Abstract
The structure and folding of a protein in solution depends on noncovalent interactions within the protein and those with surrounding ions and molecules. Decoupling these interactions in solution is challenging, which has hindered the development of accurate physics-based models for structure prediction. Investigations of proteins in the gas phase can be used to selectively decouple factors affecting the structures of proteins. Here, we use Cation to Anion Proton Transfer Reactions (CAPTR) to reduce the charge states of denatured ubiquitin ions in the gas phase, and ion mobility to probe their structures. In CAPTR, a precursor charge state is selected (P) and reacted with monoanions to generate charge-reduced product ions (C). Following each CAPTR event, denatured ubiquitin ions (13+ to 6+) yield products that rapidly isomerize to structures that have smaller collision cross sections (Ω). The Ω of CAPTR product ions depend strongly on C and very weakly on P. Pre- and post-CAPTR activation was then used to probe the potential-energy surfaces of the precursor and product ions, respectively. Post-CAPTR activation showed that ions of different P fold differently and populate different regions of the potential-energy surface of that ion. Finally, pre-CAPTR activation showed that the structures of protein ions can be indirectly investigated using ion mobility of their CAPTR product ions, even for subtle structural differences that are not apparent from ion mobility characterization of the activated precursor ions. More generally, these results show that CAPTR strongly complements existing techniques for characterizing the structures and dynamics of biological molecules in the gas phase.
Graphical abstract
Introduction
Despite progress in understanding protein folding1,2 and protein structure prediction,3,4 universal methods for predicting the tertiary and quaternary structures of proteins on the basis of their sequence remain elusive, partially due to limited experimental data.1,5–7 The folding of proteins depends on covalent interactions and noncovalent interactions within the protein and with surrounding ions and molecules. Although a great deal of progress has been made,8 how these contributions work in concert to achieve native, biologically active proteins remains difficult to predict using physics-based models.1 Decoupling these contributions is particularly challenging in the condensed phase, where all interactions contribute simultaneously.
Mass spectrometry (MS) and associated methods have emerged as robust tools for studying proteins and noncovalent complexes in the gas phase, particularly due to their ability to selectively characterize individual components of mixtures.9–11 Electrospray ionization of proteins from native conditions, e.g., pH = 7.0 and physiological ionic strengths, enables measurements of “native-like” gas-phase ions, which can retain ligands, stoichiometries, and tertiary structures from solution.9–11 Coupling mass spectrometry to ion/neutral proton transfer reactions,12–15 hydrogen/deuterium exchange reactions,14,16–20 electron-mediated dissociation techniques,21–23 and ion mobility (IM),23–29 enables the characterization of gas-phase structures, which can be compared to their solution-phase counterparts.9,29–33 For example, IM-MS measurements can be used to determine the collision cross section (Ω) of an ion with a buffer gas molecule,34 which can be compared with Ω values calculated for atomic and coarse-grained models and structures.35–38
Biomolecules can adopt multiple structures that are kinetically stable in the gas phase. For example, native-like ions typically exhibit a single feature in their IM arrival-time distributions at low energies39,40 and activation of these ions can result in the formation of multiple structures that are kinetically stable and appear at different drift times.41–44 IM-MS of protein ions generated from denaturing solutions, e.g., low pH and a large fraction of organic solvent, frequently exhibit features for multiple coexisting conformers.27,31,45 For example, IM-MS of 3+ bradykinin from different solutions shows evidence for a total of ten conformers that are all kinetically stable on the millisecond time scale of the separation.46 To investigate the relationship between these conformers, Clemmer and coworkers pioneered IM-IM-MS47 and IM-IM-IM-MS48 approaches to characterize the potential-energy surfaces of peptide ions in the gas phase. For example, six conformers of 3+ bradykinin were each mobility selected, activated, and reanalyzed using a second IM separation.49,50 At high energies, the products of these experiments all exhibited the same three populations in approximately identical ratios.49,50 They propose that these ions formed a quasi-equilibrium of structures in which the relative populations of those structures depended on their free energies after thermalization. These tandem IM experiments have established the ground work for quantitative thermodynamic measurements of potential-energy surfaces in the gas phase.50
The relationship between the structure, charge state, and original solution of protein ions remains challenging to decouple. For example, native-like ions of prion51 and intrinsically disordered52 proteins as well as proteins ions from denaturing solutions31,53,54 can exhibit a strong correlation between charge state and Ω. However, charge state39–41,55 and polarity40 can have a comparatively small effect on the Ω of native-like ions of large, globular proteins and protein complexes. One approach to probe the relationship between structure and charge state is to manipulate the charge states of protein ions in the gas phase. For example, in ion/neutral proton transfer reactions a multiply charged precursor cation is reacted with a neutral molecule, A, which has a higher gas-phase basicity:13,56
| (Reaction 1) |
Foundational results from ion/neutral proton transfer reactions in tandem with IM-MS have shown that charge reduction of mixtures of ions that have high-charge states and unfolded structures can yield product ions that have lower charge states and partially folded structures.57,58
An alternative approach to manipulate the charge states of protein ions in the gas phase is to use ion/ion proton transfer reactions, in which multiply charged protein cations react with monoanions to yield charge-reduced protein cations:
| (Reaction 2) |
Ion/ion reactions benefit from more favorable kinetics and thermodynamics relative to ion/neutral proton transfer reactions.59–62 Recently, we reported an approach for these reactions that we refer to as cation to anion proton transfer reactions (CAPTR).62 CAPTR is most similar to ion/ion proton transfer reactions pioneered by McLuckey and coworkers.60,61 However, those reactions are typically performed under pseudo first-order kinetics (effectively constant anion abundance),60,61,63 whereas the abundance of anions in CAPTR depletes during individual experiments and as a result a wide range of product ion charge states (from different numbers of sequential proton transfer events) can be formed during a single experiment.62 It differs from analogous experiments using electron transfer (“electron transfer no dissociation”)22 in that charge reduction is caused by proton transfer rather than electron transfer, which can also cause bond cleavage.64,65
Here, CAPTR of m/z-selected precursor ions are used to monitor the stepwise changes in conformation as the charge state of a protein ion is reduced. We perform CAPTR within the vacuum system of a mass spectrometer in order to reduce an individual charge state of denatured ubiquitin, IM to characterize the structural effects of removing charge, and collisional activation to probe the structures and stabilities of CAPTR precursor and product ions. IM of CAPTR product ions show that there is a strong relationship between their charge state and Ω. Energy-dependent experiments suggest that 6+ ubiquitin can adopt at least two sets of structures that are unable to interconvert under the conditions of these experiments.
Experimental Methods
Bovine ubiquitin was purchased from Sigma Aldrich and dissolved into ultrapure (18.2 MΩ) water to a concentration of ~100 μM without additional purification. This solution was then diluted with methanol and water, which were acidified to a pH of 2 using trifluoroacetic acid. The final solution contained 10 μM ubiquitin in 70%:30% water:methanol by volume. Cations were generated using nanoelectrospray ionization from borosilicate capillaries with inner diameters of 0.78 mm that were pulled to ~1 to 3 μm on one end using a Sutter Instruments P-97 micropipette puller (Novato, CA). Electrical contact with the solution was achieved by inserting a platinum wire into the wide end of the capillary. The source was maintained at 120 °C. Note that under these conditions there is some heat transfer to the sample capillary, which is discussed with Figure S1.
Experiments were performed using a Waters Synapt G2 HDMS modified with a glow-discharge ionization source66 and a radio-frequency confining drift cell (Figure 1A).67 This instrument is equipped for ion/ion reactions in the trap cell, which contains 0.08 mBar helium and is positioned before the mobility cell and the time-of-flight mass analyzer. Voltages were selected to minimize activation and maximize transmission. Monoanions for CAPTR, the fragments of perfluoro-1,3-dimethylcyclohexane (PDCH, Sigma Aldrich) at m/z 381 ([PDCH-F]−),68 were generated via glow discharge for 100 ms, quadrupole selected, and accumulated in the trap cell.62 The instrument was then switched into positive mode for 2 to 3 s, during which a single charge state of ubiquitin was quadrupole selected and trapped with [PDCH-F]− to initiate CAPTR. The CAPTR products and residual precursors were then pulsed into the RF-confining drift cell for 200 μs every 13.8 or 22 ms, depending on the maximum m/z measured. For selected experiments, ions were collisionally activated by increasing their kinetic energy (Figure 1B) prior to injection into either the trap cell (pre-CAPTR activation) or the mobility cell (post-CAPTR activation). Spontaneous charge loss following IM separation is minor relative to the other charge reduction processes in these experiments, as discussed in the Supporting Information.
Figure 1.
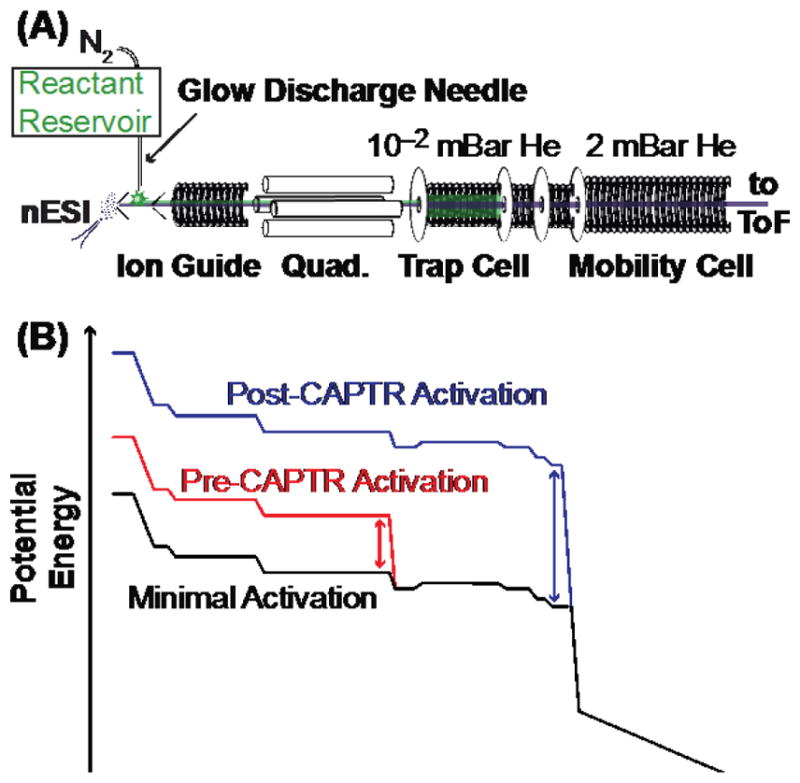
(A) Diagram of the modified Waters Synapt G2 HDMS used in these experiments. Anions (green) are generated by glow-discharge ionization66 and accumulated in the trap cell. Cations (purple) are generated by nanoelectrospray ionization (nESI) and are trapped with anions for CAPTR. Residual precursor and CAPTR product ions are separated using IM in a radio-frequency confining drift cell67 prior to mass analysis. (B) Representative potential-energy diagrams for cation transmission during minimal activation, pre-CAPTR activation, and post-CAPTR activation experiments. The double headed arrows indicate the DC bias adjusted relative to the minimal activation conditions.
IM arrival-time distributions were measured using a radio-frequency confining drift cell,67 a field of 5 V·cm−1, and 2.0 mBar helium. Ω values were determined from those distributions using a procedure discussed in the Supporting Information. Briefly, field-dependent measurements were used to determine the m/z-independent and m/z-dependent69 transport times of ions from the exit of the mobility cell to the time-of-flight mass analyzer. The experimental drift time of CAPTR ions obtained from a measurement at a single field strength were corrected for these transport times to determine their mobility (K), which was converted to Ω using the Mason-Schamp equation:34
| (Equation 1) |
where z is the charge state, e is the elementary charge, N is the number density of the drift gas, μ is the reduced mass of the ion and drift gas (helium), kB is the Boltzmann constant, and T is the temperature of the drift gas, which was 301 K for these experiments.
Results & Discussion
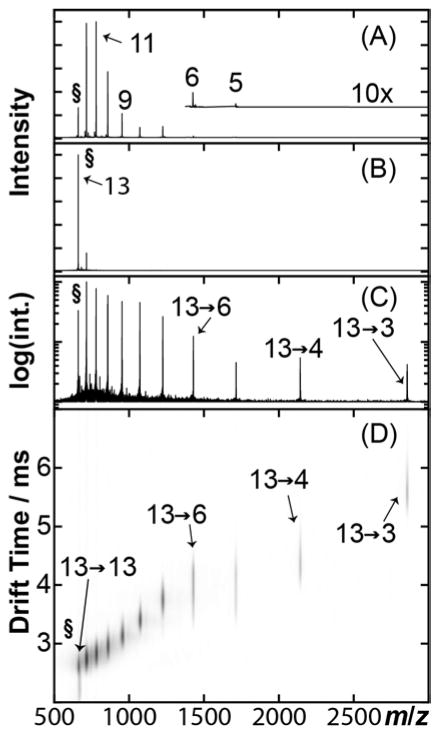
Recently, we reported experiments using CAPTR to manipulate the charge states of protein complex ions.62 Here, we couple CAPTR to reduce the charge states of protein ions generated from denaturing solutions and IM to monitor to how changes in z of protein ions affect their Ω. Figure 2A shows a nanoelectrospray ionization mass spectrum of ubiquitin from a denaturing solution of 70%:30% water:methanol at pH 2. 13+ to 5+ cations were observed, but the abundance of the 5+ cation was too weak for further analysis (Figure 2A, inset). Precursor ions were m/z selected (Figure 2B) and then subjected to CAPTR (Reaction 2) in the trap cell (Figure 2C). In this implementation, CAPTR spectra exhibit peaks for the charge-reduced product ions and a small population of unreacted precursor ions.62 The product ions and any unreacted precursor ions were then characterized using IM-MS (Figure 2D). For the remainder of the discussion, precursor ions are defined as quadrupole-selected ions that have not undergone CAPTR, and will be noted as either “P+” or “P→P”. The product ions of CAPTR are noted as “P→C”, where P is the charge state of the precursor ion and C is the charge state of the product ion.
Figure 2.
CAPTR experimental workflow. (A) Mass spectrum of ubiquitin ions generated using nanoelectrospray ionization from a denaturing solution. (B) 13+ ubiquitin (§) is m/z selected using a quadrupole mass filter. (C) Precursor ions are subjected to CAPTR; the intensity axis is plotted on a logarithmic scale to aid in visualizing the low abundance product ions. (D) CAPTR mass spectrum plotted versus the corresponding ion mobility drift times. Intensities were scaled using a nested logarithm function (log10(log10(intensity))) to aid in the visualization of low-intensity product ions.
CAPTR of Denatured Ubiquitin
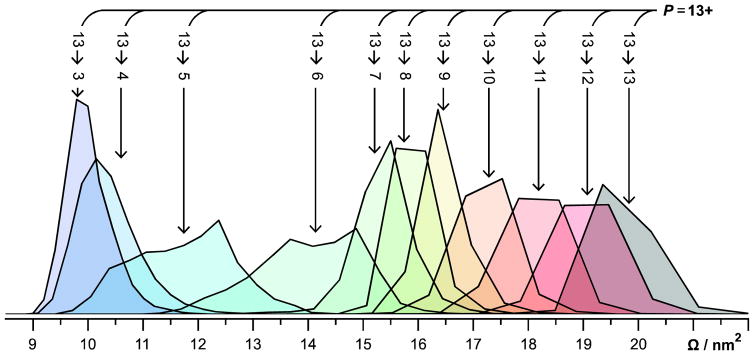
Figure 3 shows the Ω distributions for the 13→C ions. All of these ions, except 13→6 and 13→5, exhibit monomodal Ω distributions. The Ω distribution for the 13→6 ions exhibits three features, whereas that for the 13→5 ions exhibits two features. Figure S2 shows the Ω distributions for selected P→7 and P→6 ions, which exhibit monomodal and trimodal Ω distributions, respectively. The Ω distributions of the P→7 ions were fit using one Gaussian distribution and those for the P→6 ions were fit using three Gaussian distributions. Interestingly, all P→6 ions exhibit features I, II, and III, but the relative intensity of those features depend on P. Note that each Gaussian function used likely represents a family of conformers, rather than a single structure.70
Figure 3.
Normalized Ω distributions of all 13→C ions. All ions except 13→6 and 13→5 exhibit monomodal Ω distributions. 13→6 and 13→5 exhibit trimodal and bimodal Ω distributions, respectively. Ω distributions were determined using drift times measured in a radio-frequency confining drift cell67 and the Mason-Schamp equation (Equation 1), as discussed in the Supporting Information.
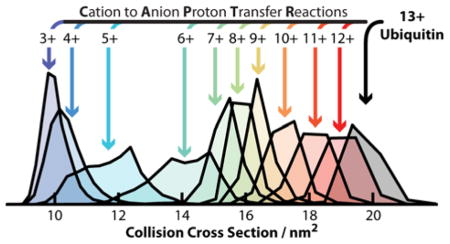
Figure 4 shows the centroids of the Gaussians fit to the features in the Ω distributions for isolated 13+ to 6+ ubiquitin cations and the corresponding CAPTR products. For each precursor charge state selected, the resulting CAPTR products exhibit smaller Ω values. This result is consistent with the product ions isomerizing to more compact conformations after charge reduction. The Ω of each feature observed for the product ions of a given charge state (C) are remarkably similar. The average root-mean-square deviation (RMSD) of the Ω for each feature is 0.04 nm2 and the greatest RMSD is 0.15 nm2 (found for feature I of the P→6 cations). These experiments show that the Ω of CAPTR product ions depend weakly on precursor charge state, regardless of whether the ion was produced via electrospray ionization or CAPTR.
Figure 4.
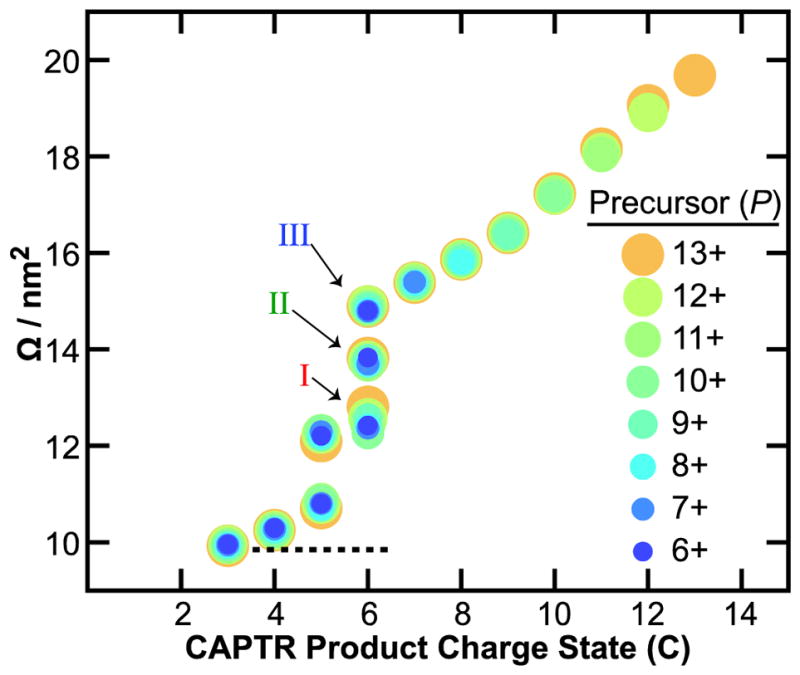
Ω of precursor (P) and CAPTR product ions (P→C) of ubiquitin. The lowest charge state product detected for each precursor ion was 3+. Precursor charge states are represented by differently colored circles, which were selected to facilitate visualization of the data. Average Ω of 4+ to 6+ ubiquitin from a native-like solution71 is shown with a dotted line for comparison.
The Ω values of native-like ubiquitin cations generated from buffered solutions of aqueous ammonium acetate are 9.72, 9.83, and 10.0 nm2 for the 4+, 5+, and 6+ cations, respectively.71 The average of those three values is 9.85 nm2 and is also shown on Figure 4.71 Features I, II, and III of the P→6 cations in these experiments range from 12.5 to 14.6 nm2 and are significantly larger than the 6+ native-like ions. In contrast, the P→3 cations in these experiments all have Ω values near 9.9 nm2, which are indistinguishable from the Ω of the native-like cations given the uncertainties in the mobility experiments.39,67 Although the native-like ions and the low-z CAPTR product ions were formed through very different processes, these results suggest that they have similar size, and may share some structural motifs. At a minimum, these results suggest that the densities of the low-z CAPTR product ions converge with those of the corresponding native-like ions.
These results demonstrate that the CAPTR products of ubiquitin adopt more compact structures with decreasing z. This finding is consistent with results from ion/neutral proton transfer reactions in which all ions generated from electrospray were reacted simultaneously with neutrals,57,58 i.e., the charge-reduced product ions originated from precursor ions that had a range of charge states. CAPTR of m/z-selected precursor ions in this study enabled the Ω to be monitored for up to ten consecutive proton transfer reactions, i.e., the products from different precursors are directly comparable to each other. Furthermore, the rates of ion/ion proton transfer reactions are expected to depend primarily on the charge state of the protein cation,59–62 whereas those for ion/neutral proton transfer reactions also depend on the structure of the protein cation.12,14,15 Previously, Badman and coworkers reported IM-MS analysis for ion/ion proton transfer reactions of two adjacent charge states each of cytochrome c72 and ubiquitin.73 Similar to the results reported here, Badman and coworkers observed that the Ω for the charge-reduced products of a mixture of 7+ and 8+ ubiquitin decreased through the 3+ product ion, and the Ω of the product ions were independent of their precursor charge state.72 In the present study, in which significantly more charge states were characterized, we can draw more general conclusions regarding the relationship between P, C, and Ω. Specifically, the results in Figure S2 show that the Ω distribution of the features observed for each C depend weakly on P, but that there are some differences in the relative intensities of those features. These results are consistent with the structure of CAPTR product ions depending strongly on C, or at least, that the structures formed as a function of C have similar Ω.
Note that each of these experiments, from ionization until time-of-flight mass analysis, take less than 44 ms. Furthermore, the widths of the arrival-time distributions are typically less than one millisecond, even though ions are formed during the entire accumulation period for the ion mobility experiment (13.8 or 22 ms). This suggests that these structural changes occur prior to IM and are therefore very fast, perhaps even sub-millisecond. For comparison, the folding time of ubiquitin in water has been estimated to be ~3 ms.74 This suggests that structural changes for proteins in the absence of solvent can occur on time scales similar to those in solution.
Post-CAPTR Activation
In order to probe the structures and stabilities of the CAPTR product ions in more detail, arrival-time distributions were measured as a function of the voltage used to inject the ions into the mobility cell (Figure 1B). The collisionally activated species will be indicated with an asterisk, i.e., P→C* indicates that the CAPTR product ions are activated following CAPTR and prior to IM separation. Figure S3 shows results for P→C*, where P = 13, 8, and 6. Interestingly, the arrival-time distributions for the 13→13*, 13→8*, and 8→8* ions (Figures S3A to S3C, respectively) are independent of the injection voltage over the range studied. These results are consistent with those ions adopting their lowest-energy structures over all energies, those ions not overcoming the barrier to structural isomerization in these experiments, or that any new structures formed have similar Ω. Note that the range of injection voltages used in these experiments is limited by competition with covalent fragmentation of CAPTR product ions.
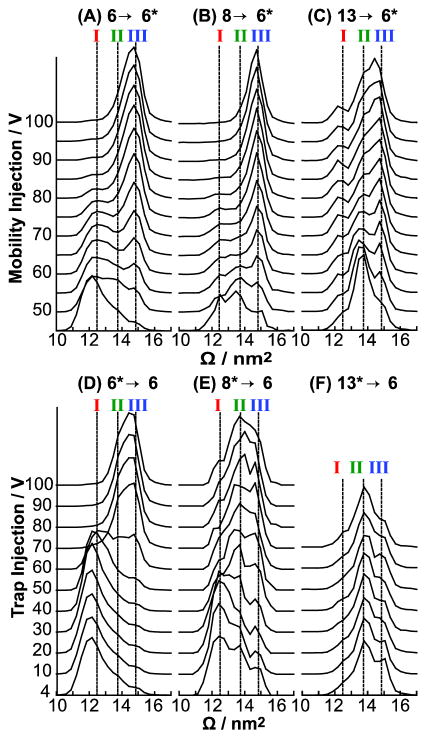
Figures 5A and 5B show the Ω distributions observed for the 6→6* and 8→6* ions, respectively, as a function of the injection voltage as they enter the mobility cell. Vertical dashed lines correspond to the average of the centroids of features I, II, and III for all P→6 ions in Figure 4. The 6→6* and 8→6* cations at low energies have intense populations corresponding to feature I. However, the 6→6* ions exhibit low-intensity peaks corresponding to features II and III, whereas the 8→6* ions exhibit intense peaks for features II and III. From 55 to 85 V the 6→6* ions convert predominantly to feature III. The 8→6* cations unfold to feature III similarly to the 6→6* cation, however it unfolds at ~10 V lower than the 6→6* ions and exhibits a slightly narrower feature at the highest energies. Therefore, in contrast to the Ω distributions for the 13→13*, 8→8*, and 13→8* ions, those for the P→6* ions depend strongly on the injection voltage.
Figure 5.
Post-CAPTR activation of (A) 6→6*, (B) 8→6*, and (C) 13→6* ubiquitin ions. Pre-CAPTR activation of (D) 6*→6, (E) 8*→6, and (F) 13*→6 ubiquitin ions. Vertical lines corresponding to the average Ω for the three features of the Ω distribution of 6+ (I to III) from Figure 4 are included for comparison. The Ω distributions obtained at the lowest energies in A to C are also shown in Figure S2. These mobility experiments used a field of 6.4 V·cm−1.
Figure 6A shows a qualitative reaction coordinate that is consistent with the results for the 6→6* ions (Figure 5A) and has three local minima corresponding to features I to III. The energies of the three features descend from I to III, consistent with features I and II converting to III with sufficient activation. For injection voltages greater than 80 V, the Ω distributions for the 6→6* ions no longer depend on energy, which is consistent with these ions establishing a quasi-equilibrium of structures50 at high energy. As previously discussed, the results for the 8→6* and 6→6* ions are similar, and therefore the qualitative reaction coordinate in Figure 6A is consistent with the results for both experiments.
Figure 6.
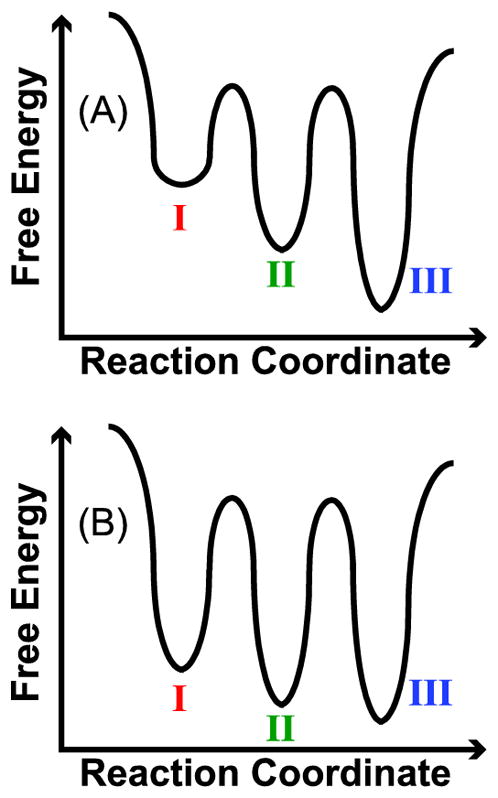
(A) Qualitative reaction coordinate of 6→6* and 8→6* ubiquitin that is consistent with the post-CAPTR activation data shown in Figures 5A and 5B. The labels for the three wells correspond to features in the Ω distributions. (B) Qualitative reaction coordinate of 13→6* ubiquitin that is consistent with the post-CAPTR activation data shown in Figure 5C. Again, the labels for the wells correspond to the features in the Ω distributions; the post-CAPTR activation experiments indicate that the structures populated in the wells in B are different than those in A.
At low injection energies, the 13→6* ions predominantly exhibit feature II, with less intense features centered at I and III (Figure 5C). With injection voltages of 45 to 70 V, feature II converts to feature III until they are roughly equal in intensity from 80 to 100 V. Interestingly, the intensity of feature I remains constant over the injection voltages studied, in contrast to that feature for the 8→6* and 6→6* ions. At high energies, the Ω distribution for the 13→6* ions (Figure 5C) are very different than those for the 6→6* and 8→6* ions (Figures 5A and 5B, respectively). These results are consistent with the 13→6* ions adopting different structures than the 6→6* and 8→6* ions. As a result, the qualitative reaction coordinate in Figure 6A alone cannot explain all the data in Figures 5A to 5C.
A qualitative reaction coordinate that is consistent with the post-CAPTR activation of the 13→6* ions is shown in Figure 6B. The qualitative reaction coordinate in Figure 6B differs from that in Figure 6A in that the local minima are more similar in energy, consistent with features I to III all being populated under quasi-equilibrium conditions (Figure 5C). Two alternative explanations are that (1) the persistence of feature I at high energies is the result of a high barrier for isomerization from that feature to the other features or (2) the 13→6* ions have a subpopulation of ions that are consistent with the qualitative reaction coordinate in Figure 6A, i.e., isomerize to feature III at high energies, and additional subpopulation that yield features I and II and are unable to interconvert to III even at very high energies.
The post-CAPTR activation results for the 6→6*, 8→6*, and 13→6* ions suggest that 6+ ubiquitin can adopt at least two sets of structures that are unable to interconvert under the conditions of these experiments, and that these experiments probe at least two different regions of the potential-energy surface of 6+ ubiquitin. The pair of qualitative reaction coordinates shown in Figure 6 are consistent with these observations. Although we do not believe that it is possible to explain all of the results for the 6→6*, 8→6*, and 13→6* ions using a single reaction coordinate with three local minima, there are other reaction coordinates that are also consistent with these results. For instance, it is possible that feature I for the 6→6* and 8→6* ions convert directly to feature III, without the formation of feature II as an intermediate.
The ions populating different regions on the potential-energy surface probed in the post-CAPTR experiments cannot interconvert and therefore have significant structural differences with barriers to isomerization that are higher than that for covalent fragmentation. One possibility is that these different regions correspond to different configurations of protonation sites. If the barriers to proton migration are sufficiently high, the protonation sites for the precursor ions may depend on the locations favored during ionization. In contrast, the protonation sites of the CAPTR product ions will also depend on the reactivity of the protonation sites for the precursor ion. Individual CAPTR events are highly exothermic,62 which will favor proton transfer through the most entropically favored channels. Therefore, the 6+ ions generated directly from electrospray and the 13→6 ions may have very different protonation sites, which may affect their structures and stabilities. These results all indicate that CAPTR product ions can have structural differences that depend on P, but that do not necessarily affect their Ω.
Pre-CAPTR Activation
To investigate the effects of the structure of the precursor ion on those of the resulting CAPTR products, ions were activated prior to CAPTR as they were injected into the trap cell (Figure 1B). Ions activated prior to CAPTR are indicated with an asterisk, i.e., P*→C. Figure S4 shows results for P*→C, where P = 13, 8, and 6. Our interpretation of these results will assume that pre-CAPTR activation occurs prior to charge reduction. This assumption is reasonable because the density of helium in the trap cell greatly exceeds that of the monoanions and the CAPTR cross section will decrease with increasing relative velocity of the cation and monoanion pair.59 Note that the range of injection voltages used in these experiments is limited by competition with covalent fragmentation.
The Ω distributions for the 6*→6 ions as a function of injection voltage are shown in Figure 5D. With increasing injection voltage, the intensity of feature I decreases and that for feature III increases. The results for the 6*→6 ions are similar to those for the 6→6* ions, although the distribution at the highest energy for the 6*→6 ions is broader than that for the 6→6* ions, suggesting that a small population of ions that yield feature II may persist. This suggests that for a given ion, high-energy injection into either the trap or mobility cells results in activation via a similar mechanism, despite differences in the efficiencies and activated ion lifetimes.
The Ω distributions of the 8*→6 ions (Figure 5E) change with increasing energy, even though the arrival-time distributions for the 8*→8 ions appear to be independent of energy (Figure S4C). The Ω distribution of the 8*→6 ions at the lowest energy studied contains features I, II and III in decreasing intensities. With activation of the 8+ precursor ion, feature I of the 8*→6 product ion becomes less abundant and features II and III become more abundant. At high energies (≥70 V), feature II is marginally more populated than feature III, which are both more intense than feature I. One explanation of these results is that the 8*→8 ions isomerize to form additional conformers with indistinguishable Ω distributions, which go on to form different structural populations of the 8*→6 ions. Thus, as the 8*→8 ions are activated and populate new conformations that yield an indistinguishable Ω distribution, those structural changes are reflected in the Ω distribution of the 8*→6 products. Therefore, CAPTR products appear to provide indirect evidence of a change in the structure of a precursor ion. Interestingly, the Ω distributions of the 8*→6 ions are indistinguishable from 70 to 100 V (Figure 5E). This result is consistent with the 8*→8 ions achieving a quasi-equilibrium of structures that go on to yield 8*→6 products with indistinguishable Ω distributions over this range of energies. Analogous experiments for the 13*→6 ions are consistent with the 13+ ubiquitin ions adopting a quasi-equilibrium of structures over all energies, and consequently, all 13*→6 ions have similar distributions of structures (Figure 5F).
Interestingly, the arrival-time distributions of the 6*→5 ion exhibit very little change over the entire voltage range (Figure S4I), even though the Ω distribution of the 6*→6 ions changes from predominantly feature I to predominantly feature III as the injection voltage is increased from 50 to 70 V (Figure 5D). This suggests that CAPTR of the 6+ precursor ions associated with features I and III both result in 5+ products that have more similar structures or at least structures that have more similar Ω.
In the preceding discussion of the 6*→6, 8*→6, and 13*→6 ions, the relative intensity of features I, II, and III for the 6+ product ions depended on both P and energy. From these experiments alone, there is not enough information to know the extent to which the qualitative reaction coordinates in Figure 6 apply to the ions formed in the P*→6 experiments, for P > 6. Future experiments using energy-dependent activation both before and after CAPTR (P*→C* experiments) may provide additional insights.
Conclusion
The experiments presented used CAPTR and IM to investigate the relationship between the Ω and charge state of a protein cation. We have shown the Ω of ubiquitin ions depend most strongly on its charge state. This conclusion is based on the observation that regardless of precursor charge state, ions of the same product ion charge state exhibit features that have similar Ω (Figure 4). This observation indicates that their structures may be determined predominantly by a delicate balance between intramolecular bonding and Coulombic repulsions. These results build upon those showing that ion/neutral charge reduction of mixtures of protein ions that have both high charge states and extended structures yield product ions with partially folded structures,57,58 by tracking this process for series of precursor ions with different charge states that were each m/z selected prior to a sequential series of CAPTR events.
The results from the pre-CAPTR and post-CAPTR activation experiments provide several new insights into the effects of charge reduction and the concomitant gas-phase folding. Despite the differences in the low-energy Ω distributions of the 6→6* and 8→6* ions, the ions isomerize to similar Ω distributions at high energies, exhibiting predominantly feature III (Figures 5A and 5B). These results are consistent with these ions forming a quasi-equilibrium of structures and the qualitative reaction coordinate in Figure 6A. In contrast, post-CAPTR activation of the 13→6* ions results in a different Ω distribution at high energy (Figure 5C), consistent with forming a different set of structures. Consequently, these data suggest that 6+ ubiquitin can adopt at least two sets of structures that are unable to interconvert under the conditions of these experiments. Therefore, these experiments probe at least two different regions of the potential-energy surface of 6+ ubiquitin.
Pre-CAPTR activation enables the exploration of the potential-energy surfaces of CAPTR precursor ions, which then is probed indirectly through Ω analysis of the corresponding CAPTR product ions. For example, the Ω distributions of 8+ ubiquitin ions do not depend on the activation energy used (Figure S4C), but the Ω distributions of 8*→6 ions do depend on the energy used to activate the 8+ precursor (Figure 5E). Together, these results show that subtle structural difference that are not apparent from IM of the precursor ions can be indirectly probed using IM of CAPTR product ions.
More generally, these results show that CAPTR complements existing techniques for characterizing the potential-energy surfaces of biological molecules in the gas phase. The final products ions in these experiments were characterized in terms of their m/z and ion mobility, but as illustrated several times in the present work, different structures can have similar ion mobilities. Future experiments will benefit greatly from complementary probes of structure, e.g., hydrogen/deuterium exchange16,17,14,18–20 and electron-mediated dissociation techniques.21,23,22 Furthermore, the energy-dependent experiments reported here used collisional activation under multicollision conditions, thus the results are qualitative. Alternative approaches in which the ions have more clearly defined internal energies and activated lifetimes may enable more quantitative analogs to the present energy-dependent experiments. Future hybrid CAPTR experiments will provide new insights into the structures and dynamics of biological molecules in the gas phase, which in turn may inform condensed-phase protein folding experiments by decoupling solvent interactions from intramolecular forces.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the American Society for Mass Spectrometry (Research Award to M. F. B.), the Amgen Foundation (scholarship to E. B. M.), the ARCS Foundation (fellowship to K. J. L.), Eli Lilly and Company (Young Investigator Award in Analytical Chemistry to M. F. B.), and the National Institute of General Medical Sciences of the National Institutes of Health under Award Number T32GM008268 (support to K. J. L.).
Footnotes
Supporting Information. The Supporting Information is available free of charge on the ACS Publications website.
Figures S1 to S4 and discussions of the effects of source temperature, spontaneous charge loss, and determination of collision cross sections.
References
- 1.Dill KA, MacCallum JL. Science. 2012;338:1042–1046. doi: 10.1126/science.1219021. [DOI] [PubMed] [Google Scholar]
- 2.Gelman H, Gruebele M. Q Rev Biophys. 2014;47:95–142. doi: 10.1017/S003358351400002X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kiss G, Çelebi-Ölçüm N, Moretti R, Baker D, Houk KN. Angew Chem. 2013;52:5700–5725. doi: 10.1002/anie.201204077. [DOI] [PubMed] [Google Scholar]
- 4.Moretti R, Fleishman SJ, Agius R, Torchala M, Bates PA, Kastritis PL, Rodrigues JPGLM, Trellet M, Bonvin AMJJ, Cui M, Rooman M, Gillis D, Dehouck Y, Moal I, Romero-Durana M, Perez-Cano L, Pallara C, Jimenez B, Fernandez-Recio J, Flores S, Pacella M, Praneeth Kilambi K, Gray JJ, Popov P, Grudinin S, Esquivel-Rodríguez J, Kihara D, Zhao N, Korkin D, Zhu X, Demerdash ONA, Mitchell JC, Kanamori E, Tsuchiya Y, Nakamura H, Lee H, Park H, Seok C, Sarmiento J, Liang S, Teraguchi S, Standley DM, Shimoyama H, Terashi G, Takeda-Shitaka M, Iwadate M, Umeyama H, Beglov D, Hall DR, Kozakov D, Vajda S, Pierce BG, Hwang H, Vreven T, Weng Z, Huang Y, Li H, Yang X, Ji X, Liu S, Xiao Y, Zacharias M, Qin S, Zhou H-X, Huang S-Y, Zou X, Velankar S, Janin J, Wodak SJ, Baker D. Proteins Struct Funct Bioinforma. 2013;81:1980–1987. [Google Scholar]
- 5.Shea JE, Brooks CLI. Annu Rev Phys Chem. 2001;52:499–535. doi: 10.1146/annurev.physchem.52.1.499. [DOI] [PubMed] [Google Scholar]
- 6.Fersht AR. Nat Rev Mol Cell Biol. 2008;9:650–654. doi: 10.1038/nrm2446. [DOI] [PubMed] [Google Scholar]
- 7.Englander SW, Mayne L. Proc Natl Acad Sci U S A. 2014;111:15873–15880. doi: 10.1073/pnas.1411798111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dill KA, Ozkan SB, Shell MS, Weikl TR. Annu Rev Biophys. 2008;37:289–316. doi: 10.1146/annurev.biophys.37.092707.153558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou M, Sandercock AM, Fraser CS, Ridlova G, Stephens E, Schenauer MR, Yokoi-Fong T, Barsky D, Leary JA, Hershey JW, Doudna JA, Robinson CV. Proc Natl Acad Sci U S A. 2008;105:18139–18144. doi: 10.1073/pnas.0801313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heck AJR. Nat Methods. 2008;5:927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- 11.Hilton GR, Benesch JLP. J R Soc Interface. 2012;9:801–816. doi: 10.1098/rsif.2011.0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cassady CJ, Wronka J, Kruppa GH, Laukien FH, Hettich R. Rapid Commun Mass Spectrom. 1994;8:394–400. doi: 10.1002/rcm.1290080511. [DOI] [PubMed] [Google Scholar]
- 13.Schnier PD, Gross DS, Williams ER. J Am Chem Soc. 1995;117:6747–6757. [Google Scholar]
- 14.Cassady CJ, Carr SR. J Mass Spectrom. 1996;31:247–254. doi: 10.1002/(SICI)1096-9888(199603)31:3<247::AID-JMS285>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 15.Valentine SJ, Counterman AE, Clemmer DE. J Am Soc Mass Spectrom. 1997;8:954–961. doi: 10.1016/S1044-0305(99)00079-3. [DOI] [PubMed] [Google Scholar]
- 16.Winger BE, Light-Wahl KJ, Rockwood AL, Smith RD. J Am Chem Soc. 1992;114:5897–5898. [Google Scholar]
- 17.Wood TD, Chorush RA, Wampler FM, Little DP, O’Connor PB, McLafferty FW. Proc Natl Acad Sci U S A. 1995;92:2451–2454. doi: 10.1073/pnas.92.7.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valentine SJ, Clemmer DE. J Am Chem Soc. 1997;119:3558–3566. [Google Scholar]
- 19.Breuker K, McLafferty FW. Proc Natl Acad Sci U S A. 2008;105:18145. doi: 10.1073/pnas.0807005105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bohrer BC, Atlasevich N, Clemmer DE. J Phys Chem B. 2011;115:4509–4515. doi: 10.1021/jp2008495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breuker K, Oh H, Horn DM, Cerda BA, McLafferty FW. J Am Chem Soc. 2002;124:6407–6420. doi: 10.1021/ja012267j. [DOI] [PubMed] [Google Scholar]
- 22.Lermyte F, Williams JP, Brown JM, Martin EM, Sobott F. J Am Soc Mass Spectrom. 2015;26:1068–1076. doi: 10.1007/s13361-015-1124-z. [DOI] [PubMed] [Google Scholar]
- 23.Harvey SR, Porrini M, Tyler RC, MacPhee CE, Volkman BF, Barran PE. Phys Chem Chem Phys. 2015;17:10538–10550. doi: 10.1039/c4cp05136j. [DOI] [PubMed] [Google Scholar]
- 24.Woenckhaus J, Mao Y, Jarrold MF. J Phys Chem B. 1997;101:847–851. [Google Scholar]
- 25.Myung S, Badman ER, Lee YJ, Clemmer DE. J Phys Chem A. 2002;106:9976–9982. [Google Scholar]
- 26.Badman ER, Myung S, Clemmer DE. J Am Soc Mass Spectrom. 2005;16:1493–1497. doi: 10.1016/j.jasms.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 27.Bohrer BC, Merenbloom SI, Koeniger SL, Hilderbrand AE, Clemmer DE. Annu Rev Anal Chem. 2008;1:293–327. doi: 10.1146/annurev.anchem.1.031207.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrison LJ, Wysocki VH. J Am Chem Soc. 2014;136:14173–14183. doi: 10.1021/ja507298e. [DOI] [PubMed] [Google Scholar]
- 29.Chen SH, Chen L, Russell DH. J Am Chem Soc. 2014;136:9499–9508. doi: 10.1021/ja5047878. [DOI] [PubMed] [Google Scholar]
- 30.Shoemaker GK, van Duijn E, Crawford SE, Uetrecht C, Baclayon M, Roos WH, Wuite GJL, Estes MK, Prasad BVV, Heck AJR. Mol Cell Proteomics. 2010;9:1742–1751. doi: 10.1074/mcp.M900620-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wyttenbach T, Bowers MT. J Phys Chem B. 2011;115:12266–12275. doi: 10.1021/jp206867a. [DOI] [PubMed] [Google Scholar]
- 32.Shi H, Pierson NA, Valentine SJ, Clemmer DE. J Phys Chem B. 2012;116:3344–3352. doi: 10.1021/jp210797x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi H, Atlasevich N, Merenbloom SI, Clemmer DE. J Am Soc Mass Spectrom. 2014;25:2000–2008. doi: 10.1007/s13361-014-0834-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mason EA, McDaniel W. Transport properties of ions in gases. Wiley; 1988. [Google Scholar]
- 35.Bernstein SL, Dupuis NF, Lazo ND, Wyttenbach T, Condron MM, Bitan G, Teplow DB, Shea JE, Ruotolo BT, Robinson CV, Bowers MT. Nat Chem. 2009;1:326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jurneczko E, Barran PE. Analyst. 2011;136:20–28. doi: 10.1039/c0an00373e. [DOI] [PubMed] [Google Scholar]
- 37.Benesch JLP, Ruotolo BT. Curr Opin Struct Biol. 2011;21:641–649. doi: 10.1016/j.sbi.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wyttenbach T, Bleiholder C, Bowers MT. Anal Chem. 2013;85:2191–2199. doi: 10.1021/ac3029008. [DOI] [PubMed] [Google Scholar]
- 39.Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT. Anal Chem. 2010;82:9557–9565. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- 40.Allen SJ, Schwartz AM, Bush MF. Anal Chem. 2013;85:12055–12061. doi: 10.1021/ac403139d. [DOI] [PubMed] [Google Scholar]
- 41.Hall Z, Politis A, Bush MF, Smith LJ, Robinson CV. J Am Chem Soc. 2012;134:3429–3438. doi: 10.1021/ja2096859. [DOI] [PubMed] [Google Scholar]
- 42.Hopper JTS, Oldham NJ. J Am Soc Mass Spectrom. 2009;20:1851–1858. doi: 10.1016/j.jasms.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 43.Ruotolo BT, Hyung SJ, Robinson PM, Giles K, Bateman RH, Robinson CV. Angew Chem. 2007;119:8147–8150. doi: 10.1002/anie.200702161. [DOI] [PubMed] [Google Scholar]
- 44.Freeke J, Bush MF, Robinson CV, Ruotolo BT. Chem Phys Lett. 2012;524:1–9. [Google Scholar]
- 45.Liu FC, Kirk SR, Bleiholder C. Analyst. 2016;141:3722–3730. doi: 10.1039/c5an02399h. [DOI] [PubMed] [Google Scholar]
- 46.Pierson NA, Chen L, Valentine SJ, Russell DH, Clemmer DE. J Am Chem Soc. 2011;133:13810–13813. doi: 10.1021/ja203895j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koeniger SL, Merenbloom SI, Valentine SJ, Jarrold MF, Udseth HR, Smith RD, Clemmer DE. Anal Chem. 2006;78:4161–4174. doi: 10.1021/ac051060w. [DOI] [PubMed] [Google Scholar]
- 48.Merenbloom SI, Koeniger SL, Valentine SJ, Plasencia MD, Clemmer DE. Anal Chem. 2006;78:2802–2809. doi: 10.1021/ac052208e. [DOI] [PubMed] [Google Scholar]
- 49.Pierson NA, Valentine SJ, Clemmer DE. J Phys Chem B. 2010;114:7777–7783. doi: 10.1021/jp102478k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pierson NA, Clemmer DE. Int J Mass Spectrom. 2015;377:646–654. doi: 10.1016/j.ijms.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hilton GR, Thalassinos K, Grabenauer M, Sanghera N, Slade SE, Wyttenbach T, Robinson PJ, Pinheiro TJT, Bowers MT, Scrivens JH. J Am Soc Mass Spectrom. 2010;21:845–854. doi: 10.1016/j.jasms.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 52.Beveridge R, Covill S, Pacholarz KJ, Kalapothakis JMD, MacPhee CE, Barran PE. Anal Chem. 2014;86:10979–10991. doi: 10.1021/ac5027435. [DOI] [PubMed] [Google Scholar]
- 53.Clemmer DE, Hudgins RR, Jarrold MF. J Am Chem Soc. 1995;117:10141–10142. [Google Scholar]
- 54.Faull PA, Korkeila KE, Kalapothakis JM, Gray A, McCullough BJ, Barran PE. Int J Mass Spectrom. 2009;283:140–148. [Google Scholar]
- 55.Hogan CJ, Ruotolo BT, Robinson CV, Fernandez de la Mora J. J Phys Chem B. 2011;115:3614–3621. doi: 10.1021/jp109172k. [DOI] [PubMed] [Google Scholar]
- 56.McLuckey SA, Goeringer DE. Anal Chem. 1995;67:2493–2497. doi: 10.1021/ac00110a026. [DOI] [PubMed] [Google Scholar]
- 57.Shelimov KB, Jarrold MF. J Am Chem Soc. 1996;118:10313–10314. [Google Scholar]
- 58.Shelimov KB, Jarrold MF. J Am Chem Soc. 1997;119:2987–2994. [Google Scholar]
- 59.McLuckey SA, Stephenson JL, Asano KG. Anal Chem. 1998;70:1198–1202. doi: 10.1021/ac9710137. [DOI] [PubMed] [Google Scholar]
- 60.McLuckey SA, Stephenson JL. Mass Spectrom Rev. 1998;17:369–407. doi: 10.1002/(SICI)1098-2787(1998)17:6<369::AID-MAS1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 61.Pitteri SJ, McLuckey SA. Mass Spectrom Rev. 2005;24:931–958. doi: 10.1002/mas.20048. [DOI] [PubMed] [Google Scholar]
- 62.Laszlo KJ, Bush MF. J Am Soc Mass Spectrom. 2015;26:2152–2161. doi: 10.1007/s13361-015-1245-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stephenson JL, Jr, Van Berkel GJ, McLuckey SA. J Am Soc Mass Spectrom. 1997;8:637–644. [Google Scholar]
- 64.Zhang H, Cui W, Wen J, Blankenship RE, Gross ML. Anal Chem. 2011;83:5598–5606. doi: 10.1021/ac200695d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lermyte F, Konijnenberg A, Williams JP, Brown JM, Valkenborg D, Sobott F. J Am Soc Mass Spectrom. 2014;25:343–350. doi: 10.1007/s13361-013-0798-3. [DOI] [PubMed] [Google Scholar]
- 66.Williams JP, Brown JM, Campuzano I, Sadler PJ. Chem Commun. 2010;46:5458–5460. doi: 10.1039/c0cc00358a. [DOI] [PubMed] [Google Scholar]
- 67.Allen S, Giles K, Golbert T, Bush M. Analyst. 2016;141:884–891. doi: 10.1039/c5an02107c. [DOI] [PubMed] [Google Scholar]
- 68.Stephenson JL, McLuckey SA. Anal Chem. 1996;68:4026–4032. doi: 10.1021/ac9605657. [DOI] [PubMed] [Google Scholar]
- 69.Ruotolo BT, Benesch JL, Sandercock AM, Hyung SJ, Robinson CV. Nat Protoc. 2008;3:1139–1152. doi: 10.1038/nprot.2008.78. [DOI] [PubMed] [Google Scholar]
- 70.Koeniger SL, Merenbloom SI, Clemmer DE. J Phys Chem B. 2006;110:7017–7021. doi: 10.1021/jp056165h. [DOI] [PubMed] [Google Scholar]
- 71.Salbo R, Bush MF, Naver H, Campuzano I, Robinson CV, Pettersson I, Jørgensen TJD, Haselmann KF. Rapid Commun Mass Spectrom. 2012;26:1181–1193. doi: 10.1002/rcm.6211. [DOI] [PubMed] [Google Scholar]
- 72.Zhao Q, Schieffer GM, Soyk MW, Anderson TJ, Houk RS, Badman ER. J Am Soc Mass Spectrom. 2010;21:1208–1217. doi: 10.1016/j.jasms.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 73.Zhao Q, Soyk MW, Schieffer GM, Fuhrer K, Gonin MM, Houk RS, Badman ER. J Am Soc Mass Spectrom. 2009;20:1549–1561. doi: 10.1016/j.jasms.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 74.Piana S, Lindorff-Larsen K, Shaw DE. Proc Natl Acad Sci U S A. 2013;110:5915–5920. doi: 10.1073/pnas.1218321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.