Abstract
The LATERAL ORGAN BOUNDARIES DOMAIN (LBD) gene family has been well-studied in Arabidopsis and play crucial roles in the diverse growth and development processes including establishment and maintenance of boundary of developmental lateral organs. In this study we identified and characterized 38 LBD genes in Lotus japonicus (LjLBD) and 57 LBD genes in Medicago truncatula (MtLBD), both of which are model legume plants that have some specific development features absent in Arabidopsis. The phylogenetic relationships, their locations in the genome, genes structure and conserved motifs were examined. The results revealed that all LjLBD and MtLBD genes could be distinctly divided into two classes: Class I and II. The evolutionary analysis showed that Type I functional divergence with some significantly site-specific shifts may be the main force for the divergence between Class I and Class II. In addition, the expression patterns of LjLBD genes uncovered the diverse functions in plant development. Interestingly, we found that two LjLBD proteins that were highly expressed during compound leaf and pulvinus development, can interact via yeast two-hybrid assays. Taken together, our findings provide an evolutionary and genetic foundation in further understanding the molecular basis of LBD gene family in general, specifically in L. japonicus and M. truncatula.
Introduction
LATERAL ORGAN BOUNDARIES DOMAIN (LBD) proteins, a plant-specific transcription factor family, possess a characteristic N-terminal LOB domain and play important roles in many aspects of plant development. The LBD protein typically contains four highly conserved cysteine (C) residues in a CX2CX6CX3C zinc finger-like motif (also called C block, where X represents variable residues) that is suggested to play crucial role in DNA binding. In addition, two other conserved features are found in the N-terminal half of the LBD: an invariant glycine residue and a leucine-zipper-like sequence LX6LX3LX6L [1,2]. Usually, the LBD gene family can be divided into two classes (class I and II) based on conserved motif number and structural features. Typically, class I members contain a CX2CX6CX3C motif and a leucine-zipper-like motif, while class II members contain only a CX2CX6CX3C motif. As transcription factors, LBD proteins function in the nucleus and bind to the conserved nucleotide consensus sequence GCGGCG. In addition, there is evidence that the interaction between LBD and bHLH proteins can reduce the DNA-binding affinity of LBDs [3].
Studies show that LBD genes usually exhibit temporal or tissue-specific expression patterns [1]. For example LBD genes are expressed in specialized regions such as the adaxial base of lateral organs, shoot apical meristem and boundary between lateral organs, indicating their important function in plant lateral organ development [4,5]. In Arabidopsis, LBD genes are found to involve in various tissues development such as leaf development [6] and lateral root initiation [7–9], as well as signaling transduction such as cytokinin [10] and gibberellin pathways [11]. Also, three LBD genes in Arabidopsis including AtLBD37, AtLBD38 and AtLBD39, are implicated in anthocyanin biosynthesis and nitrate metabolism [12]. In other species, a panel of LBD genes were also well studied and characterized. For instance, the maize Ramosa2 gene plays a role in regulation of inflorescence architecture [13]. Two maize LBD family genes RTCS and RTCL cooperatively act in shoot-borne root formation [14]. The rice auxin-inducible ARL1 gene encodes a LBD protein that promotes adventitious root formation [15].
With the advent of high-throughput sequencing techniques, genome-wide identification and characterization of LBD genes have been conducted in Arabidopsis [1], rice [16], maize [17] and apple [18]. However, little is known about LBD genes in legumes to date. Legumes are important crop plants that not only have the unique ability to fix nitrogen from the atmosphere, but also are rich sources of protein and oil for the human diet. The genomes of many legume plants have recently been sequenced, providing an opportunity for detailed characterization of LBD genes in this important family of plants. Among legumes, Lotus japonicus and Medicago truncatula are used as model legumes due to their short life cycles, self-fertility, and relatively small diploid genome. In particular, these two species provide an excellent system for study of some specific development features including compound leaf development, motor organ specification, and root nodule formation that are absent in Arabidopsis. For example, ELP1 (Elongated Petiolule1; also called MtLBD13 in this study) and SLP (Sleepless, also called LjLBD6 in this study) were isolated from M. truncatula and L. japonicus, respectively, and found that they are involved in specification of so-called motor organ or pulvinus identity, uncovering a novel function for LBD genes in the determination of motor organs and the control of plant movement in legumes [5]. In addition, three LBD genes (LjLOB1, LjLOB3 and LjLOB4) have been isolated from L. japonicus, and their specific expression patterns strongly showed that LjLOB1 and LjLOB3 (also called LjLBD11 and LjLBD6 in this study) might have important roles in determining compound leaf development, and that another gene LjLOB4 (also called LjLBD22 in this study) may be involved in floral development [4]. Thus, investigation of LBD genes at genome-wide level in legumes would likely provide new insights into the regulatory mechanisms of plant growth and development, especially, motor organs and compound leaves development.
In the present study, we identified and characterized the LBD gene family in L. japonicus and M. truncatula on the genome-wide scale. In total, we found 38 putative LBD genes in L. japonicus and 57 members in M. truncatula, respectively. Their motif distribution, evolutionary relationship and expression profiles were further characterized in detail. Importantly, we found that two LBD members LjLBD11 (LjLOB1) and LjLBD6 (LjLOB3, SLP1) were highly expressed in compound leaf and can form complex by proteins interaction, implying that they may work together in controlling compound leaf development in L. japonicus. These results obtained from this study provide global information important for further understanding the molecular functions of the LBD gene family in legumes.
Materials and Methods
Screening of LBD gene members
All Arabidopsis LBD proteins were retrieved from the DATF database (http://datf.cbi.pku.edu.cn) based on the previous study [1]. All protein sequences of L. japonicus and M. truncatula were downloaded from the database (http://www.plantgdb.org/LjGDB/) and Phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Mtruncatula), respectively. Using BLASTP program, all Arabidopsis LBD protein sequences were used as query against corresponding protein database. The hits with a significant E-value (< lE-3) and more than 70% identity were collected and considered as candidate proteins. Subsequently, redundant sequences or incomplete ORF sequences were removed from our gene set. Finally, all candidate proteins were further subjected to SMART (http://smart.embl-heidelberg.de/) and Pfam (http://www.pfam.sanger.ac.uk/) to confirm the presence of LOB domain.
Chromosomal localization and gene structure analysis
To determine the distribution of LBD genes on chromosomes, position information of each LBD gene was obtained from genome annotation file (downloaded from website: http://www.plantgdb.org/LjGDB/ and http://www.jcvi.org/medicago/, respectively). A custom MATLAB script was used to draw the location information of LBD genes in chromosome or scaffold. According to the alignment between full-length CDS and corresponding gene sequence, exon-intron organization for individual LBD gene was illustrated by using Gene structure display server software (GSDS, http://gsds.cbi.pku.edu.cn/)
Phylogenetic tree, evolutionary analysis and conserved motifs
Multiple sequence alignment of amino acid sequences was carried out using Clustalx v2.1 (http://www.clustal.org/clustal2/), and phylogenetic trees were then generated using the neighbor-joining (NJ) method with 10,000 bootstrap replicates in MEGA5.0 [19]. The functional divergence for classes or subclasses was estimated using the DIVERGE v3.0 package [20]. The coefficients of type-I (θI) and type-II (θII) functional divergence between any two classes were estimated based on the ML algorithm. The Type-I functional divergence means that some significantly site-specific changes mainly occurs between two classes. The Type-II means that functional divergence between two classes were mainly due to some site-specific shifts in amino acid properties. The coefficients of Type I or type II functional divergence with greater than zero were considered significant. Additionally, to identify which sites are critical for functional divergence between two classes, a site-specific posterior probability were predicted with a cut-off value more than 0.9 to reduce possible false positives.
Besides, conserved motifs of each LBD protein were identified using MEME Suite (http://meme-suite.org/) with parameters set: optimum width 6–200 amino acids and maximum number of motifs 15, and then visualized by MAST (Motif Alignment and Search Tool). In addition, the molecular weight (MW, Da) and isoelectric point (pI) of each protein were estimated by online ExPASy programs (http://www.expasy.org/tools/).
Expression profiles of LBD genes in L. japonicus
The expression profiles of all LjLBD genes were investigated among different tissues or developmental stages using L. japonicus gene expression altas (LjGEA, http://ljgea.noble.org/v2/) [21]. Briefly, probe ID of each LjLBD gene was firstly obtained by gene sequence BLASTN program that is available at LjGEA website. If a given gene has more than one probe ID, we choose the probe ID showing the best e-value and higher identity (S1 Table). Expression level of each gene was obtained from different tissues by normalizing probe count, and then a global gene expression profiles were visualized by using MultiExperiment Viewer (MeV) software (v4.8.1). Some LBD genes with high expression level in leaf were confirmed by quantitative RT-PCR (qRT-PCR) with three independent biological replications.
Yeast Two-Hybird Assay
Yeast two-hybrid analysis was performed in Saccharomyces cerevisiae strain Y2H gold according to the manufacturer’s instructions (http://www.clontech.com/). The full-length of LjLBD11 (LBD11-Fw: CCGGggatccATGAAGGGTTATGAACCACG; LBD11-Rv: CCGGgtcgacTCAAAATATATATGGGATTTGA) and LjLBD6 (LBD6-Fw: CCGGggatccATGGCATCATCAAGCGCTTA; LBD6-Rv: CCGGgtcgacTCATAAATTACCTCCTCCTTCAC) were cloned into the prey vector pGADT7 or the bait vector pGBKT7. The yeast cells were co-transformed with the prey vector and the bait vector using Yeast Transformation II Kit (The Epigenetics CompanyTM). Quadruple dropout medium (without adenine, histidine, leucine and trptophan) containing 200 ng/ml Aureobasidin and 40 mg/ml x-a-gal was used to test the expression of reported genes AUR1-C and MEL1.
Results
Identification and physical locations of LBD genes
A total of 45 candidate LBD genes were identified in L. japonicus genome based on an extensive search. Unfortunately, seven LBD proteins were excluded due to the lack of typical LOB domain or C motif (CX2CX6CX3C) in the N-terminus. Thus, we finally identified 38 LjLBD candidate proteins in the L. Japonicus, ranging from 95 aa to 349 aa in length. The molecular weight of LjLBD proteins varied from 10.36 kDa to 39.96 kDa, and protein pI ranged from 4.40 to 10.12 (see Table 1). For the better description in subsequent analyses, the 38 non-redundant LBD genes in L. japonicus were designated LjLBD1–38 according to their positions from the top to the bottom on the chromosomes or scaffolds (Table 1). In addition, we also identify 57 LBD proteins in M. truncatula (named MtLBD1-57) based on the same criteria and found similar protein lengths and molecular weights (S1 Table). The number of LBD genes in these two model legumes was comparable to their homologs in other plants such as Arabidopsis (43 members) [1], rice (35 members) [16], maize (43 members) [17] and apple (58 members) [18].
Table 1. Information of LBD gene family identified in Lotus japonicus.
| Gene identifier | Gene name | Genomic position | Size(aa) | MW(Da) | pI |
|---|---|---|---|---|---|
| chr1.CM0001.80.r2.a | LjLBD1 | chr1:44897181–44897675 | 164 | 18162.3 | 6.25 |
| chr1.CM0017.40.r2.m | LjLBD2 | chr1:39247040–39248087 | 316 | 34117.4 | 8.65 |
| chr1.CM0088.550.r2.d | LjLBD3 | chr1:201658–202206 | 156 | 16733.2 | 8.3 |
| chr1.CM0104.3290.r2.d | LjLBD4 | chr1:50334865–50335833 | 234 | 25236.3 | 7.86 |
| chr1.CM0125.620.r2.d | LjLBD5 | chr1:16763699–16764628 | 190 | 20829.9 | 9.03 |
| chr1.CM0171.410.r2.m | LjLBD6 | chr1:3870813–3871385 | 190 | 20834.6 | 7.83 |
| chr1.CM0375.610.r2.a | LjLBD7 | chr1:56389849–56391175 | 205 | 22398.4 | 8.11 |
| chr1.CM0375.630.r2.a | LjLBD8 | chr1:56398498–56399492 | 222 | 24576.6 | 6.49 |
| chr1.CM1255.330.r2.m | LjLBD9 | chr1:31734423–31736630 | 184 | 20117.9 | 8.17 |
| chr1.CM1255.380.r2.m | LjLBD10 | chr1:31764240–31767714 | 247 | 25610 | 8.41 |
| chr2.CM0002.640.r2.m | LjLBD11 | chr2:37239732–37240286 | 184 | 20456.2 | 7.06 |
| chr2.CM0008.840.r2.d | LjLBD12 | chr2:23416613–23417729 | 220 | 24392.4 | 5.59 |
| chr2.CM0081.1790.r2.a | LjLBD13 | chr2:16470717–16471499 | 260 | 29606.1 | 7.97 |
| chr2.CM0263.240.r2.m | LjLBD14 | chr2:15129929–15130414 | 161 | 17933.5 | 7.83 |
| chr2.LjB15M17.20.r2.m | LjLBD15 | chr2:40232463–40234079 | 349 | 39956.8 | 7.06 |
| chr2.LjT16G06.10.r2.d | LjLBD16 | chr2:28013987–28014913 | 308 | 33953.9 | 7.22 |
| chr3.CM0049.340.r2.d | LjLBD17 | chr3:35912062–35912889 | 275 | 31115.4 | 6.63 |
| chr3.CM0135.10.r2.m | LjLBD18 | chr3:45395048–45395762 | 136 | 15523.8 | 8.15 |
| chr3.CM0160.1010.r2.m | LjLBD19 | chr3:37974603–37975202 | 199 | 23215.5 | 7.83 |
| chr3.CM0176.110.r2.m | LjLBD20 | chr3:3033466–3038666 | 182 | 20201.9 | 8.38 |
| chr3.CM0246.630.r2.m | LjLBD21 | chr3:30927790–30929651 | 203 | 22258.2 | 5.21 |
| chr3.CM0649.90.r2.d | LjLBD22 | chr3:41206303–41206659 | 119 | 13314.6 | 8.69 |
| chr3.CM1488.110.r2.d | LjLBD23 | chr3:1617981–1618478 | 165 | 18510.2 | 8.23 |
| chr4.CM0128.420.r2.m | LjLBD24 | chr4:12578313–12579139 | 231 | 25041.4 | 8.42 |
| chr4.CM0229.130.r2.m | LjLBD25 | chr4:11249399–11252380 | 174 | 19080.7 | 7.84 |
| chr4.CM0432.3340.r2.d | LjLBD26 | chr4:8660066–8660599 | 177 | 19674 | 5.35 |
| chr5.CM0200.2670.r2.d | LjLBD27 | chr5:33419174–33420445 | 199 | 21285 | 8 |
| chr5.CM0494.160.r2.d | LjLBD28 | chr5:19260103–19260417 | 104 | 11165.8 | 8.5 |
| chr5.CM1667.70.r2.a | LjLBD29 | chr5:29053657–29055208 | 223 | 24304.9 | 7.34 |
| chr5.LjT39A22.130.r2.a | LjLBD30 | chr5:31524745–31525388 | 130 | 14961 | 8.03 |
| chr6.CM0738.260.r2.d | LjLBD31 | chr6:5288434–5290516 | 167 | 18616.1 | 5.14 |
| chr6.LjB02K20.100.r2.m | LjLBD32 | chr6:5570935–5571474 | 179 | 20164 | 6.77 |
| LjSGA_002536 | LjLBD33 | Scaffold:2417–6865 | 231 | 24409.8 | 7.73 |
| LjSGA_010274 | LjLBD34 | Scaffold:3488–3787 | 99 | 11252.3 | 10.12 |
| LjSGA_023437 | LjLBD35 | Scaffold:60–1891 | 251 | 27916.9 | 8.28 |
| LjSGA_054888 | LjLBD36 | Scaffold:853–1140 | 95 | 10354.9 | 8.49 |
| LjSGA_076325 | LjLBD37 | Scaffold:357–1193 | 278 | 30656 | 4.4 |
| LjSGA_080276 | LjLBD38 | Scaffold:564–1165 | 199 | 22252.7 | 8.71 |
Among the 38 LjLBD genes, most members (84.2%) were successfully located to the six L. japonicus chromosomes, while the remaining members, LjLBD33-38, were localized to the six scaffolds that were not assembled into chromosomes (S1 Fig). It is obvious that the number of LjLBD genes on each chromosome was uneven. For instance, ten LjLBD members were detected on chromosome 1 followed by seven members in chromosome 3 and six in chromosome 2, whereas the fewest were found on chromosome 6 (only 2 members). Within chromosomes 4 and 5, there were only three and four LjLBD genes, respectively. In Medicago truncatula, the LBD genes were distributed unevenly among 8 chromosomes, and chromosome 2 contained the fewest LBD members (only 3 genes, S1 Fig).
Sequence alignment and phylogenetic analyses
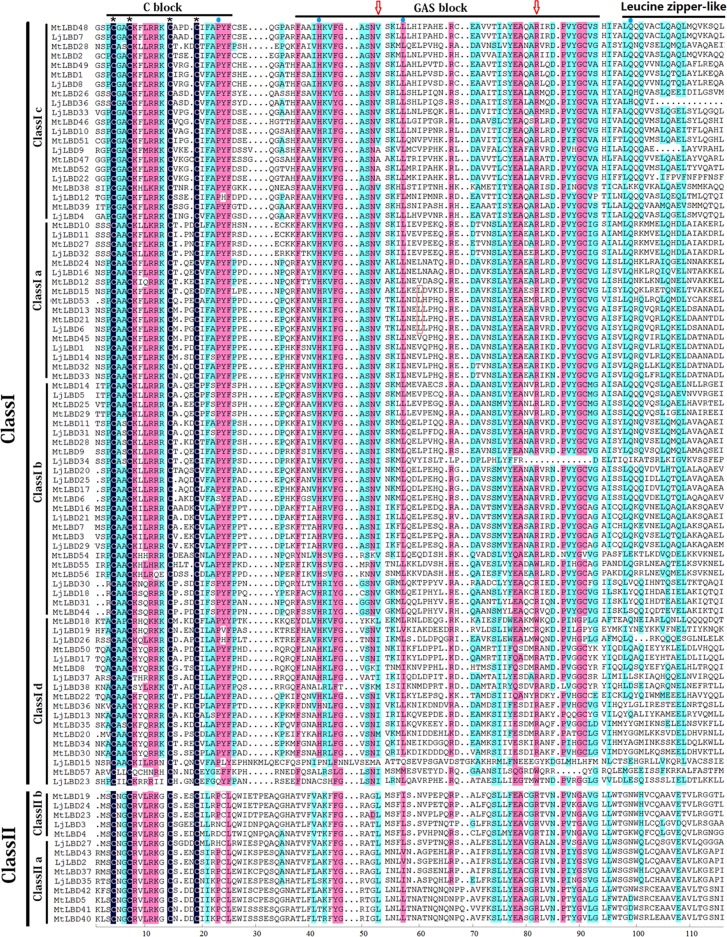
To identify conserved amino acid residues of LBD proteins and classify LBD members into the two classes as previously defined, we performed a sequence alignment using all of the LjLBD and MtLBD proteins. As a result, we found that all LBD proteins including 38 LjLBD and 57 MtLBD had a completely conserved CX2CX6CX3C motif (a C block, see Fig 1). It should be noted that the GAS block between the C block and the leucine zipper-like motif (LX6LX3LX6L) was also highly conserved in the LOB domain region (Fig 1). Based on the presence or absence of the leucine zipper-like motif, we further identified 33 and 48 class I LBD members, 5 and 9 class II members in L. japonicus and M. truncatula, respectively. In addition, recent study evidenced that a valine (V) residue and a leucine (L) in the GAS block, and a glutamine (Q) residue in the leucine-zipper-like motif is required for motor organ specification in pea [5]. These amino acid resides were also highly conserved in L. japonicus and M. truncatula (Fig 1). Also, an arginine (R) in GAS block region, which is very conserved in L. japonicus and M. truncatula (Fig 1), mutated into cysteine (C) in L. japonicus can lead to the defect in motor organ [5].
Fig 1. Amino acid sequence alignments of LOB domain region from Lotus japonicus and Medicago truncatula.
The N-terminal LOB domain includes cysteine C block, GAS block and leucine-zipper-like regions is displayed. The valine (V) and leucine (L) residues required for motor organ specification in pea were denoted by red arrow and red frame. An arginine (R) in the GAS block required for motor organ specification was denoted by red arrow.
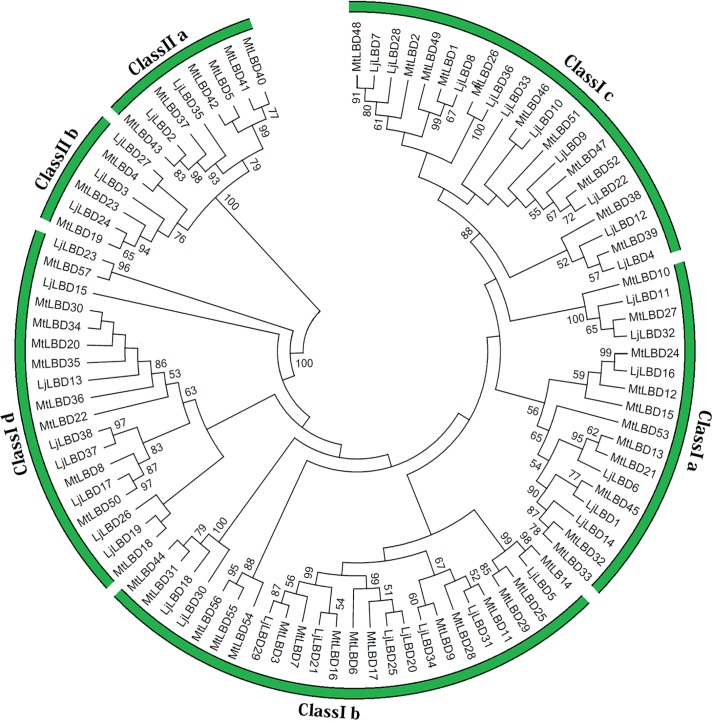
According to the protein sequence alignments, we investigated the evolutionary relationships among LBD members from L. japonicus and M. truncatula. An un-rooted phylogenetic tree was constructed using the neighbor-joining method with 10,000 bootstrap replicates. Two distinct clusters were formed corresponding to the two classes in the LBD gene family with well supported bootstrap values as shown in Fig 2, consistent with the classification described above. Among class I, members could be further grouped into four sub-classes (Ia-Id), and class II members were divided into two sub-classes (IIa and IIb). Although three members (LjLBD15, 23 and MtLBD57) were not clustered into any subclass, we classified them into class Id subclass due to the closest genetic distances. As shown in Fig 2, two known motor organ determined genes (MtELP1, also called MtLBD13 and LjSLP, also called LjLBD6) belonging to the class Ia. In addition, we found three pairs of genes duplication in M. truncatula genome including MtLBD7 and MtLBD16, MtLBD10 and MtLBD27, MtLBD19 and MtLBD23 [22]. The tree topology of LjLBD and MtLBD proteins was quite similar to the tree reported in Arabidopsis and rice [23]. Moreover, counterparts between L. japonicus and M. truncatula were clustered into all major clades and subclades, suggesting the functional conservation and similarity of LBD proteins in these clusters.
Fig 2. The phylogenetic tree of LBD proteins form Lotus japonicus and Medicago truncatula.
The amino acid sequences of the LBD proteins were aligned with Clustal X, and the phylogenetic tree was constructed using the neighbor-joining method of MEGA 5.0 software.
To detect the evolutionary relationships in the LBD family between L. japonicus and Arabidopsis, another un-rooted phylogenetic tree was constructed (S2 Fig). Similarly, LBD proteins from L. japonicus and Arabidopsis were clustered into two main clades (corresponding to class I and II), and each clade or subclade comprised members of both species, suggesting that LjLBD transcription factors were homologous with those in Arabidopsis. According to previously functional researches in Arabidopsis [23], five members from class I including AtLBD3 (ASL9), AtLBD12 (ASL5), AtLBD6 (ASL2), AtLBD36 (ASL1), AtLOB1 (ASL4) are involved in lateral organ (leaf and flower) development. Three genes AtLBD16 (ASL18), AtLBD18 (ASL20) and AtLBD29 (ASL16) participate in the auxin signal transduction cascade that leads to the formation of lateral roots in Arabidopsis. As for the characterized class II LBD genes, three genes AtLBD37 (ASL39), AtLBD38 (ASL40), AtLBD39 (ASL41) are involved in metabolism, acting as repressors of anthocyanin synthesis and N availability signals in the plant. AtLBD37 (ASL40), another class II LBD gene, is reported to be downregulated by gibberellin and upregulated by DELLA proteins [23]. The phylogenetic analysis indicated that the homologs in L. japonicus may have a similar function as described above.
Functional divergence for classes and subclasses
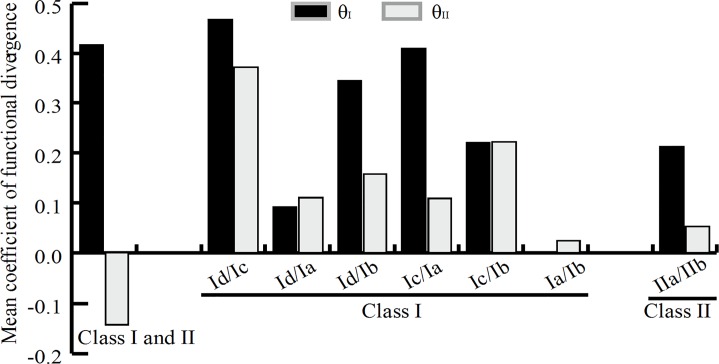
To examine whether the functional divergence between classes or subclasses was underwent in the conserved LOB domain region, we calculated the coefficients of Type-I (θI) and Type-II (θII) functional divergence as described in materials and methods. As shown in Fig 3, we found that the coefficient of Type-I functional divergence (θI = 0.42) between class I and class II was significant greater than 0, but the Type-II coefficient θII was negative, suggesting that significant type I functional divergence was underwent between these two classes. Moreover, we calculated the posterior probability (Qk) value to detect the potential amino acid residues that occurred significant changes between class I and II members. We considered 0.9 as the Qk cutoff in this study and found four sites including 23P, 42H, 57L and 99Q were markedly changed, in which these four amino acid residues were highly conserved within class I but highly variable within class II (see Fig 1). Within class I, the coefficients of both θI and θII were significant between pairs Id/Ic, Id/Ia, Id/Ib, Ic/Ia and Ic/Ib, suggesting that these subclasses, particularly Id/Ic, might have experienced both Type-I and Type-II functional divergence. However, the functional divergence between Ia and Ib may be caused by the Type-II functional divergence (Fig 3). Similarly, within class II, Type-I and Type-II functional divergence were detected in the pair IIa/IIb (Fig 3).
Fig 3. The functional divergence analysis between classes or subclasses.
The estimated mean coefficients of Type-I (θI) and type-II (θII) functional divergence based on the aligned LOB domain sequences from Lotus japonicus and Medicago truncatula.
Conserved motifs and gene structure analyses
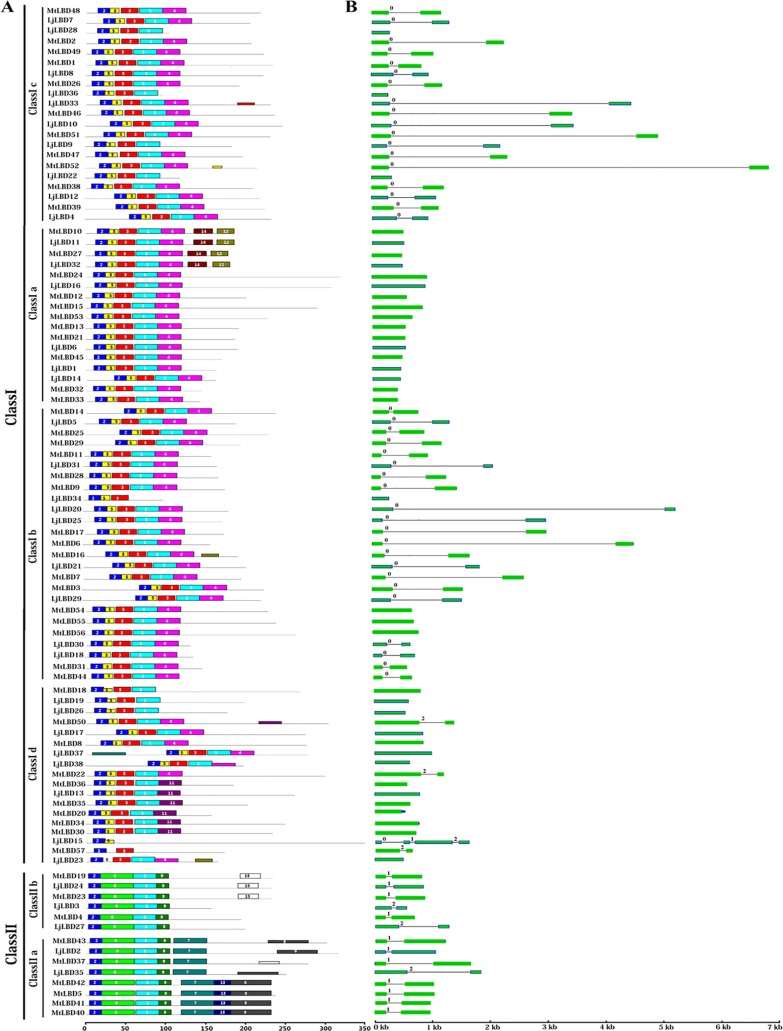
Conserved motifs and exon/intron structures could further explore the possible evolutionary relationship of related LBD proteins. To identify the potential conserved motifs, all LBD proteins from L. japonicus and M. truncatula were subjected to the MEME suite (Fig 4A). Totally, we detected 15 high-confidence motifs (designed as motif 1–15) with significant p value (p < 0.001), and the consensus sequence of motifs were listed in S2 Table. Of them, motif 2 was embedded in the LOB domain that was present in all LBD proteins. Notably, the class I and class II LBD proteins harbored distinct sets of motifs. For instance, all class I proteins possessed motifs 1–5, whereas class II proteins included motifs 1, 2, 6 and 9. Further, class II proteins were split into two groups based on differential distribution of motifs 7, 13 and 8. Most of members from class I shared common motifs. In addition, some motifs were nested in specific clades. For instance, motifs 12 and 14 were shared by four members (MtLBD10, 27 and LjLBD11, 32) in class Ia; motif 11 was uniquely found in six proteins (MtLBD20, 30, 34, 35, 36 and LjLBD13) from class Id; motifs 7, 13, and 8 were specific to class IIa proteins. These findings suggest that LBD proteins with the same motifs are likely to have similar functions in plant development.
Fig 4. Distribution of conserved motifs and gene structure analysis of LBD gene family from Lotus japonicus and Medicago truncatula.
(A) Conserved motifs analysis by MEME suite. The colorful boxes represent the different motifs 1–15. (B) The exons and introns splicing patterns. The green boxes and black lines represent the exon ans intron, respectively. The numbers indicate the intron phase. The motifs and gene sizes are indicated at the bottom of the figure.
Additionally, gene structure analysis showed that almost all of the LjLBD genes had either one or two exons, except LjLBD15 with four exons (see Fig 4B). We found that 20 genes included two exons, and 17 genes contained just one exon. Specifically, all LBD members in class Ia and most class Id members were intronless. Interestingly, while inspecting the pattern of intron insertion and splicing phase for those genes containing introns, we found that most of class I members shared splicing phase 0 (splicing occurred after the third nucleotide of the codon), whereas most of members from class II shared splicing phase 1 (splicing occurred after the first nucleotide of the codon) except three members with phase 2 (splicing occurred after the second nucleotide of the codon). Collectively, these results from conserved motifs and gene structure analyses provided additional evidence to confirm our phylogeny-based groupings.
Expression profiles of LBD genes in L. japonicus
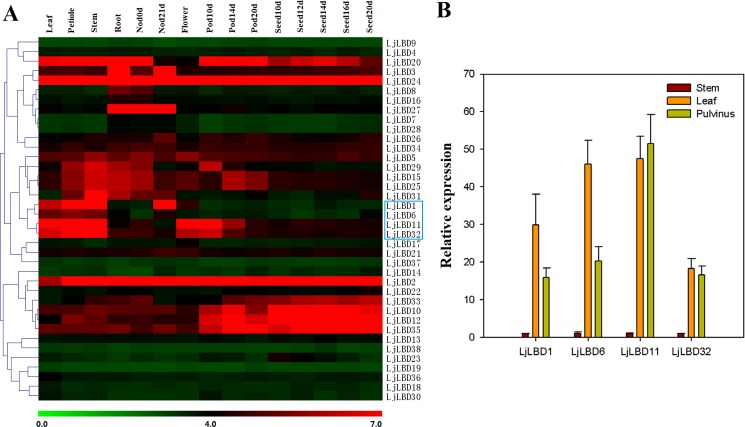
To gain insights in understanding the potential function of LBD genes in L. japonicus, we investigated their expression patterns in various tissues/organs/developmental stages (Fig 5A). Transcripts for all 38 LjLBD genes could be detected and about half of the genes displayed high expression in at least one tissue tested. With the exception of LjLBD2 and LBD24 that were highly transcribed in all tissues, the remaining LjLBD genes exhibited tissue/organ-specific expression. Two class II genes (LjLBD3 and LjLBD27) were specifically expressed in root and nodules; five genes (LjLBD10, 12, 20, 33, 35) showed high transcript abundance throughout pod and seed development. Also, we detected ten genes (LjLBD1, 2, 5, 6, 10, 11, 20, 24, 32 and 35) that were highly expressed in compound leaf and 14 members (LjLBD1, 2, 3, 5, 10, 15, 20, 25, 26, 27, 29, 31, 33, 35) that were highly expressed in root nodule. These genes represented candidate genes involved in compound leaf development or root nodule formation, respectively, in which these two traits in L. japonicus are distinct phenotypic features from Arabidopsis. In addition, we performed hierarchical clustering based on these expression data to test whether the LBD members placed in same phylogenetic clade had similar expression patterns. Unfortunately, the result showed no clade-specific expression pattern for all LjLBD members.
Fig 5. Expression profiles of LBD genes in Lotus japonicus.
(A) Heatmap showing LBD gene expression patterns in different tissues/organs/development stages. The scale at the bottom represents log2 value. (B) Some genes highly expressed in leaf (the blue box) were confirmed by quantitative RT-PCR. The expression level of stem sample was normalized to 1.
One of main interests in this study is to identify potential LBD transcription factors involved in compound leaf or motor organ development in L. japonicus. Thus, we selected a set of genes that were highly expressed in leaf but not constitutively expressed, and examined their expression levels in stem, compound leaf and pulvinus using qRT-PCR technique. Results showed that all selected four genes including LjLBD1,6,11 and 32 had high expression levels in leaf and pulvinus tissue but were quite low expression in stem (Fig 5B), suggesting the potential roles in compound leaf and pulvinus development.
The interaction of LjLBD11 and LjLBD6
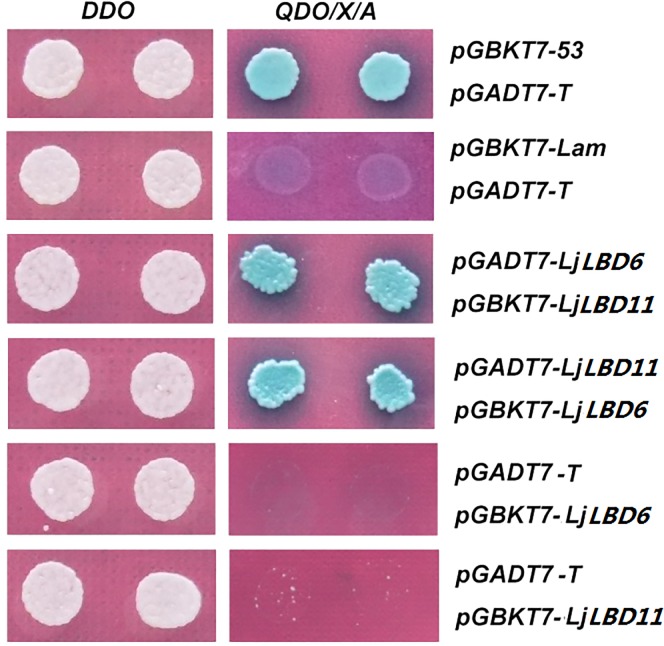
Based on the expression analysis in this study and previous study [4], LjLBD6 and LjLBD11 (responding to LjLOB3 and LjLOB1) had a strong expression at the bases of leaflet primordia and considered likely to have roles in compound leaf development. Moreover, mutation of LjLBD6 results in loss of motor organs in L. Japonicus [5] that was in good agreement with it expression in pulvinus (Fig 5). It is should be noted that LjLBD11 gene exhibited more high expression level in compound leaf and pulvinus relative to LjLBD6. However, the functional roles of LjLBD11 in compound development and motor organs formation remained unknown. In addition, previous studies uncovered that LBD proteins can usually form the Homo- or heter-dimerization through the leucine-zipper-like motif located at the C-terminus of the LBD domain [3]. Therefore, we speculated that LjLBD6 and LjLBD11 might exert their function via a protein complex. To test this, the protein interaction between LjLBD6 and LjLBD11 was performed by yeast two-hybird experiment. The yeast strain was co-transformed with the indicated combinations of LjLBD6 and LjLBD11 fused to the GAL4 activition domain (AD) and GAL4 DNA-binding domain (BD), respectively. Among the clones grown on the dropout medium lacking tryptophan, leucine, histidine and adenine, we found that clones containing LjLBD6 and LjLBD11 showed growth (see Fig 6). This result showed that LjLBD11 protein can interact with LjLBD6 protein, suggesting that the LjLBD11 may play a similar function of LjLBD6 involved in compound leaf development or specification of motor organ identity, but the detail biological function remains to be determined.
Fig 6. Interaction between LjLBD11 and LjLBD6 protein by yeast two-hybird assay.
pGBKT7-53 in combination with pGADT7-T was used as a positive control and pGBKT7-Lam with pGADT7-T was used as negative control. Yeast grew on DDO medium to select for both the bait and prey proteins (left). QDO/X/A media allow the growth of only positively interacting clones (right).
Discussion
The plant-specific LBD (Lateral Organ Boundaries domain) transcription factors play critical roles in the control of plant development, in particular in lateral organ development. The LBD gene family has been extensively studied in diverse species [1,16–18] but very little is known in legumes. Legumes are models for studying development of some specific features absent in other model plants such as Arabidopsis, including compound leaves, motor organs, and root nodules. Thus, studies on LBD genes in legumes would provide the basis for unraveling mechanisms of plant development that are currently not understood.
In this study, we identified 38 and 57 putative LBD transcription factors in L. japonicus and M. truncatula respectively, both of which are important model plants in legumes. Compared with other plants, L. japonicus and M. truncatula apparently harbored more LBD members in its genome, probably due to genome duplication that resulted in gene family expansion during evolution. All LBD proteins identified in this study contained a highly conserved CX2CX6CX3C zinc finger-like domain, implying its structural and functional necessity. Among class I members, most proteins contained an additional motif (LX6LX3LX6L) in the C-terminus, which has been demonstrated to function in protein-protein interaction [2]. Interestingly, analysis of conserved motifs outside the LOB domain found that members of the two classes (class I and II) harbored different motifs. The phylogenetic tree of LBD transcription factors from L. japonicus, M. truncatula and Arabidopsis was distinctly split into two clusters, corresponding to class I and class II members. Gene structure and motif analyses further supported phylogenetic tree analysis similar to the previous reports in Arabidopsis, rice, maize and apple [17,18,23], indicating that the LBD gene family may be highly conserved among species. In addition, functional divergence analysis showed that significant type I functional divergence was detected between classes I and II, and four amino acid residues with site-specific changes in evolutionary rates may have a main contribution.
To understand the functions of LBD genes in L. japonicus, we examined their expression profiles in different tissues/organ/developmental stages. For example, AtLBD37 (AT5G67420), AtLBD38 (AT3G49940) and AtLBD39 (AT4G37540) function in the regulation of plant basic metabolism in Arabidopsis [12], and their homologous gene LjLBD24 in L. japonicus exhibited constitutive expression, suggesting that it may also have a basic function in plant growth and development. For three genes (LjLBD7, LjLBD8 and LjLBD28) with relatively high expression in root, their homologs in Arabidopsis (AtLBD16, AtLBD18 and AtLBD29) represent a set of auxin-regulated genes that also display root-specific expression [7,8,9], strongly implying that these three genes may be involved in lateral root formation in L. japonicus. The LjLBD2 was expressed in all tissues tested in this study, while its homolog AtLBD40 is reported to respond to gibberellin [11]. Arabidopsis AtLBD30 is thought to regulate embryogenesis and floral development, and its homologs LjLBD10 and LjLBD33 were had abundant transcripts in flower and seed developmental stages [24,25], suggesting that they probably have similar functions. In addition, we noticed that both LjLBD11 and LjLBD6 were strongly expressed in compound leaf, in good agreement with previous results from RNA in situ hybridization. Previous study showed that LjLBD6 may play a conserved role in genetic determinant of motor organ identity in legumes [5]. By the Yeast two-hybrid experiment, we further found that LjLBD6 can interact with LjLBD11 at the protein level, indicating that LjLBD11 protein may have similar important function in the control of compound leaf development and determination of motor organ identity. Due to the lack of a LjLBD11 mutant in L. japonicus, the detailed function of LjLBD11 gene remains unknown, but worth further inquiry using other methods such as siRNA-mediated silencing.
In summary, the current study defined in detail the LBD gene family in legumes based on genome sequences. The gene structure, conserved motif, and phylogenetic analyses indicated that the functions of LBD genes are likely conserved among angiosperms. In addition, our results indicate that the Type I functional divergence with some site-specific shifts may be the main force for between class I and II. Importantly, The expression patterns of LjLBD11 and its interaction with LjLBD6 provided the molecular basis for the mechanisms underlying LjLBD11 gene in compound leaf development, motor organ specification in L. japonicus, even more generally in legumes.
Supporting Information
The chromosome number is indicated at the bottom of each chromosome. Genes without intron are marked with red asterisk. Segmental duplication genes in M. truncatula are linked by red dash lines.
(TIF)
The amino acid sequences of the LBD proteins were aligned with Clustal X, and the phylogenetic tree was constructed using the neighbor-joining method of MEGA 5.0 software. The red clade represents the Class II members. The LBD proteins in bracket meant that they have been investigated in other studies.
(TIF)
(DOCX)
(DOCX)
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31500989).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
National Natural Science Foundation of China (31500989). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Shuai B, Reynaga-Peña CG, Springer PS. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002; 129(2): 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsumura Y, Iwakawa H, Machida Y, Machida C. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J. 2009; 58(3): 525–537. 10.1111/j.1365-313X.2009.03797.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Husbands A, Bell EM, Shuai B, Smith HM, Springer PS. LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 2007; 35(19): 6663–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo JH, Weng L, Luo D. Isolation and expression pattern of LATERAL ORGAN BOUNDARIES-like genes in Lotus japonicas. J Plant Physiol Mol Biol. 2006; 32(2): 202–208. [PubMed] [Google Scholar]
- 5.Chen J, Moreau C, Kawaquchi M, Hofer J, Ellis N, Chen R. Conserved genetic determinant of motor organ identity in Medicago truncatula and related legumes. Proc Natl Acad Sci USA. 2012; 109(29): 11723–11728. 10.1073/pnas.1204566109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Semiarti E, Ueno Y, Tsukaya H, Iwakawa H, Machida C, Machida Y. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development. 2001; 128(10): 1771–1783. [DOI] [PubMed] [Google Scholar]
- 7.Lee HW, Kim NY, Lee DJ, Kim J. LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 2009; 151(3): 1377–1389. 10.1104/pp.109.143685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005; 17(2): 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007; 19(1): 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naito T, Yamashino T, Kiba T, Koizumi N, Kojima M, Sakakibara H et al. A link between cytokinin and ASL9 (ASYMMETRIC LEAVES 2 LIKE 9) that belongs to the AS2/LOB (LATERAL ORGAN BOUNDARIES) family genes in Arabidopsis thaliana. Biosci Biotechnol Biochem. 2007; 71(5): 1269–1278. [DOI] [PubMed] [Google Scholar]
- 11.Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, et al. Global analysis of DELLA direct targets in early gibberellins signaling in Arabidopsis. Plant Cell. 2007; 19(10): 3037–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell. 2009; 21(11): 3567–3584. 10.1105/tpc.109.067041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bortiri E, Chuck G, Vollbrecht E, Rocheford T, Martienssen R, Hake S. ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell. 2006; 18(3): 574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu C, Tai H, Saleem M, Ludwig Y, Majer C, Berendzen KW et al. Cooperative action of the paralogous maize lateral organ boundaries (LOB) domain proteins RTCS and RTCL in shoot-borne root formation. New Phytol. 2015; 207(4): 1123–1133. 10.1111/nph.13420 [DOI] [PubMed] [Google Scholar]
- 15.Liu H, Wang S, Yu X, Yu J, He X, Zhang S, et al. ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 2005; 43(1); 47–56. [DOI] [PubMed] [Google Scholar]
- 16.Yang Y, Yu X, Wu P. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol Phylogenet Evol. 2006; 39(1): 248–262. [DOI] [PubMed] [Google Scholar]
- 17.Zhang YM, Zhang SZ, Zheng CC. Genomewide analysis of LATERAL ORGAN BOUNDARIES Domain gene family in Zea mays. J Genet. 2014; 93(1): 79–91. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Zhang S, Su L, Liu X, Hao Y. A genome-wide analysis of the LBD (LATERAL ORGAN BOUNDARIES domain) gene family in Malus domestica with a functional characterization of MdLBD11. PLoS One. 2013; 8(2): e57044 10.1371/journal.pone.0057044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28(10): 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu X, Zou Y, Su Z, Huang W, Zhou Z, Arendsee Z, et al. An update of DIVERGE software for functional divergence analysis of protein family. Mol Biol Evol. 2013; 30(7): 1713–1719. 10.1093/molbev/mst069 [DOI] [PubMed] [Google Scholar]
- 21.Verdier J, Torres-Jerez I, Wang M, Andriankaja A, Allen SN, He J, et al. Establishment of the Lotus japonicus Gene Expression Atlas (LjGEA) and its use to explore legume seed maturation. Plant J. 2013; 74(2): 351–362. 10.1111/tpj.12119 [DOI] [PubMed] [Google Scholar]
- 22.Young ND, Debellé F, Oldroyd GE, Geurts R, Cannon SB, Udvardi MK, et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011; 480(7378): 520–524. 10.1038/nature10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Majer C, Hochholdinger F. Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci. 2011; 16(1): 47–52. 10.1016/j.tplants.2010.09.009 [DOI] [PubMed] [Google Scholar]
- 24.Borghi L, Bureau M, Simon R. Arabidopsis JAGGED LATERAL ORGANS is expressed in boundaries and coordinates KNOX and PIN activity. Plant Cell. 2007; 19(6): 1795–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soyano T, Thitamadee S, Machida Y, Chua NH. ASYMMETRIC LEAVES2–LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell. 2008; 20(12): 3359–3373. 10.1105/tpc.108.061796 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The chromosome number is indicated at the bottom of each chromosome. Genes without intron are marked with red asterisk. Segmental duplication genes in M. truncatula are linked by red dash lines.
(TIF)
The amino acid sequences of the LBD proteins were aligned with Clustal X, and the phylogenetic tree was constructed using the neighbor-joining method of MEGA 5.0 software. The red clade represents the Class II members. The LBD proteins in bracket meant that they have been investigated in other studies.
(TIF)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.