Abstract
The productivity of maize (Zea mays L.) depends on the development of chloroplasts, and G2-like transcription factors play a central role in regulating chloroplast development. In this study, we identified 59 G2-like genes in the B73 maize genome and systematically analyzed these genes at the molecular and evolutionary levels. Based on gene structure character, motif compositions and phylogenetic analysis, maize G2-like genes (ZmG1- ZmG59) were divided into seven groups (I-VII). By synteny analysis, 18 collinear gene pairs and strongly conserved microsyntny among regions hosting G2-like genes across maize and sorghum were found. Here, we showed that the vast majority of ZmG gene duplications resulted from whole genome duplication events rather than tandem duplications. After gene duplication events, some ZmG genes were silenced. The functions of G2-like genes were multifarious and most genes that are expressed in green tissues may relate to maize photosynthesis. The qRT-PCR showed that the expression of these genes was sensitive to low temperature and drought. Furthermore, we analyzed differences of ZmGs specific to cultivars in temperate and tropical regions at the population level. Interestingly, the single nucleotide polymorphism (SNP) analysis revealed that nucleotide polymorphism associated with different temperature zones. Above all, G2-like genes were highly conserved during evolution, but polymorphism could be caused due to a different geographical location. Moreover, G2-like genes might be related to cold and drought stresses.
Introduction
The chloroplasts of higher plants are believed to have originated from aquatic single-celled cyanobacteria more than 2.5 billion years ago [1–3]. Chloroplasts contain the green pigment chlorophyll and are responsible for the light-powered reactions of photosynthesis, upon which essentially all life depends [2–4]. Recent studies supported the view that chloroplasts were derived from a primary endosymbiotic event involving such cyanobacteria [5–8]. As a result, the regulation of chloroplast biogenesis involves cooperation between the nucleus and chloroplast. Plastids play a variety of roles in addition to functioning in photosynthesis, including roles in the synthesis of amino acids, fatty acids, purine and pyrimidine bases, terpenoids and various pigments and hormones, as well as functioning in key aspects of nitrogen and sulfur assimilation [2,9–11].
Proplastids in subepidermal meristematic cells (or etioplasts in dark-grown cotyledons) are transformed into mesophyll chloroplasts upon exposure to light [1,12]. Approximately 50% of genes are differentially expressed during this transformation [1,13–15] and transcription factors play key regulation in this process. Members of Golden 2-like (G2-like) family have been characterized with roles of regulating the formation of chloroplasts during the transition and early maturation phases [1,16]. And G2-like genes are indispensable for chloroplast development in angiosperms.
G2-like genes are members of the recently categorized GARP superfamily of transcription factors [17,18]. Within the GARP family, G2-like genes are monophyletic, while interestingly, gene duplications have occurred independently in monocots and eudicots [19]. Most G2-like genes have two domains, including a Myb-DNA binding domain (DBD; containing an HLH motif) and a C-terminal domain (containing a conserved GCT box). Several chloroplast development-related transcription factors have been reported in plants. For example, ACCUMULATION AND REPLICATION OF CHLOROPLASTS5 encodes a cytoplasmically localized dynamin-like protein that regulates chloroplast division in Arabidopsis thaliana [20]. Two transposase-derived transcription factors, FAR-RED ELONGATED HYPOCOTYL3 (FHY3) and FAR-RED IMPAIRED RESPONSE1 (FAR1), are positive regulators of chlorophyll biosynthesis in Arabidopsis [21]. Golden2-like (GLK) genes regulate chloroplast development in the monocot maize (Zea mays L.) and in the eudicot Arabidopsis [4,21–24]. GLK genes help coregulate and synchronize the expression of a suite of nucleus photosynthetic genes and thus act to optimize photosynthetic capacity under varying environmental and developmental conditions [25]. GLK overexpression enhances the expression of genes related to fruit photosynthesis and chloroplast development, leading to elevated carbohydrate and carotenoid levels in ripe fruit [26]. SlGLK2 influences photosynthesis in developing fruit, contributing to mature fruit characteristics [26]. ZmGLK1 is thought to regulate mesophyll cell chloroplast development in C4 tissues, and GLK gene pairs act redundantly to promote chloroplast development in C3 species [4,23]. In addition, overexpression of AtGLK1 (35S:AtGLK1) in Arabidopsis also confers resistance to the cereal pathogen Fusarium graminearum [27]. Maize is a model genetic system and one of the world’s highest valued crops, accounting for billions of dollars of annual agricultural revenue [28]. To further increase crop productivity, one way is to improve the stress and disease resistance. G2-like genes might have a function in stress and disease resistance. Although G2-like transcription factors were first characterized in maize [24], a systematic analysis and comparison of G2-like genes in maize has not previously been reported. In this study, systems analysis and population SNP analysis were performed to gain insight into the evolutionary trajectory of G2-like gene in maize expression patterns of some important ZmG genes were also investigated under cold and drought conditions.
Materials and Methods
Identification of G2-like genes
Maize protein and nucleic acid sequences were obtained from maize genome database (http://www.maizesequence.org). Sorghum data were downloaded from Phytozome (v9.1). DNATOOLS software was used to construct a local database of maize nucleotide and protein sequences. Previously reported sequences of Arabidopsis G2-like protein sequences were used as queries to search against the maize and sorghum protein database with BLASTP (E-values below 0.001). All candidate sequences that met the standards were confirmed to be G2-like proteins using Pfam (http://pfam.xfam.org/) and SMART (http://smart.embl-heidelberg.de/). Finally, sequences of all confirmed proteins were aligned using MEGA6 software [29], and redundant sequences were removed manually (all sequence data in supplementary file). Physical parameters of these proteins including molecular mass (kDa), and isoelectric point (pI) were estimated using the compute pI/Mw tool in ExPASy (http://web.expasy.org/compute_pi/). Protein subcellular localization was predicted by online softwares: TargetP online server (http://www.expasy.org/), SubLoc online server (http://www.bioinfo.tsinghua.edu.cn/SubLoc/) and WoLF PSORT online server (http://www.genscript.com/).
Phylogenetic and motif analysis of G2-like genes
A neighbor-joining phylogenetic tree of G2-like genes from maize and sorghum was generated with MEGA6 software [29]. The confidence limits of each branch were assessed by 1,000 bootstrap replications and expressed as percentage values.
Conserved motifs among maize G2-like genes were examined using MEME software (http://meme-suite.org/) [30]. The width of each motif was limited to 6–50, and the maximum number of motifs was set to 20. In addition, structural motif annotation was performed using Pfam (http://pfam.xfam.org/) and SMART (http://smart.embl-heidelberg.de/) online service.
Chromosomal locations, gene structure and duplication events of G2-like genes
G2-like genes were mapped to the maize chromosomes according to their position information from maize genomic database (http://www.maizesequence.org). MapInspect software (http://www.plantbreeding.wur.nl/uk/software_mapinspect.html) was subsequently used to graphically portray G2-like genes from maize. Introns and exons were predicted via the online tool Gene Structure Display Server 2.0 (http://gsds.cbi.pku.edu.cn/) [31].The duplication pattern for each G2-like genes was analyzed by MCScanX softaware (http://chibba.pgml.uga.edu/mcscan2/) [32]. Then, whole-genome BLASTP analysis of maize and sorghum were performed using local blast software with e-value under 1e-5. Blast outputs and position of all protein-coding genes were imported into Circos software (http://circos.ca/) [33] and genes were classified into various types of duplications.
The Ka and Ks were calculated by DnaSPv5.0 [34]. The ratio of nonsynonymous to synonymous nucleotide substitutions (Ka/Ks ratios between paralogs) was analyzed to detect the mode of selection. Values of Ka/Ks >1, = 1 and <1 represent positive selection, neutral selection and purifying selection, respectively [35].
Expression pattern analysis
Gene expression data from Maize B73 transcriptomes were used to draw a heat map, including germinating seeds, primary roots, stems, SAMs (shoot apical meristem), leaves, endosperm, embryos and whole seeds [36,37]. The expression data were used to generate a heatmap using R/Bioconductor (http://www.bioconductor.org/).
RNA extraction and qRT-PCR analysis
The maize inbred line B73 was grown in a greenhouse (16h light/ 8h dark, 28±2°C). After 3 weeks, seedlings were treated with cold and drought stresses, respectively. For cold stress, leaves were sampled at 0h, 6h, 12h and 24h after cold stress (4°C) treatment. For drought stress, leaves were collected at 0h, 0.5h, 1h, 2h, 3h and 6h after treated with 20% PEG6000. Total RNAs of all the samples collected in this study were extracted using the RNAiso plus (TaKaRa) accords to the manufacturer’s instructions. The DNase-treated RNA was reverse-transcribed using First-Strand cDNA Synthesis Kit (TRANSGEN). Reaction was used SYBR Green Master Mix reagent (Roche). The qRT-PCR was performed in a 20 μl volume, which contained 10 μl of 2×SYBR Green Master Mix, 2 μl diluted cDNA template, 1 μl of each specific primer, and 6 μl ddH2O and then performed on ABI 7300 real time system (Applied Biosystems). The qRT-PCR program was used as follows: 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. The gene-specific primers designed by Primer 6.0 software were employed to amplify 120–300 bp PCR products unique to each gene (S7 Table). Expression level of the maize Actin 1 gene was used as an internal control. The relative expression level of each gene was calculated as 2-ΔΔCT [38] compared to that of untreated control plant which was set as 1. The qRT-PCR assays were performed with three biological and three technical replicates.
Nucleotide diversity of G2-like genes in 85 maize inbreed lines
To further investigate G2-like gene evolution in natural populations of maize, G2-like genes sequences were examined from 85 maize inbred lines grown in tropical and temperate regions combined with whole genome sequencing SNP data (unpublished data). DNASP 5.0 software was used to analyze sequence nucleotide polymorphism and gene flow. The nucleotide diversity parameter Pi (π) was estimated, where Pi is the average number of nucleotide differences per site between any two DNA sequences. In addition, in gene flow analysis, Gst is the genetic differentiation coefficient and Fst is the fixed coefficient. χ2 assessment was used to test the significance of genetic differences among groups.
Results
Identification and analysis of G2-like genes in maize
After extensive searches and alignment of the maize genome database using previously reported Arabidopsis G2-like proteins AtGLK1 (AT2G20570) and AtGLK2 (AT5G44190) [23] as BLASTP queries, a total of 59 G2-like genes (designated ZmG1 to ZmG59) were identified (S1 Table and S1 Text). Basic information about maize G2-like genes is presented in S1 Table. The exon number of these 59 genes ranges from 1 to 8. The predicted molecular weights (MW) of ZmG proteins range from 16.68 kDa to 59.22 kDa, while their lengths range from 145 to 554 amino acids. ZmG10 has the shortest sequence while ZmG37 has the longest. In addition, the proteins could be divided into two groups of roughly equal size based on isoelectric point, comprising 30 acidic proteins and 29 basic proteins. Moreover, the subcellular localizations of 59 genes predicted by the three online server TargetP online server (http://www.expasy.org/), SubLoc online server (http://www.bioinfo.tsinghua.edu.cn/SubLoc/) and WoLF PSORT online server (http://www.genscript.com/). Combined with results from the three online server analyses, we found that most of ZmGs were predicted in the nucleus.
Phylogenetic and motif analysis of maize G2-like genes
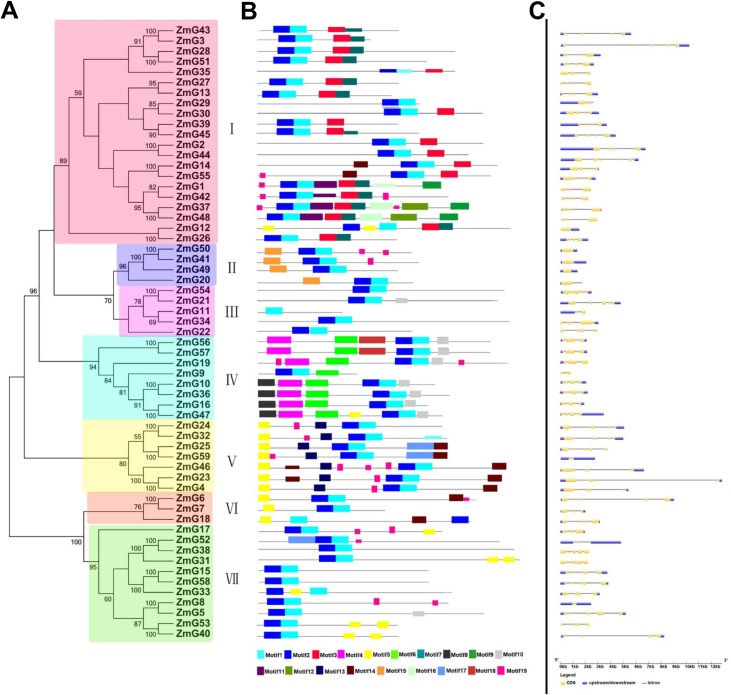
The phylogenetic relationships and evolutionary history of maize G2-like gene family were inferred by constructing a phylogenetic tree based on aligned G2-like protein sequences (Fig 1A). The G2-like family was classed into seven groups (I–VII) based on evolutionary relationships and motif analysis; Groups I to VII contain 21, 4, 5, 8, 7, 3 and 11 genes, respectively.
Fig 1. Neighbor-joining phylogenetic tree, motif and structure of the 59 predicted maize G2-like genes.
(A) The phylogenetic tree, constructed with MEGA6.0, was generated based on amino acid sequences. G2-like proteins were divided into seven groups (I-VII) based on high bootstrap values (>50). (B) All motifs were identified by MEME using the complete amino acid sequences of 59 maize G2-like proteins. Different motifs are indicated by different-colored numbers (1–20) as shown in the scale at the bottom of the Fig (C) Exons and introns are indicated by yellow boxes and single lines, respectively. Thick blue lines represent untranslated regions (UTRs). The length of each G2-like gene can be estimated based on the scale at the bottom.
Motif analysis of 59 maize G2-like genes also proved phylogenetic kinship. Conserved motifs (Fig 1B and S2 Table) were examined using MEME software and the motif sequences were shown in S2 Table. In addition to a conserved G2-like Myb DNA-binding domain, each group has unique motifs that might represent their functional diversity. Group I possess a Myb-CC-LHEQLE domain (a type of MYB-like domain). MYB transcription factors play diverse roles in plant development and in response to abiotic stress [39]. Besides, in gene structure analysis diverse distribution of intronic regions (from 1 to 8 in numbers) was found among ZmG genes (Fig 1C). In general, ZmGs clustered in the same group exhibit similar exon/intron structure (Fig 1C) and all ZmGs genes in group Ⅱ have no intron.
Multiple sequence alignment of ZmGs and other identified G2-like proteins of rice OsGLK1/2 (LOC_Os06g24070/ LOC_Os01g13740) and Arabidopsis AtGLK1/2 (AT2G20570/ AT5G44190) [23] (S1 Fig) demonstrated that these sequences are particularly conserved across two regions of a putative DNA binding domain with an HLH structure (The first helix contains initial sequences PELHRR and invariably comprises 14 amino acids and the second helix contains an initial NI/VASHLQ motif. These helices are separated by a 22 amino acid loop). In a number of well-characterized transcriptional regulators, HLH domains bind DNA and mediate dimerization [40,41].
Moreover, to investigate the potential function of maize G2-like genes, further evolutionary relationship among ZmGs, AtGLK1, AtGLK2, OsGLK1 and OsGLK2 was investigated. As showed in S2 Fig, AtGLK1, AtGLK2, OsGLK1 and OsGLK2 were classed into one group (ZmG54 classed with OsGLK1, and ZmG21 classed with OsGLK2).
Physical locations and duplication events of G2-like genes in maize
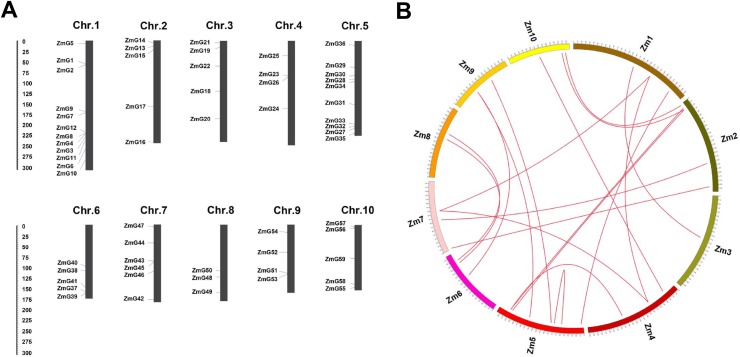
Chromosome locations of maize 59 G2-like genes revealed that 11, 5, 5, 4, 10, 5, 6, 3, 5 and 5 genes are distributed on chromosomes 1 to 10, respectively (Fig 2A). Although G2-like genes are distributed on every maize chromosome, their distribution and gene groups are nonrandom. For example, chromosome 1 not only contains the greatest number of G2-like genes (11), but it also exhibits the greatest variation of gene groups, whereas only three G2-like genes are located on chromosome 8. In addition, 18 duplicated pairs of ZmG genes were identified in syntney map (Fig 2B), and all of them were segmental duplication events. ZmGs duplication events might have resulted from ancient tetraploidy processes during the course of evolution. All chromosomes were involved in these duplication events.
Fig 2. Locations and duplication events of maize G2-like genes.
(A) Chrosome locations of maize G2-like genes. The scale represents megabases (Mb). The chromosome numbers are indicated above of each bar. (B) Synteny of maize G2-like gene. Numbers along each chromosome box indicate sequence lengths in megabases. All the syntenic genes were located in map, and linked by red lines.
The Ka/Ks ratio (ratio of nonsynonymous to synonymous nucleotide substitutions) is used as an indicator of selective pressure acting on a protein-coding gene [42]. The Ka (number of nonsynonymous substitutions per nonsynonymous site) and Ks (number of synonymous substitution per synonymous site of duplicated genes) values of 18 ZmGs pairs were calculated (Table 1), and results showed that five pairs genes were undergone positive selection (Ka/Ks> 1), and others were purifying selection (Ka/Ks< 1).
Table 1. Ka/Ks and divergence analysis of paralogous maize G2-like genes.
| Paralogoue pair | Ka | Ks | ka/ks | Selection pressure | Duplicate type |
|---|---|---|---|---|---|
| ZmG55/13 | 1.01 | 1.01 | 1.01 | Positive selection | Segmental duplication |
| ZmG50/27 | 0.99 | 1.07 | 1.08 | Positive selection | Segmental duplication |
| ZmG45/4 | 0.96 | 1.15 | 1.19 | Positive selection | Segmental duplication |
| ZmG35/9 | 0.75 | 0.94 | 1.26 | Positive selection | Segmental duplication |
| ZmG4/22 | 0.69 | 0.99 | 1.43 | Positive selection | Segmental duplication |
| ZmG12/26 | 1.06 | 0.90 | 0.85 | Purifying selection | Segmental duplication |
| ZmG36/47 | 0.56 | 0.46 | 0.82 | Purifying selection | Segmental duplication |
| ZmG28/29 | 1.18 | 0.92 | 0.78 | Purifying selection | Segmental duplication |
| ZmG27/7 | 0.92 | 0.85 | 0.93 | Purifying selection | Segmental duplication |
| ZmG23/31 | 1.00 | 0.91 | 0.91 | Purifying selection | Segmental duplication |
| ZmG58/14 | 1.23 | 0.99 | 0.80 | Purifying selection | Segmental duplication |
| ZmG32/14 | 0.96 | 0.77 | 0.80 | Purifying selection | Segmental duplication |
| ZmG22/45 | 1.39 | 0.97 | 0.70 | Purifying selection | Segmental duplication |
| ZmG15/46 | 0.99 | 0.88 | 0.89 | Purifying selection | Segmental duplication |
| ZmG24/59 | 0.69 | 0.58 | 0.83 | Purifying selection | Segmental duplication |
| ZmG37/51 | 2.09 | 0.58 | 0.28 | Purifying selection | Segmental duplication |
| ZmG49/40 | 1.02 | 0.95 | 0.93 | Purifying selection | Segmental duplication |
Phylogenetic comparison of G2-like genes between maize and sorghum
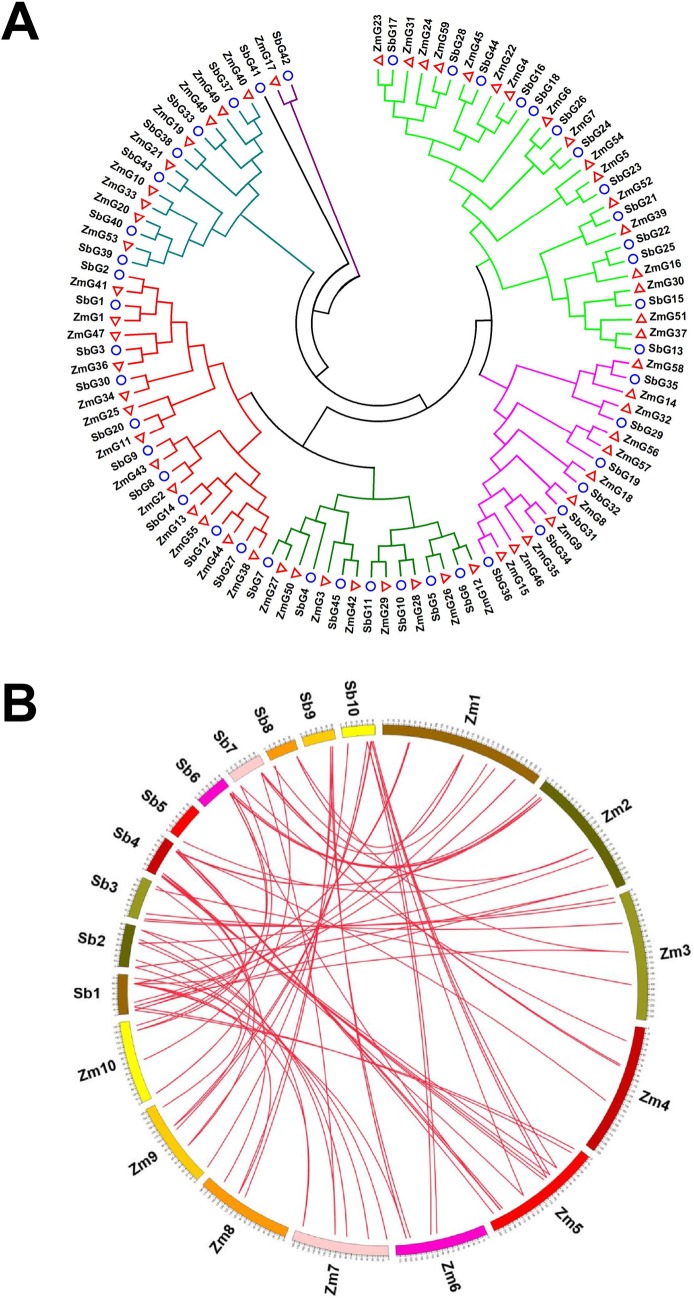
To further analyze evolutionary relationship of G2-like family, ZmGs and SbGs (G2-like genes of sorghum) were subjected to a comprehensive phylogenetic analysis. 45 sorghum G2-like genes (SbG1-SbG45) were identified (S3 Table and S2 Text), and 11 collinear gene pairs were found (S4 Table), distributing in 9 chromosomes (except chromosome 3) (S3 Fig). Then the unrooted phylogenetic tree between ZmGs and SbGs was constructed using the full-length protein sequences (Fig 3A). The phylogenetic analysis classified the ZmGs into several groups together with their sorghum orthologs. To identify the evolutionary orthologous relationships within G2-like genes of maize and sorghum, a synteny map was plotted between maize and sorghum (Fig 3B). A total of 75 orthologous gene pairs between maize and sorghum were found (S5 Table). Across maize and sorghum, strongly conserved microsyntny among regions hosting G2-like genes were observed, especially in Zm1 and Sb1 (6 synteny genes), Zm3 and Sb3 (4 synteny genes), Zm5 and Sb10 (4 synteny genes), Zm5 and Sb4 (9 synteny genes), Zm7 and Sb2 (6 synteny genes), Zm9 and Sb10 (4 synteny genes).
Fig 3. Phylogenetic analysis and duplication events of G2-like proteins between maize and sorghum.
(A) Phylogenetic tree of ZmGs and SbGs. The tree was constructed using neighbor-joining method with MEGA 6.0. Maize G2-like proteins were presented by hollow red triangle, and sorghum G2-like proteins were presented hollow blue circular. (B) Synteny of maize and sorghum G2-like genes regions. Numbers along each chromosome box indicate sequence lengths in megabases. Maize and sorghum chromosome were presented by Zm1-Zm10 and Sb1-Sb10, respectively. All the syntenic genes across maize and sorghum were linked by red lines.
Digital expression analysis
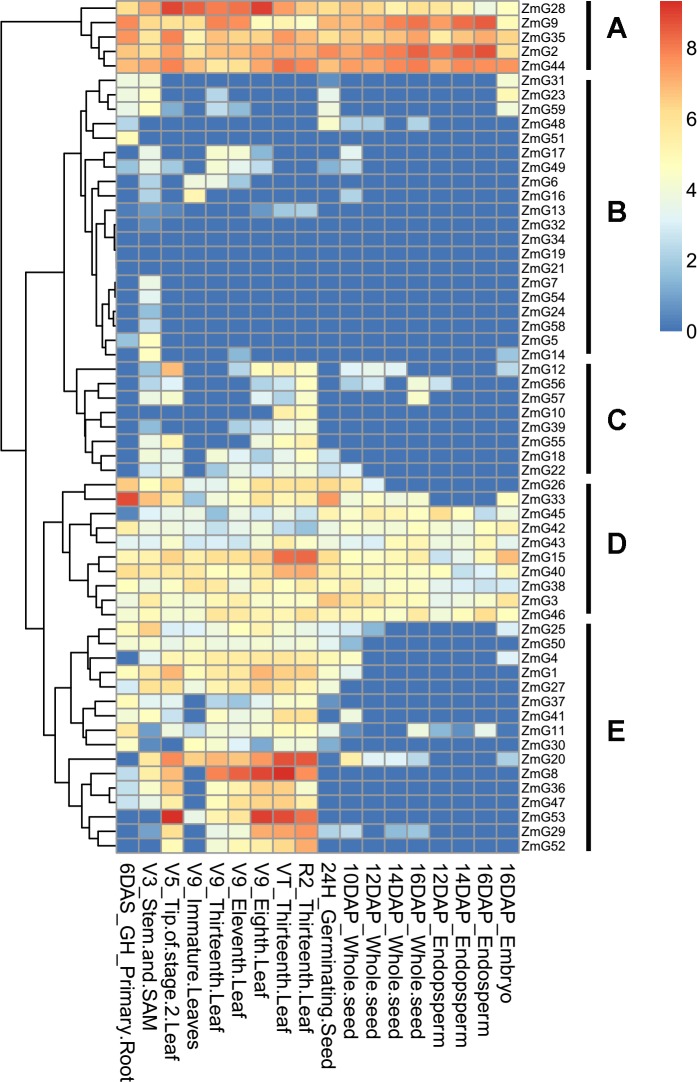
Gene expression patterns in different tissues provide important information for studying gene function. Results of expression pattern analysis (Fig 4) revealed that most G2-like genes were expressed in green tissues (e.g., stems and leaves), but some genes were expressed in non-green tissue (e.g., seeds, roots endosperm and embryos). The gene family consisted of five groups based on expression patterns (A–E). Pattern A and pattern D genes were expressed during almost all periods in seed, root, stem, leaf, endosperm and embryo tissue, with high levels of expression, especially for pattern A genes. Pattern B genes exhibited little or no expression in any tissue or organ, with only a few genes expressed in stems and leaves during some periods. Pattern C and E genes were mainly expressed in leaves, with high expression levels. We also examined the expression patterns of duplicated groups of G2-like genes. Pattern B contains the most duplicated genes (11 of 30 duplication genes). Pattern A, C and D include only 1, 2 and 3 duplicated genes, respectively, while pattern E contains 8 duplicated genes. Among these duplicated gene pairs, eight pairs (16 genes) shared the same expression patterns, including the following: ZmG9 and ZmG35 (pattern A); ZmG23 and ZmG31, ZmG24 and ZmG59, ZmG58 and ZmG14, ZmG32 and ZmG58 (pattern B); ZmG15 and ZmG46 (pattern D); and ZmG36 and ZmG47, ZmG50 and ZmG27 (pattern E).
Fig 4. Heat map of maize G2-like genes.
The expression level of each G2-like gene can be estimated based on the scale to the right. Red, yellow and blue indicate high, medium and low levels of gene expression, respectively. H, hours; DAS, days after sowing; GH, greenhouse; V, vegetative; DAP, days after pollination; VT, vegetative tasseling; R, reproductive.
Expression levels of maize G2-like genes in response to cold and drought stress
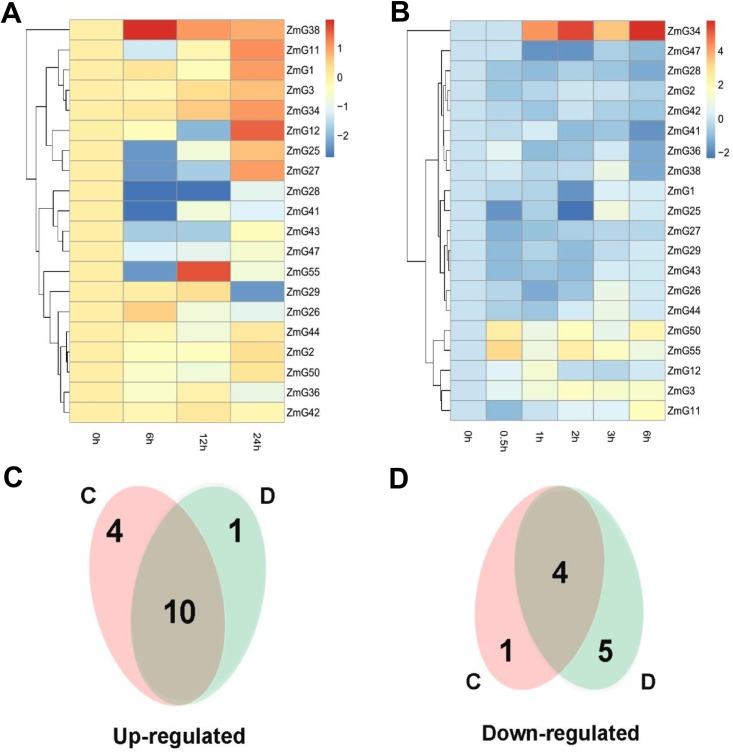
In this study, we investigated possible stress-responsive cis-elements in the promoter regions of these genes. Two abiotic relative cis-elements DRE (dehydration-responsive element) and LTRE (low-temperature responsive element) were detected in these genes 2000 bp promoter sequences, which may be responsive to their stress responsiveness (Table 2). Then we further investigated expression levels of some G2-like genes in response to abiotic stress by subjecting three-week-old seedling leaves to drought (20% PEG6000) and cold (4°C) treatments. We choose some ZmG genes for the followed verification experiment-qRT-PCR, randomly. Detailed expression profiles of these G2-like genes under cold and drought stress conditions were presented in S4 Fig. Heat map representation for transcript expression fold change in response to these two abiotic stresses was shown in Fig 5. Ten genes (ZmG3, ZmG11, ZmG12, ZmG25, ZmG26, ZmG34, ZmG38, ZmG44, ZmG50 and ZmG55) were up-regulated by both drought (Fig 5A) and cold stresses (Fig 5B), while four genes (ZmG28, ZmG41, ZmG43 and ZmG47) were down-regulated under both two conditions. Moreover, the expression of five genes (ZmG1, ZmG2, ZmG27, ZmG29 and ZmG42) were induced (Except ZmG42) by drought stress (Fig 5A) but repressed by cold (Fig 5B), and only one gene (ZmG36) was induced by cold but repressed by drought.
Table 2. Cis-elements in the promoter regions of stress-responsive G2-like genes.
| Genes No. | Promotor Elements | Genes No. | Promotor Elements | ||
|---|---|---|---|---|---|
| DRE | LTRE | DRE | LTRE | ||
| ZmG1 | 2 | 3 | ZmG31 | 0 | 2 |
| ZmG2 | 2 | 2 | ZmG32 | 1 | 1 |
| ZmG3 | 0 | 6 | ZmG33 | 0 | 0 |
| ZmG4 | 7 | 9 | ZmG34 | 1 | 2 |
| ZmG5 | 0 | 1 | ZmG35 | 0 | 1 |
| ZmG6 | 0 | 0 | ZmG36 | 3 | 3 |
| ZmG7 | 8 | 7 | ZmG37 | 0 | 1 |
| ZmG8 | 0 | 2 | ZmG38 | 0 | 1 |
| ZmG9 | 0 | 9 | ZmG39 | 3 | 4 |
| ZmG10 | 0 | 1 | ZmG40 | 2 | 2 |
| ZmG11 | 2 | 2 | ZmG41 | 0 | 0 |
| ZmG12 | 9 | 0 | ZmG42 | 4 | 4 |
| ZmG13 | 2 | 4 | ZmG43 | 1 | 2 |
| ZmG14 | 1 | 3 | ZmG44 | 2 | 6 |
| ZmG15 | 1 | 1 | ZmG45 | 0 | 2 |
| ZmG16 | 2 | 4 | ZmG46 | 0 | 0 |
| ZmG17 | 1 | 1 | ZmG47 | 1 | 4 |
| ZmG18 | 2 | 2 | ZmG48 | 0 | 0 |
| ZmG19 | 0 | 0 | ZmG49 | 4 | 3 |
| ZmG20 | 6 | 5 | ZmG50 | 0 | 2 |
| ZmG21 | 0 | 0 | ZmG51 | 0 | 0 |
| ZmG22 | 3 | 4 | ZmG52 | 1 | 1 |
| ZmG23 | 1 | 1 | ZmG53 | 0 | 3 |
| ZmG24 | 1 | 2 | ZmG54 | 7 | 6 |
| ZmG25 | 1 | 2 | ZmG55 | 1 | 0 |
| ZmG26 | 8 | 6 | ZmG56 | 0 | 0 |
| ZmG27 | 1 | 3 | ZmG57 | 0 | 4 |
| ZmG28 | 0 | 1 | ZmG58 | 0 | 5 |
| ZmG29 | 0 | 1 | ZmG59 | 0 | 0 |
| ZmG30 | 0 | 1 | |||
Fig 5. Expression of twenty maize stress-responsive G2-like genes under stress treatments.
Three-week-old seedlings were treated with 20% PEG6000 (A) and 4°C (B), respectively. Relative expression levels of the ZmG genes were analyzed by quantitative qPCR, and log2-transformed fold-change values were used for creating the heatmap (original data were shown in S4 Fig). Venn diagram illustrated the distribution of the up-regulated (C) or down-regulated (D) ZmG genes response to cold and drought treatments. The common subset of genes regulated by the two stressed was marked by the overlapping circle.
Nucleotide diversity of G2-like genes in 85 maize inbreed lines
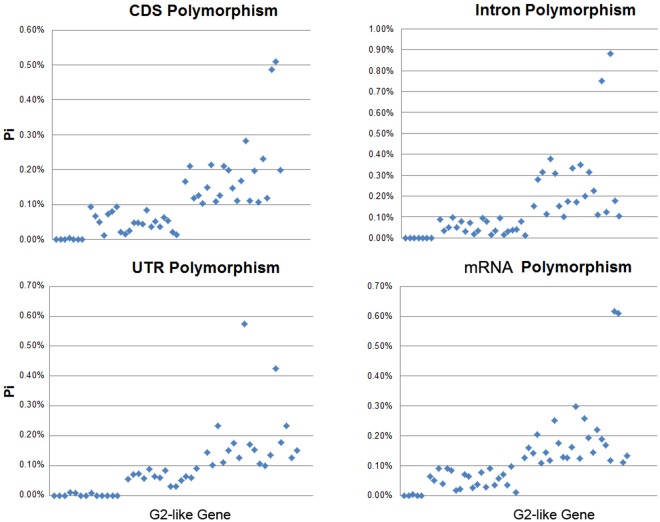
The polymorphism of G2-like genes in different regions (CDS, UTR, introns and mRNA) among 85 maize inbreed lines were further analyzed (Fig 6). Categories of ZmGs polymorphism on all regions were divided depending on Pi value (Pi is the average number of nucleotide differences per site between any two DNA sequences). Results showed that these four regions could be classified into similar groups: Pi ≤ 0.01%, 0.01% < Pi ≤ 0.1% and Pi > 0.1%, which we designated Type I, Type II and Type III, respectively. Type III includes the largest genes, while Type I, harboring a low level of nucleotide variation, and includes the fewest genes. We analyzed the Ka/Ks ratios of highly diverse genes among 85 maize lines (Table 3) to explore the selection pressure they are undergoing. Among the 23 genes examined, only one gene (ZmG34) was under positive selection.
Fig 6. Polymorphism of G2-like genes.
The polymorphism in different conserved regions (CDS, UTR, intron and mRNA) is shown. Vertical axis shows the nucleotide polymorphism Pi, and the horizontal axis represents G2-like genes in maize.
Table 3. Ka/Ks and selection pressure on diverse G2-like genes.
| Gene ID | Ka | Ks | Ka/Ks | selection Pressure |
|---|---|---|---|---|
| ZmG1 | 0.000796 | 0.002883 | 0.275923 | Negative selection |
| ZmG5 | 0.001525 | 0.003635 | 0.419355 | Negative selection |
| ZmG6 | 0.000968 | 0.003109 | 0.311423 | Negative selection |
| ZmG9 | 0.001677 | 0.006709 | 0.249946 | Negative selection |
| ZmG11 | 0.000879 | 0.001751 | 0.50206 | Negative selection |
| ZmG14 | 0.000763 | 0.002921 | 0.261349 | Negative selection |
| ZmG15 | 0.001667 | 0.00301 | 0.553781 | Negative selection |
| ZmG17 | 0.001898 | 0.003724 | 0.509689 | Negative selection |
| ZmG24 | 0.000871 | 0.001564 | 0.556699 | Negative selection |
| ZmG26 | 0.001311 | 0.004799 | 0.273232 | Negative selection |
| ZmG27 | 0.000316 | 0.006472 | 0.048882 | Negative selection |
| ZmG30 | 0.002397 | 0.01413 | 0.169648 | Negative selection |
| ZmG33 | 0.000548 | 0.002577 | 0.212772 | Negative selection |
| ZmG34 | 0.005853 | 0.002575 | 2.273277 | positive selection |
| ZmG35 | 0.001462 | 0 | ||
| ZmG37 | 0.00091 | 0.002306 | 0.394459 | Negative selection |
| ZmG38 | 0.00016 | 0.007295 | 0.021897 | Negative selection |
| ZmG39 | 0.001992 | 0 | ||
| ZmG41 | 0.000674 | 0.005051 | 0.133499 | Negative selection |
| ZmG46 | 0.00076 | 0.006038 | 0.125846 | Negative selection |
| ZmG53 | 0.000275 | 0.003129 | 0.08802 | Negative selection |
| ZmG54 | 0.000958 | 0.002034 | 0.470698 | Negative selection |
| ZmG58 | 0.001525 | 0.003635 | 0.419355 | Negative selection |
Comparison of G2-like genes genetic diversity between tropical and temperate lines
Nucleotide polymorphism of G2-like genes among 85 tropical and temperate maize inbred lines were also examined (S6 Table). The single nucleotide polymorphisms (SNP) between tropical and temperate lines were not significantly different, whereas, genetic variance analysis between these lines revealed that 14 genes (27.45%) exhibited differences between tropical and temperate lines, among which 11 genes (ZmG37, ZmG47, ZmG14, ZmG39, ZmG32, ZmG11, ZmG15, ZmG22, ZmG17, ZmG30 and ZmG5) exhibited significant differences (P < 0.01). We further analyzed the haplotype diversity between tropical and temperate lines (S6 Table), revealing that the haplotype diversity of tropical lines is greater than temperate. Seven of the 11 genes had moderate genetic differentiation, with Fst values (fixation index of the subpopulation within the total) [43] between 0.05 and 0.15, while 4 other genes (ZmG22, ZmG11, ZmG32 and ZmG47) had a high level of genetic differentiation, with Fst values greater than 0.15.
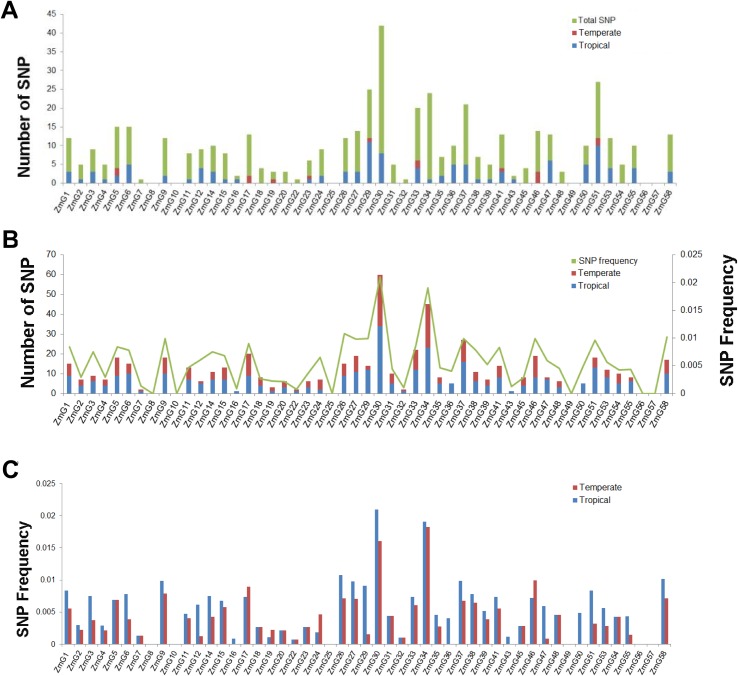
We compared the number of unique fixed SNPs in each G2-like gene in the tropical and temperate lines, respectively (Fig 7A). Results showed that G2-like genes of tropical lines have much more unique fixed SNP sites compared to temperate lines. Only five genes (ZmG5, ZmG23, ZmG17, ZmG19 and ZmG46) have as many or more unique fixed SNP sites in temperate versus tropical lines. However, the total number of fixed SNP statistics showed that ZmG genes shared more same fixed SNPs than their unique fixed SNPs in both tropical and temperate lines (Fig 7A), suggesting that these SNPs may be not selected based on the environment relative to temperature. Furthermore, the total number of fixed SNPs in tropical and temperate lines was analyzed (Fig 7B), revealing that more fixed SNPs of tropical lines are present than temperate lines. The frequency of fixed SNP exhibited variation that is always proportional to the number of SNPs per gene (the SNP frequency is the ratio of the total SNP number of each gene with their corresponding coding sequence length) (Fig 7B). However, 42 of 59 ZmG genes in tropical lines have higher fixed SNP frequency than temperate lines (Fig 7C).
Fig 7. Number of SNPs in each G2-like gene (CDS sequence) among 85 maize inbred lines.
Blue boxes represent the number of SNPs in tropical lines, red boxes represent the number of SNPs in temperate lines, green boxes represent the total number of SNPs and green lines indicate SNP frequency. (A) Number of tropical and temperate fixed SNPs and total number of SNPs in 85 maize inbred lines. (B) Number of SNPs in G2-like genes in tropical and temperate lines and SNP frequency. (C) Comparisons of tropical and temperate lines SNP frequency.
Discussion
Interestingly, in this study, 59 G2-like genes were identified in maize and a same set of G2-like gene was identified in GRASSIUS (http://grassius.org/grasstfdb.html) database ever (S3 Text). And ZmG genes in this study matched the list in GRASSIUS, except for one (ZmG34 and ZmGLK12). We checked these two genes, and found that ZmGLK12 does not contain Myb_DNA-binding domain, but ZmG34 contains, indicating ZmGLK12 was not G2-like gene.
In this study, numerous segmental duplications of maize G2-like genes were identified, indicating that segmental duplication was the main contributor to the expansion of maize G2-like genes. In tissues expression patterns, Pattern B contains the most duplicated genes (11 of 30 duplication genes), exhibited little or no expression in any tissue or organ, indicating some of these genes are pseudogenes or silenced paralogs. In general, segmental duplications are thought to occur regularly in more slowly evolving gene families. Thus we speculated that G2-like genes have been considerably conserved during the process of evolution. Since maize underwent whole genome duplication (WGD) event after diverging from sorghum, there ought to be twice as many GLK genes, but it is not double, clearly some have been lost 45 in sorghum versus 59 in maize. A phylogenetic comparison of G2-like genes between maize and sorghum divided 104 G2-like members into six clades, with ZmGs and SbGs appear to be more closely to each other. Moreover, synteny analysis showed, strongly conserved microsyntny among regions hosting G2-like genes across maize and sorghum was observed, further illustrated these genes derived from gene duplication.
The evolution of maize G2-like genes may have occurred during maize genome evolution. G2-like genes were amplified through duplication, which enabled C4 plants to undergo subfunctionalization, in turn enabling the development of cell-specific functions in dimorphic chloroplasts [44]. Current evidence suggests that the ancestral state was a single G2-like gene and that G2-like gene duplication enabled subfunctionalization [45]. Moreover, G2-like gene duplication preconditioned the evolution of chloroplast dimorphism. According to classical theory, when gene duplication occurs, each copy of the gene has two possible fates: (1) in most cases, a copy retains the original features, which are stabilized through negative selection; (2) the remaining copy does not undergo selection and becomes pseudogenic. This theory helps explain the slow evolution of G2-like genes and a certain level of total number of genes, further illustrating the highly conserved evolution of chloroplast. In rare cases, one copy of gene undergoes more adaptation, drive the evolution of gene function [46]. Based on analysis of G2-like gene family, we proposed that this family belongs to the former category, that is, the function of this gene family has been stabilized by duplication.
Furthermore, SNPs analysis of maize ZmG genes in natural population revealed the influences of temperature and humidity on ZmGs. Polymorphism analysis of G2-like genes in different conserved regions (Fig 6) revealed that highly diverse Type III includes the largest number of genes, which might have quickly evolved to adapt to environment, while highly conserved Type I includes the fewest genes, which may play an important role in basic functions rather than adaptation to temperature or humidity conditions. Selection pressure was revealed for genes in Type III, while only one gene (ZmG34) was under positive selection (Table 3), indicating that this family tends to retain vary in order to maintain stability. However, the variation retained in ZmG34 might result from its significant role in growth and development in maize. And the expression pattern of ZmG34 also suggested its importance. Genetic diversity analysis of G2-like genes between tropical and temperate lines (based on CDS) revealed 11 genes with Pi values lower than 0.01, which may be related to their role in the response to temperature and light conditions. These genes will be a focus for future research aimed at elucidating the influence of temperature and humidity conditions on maize growth. In these genes, ZmG47 had a high level of genetic differentiation, and a high level expression in all tissues, we hypothesized that this gene may have significant roles in the formation and evolution of chloroplast.
In addition to total SNPs number, compared to temperate lines, more SNPs of ZmG genes were fixed in tropical lines. It suggested that these SNPs were selected based on the environment, which may relate to temperature and humidity. Our promoter element analysis results support this view, as promoters of these genes include DRE, ABRE, and LTRE resistance elements [47,48], and qRT-PCR results supported this view, too. In addition, a recent report suggested that G2-like genes play a role in the temperature stress response, as increased accumulation of G2-like1 was observed in frost-tolerant transgenic Brassica napus overexpressing two transcripts harboring DREB1/CBFs [49]. These stress-related genes are induced in order to adapt to environmental stresses. Overexpression of AtGLK1 (35S: AtGLK1) in Arabidopsis confers resistance to the cereal pathogen Fusarium graminearum [27]. Thus, we speculate that maize G2-like gene family might have a function in stress and disease resistance. In addition, as ZmG34 was significantly up-regulated by low temperature and drought stress, moreover, it was subjected to positive selection in natural population lines; we convinced more on this gene’s importance in crop breeding.
Due to highly conserved evolution of G2-like genes, we speculated these genes might play vital roles in maize growth and development. Moreover, both of these genes polymorphism and expression were sensitive for environment; further exploration of them would serve as useful information for maize culture in drought or cold environment.
Supporting Information
Pileup multiple sequence alignment of maize ZmG1–59, rice OsGLK1–2 and Arabidopsis AtGLK1–2 GLK proteins. The putative DNA binding domain folds into an HLH structure. Black horizontal bars indicate the predicted α-helix segments conserved in all proteins. The GCT box is delimited, marked by red horizontal bars.
(TIF)
(TIF)
Numbers along each chromosome box indicate sequence lengths in megabases. All the syntenic genes were located in sorghum chromosome, and linked by red lines.
(TIF)
The Y-axis is the scale of relative expression levels. The X-axis is time courses of stress treatments. Error bars, +SE. (A) Relative expression levels of the twenty stress-responsive G2-like genes in responsive to drought stress. Seedlings were sampled at 0 h (CK), 6 h, 12 h, and 24 h after drought treatment. (B) Relative expression levels of the twenty stress- responsive G2-like genes in response to low temperature treatment (4°C). Seedlings were sampled at 0 h (CK), 0.5 h, 1 h, 2 h, 3 h and 6 h after 4°C treatment.
(TIF)
(DOC)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(TXT)
(TXT)
(TXT)
Acknowledgments
We thank all the members of the Key Laboratory of Crop Biology of Anhui province for their assistance in this study.
Data Availability
The 85 maize inbred lines sequencing SNP data are available upon request to Chuanxiao Xie (xiechuanxiao@caas.cn) National Key Facility for Crop Gene Resources and Genetic Improvement, Institute of Crop Science, Chinese Academy of Agricultural Sciences. Other remaining data were provided as a Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Jarvis P, Lopez-Juez E (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nature Reviews Molecular Cell Biology 14: 787–802. 10.1038/nrm3702 [DOI] [PubMed] [Google Scholar]
- 2.Jarvis P, Lopez-Juez E (2014) Biogenesis and homeostasis of chloroplasts and other plastids (vol 14, pg 787, 2013). Nature Reviews Molecular Cell Biology 15: 147–147. [DOI] [PubMed] [Google Scholar]
- 3.Eberhard S, Finazzi G, Wollman FA (2008) The Dynamics of Photosynthesis. Annual Review of Genetics 42: 463–515. 10.1146/annurev.genet.42.110807.091452 [DOI] [PubMed] [Google Scholar]
- 4.Waters MT, Langdale JA (2009) The making of a chloroplast. Embo Journal 28: 2861–2873. 10.1038/emboj.2009.264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker SFS, Mayor R, Kashef J (2013) Cadherin-11 Mediates Contact Inhibition of Locomotion during Xenopus Neural Crest Cell Migration. Plos One 8: e85717 10.1371/journal.pone.0085717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parente DJ, Swint-Kruse L (2013) Multiple Co-Evolutionary Networks Are Supported by the Common Tertiary Scaffold of the LacI/GalR Proteins. Plos One 8: e84398 10.1371/journal.pone.0084398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Douglas SE (1998) Plastid evolution: origins, diversity, trends. Curr Opin Genet Dev 8: 655–661. [DOI] [PubMed] [Google Scholar]
- 8.Moreira D, Le Guyader H, Philippe H (2000) The origin of red algae and the evolution of chloroplasts. Nature 405: 69–72. [DOI] [PubMed] [Google Scholar]
- 9.Lopez-Juez E, Pyke KA (2005) Plastids unleashed: their development and their integration in plant development. International Journal of Developmental Biology 49: 557–577. [DOI] [PubMed] [Google Scholar]
- 10.Park J, Werley CA, Venkatachalam V, Kralj JM, Dib-Hajj SD, Waxman SG, et al. (2013) Screening Fluorescent Voltage Indicators with Spontaneously Spiking HEK Cells. Plos One 8: e85221 10.1371/journal.pone.0085221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neuhaus HE, Emes MJ (2000) Nonphotosynthetic metabolism in plastids. Annual Review of Plant Physiology and Plant Molecular Biology 51: 111–140. [DOI] [PubMed] [Google Scholar]
- 12.Charuvi D, Kiss V, Nevo R, Shimoni E, Adam Z, Reich Z, et al. (2012) Gain and Loss of Photosynthetic Membranes during Plastid Differentiation in the Shoot Apex of Arabidopsis. Plant Cell 24: 1143–1157. 10.1105/tpc.111.094458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez-Juez E, Dillon E, Magyar Z, Khan S, Hazeldine S, de Jager SM, et al. (2008) Distinct light-initiated gene expression and cell cycle programs in the shoot apex and cotyledons of Arabidopsis. Plant Cell 20: 947–968. 10.1105/tpc.107.057075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seu L, Ortiz GM, Epling L, Sinclair E, Swainson LA, Bajpai UD, et al. (2013) Higher CD27(+)CD8(+) T Cells Percentages during Suppressive Antiretroviral Therapy Predict Greater Subsequent CD4(+) T Cell Recovery in Treated HIV Infection. Plos One 8: e84091 10.1371/journal.pone.0084091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiao YL, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nature Reviews Genetics 8: 217–230. [DOI] [PubMed] [Google Scholar]
- 16.Li PH, Ponnala L, Gandotra N, Wang L, Si YQ, Tausta SL, et al. (2010) The developmental dynamics of the maize leaf transcriptome. Nature Genetics 42: 1060–U1051. 10.1038/ng.703 [DOI] [PubMed] [Google Scholar]
- 17.La Marca M, Beffy P, Pugliese A, Longo V (2013) Fermented Wheat Powder Induces the Antioxidant and Detoxifying System in Primary Rat Hepatocytes. Plos One 8: e83538 10.1371/journal.pone.0083538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, et al. (2000) Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110. [DOI] [PubMed] [Google Scholar]
- 19.Yasumura Y, Moylan EC, Langdale JA (2005) A conserved transcription factor mediates nuclear control of organelle biogenesis in anciently diverged land plants. Plant Cell 17: 1894–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao HB, Kadirjan-Kalbach D, Froehlich JE, Osteryoung KW (2003) ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proceedings of the National Academy of Sciences of the United States of America 100: 4328–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang WJ, Wang WQ, Chen DQ, Ji Q, Jing YJ, Wang HY, et al. (2012) Transposase-Derived Proteins FHY3/FAR1 Interact with PHYTOCHROME-INTERACTING FACTOR1 to Regulate Chlorophyll Biosynthesis by Modulating HEMB1 during Deetiolation in Arabidopsis. Plant Cell 24: 1984–2000. 10.1105/tpc.112.097022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossini L, Cribb L, Martin DJ, Langdale JA (2001) The maize Golden2 gene defines a novel class of transcriptional regulators in plants. Plant Cell 13: 1231–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA (2002) GLK gene pairs regulate chloroplast development in diverse plant species. Plant Journal 31: 713–727. [DOI] [PubMed] [Google Scholar]
- 24.Hall LN, Rossini L, Cribb L, Langdale JA (1998) GOLDEN 2: a novel transcriptional regulator of cellular differentiation in the maize leaf. Plant Cell 10: 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA, et al. (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128. 10.1105/tpc.108.065250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Powell AL, Nguyen CV, Hill T, Cheng KL, Figueroa-Balderas R, Aktas H, et al. (2012) Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 336: 1711–1715. 10.1126/science.1222218 [DOI] [PubMed] [Google Scholar]
- 27.Tan XP, Meyers BC, Kozik A, Al West M, Morgante M, St Clair DA, et al. (2007) Global expression analysis of nucleotide binding site-leucine rich repeat-encoding and related genes in Arabidopsis. Bmc Plant Biology 7: 56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kretschmer M, Reiner E, Hu GG, Tam N, Oliveira DL, Caza M, et al. (2014) Defects in Phosphate Acquisition and Storage Influence Virulence of Cryptococcus neoformans. Infection and Immunity 82: 2697–2712. 10.1128/IAI.01607-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailey TL, Elkan C (1995) The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol 3: 21–29. [PubMed] [Google Scholar]
- 31.Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G, et al. (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31: 1296–1297. 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, et al. (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40: e49 10.1093/nar/gkr1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 35.Swanson WJ, Yang Z, Wolfner MF, Aquadro CF (2001) Positive Darwinian selection drives the evolution of several female reproductive proteins in mammals. Proc Natl Acad Sci U S A 98: 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sekhon RS, Briskine R, Hirsch CN, Myers CL, Springer NM, Buell CR, et al. (2013) Maize Gene Atlas Developed by RNA Sequencing and Comparative Evaluation of Transcriptomes Based on RNA Sequencing and Microarrays. Plos One 8: e61005 10.1371/journal.pone.0061005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sekhon RS, Lin HN, Childs KL, Hansey CN, Buell CR, de Leon N, et al. (2011) Genome-wide atlas of transcription during maize development. Plant Journal 66: 553–563. 10.1111/j.1365-313X.2011.04527.x [DOI] [PubMed] [Google Scholar]
- 38.Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- 39.Murmu J, Wilton M, Allard G, Pandeya R, Desveaux D, Singh J, et al. (2014) Arabidopsis GOLDEN2-LIKE (GLK) transcription factors activate jasmonic acid (JA)-dependent disease susceptibility to the biotrophic pathogen Hyaloperonospora arabidopsidis, as well as JA-independent plant immunity against the necrotrophic pathogen Botrytis cinerea. Mol Plant Pathol 15: 174–184. 10.1111/mpp.12077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cinquin O, Demongeot J (2005) High-dimensional switches and the modelling of cellular differentiation. Journal of Theoretical Biology 233: 391–411. [DOI] [PubMed] [Google Scholar]
- 41.Massari ME, Murre C (2000) Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Molecular and Cellular Biology 20: 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang ZF, Zhou Y, Wang XF, Gu SL, Yu JM, Liang G H, et al. (2008) Genomewide comparative phylogenetic and molecular evolutionary analysis of tubby-like protein family in Arabidopsis, rice, and poplar. Genomics 92: 246–253. 10.1016/j.ygeno.2008.06.001 [DOI] [PubMed] [Google Scholar]
- 43.Hudson RR (1990) Genetic Data Analysis. Methods for Discrete Population Genetic Data. Bruce S. Weir. Sinauer, Sunderland, MA, 1990. xiv, 377 pp., illus. $48; paper, $27. Science 250: 575. [DOI] [PubMed]
- 44.Bravo-Garcia A, Yasumura Y, Langdale JA (2009) Specialization of the Golden2-like regulatory pathway during land plant evolution. New Phytologist 183: 133–141. 10.1111/j.1469-8137.2009.02829.x [DOI] [PubMed] [Google Scholar]
- 45.Wang P, Fouracre J, Kelly S, Karki S, Gowik U, Aubry S, et al. (2013) Evolution of GOLDEN2-LIKE gene function in C-3 and C-4 plants. Planta 237: 481–495. 10.1007/s00425-012-1754-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prince VE, Pickett FB (2002) Splitting pairs: The diverging fates of duplicated genes. Nature Reviews Genetics 3: 827–837. [DOI] [PubMed] [Google Scholar]
- 47.Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, et al. (2003) Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant Journal 34: 137–148. [DOI] [PubMed] [Google Scholar]
- 48.Yamaguchi-Shinozaki K, Shinozaki K (2005) Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends in Plant Science 10: 88–94. [DOI] [PubMed] [Google Scholar]
- 49.Savitch LV, Allard G, Seki M, Robert LS, Tinker NA, Huner NPA, et al. (2005) The effect of overexpression of two Brassica CBF/DREB1-like transcription factors on photosynthetic capacity and freezing tolerance in Brassica napus. Plant and Cell Physiology 46: 1525–1539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pileup multiple sequence alignment of maize ZmG1–59, rice OsGLK1–2 and Arabidopsis AtGLK1–2 GLK proteins. The putative DNA binding domain folds into an HLH structure. Black horizontal bars indicate the predicted α-helix segments conserved in all proteins. The GCT box is delimited, marked by red horizontal bars.
(TIF)
(TIF)
Numbers along each chromosome box indicate sequence lengths in megabases. All the syntenic genes were located in sorghum chromosome, and linked by red lines.
(TIF)
The Y-axis is the scale of relative expression levels. The X-axis is time courses of stress treatments. Error bars, +SE. (A) Relative expression levels of the twenty stress-responsive G2-like genes in responsive to drought stress. Seedlings were sampled at 0 h (CK), 6 h, 12 h, and 24 h after drought treatment. (B) Relative expression levels of the twenty stress- responsive G2-like genes in response to low temperature treatment (4°C). Seedlings were sampled at 0 h (CK), 0.5 h, 1 h, 2 h, 3 h and 6 h after 4°C treatment.
(TIF)
(DOC)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(TXT)
(TXT)
(TXT)
Data Availability Statement
The 85 maize inbred lines sequencing SNP data are available upon request to Chuanxiao Xie (xiechuanxiao@caas.cn) National Key Facility for Crop Gene Resources and Genetic Improvement, Institute of Crop Science, Chinese Academy of Agricultural Sciences. Other remaining data were provided as a Supporting Information files.