 51 (42.5) | 1.000 (reference) | ||
| AA | 2R3R+2R2R | 26 (20.8) | 27 (22.5) | 1.087 (0.559―2.114) | 0.805 | 0.805 |
| AC+CC | 3R3R | 29 (23.2) | 30 (25.0) | 1.123 (0.589―2.142) | 0.725 | 0.805 |
| AC+CC | 2R3R+2R2R | 17 (13.6) | 12 (10.0) | 0.738 (0.319―1.708) | 0.478 | 0.805 |
| MTHFR 1298A>C | TS 1494 0bp/6bp | |||||
| AA | 0bp0bp | 47 (37.6) | 34 (28.3) | 1.000 (reference) | ||
| AA | 0bp6bp+6bp6bp | 32 (25.6) | 44 (36.7) | 1.945 (1.024―3.694) | 0.042 | 0.126 |
| AC+CC | 0bp0bp | 23 (18.4) | 25 (20.8) | 1.484 (0.718―3.066) | 0.287 | 0.431 |
| AC+CC | 0bp6bp+6bp6bp | 23 (18.4) | 17 (14.2) | 1.012 (0.466―2.198) | 0.975 | 0.975 |
| TSER 2R/3R | TS 1494 0bp/6bp | |||||
| 3R3R | 0bp0bp | 53 (42.4) | 45 (37.5) | 1.000 (reference) | ||
| 3R3R | 0bp6bp+6bp6bp | 29 (23.2) | 36 (30.0) | 1.530 (0.809―2.892) | 0.191 | 0.573 |
| 2R3R+2R2R | 0bp0bp | 17 (13.6) | 14 (11.7) | 0.955 (0.423―2.158) | 0.912 | 0.912 |
| 2R3R+2R2R | 0bp6bp+6bp6bp | 26 (20.8) | 25 (20.8) | 1.166 (0.589―2.309) | 0.660 | 0.912 |
Adjusted by age of female participants.
RIF, recurrent implantation failure; SNP, single nucleotide polymorphism; AOR, adjusted odds ratio
To investigate the allele combinations of the MTHFR 677, MTHFR 1298, TS 1494, and TSER polymorphisms, we carried out gene-gene interaction analysis using the haplotype-based MDR method (Table 4). The results of the MDR analysis revealed that allele combinations increased the relative risk of RIF. The MTHFR 677/MTHFR 1298/TSER/TS 1494 allele combination (C-A-3R-6bp and T-A-2R-6bp) was significantly higher in RIF patients than in control subjects. The OR of the combined polymorphisms (C-A-3R-6bp) was 3.678 (95% CI, 1.363–9.929) and the OR of T-A-2R-6bp was 2.885 (95% CI, 1.222–6.814) when the reference combination was C-A-3R-0bp. In addition, the MTHFR 677/MTHFR 1298/TSER 238, MTHFR 677/MTHFR 1298/TS 1494, MTHFR 677/TSER 238/TS 1494, and MTHFR 1298/TSER 238/TS 1494 allele combinations increased the risk of RIF. The OR of the combined alleles (T-A-2R) was 2.407 (95% CI, 1.166–4.970) in the MTHFR 677/MTHFR 1298/TSER 238 combination. As well, the T-A-0bp (MTHFR 677/MTHFR 1298/TS 1494) increased the risk of RIF (OR, 1.964; 95% CI, 1.109–3.478). Further, the T-2R-6bp (MTHFR 677/TSER 238/TS 1494) and the A-3R-6bp (MTHFR 1298/TSER 238/TS 1494) allele combinations had higher ORs of 2.395 (95% CI, 1.036–5.534) and 1.788 (95% CI, 1.029–3.107), respectively. In particular, the risk of RIF in patients carrying the combined MTHFR 677/MTHFR 1298/TSER 238/TS 1494 (C-A-3R-6bp) or MTHFR 677/MTHFR 1298/TSER 238 (T-A-2R) alleles increased 3.7 and 2.4 times, respectively, compared to women with the C-A-3R-0bp or C-A-3R reference alleles. The risk of RIF was not altered when the reference was the value combined, except in the self in haplotype-based MDR analysis. In contrast, the MTHFR 677/MTHFR 1298/TSER 238/TS 1494 (C-A-2R-6bp) and MTHFR 677/MTHFR 1298/TS 1494 (C-A-2R) allele combinations exhibited protective effects compared to the reference allele combinations. The ORs of C-A-2R-6bp and C-2R-6bp were 0.045 (95% CI, 0.003–0.765) and 0.345 (95% CI, 0.147–0.812), respectively (Table 4), which suggested that patients with C-A-2R-6bp allele combination reduced risk of RIF. In addition, the MTHFR 677/TSER 238 (C-2R and T-2R) was significantly association with RIF risk (S5 Table).
Table 4. Allelic gene-gene interaction of one-carbon metabolism-related gene polymorphisms between controls and RIF patients.
| Haplotype | Controls (2n = 250) | RIF patients (2n = 240) | OR (95% CI) | P | FDR-P |
|---|---|---|---|---|---|
| MTHFR 677/MTHFR 1298/TSER 238/TS 1494 | |||||
| C-A-3R-0bp | 74 (29.7) | 57 (23.8) | 1.000 (reference) | ||
| C-A-3R-6bp | 6 (2.5) | 17 (7.1) | 3.678 (1.363–9.929) | 0.012 | 0.055 |
| C-A-2R-0bp | 13 (5.2) | 10 (4.2) | 0.999 (0.408–2.442) | 1.000 | 1.000 |
| C-A-2R-6bp | 14 (5.4) | 0 (0.0) | 0.045 (0.003–0.765) | 0.001 | 0.011 |
| C-C-3R-0bp | 24 (9.7) | 32 (13.3) | 1.731 (0.920–3.257) | 0.110 | 0.242 |
| C-C-3R-6bp | 13 (5.0) | 6 (2.7) | 0.599 (0.215–1.674) | 0.457 | 0.628 |
| C-C-2R-0bp | 9 (3.5) | 4 (1.6) | 0.577 (0.169–1.970) | 0.558 | 0.682 |
| C-C-2R-6bp | 4 (1.4) | 4 (1.7) | 1.298 (0.311–5.418) | 0.730 | 0.803 |
| T-A-3R-0bp | 64 (25.6) | 61 (25.2) | 1.237 (0.756–2.025) | 0.452 | 0.628 |
| T-A-3R-6bp | 20 (7.9) | 23 (9.7) | 1.493 (0.748–2.982) | 0.292 | 0.535 |
| T-A-2R-0bp | 1 (0.4) | 6 (2.4) | 7.789 (0.911–66.57) | 0.047 | 0.129 |
| T-A-2R-6bp | 9 (3.7) | 20 (8.5) | 2.885 (1.222–6.814) | 0.015 | 0.055 |
| MTHFR 677/MTHFR 1298/TSER 238 | |||||
| C-A-3R | 82 (32.8) | 76 (31.8) | 1.000 (reference) | ||
| C-A-2R | 25 (10.0) | 8 (3.2) | 0.345 (0.147–0.812) | 0.013 | 0.058 |
| C-C-3R | 38 (15.3) | 39 (16.1) | 1.107 (0.642–1.910) | 0.781 | 0.781 |
| C-C-2R | 11 (4.3) | 7 (3.1) | 0.687 (0.253–1.863) | 0.619 | 0.781 |
| T-A-3R | 81 (32.3) | 81 (33.8) | 1.079 (0.696–1.673) | 0.739 | 0.781 |
| T-A-2R | 13 (5.3) | 29 (12.1) | 2.407 (1.166–4.970) | 0.023 | 0.058 |
| MTHFR 677/MTHFR 1298/TS 1494 | |||||
| C-A-0bp | 85 (34.0) | 68 (28.3) | 1.000 (reference) | ||
| C-A-6bp | 22 (8.8) | 16 (6.7) | 0.909 (0.443–1.865) | 0.856 | 0.856 |
| C-C-0bp | 35 (13.8) | 36 (14.8) | 1.286 (0.731–2.260) | 0.392 | 0.675 |
| C-C-6bp | 14 (5.8) | 10 (4.4) | 0.893 (0.373–2.136) | 0.829 | 0.856 |
| T-A-0bp | 66 (26.2) | 65 (27.3) | 1.231 (0.771–1.966) | 0.405 | 0.675 |
| T-A-6bp | 28 (11.4) | 44 (18.5) | 1.964 (1.109–3.478) | 0.023 | 0.115 |
| MTHFR 677/TSER 238/TS 1494 | |||||
| C-3R-0bp | 97 (39.0) | 90 (37.5) | 1.000 (reference) | ||
| C-3R-6bp | 19 (7.6) | 22 (9.0) | 1.248 (0.634–2.458) | 0.606 | 0.707 |
| C-2R-0bp | 23 (9.1) | 13 (5.4) | 0.609 (0.291–1.275) | 0.205 | 0.359 |
| C-2R-6bp | 17 (6.8) | 6 (2.3) | 0.380 (0.144–1.008) | 0.049 | 0.114 |
| T-3R-0bp | 65 (26.0) | 60 (25.1) | 0.995 (0.632–1.565) | 1.000 | 1.000 |
| T-3R-6bp | 20 (7.8) | 24 (10.1) | 1.293 (0.669–2.501) | 0.504 | 0.706 |
| T-2R-0bp | 0 (0.0) | 6 (2.5) | 14.01 (0.777–252.3) | 0.014 | 0.098 |
| T-2R-6bp | 9 (3.8) | 20 (8.2) | 2.395 (1.036–5.534) | 0.046 | 0.114 |
| MTHFR 1298/TSER 238/TS 1494 | |||||
| A-3R-0bp | 136 (54.4) | 117 (48.7) | 1.000 (reference) | ||
| A-3R-6bp | 26 (10.5) | 40 (16.5) | 1.788 (1.029–3.107) | 0.039 | 0.273 |
| A-2R-0bp | 16 (6.3) | 16 (6.7) | 1.162 (0.557–2.426) | 0.711 | 0.749 |
| A-2R-6bp | 23 (9.2) | 22 (9.0) | 1.112 (0.589–2.098) | 0.749 | 0.749 |
| C-3R-0bp | 26 (10.5) | 34 (14.0) | 1.520 (0.862–2.681) | 0.154 | 0.539 |
| C-3R-6bp | 12 (5.0) | 6 (2.5) | 0.581 (0.212–1.597) | 0.335 | 0.620 |
| C-2R-0bp | 7 (2.8) | 3 (1.1) | 0.498 (0.126–1.971) | 0.354 | 0.620 |
| C-2R-6bp | 3 (1.3) | 4 (1.6) | 1.550 (0.340–7.069) | 0.708 | 0.749 |
RIF, recurrent implantation failure; OR, odds ratio; p-value Fisher’s exact test.
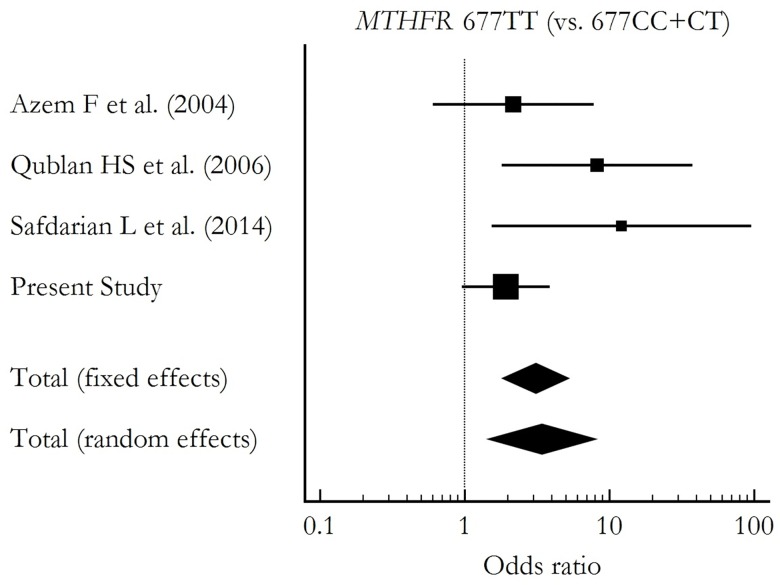
The results of the MTHFR 677C>T screening is summarized in Fig 1. We were search association study between MTHFR 677C>T polymorphism and RIF, found 3 studies in PubMed database (http://www.ncbi.nlm.nih.gov/pubmed). A total of 364 controls and 351 patients were positive for the MTHFR 677C>T polymorphism. Overall, the results indicate an association of this polymorphism with risk of RIF (OR, 3.394; 95% CI, 1.451–7.938; Fig 1, S6 Table).
Fig 1. A meta-analysis of MTHFR 677C>T in RIF.
A meta-analysis of the association between carriers of the T allele (individuals with TT genotype) in the MTHFR 677C>T polymorphism and recurrent implantation failure (RIF). The fixed and random effects models were used to calculate the pooled weighted odds ratios (ORs).
Discussion
At present, the relationship between RIF and polymorphisms in genes that encode major folate metabolism enzymes remains unclear. However, in this study, we demonstrated that patients carrying the MTHFR 677 and MTHFR 1298 alleles in combination with the TS 1494 polymorphism had a significantly higher risk of experiencing RIF.
Successful implantation begins with proper implantation of the embryo in a receptive uterine endometrium. Afterward, the endometrium undergoes dynamic morphological and functional changes to become receptive to the embryo during early implantation [12]. In addition, the uterus undergoes cellular processes, including DNA synthesis and angiogenesis, which are required for cell proliferation and decidualization, and for orchestrating complicated implantation processes. Hence, DNA synthesis and homeostasis are important during this process. As well, folate acts as a methyl group carrier for the targets and is required for the homeostasis of the one-carbon metabolism pathway. One of the forms of folate is 5,10-MTHF, which can be used for the synthesis of either methionine or dTMP. Methionine is synthesized by the transfer of a 5-MTHF methyl group to homocysteine, whereas dTMP is generated by the transfer of a 5-MTHF methyl group to deoxyuridine monophosphate (dUTP) [13].
The one-carbon metabolism pathway is precisely regulated by two critical enzymes, MTHFR and TS. Further, MTHFR generates 5-MTHF from 5,10-MTHF [13,14], and is located on the short arm of chromosome 1 (1p36.3[15]. MTHFR 677C>T, a polymorphism of MTHFR, leads to the conversion of the amino acid alanine to valine, resulting in decreased MTHFR activity and impaired enzyme activity [16]. Also, the MTHFR 677C>T polymorphism is associated with various diseases, including stroke, hypertension, and cancer [reviewed in [9,17]]. The A and C nucleotides are involved in the MTHFR 1298 polymorphism, but the MTHFR 1298A allele is more common than the 1298C allele. The MTHFR 1298A>C polymorphism leads to the substitution of alanine for glutamine at amino acid 429, but the mutation does not affect the thermolability or FAD release activity functions of the MTHFR protein, or the protective effects of 5-methyl-THF [18]. Several recent reports have demonstrated an association between the one-carbon metabolism enzymes and recurrent spontaneous abortion [19–21]. In addition, idiopathic infertile women exhibit an increased frequency of MTHFR 677C>T polymorphisms compared to control women [22]. Safdarian et al. reported association between MTHFR 677C>T and RIF risk in term of hereditary thrombophilia [23].
Thymidylate synthetase catalyzes the conversion of dUMP to dTMP by oxidation of 5,10-MTHF [24]. As well, dTMP is required for de novo DNA synthesis. Further, two TSER and TS 1494 polymorphisms affect the transcription and translation of the TS gene [25]. The 5’-untranslated region (UTR) of TSER contains 2R and 3R repeats of 28 bp sequences, and the 3’-UTR of TS 1494 has either a 6 bp deletion or insertion, which results in the modulation of TS expression and stability [25–28]. However, little is known regarding the effect of TSER and TS polymorphisms on RIF. Recently, we reported that the TSER 2R2R and TS 6bp6bp combined genotype was associated with cancer [29], which suggests that the presence of these combinations might affect susceptibility to RIF.
The results of our previous studies established an association between polymorphisms in folate metabolism-related genes (MTHFR 677C>T, 1298A>C, TSER 2R/3R, and TS 1494 0bp/6bpins/del) and increased risk of reproductive diseases, including RPL, premature ovarian failure, and spontaneously aborted embryos in the Korean population [30–34]. However, these studies only showed the difference in genotype frequencies between control subjects and patient groups. In addition, we have identified an indirect effect of the MTHFR 677C>T, 1298A>C, and TSER 2R/3R polymorphisms on RPL [35]. In previous study, MTHFR gene polymorphisms (677C>T and 1298A>C) were reported association between maternal, fetal and paternal in RPL risk by meta-analysis [36]. These meta-analyses of RPL and our screening data of RIF were suggestion that MTHFR 677C>T was considered to crucial genetic factor during implantation, or maintaining pregnancy. In this study, one interesting result was the indication that the MTHFR 677TT/1298AA and MTHFR 677TT/TS 1494 0bp6bp+6bp6bp combinations conveyed increased risk of RIF occurrence. In addition, we identified relationships between the MTHFR/TSER/TS genetic polymorphisms and risk of RIF in Korean women. The genetic combinations of MTHFR 677/MTHFR 1298/TSER, TS 1494 (C-A-3R-6bp), and MTHFR 677/MTHFR 1298/TS 1494(C-A-6bp) increased the risk of RIF compared to the risk associated with each reference combination.
We also performed a screening of published studies to investigate the genetic association between MTHFR 677C>T and the risk of RIF, which revealed that MTHFR 677C>T increased the risk of RIF. However, this study has some limitations. First, the lack of clinical parameters, such as vitamin B6, inflammatory cytokine and hormone levels in RIF women remains to be investigated. Second, the sample size (number of included studies) was so small; therefore, we cannot rule out the possibility of the results being biased, although no significant publication bias was found.
In conclusion, these interesting findings indicated that the combined MTHFR, TSER, and TS genotypes could potentially be novel diagnostic markers for evaluating the risk of experiencing RIF. However, due to the small number of patients and clinical insufficiency, further studies are required to confirm our conclusions. Nonetheless, the results of the current study provided us with a better understanding of idiopathic RIF and the relationship between RIF and polymorphisms in genes that encode major folate metabolism enzymes.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Support was provided by: HI15C1972010015: [https://www.htdream.kr/]; 2009-0093821: [http://www.nrf.re.kr/]; 2014R1A2A2A01003994: [http://www.nrf.re.kr/]; 2015R1 D1A1A09057432: [http://www.nrf.re.kr/]. This study was partly supported by a grant of the Korea Healthcare technology R&D Project (HI15C1972010015), Ministry for Health, Welfare & Family Affairs, Republic of Korea and by Basic Science Research Program through National Research Foundation of Korea Grants funded by the Korean Government (2009-0093821, 2014R1A2A2A01003994 and 2015R1 D1A1A09057432), Republic of Korea.
References
- 1.Stephenson MD, Fluker MR (2000) Treatment of repeated unexplained in vitro fertilization failure with intravenous immunoglobulin: a randomized, placebo-controlled Canadian trial. Fertil Steril 74: 1108–1113. [DOI] [PubMed] [Google Scholar]
- 2.Laufer N, Simon A (2012) Recurrent implantation failure: current update and clinical approach to an ongoing challenge. Fertil Steril 97: 1019–1020. 10.1016/j.fertnstert.2012.03.033 [DOI] [PubMed] [Google Scholar]
- 3.Simon A, Laufer N (2012) Repeated implantation failure: clinical approach. Fertil Steril 97: 1039–1043. 10.1016/j.fertnstert.2012.03.010 [DOI] [PubMed] [Google Scholar]
- 4.Margalioth EJ, Ben-Chetrit A, Gal M, Eldar-Geva T (2006) Investigation and treatment of repeated implantation failure following IVF-ET. Hum Reprod 21: 3036–3043. [DOI] [PubMed] [Google Scholar]
- 5.Wang H, Dey SK (2006) Roadmap to embryo implantation: clues from mouse models. Nat Rev Genet 7: 185–199. [DOI] [PubMed] [Google Scholar]
- 6.Ashworth CJ, Antipatis C (2001) Micronutrient programming of development throughout gestation. Reproduction 122: 527–535. [PubMed] [Google Scholar]
- 7.Taparia S, Gelineau-van Waes J, Rosenquist TH, Finnell RH (2007) Importance of folate-homocysteine homeostasis during early embryonic development. Clin Chem Lab Med 45: 1717–1727. [DOI] [PubMed] [Google Scholar]
- 8.Yajnik CS, Deshmukh US (2012) Fetal programming: maternal nutrition and role of one-carbon metabolism. Rev Endocr Metab Disord 13: 121–127. 10.1007/s11154-012-9214-8 [DOI] [PubMed] [Google Scholar]
- 9.Nazki FH, Sameer AS, Ganaie BA (2014) Folate: Metabolism, genes, polymorphisms and the associated diseases. Gene 533: 11–20. 10.1016/j.gene.2013.09.063 [DOI] [PubMed] [Google Scholar]
- 10.Peters GJ, Backus HH, Freemantle S, van Triest B, Codacci-Pisanelli G, van der Wilt CL, et al. (2002) Induction of thymidylate synthase as a 5-fluorouracil resistance mechanism. Biochim Biophys Acta 1587: 194–205. [DOI] [PubMed] [Google Scholar]
- 11.Horie N, Aiba H, Oguro K, Hojo H, Takeishi K (1995) Functional analysis and DNA polymorphism of the tandemly repeated sequences in the 5'-terminal regulatory region of the human gene for thymidylate synthase. Cell Struct Funct 20: 191–197. [DOI] [PubMed] [Google Scholar]
- 12.Noyes RW, Hertig AT, Rock J (1975) Dating the endometrial biopsy. Am J Obstet Gynecol 122: 262–263. [DOI] [PubMed] [Google Scholar]
- 13.Smith DE, Kok RM, Teerlink T, Jakobs C, Smulders YM (2006) Quantitative determination of erythrocyte folate vitamer distribution by liquid chromatography-tandem mass spectrometry. Clin Chem Lab Med 44:450–459 [DOI] [PubMed] [Google Scholar]
- 14.Ghandour H, Chen Z, Selhub J, Rozen R (2004) Mice deficient in methylenetetrahydrofolate reductase exhibit tissue-specific distribution of folates. J Nutr 134: 2975–2978. [DOI] [PubMed] [Google Scholar]
- 15.Hart DJ, Finglas PM, Wolfe CA, Mellon F, Wright AJ, Southon S (2002) Determination of 5-methyltetrahydrofolate (13C-labeled and unlabeled) in human plasma and urine by combined liquid chromatography mass spectrometry. Anal Biochem 305: 206–213. [DOI] [PubMed] [Google Scholar]
- 16.Goyette P, Sumner JS, Milos R, Duncan AM, Rosenblatt DS, Matthews RG, et al. (1994) Human methylenetetrahydrofolate reductase: isolation of cDNA mapping and mutation identification. Nat Genet 7: 551. [PubMed] [Google Scholar]
- 17.Rozen R (1997) Genetic predisposition to hyperhomocysteinemia: deficiency of methylenetetrahydrofolate reductase (MTHFR). Thromb Haemost 78: 523–526. [PubMed] [Google Scholar]
- 18.Reilly R, McNulty H, Pentieva K, Strain JJ, Ward M (2013) MTHFR 677TT genotype and disease risk: is there a modulating role for B-vitamins? Proc Nutr Soc: 1–10. [DOI] [PubMed] [Google Scholar]
- 19.Yamada K, Chen Z, Rozen R, Matthews RG (2001) Effects of common polymorphisms on the properties of recombinant human methylenetetrahydrofolate reductase. Proc Natl Acad Sci U S A 98: 14853–14858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobashi G, Kato EH, Morikawa M, Shimada S, Ohta K, Fujimoto S, et al. (2005) MTHFR C677T Polymorphism and factor V Leiden mutation are not associated with recurrent spontaneous abortion of unexplained etiology in Japanese women. Semin Thromb Hemost 31: 266–271. [DOI] [PubMed] [Google Scholar]
- 21.Poursadegh Zonouzi A, Chaparzadeh N, Asghari Estiar M, Mehrzad Sadaghiani M, Farzadi L, Ghasemzadeh A, et al. (2012) Methylenetetrahydrofolate Reductase C677T and A1298C Mutations in Women with Recurrent Spontaneous Abortions in the Northwest of Iran. ISRN Obstet Gynecol 2012: 945486 10.5402/2012/945486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rotondo JC, Bosi S, Bazzan E, Di Domenico M, De Mattei M, Selvatici R, et al. (2012) Methylenetetrahydrofolate reductase gene promoter hypermethylation in semen samples of infertile couples correlates with recurrent spontaneous abortion. Hum Reprod 27: 3632–3638. 10.1093/humrep/des319 [DOI] [PubMed] [Google Scholar]
- 23.Safdarian L, Najmi Z, Aleyasin A, Aghahosseini M, Rashidi M, Asadollah S (2014) Recurrent IVF failure and hereditary thrombophilia. Iran J Reprod Med 12:467–470. [PMC free article] [PubMed] [Google Scholar]
- 24.Coulam CB, Jeyendran RS (2009) Thrombophilic gene polymorphisms are risk factors for unexplained infertility. Fertil Steril 91: 1516–1517. 10.1016/j.fertnstert.2008.07.1782 [DOI] [PubMed] [Google Scholar]
- 25.Hardy LW, Finer-Moore JS, Montfort WR, Jones MO, Santi DV, Stroud RM (1987) Atomic structure of thymidylate synthase: target for rational drug design. Science 235: 448–455. [DOI] [PubMed] [Google Scholar]
- 26.Zhou JY, Shi R, Yu HL, Zeng Y, Zheng WL, Ma WL (2012) The association between two polymorphisms in the TS gene and risk of cancer: a systematic review and pooled analysis. Int J Cancer 131: 2103–2116. 10.1002/ijc.27465 [DOI] [PubMed] [Google Scholar]
- 27.Chu E, Koeller DM, Casey JL, Drake JC, Chabner BA, Elwood PC, et al. (1991) Autoregulation of human thymidylate synthase messenger RNA translation by thymidylate synthase. Proc Natl Acad Sci U S A 88: 8977–8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ulrich CM, Bigler J, Velicer CM, Greene EA, Farin FM, Potter JD (2000) Searching expressed sequence tag databases: discovery and confirmation of a common polymorphism in the thymidylate synthase gene. Cancer Epidemiol Biomarkers Prev 9: 1381–1385. [PubMed] [Google Scholar]
- 29.Yim DJ, Kim OJ, An HJ, Kang H, Ahn DH, Hwang SG, et al. (2010) Polymorphisms of thymidylate synthase gene 5'- and 3'-untranslated region and risk of gastric cancer in Koreans. Anticancer Res 30: 2325–2330. [PubMed] [Google Scholar]
- 30.Kim NK, Choi YK, Kang MS, Choi DH, Cha SH, An MO, et al. (2006) Influence of combined methylenetetrahydrofolate reductase (MTHFR) and thymidylate synthase enhancer region (TSER) polymorphisms to plasma homocysteine levels in Korean patients with recurrent spontaneous abortion. Thromb Res 117:653–658. [DOI] [PubMed] [Google Scholar]
- 31.Kim JH, Jeon YJ, Lee BE, Kang H, Shin JE, Choi DH, et al. (2013) Association of methionine synthase and thymidylate synthase genetic polymorphisms with idiopathic recurrent pregnancy loss. Fertil Steril 99:1674–1680. 10.1016/j.fertnstert.2013.01.108 [DOI] [PubMed] [Google Scholar]
- 32.Rah H, Jeon YJ, Choi Y, Shim SH, Yoon TK, Choi DH, et al. (2012) Association of methylenetetrahydrofolate reductase (MTHFR 677C>T) and thymidylate synthase (TSER and TS 1494del6) polymorphisms with premature ovarian failure in Korean women. Menopause 19:1260–1266. 10.1097/gme.0b013e3182556b08 [DOI] [PubMed] [Google Scholar]
- 33.Park HM, Shin SJ, Choi DH, Oh D, Lee S, Kim NK (2008) Association between folate metabolism-related gene polymorphisms and methylation of p16(INK4A) and hMLH1 genes in spontaneously aborted embryos with normal chromosomal integrity. Fertil Steril 90:1605–1610. 10.1016/j.fertnstert.2007.09.046 [DOI] [PubMed] [Google Scholar]
- 34.Bae J, Shin SJ, Cha SH, Choi DH, Lee S, Kim NK (2007) Prevalent genotypes of methylenetetrahydrofolate reductase (MTHFR C677T and A1298C) in spontaneously aborted embryos. Fertil Steril 87:351–355. [DOI] [PubMed] [Google Scholar]
- 35.Bae J, Choi DH, Kang MS, Cha SH, Oh D, Kim NK (2009) Effect of methylenetetrahydrofolate reductase and thymidylate synthase enhancer region polymorphisms on the risk of idiopathic recurrent spontaneous abortion in a Korean population. Fertil Steril 91:1560–1562. 10.1016/j.fertnstert.2008.09.060 [DOI] [PubMed] [Google Scholar]
- 36.Yang Y, Luo Y, Yuan J, Tang Y, Xiong L, Xu M, et al. (2016) Association between maternal, fetal and paternal MTHFR gene C677T and A1298C polymorphisms and risk of recurrent pregnancy loss: a comprehensive evaluation. Arch Gynecol Obstet 293:1197–1211. 10.1007/s00404-015-3944-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.