Abstract
More than 270 different types of papillomaviruses have been discovered in a wide array of animal species. Despite the great diversity of papillomaviruses, little is known about the evolutionary processes that drive host tropism and the emergence of oncogenic genotypes. Although host defense mechanisms have evolved to interfere with various aspects of a virus life cycle, viruses have also coevolved copious strategies to avoid host antiviral restriction. Our and other studies have shown that the cytidine deaminase APOBEC3 family members edit HPV genomes and restrict virus infectivity. Thus, we hypothesized that host restriction by APOBEC3 served as selective pressure during papillomavirus evolution. To test this hypothesis, we analyzed the relative abundance of all dinucleotide sequences in full-length genomes of 274 papillomavirus types documented in the Papillomavirus Episteme database (PaVE). Here, we report that TC dinucleotides, the preferred target sequence of several human APOBEC3 proteins (hA3A, hA3B, hA3F, and hA3H), are highly depleted in papillomavirus genomes. Given that HPV infection is highly tissue-specific, the expression levels of APOBEC3 family members were analyzed. The basal expression levels of all APOBEC3 isoforms, excluding hA3B, are significantly higher in mucosal skin compared with cutaneous skin. Interestingly, we reveal that Alphapapillomaviruses (alpha-PVs), a majority of which infects anogenital mucosa, display the most dramatic reduction in TC dinucleotide content. Computer modeling and reconstruction of ancestral alpha-PV genomes suggest that TC depletion occurred after the alpha-PVs diverged from their most recent common ancestor. In addition, we found that TC depletion in alpha-PVs is greatly affected by protein coding potential. Taken together, our results suggest that PVs replicating in tissues with high APOBEC3 levels may have evolved to evade restriction by selecting for variants that contain reduced APOBEC3 target sites in their genomes.
Keywords: papillomavirus, APOBEC3, restriction factor, coevolution
1. Introduction
Papillomaviruses (PVs) are non-enveloped, double-stranded DNA viruses that infect keratinocytes and lead to benign and malignant tumors. Over 270 types of PVs have been discovered in various host species including mammals, reptiles, and birds (Van Doorslaer et al., 2013). It is believed that PVs have evolved for hundreds of millions of years and successfully established the largest and most diversified family of vertebrate viruses.
Of about 180 different types of human papillomaviruses (HPVs) identified so far, high-risk genotypes such as HPV16 and HPV18 are causally associated with multiple human cancers including cervical and head and neck cancers. All high-risk HPV types are classified in a single genus, Alphapapillomaviridae, which diverged from a common ancestor about 75 million years ago (Rector et al. 2007; Van Doorslaer 2013). This implies that the Alphapapillomaviruses (alpha-PVs) diverged well before the emergence of Homo sapiens in the primate lineage (Van Doorslaer 2013; Ong et al., 1993). However, little is known about factors contributing to PV evolution (Van Doorslaer 2013; Bravo and Félez-Sánchez 2015).
The apolipoprotein B mRNA-editing, enzyme-catalytic, polypeptide-like 3 (APOBEC3) family of proteins have recently been discovered as host restriction factors against various viruses and endogenous retroelements (Sheehy et al. 2002; Zheng et al. 2004; Bogerd et al. 2006; Hultquist et al. 2011; Suspène et al. 2011; Ooms et al. 2012; Liang et al. 2013; Minkah et al. 2014). APOBEC3 proteins deaminate cytosine (C) residues to uracil (U) in single-stranded DNA (Yu et al. 2004; Chiu et al. 2005). These mutations can lead to defects in gene transcription, genome replication and chromosomal integration thereby restricting the viral lifecycle. Of the multiple potential targets of human APOBEC3 (hA3), retroviruses, endogenous retroelements, DNA viruses and surprisingly, host genomic DNA, have been identified as common substrates (Harris and Liddament 2004; Turelli et al. 2004; Esnault et al. 2005; Bogerd et al. 2006; Chen et al. 2006; Burns et al. 2013; Burns, Temiz, and Harris 2013).
The human and non-human primate APOBEC3 family consists of seven members (A3A, A3B, A3C, A3D, A3F, A3G, and A3H), while other mammals encode fewer isoforms (Jarmuz et al. 2002; Münk et al. 2008; LaRue et al. 2009). Substrate specificity varies amongst the seven hA3 family members with each having slightly different target sequences based on the dinucleotide context of the cytosine residue (Zheng et al. 2004; Hultquist et al. 2011; Pham et al. 2003; Liddament et al. 2004; Yu et al. 2004; Jern et al. 2009; Nik-Zainal et al. 2012). In addition to differences in dinucleotide preferences, hA3s differ in their subcellular distribution during interphase and mitosis (Lackeyet al. 2013). These differences in localization impact the class of virus targeted by a particular hA3. For example, the predominantly cytoplasmic hA3F and hA3G are important inhibitors of productive replication of viruses whose replication cycle proceeds through obligatory reverse transcribed cDNA intermediates (Turelli et al. 2004; Chiu et al. 2005; Cullen 2006). In contrast, genetic editing of DNA viruses by nuclear hA3s (hA3A, hA3B, and hA3C) has been reported (Chen et al. 2006; Vartanian et al. 2008; Suspène et al. 2011; Lackeyet al. 2013; Wang et al. 2014).
It has been demonstrated that episomal HPV genomes are edited by hA3A and hA3B in cervical lesions and keratinocytes (Vartanian et al. 2008; Wang et al. 2014). Recent work by us and others revealed that HPV induced hA3A expression restricts HPV16 infectivity in human keratinocytes (Ahasan et al. 2015; Warren et al. 2015). If deamination by hA3s represents a bona fide host restriction mechanism, PVs may have evolved to limit APOBEC3 recognition motifs in their genomes through mutation and selection. Thus, we hypothesized that the host restriction function of APOBEC3 proteins contributed to PV genome evolution by selecting for depletion of specific dinucleotide sequences within PV genomes. To test this hypothesis, the relative abundance of dinucleotide sequence patterns in the full genome sequences of all known PVs was analyzed.
In this study, we investigated the link between APOBEC3 expression levels, tissue tropism of various HPV genotypes, and the extent of dinucleotide depletion in PV genomes. We propose a novel model suggesting that PV types replicating in tissues with high APOBEC3 levels have evolved to evade host restriction by selecting for variants containing reduced APOBEC3 target sites in their genomes. Our results provide important insight into the role of host restriction factors during PV evolution.
2 Materials and methods
2.1 Papillomavirus genomes
We retrieved full-length reference sequences of all 274 PV types archived in the PV database (PaVE; pave.niaid.nih.gov, accessed on 1 August 2014).
2.2 Calculation of dinucleotide frequencies
Dinucleotide frequencies were determined using three complementary approaches. First, in order to determine a single observed vs. expected (O/E) dinucleotide ratio across the entire viral genome, a custom python script was used (available from a public computer code repository [http://www.github.com/Van-Doorslaer/Warren-et-al.]). The script uses the CompSeq program from Emboss. The expected frequencies of dinucleotide ‘words’ were estimated based on the observed frequency of single bases in the sequences. Alternatively, to assess local differences in dinucleotide content across the length of the viral genome, the frequency of each motif was counted within a 1 kb sliding window with a 100 bp overlap (Minkah et al. 2014). In order to generate a null model, the word occurrence was enumerated in 1,000 randomly shuffled versions of each 1 kb window. If the actual frequency fell within the lowest or highest fifth percentile, the word was considered to be under- or over-represented, respectively. The data are presented as percent 1 kb windows that fall in each category. Finally, the above-mentioned CompSeq based approach does not control for potential effects of coding restraints. To analyze this, the E1, E2, L1, and L2 ORFs were concatenated and the occurrence of each dinucleotide was counted in relationship to the position in a triplet (for coding sequences this triplet equals a codon). As a control, the non-coding upstream regulatory region (URR) was analyzed. In order to obtain the expected distribution, the sequences were randomly shuffled and dinucleotides were counted as earlier. This process was repeated 1,000 times.
2.3 Tissue-specific gene expression analysis
Correlation of tissue-specific APOBEC3 gene expression analysis was performed using publicly available RNAseq data from the Genotype-Tissue Expression (GTEx) project (gtexportal.org) (Lonsdale et al. 2013). DESeq2 (v 1.6.3) in R (v 3.1.0) was used for analysis of differentially expressed genes using DESeq2’s negative binomial distribution analysis model (Love, Huber, and Anders 2014).
2.4 Statistical analysis
Student’s t test and one- or two-way analysis of variance (ANOVA) were used where appropriate. Data are presented as box-and-whisker plots with Tukey’s method for outliers noted as distinct data points. All graphs were generated using Prism software. Results were considered statistically significant at a P-value of <0.05.
2.5 Phylogenetic analysis
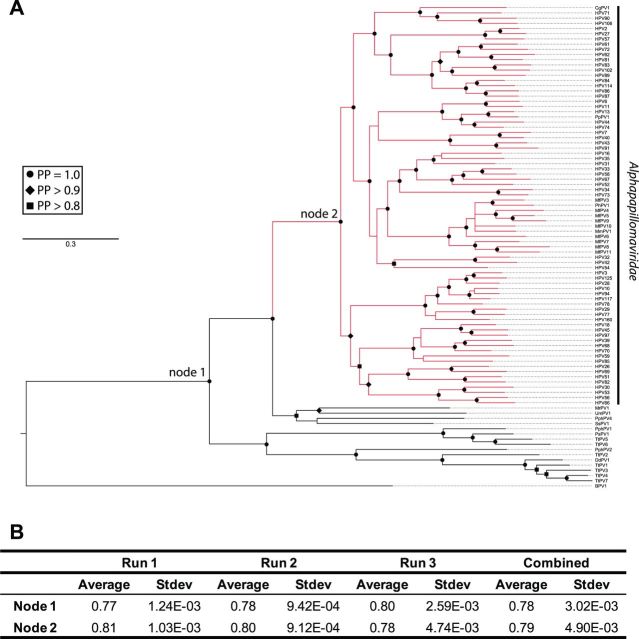
The E1, E2, L1, and L2 sequences from 274 PVs were downloaded from PaVE. The nucleotide sequences were translated into amino acids and aligned using the MAFFT algorithm (Katoh and Standley 2013). The resulting alignment was back translated to nucleotides. The individual gene alignments were concatenated into a single supermatrix. A maximum likelihood tree was constructed using RaxML (Stamatakis 2006) implementing a gamma model allowing for among-site rate variation and variable substitution rates GTR + I + G; model selected using jModeltest (Darriba et al. 2012). The phylogenetic tree shown in Figure 3 was annotated using EvolView (Zhang et al. 2012). All computational analyses were performed on the CIPRES Science Gateway (Miller, Pfeiffer, and Schwartz 2010).
Figure 3.
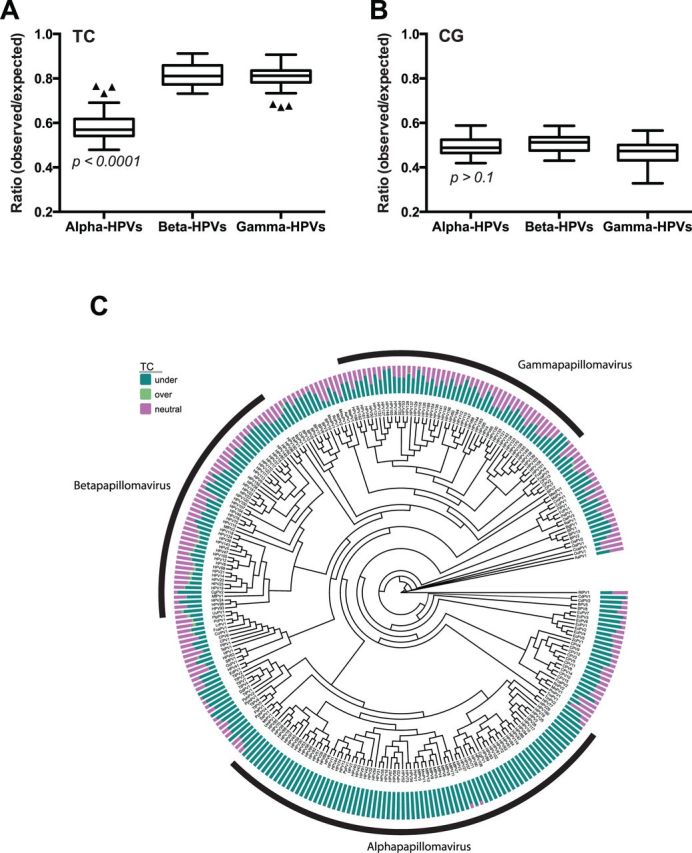
TC content is significantly lower in alpha-PVs, compared with beta- and gamma-PVs. The O/E ratios of TC (A) and CG (B) dinucleotides were compared between alpha- (n = 64), beta- (n = 44), and gamma- (n = 50) HPVs. Box-and-whisker plots are shown with the outliers (black triangles) identified using Tukey’s method. P-values were determined by one-way ANOVA analysis using Prism software. (C) A maximum likelihood phylogenetic tree is shown comparing the O/E ratios of TC dinucleotides from all PV genomes (n = 274). The alpha-, beta-, and gamma-PV genera are indicated. Bars indicate percent of 1 kb windows in which TC is under-represented (green), overrepresented (light green), and neutral (pink). Details are available in ‘Materials and Methods’ section.
2.6 Ancestral state reconstruction
The E1, E2, L1, and L2 nucleotide sequences belonging to the alpha, omega, upsilon, omicron, and dyodelta-PV genera were downloaded from PaVE. The nucleotide sequences were translated into amino acids and aligned using the MAFFT algorithm (Katoh and Standley 2013). The resulting alignment was reverse translated to nucleotides. Aligned sequences were concatenated and the resulting supermatrix was used to estimate a Bayesian phylogenetic tree using MrBayes (version 3; Cipres science gateway) implementing a GTR + I + G model of evolution. The MCMC-chain length was set to 1 × 107 while the chain was sampled every 1,000th generation. Chain convergence was analyzed using AWTY (Nylander et al. 2008). Bovine papillomavirus type 1 (BPV1), a member of the Deltapapillomavirs genus, was used to root the phylogenetic tree. Figure 5 shows the majority rule tree. Ancestral states were estimated using a Bayesian approach within BayesTraits (Barker, Meade, and Pagel 2007). The applied General Least squares approach removes the need for phylogenetic correction when comparing continuous traits between evolutionarily related species. Furthermore, in order to account for phylogenetic uncertainty during the tree building process, 500 trees were randomly sampled from the post-burn-in posterior sample of MrBayes trees and used for further analysis. Within BayesTraits, the MCMC analysis was run for 1 × 109 iterations. After a burn-in of 1 × 106 iterations, the chains were sampled every 10,000th iteration. The alpha (estimated root of the tree) and beta (directional change parameter of the directional model) parameters were estimated based on a uniform prior distribution. BayesTraits keeps a running tally of the logarithm of the harmonic mean of the likelihoods. When the analysis is completed, the final harmonic means for both models are compared using Bayes Factors (logBF = 2 (log[harmonic mean(complex model)]–log[harmonic mean(simple model)]). Kappa, lambda, and delta scaling parameters were estimated under both the ‘random walk’ and ‘directional walk’ models (Pagel 1999). These parameters test the tempo, mode, and phylogenetic associations of trait evolution, respectively. Following selection of appropriate model parameters, these parameters were used to estimate ancestral TC O/E ratios at indicated nodes. To confirm the robustness of the results, the analysis was performed three independent times.
Figure 5.
TC depletion represents an apomorphy of the alpha-PVs. (A) The Bayesian phylogenetic tree is constructed based on a concatenated E1-E2-L2-L1 nucleotide sequence alignment. The majority-rule consensus phylogenetic tree shows the evolutionary relationship between the alpha-PVs and the members of the omega, upsilon, omicron, and dyodelta-PV genera. BPV1, a delta-PV was used to root the tree. The solid branches indicate the optimized branch lengths. Dotted lines were added to the branches to facilitate visual inspection of the tree. The alpha-PV clade is highlighted in red. Posterior probability values are indicated using symbols at the nodes (circle = 1, diamond > 0.90, square > 0.80). The nodes for which the ancestral states were estimated are indicated in the figure. (B) Table showing the average (and SD) of the estimates for the ancestral TC ratios at the indicated nodes. The reconstruction process was repeated three times to ensure repeatability.
3 Results
3.1 TC dinucleotides are significantly depleted in PV genomes
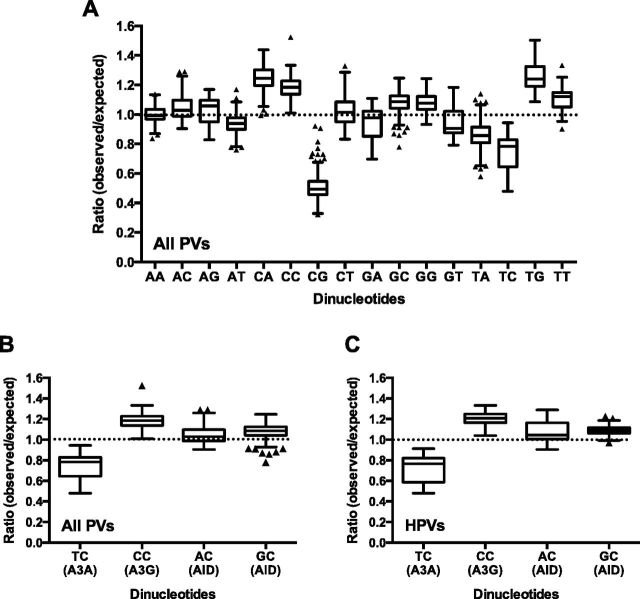
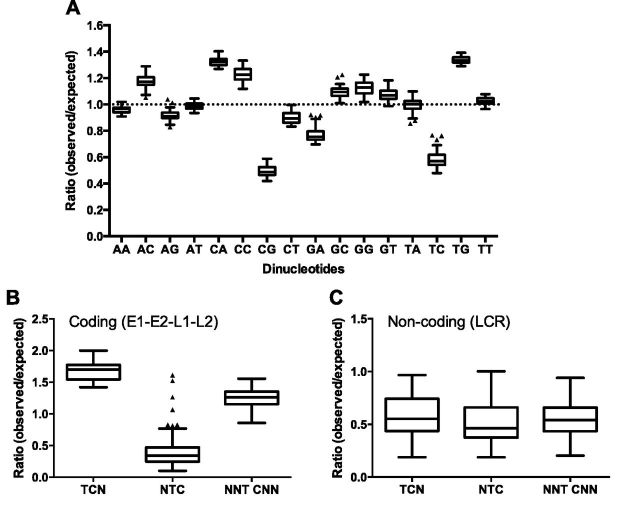
To determine whether there are preferred dinucleotide sequence patterns in PV genomes, we analyzed the relative abundance of all possible dinucleotide combinations in the full-length genomes of all 274 PVs deposited in the Papillomavirus Episteme database (Van Doorslaer et al. 2013) (PaVE; pave.niaid. nih.gov; accessed on 1 August 2014). The ratio of observed vs. expected (O/E) counts of all dinucleotides were calculated and this O/E ratio was used to determine whether dinucleotides were over-represented (ratio > 1), neutral (ratio = 1), or under-represented (ratio < 1) in these genome sequences. Among all dinucleotide combinations, CG and TC dinucleotides are the most underrepresented dinucleotides in PV genomes (Fig. 1A and Supplementary Table S1; depletion of the CG or TC dinucleotide compared with any other dinucleotide, P < 0.0001 by two-way ANOVA with Tukey correction). CG depletion has been previously noted for several small DNA viruses (Karlin, Doerfler, and Cardon 1994; Hoelzer, Shackelton, and Parrish 2008; Upadhyay et al. 2013), including PVs (Shackelton, Parrish, and Holmes 2006). Given that TC dinucleotides are preferred or specific targets of APOBEC3 DNA editing, these results imply that the PV genomes may be greatly affected by the host restriction functions of APOBEC3.
Figure 1.
TC and CG dinucleotide sequences are significantly depleted in PV genomes. (A) The observed vs. expected (O/E) ratios of each dinucleotide in PV genome sequences from all PV genotypes (n = 274) were calculated using a custom wrapper around the CompSeq program from the EMBOSS software suite. (B) The O/E ratios of TC, CC, AC, and GC dinucleotides, preferred editing sites of A3A, A3G, and AID, respectively, were analyzed with the genome sequences of all PVs (A; n = 274) and HPVs (B; n = 161). Box-and-whisker plots are shown with the outliers (black triangles) identified using Tukey's method. CG and TC dinucleotide are the most depleted cytidine containing dinucleotides in all PVs and HPVs (depletion of CG or TC compared with depletion of any other dinucleotide, P < 0.0001 by two-way ANOVA with Tukey correction).
Each APOBEC3 family member preferably targets different dinucleotide sequences for cytidine deamination. For example, the TC dinucleotide is preferably targeted by A3A, A3B, A3C, A3F, and A3H (Liddament et al. 2004; Yu et al. 2004; Zheng et al. 2004; Hultquist et al. 2011; Nik-Zainal et al. 2012), while CC and RC (where R represents any pyrimidine) dinucleotides are preferably deaminated by A3G (Jern et al. 2009) and AID (Pham et al. 2003), respectively. Interestingly, among cytidine-containing dinucleotides, only CG and TC residues are significantly depleted in all papillomavirus genomes, including HPVs (Fig. 1A–C and Supplementary Fig. S1; P < 0.0001 by two-way ANOVA with Tukey correction). Among the APOBEC3 family members preferably editing TC dinucleotides, hA3A, hA3B, and hA3H are known to restrict DNA viruses including HPVs and herpesviruses (Vartanian et al. 2008; Suspène et al. 2011; Minkah et al. 2014; Warren et al. 2015). Therefore, these results suggest that TC depletion may be influenced by host restriction factors, A3A, A3B, and/or A3H.
3.2 APOBEC3 family members are differentially expressed at mucosal and cutaneous sites.
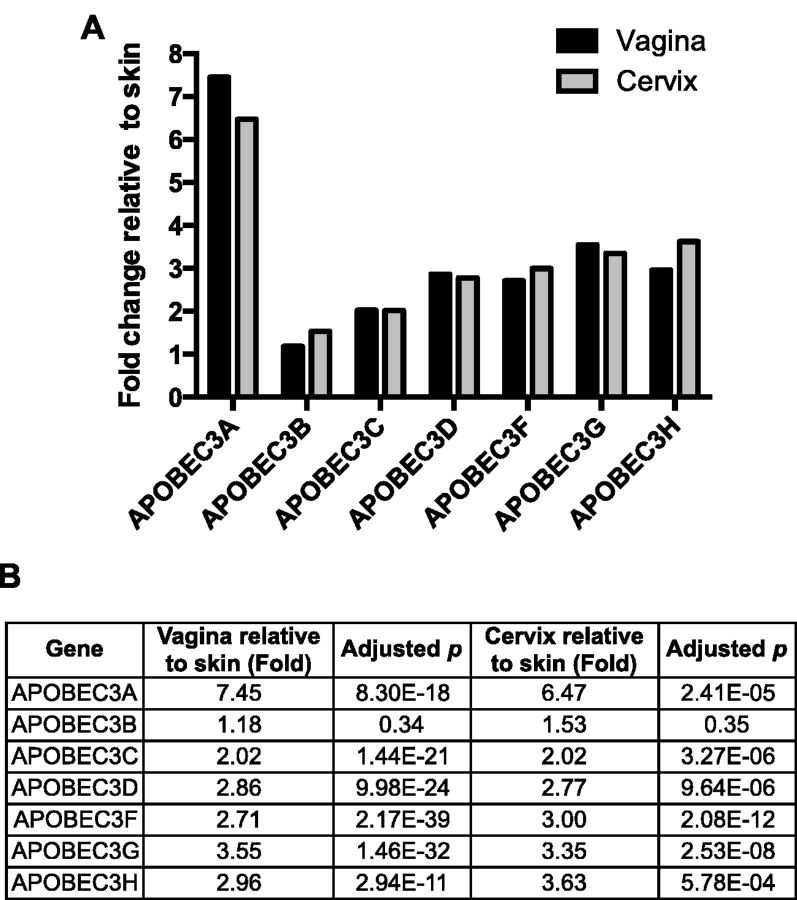
HPVs can roughly be categorized into two groups based on tissue tropism, those that infect cutaneous skin, which mainly include beta- and gamma-PVs, and those that infect the mucosa, which predominantly include alpha-PVs (de Villiers et al. 2004). Because TC depletion is significant amongst all HPV types, we sought to determine whether the expression levels of all human APOBEC3 family members differed in mucosal vs. cutaneous skin. RNA sequencing data from the GTEx project (Lonsdale et al. 2013) was used to compare the expression levels of each human APOBEC3 gene in cervix and vagina to cutaneous skin using DESeq2 (Love, Huber, and Anders 2014). Of note, no strong evidence of potential batch effects was observed from PCA plots and hierarchical clustering. High APOBEC3 expression was observed for all isoforms, except A3B, when mucosal tissues (cervix and vagina) were compared with cutaneous skin (Fig. 2A and B).
Figure 2.
The expression levels of APOBEC3s are different between cervicovaginal and cutaneous skin. (A) Gene expression levels of APOBEC3s were analyzed using RNA sequencing data retrieved from the Genotype-Tissue Expression project database (gtexportal.org) (Lonsdale et al. 2013) and analyzed using DESeq2. Data are presented as fold change in cervix (n = 6) or vagina (n = 34) compared with cutaneous skin (n = 41). (B) Table showing the fold change and P-values adjusted for repeated measurements.
3.3 TC content is significantly lower in alpha-PVs, compared with beta- and gamma-PVs
Gene expression analysis revealed increased basal expression of APOBEC3 family members at mucosal sites when compared with cutaneous skin. Because increased basal APOBEC3 expression may pose an intrinsic barrier to viral infection, it is likely that PVs infecting these tissues would evolve to evade APOBEC3 restriction pressure. Thus, we hypothesized that the TC content in mucosotropic PVs would be significantly lower than PVs with a cutaneous tropism. The TC dinucleotide ratio in the genomes of human alpha- (n = 64), beta- (n = 44), and gamma- (n = 50) PVs was calculated as described earlier. Notably, TC dinucleotides are significantly depleted in the genomes of alpha-PVs (mean O/E ratio <0.6), compared with the TC contents of beta- and gamma-PVs (mean O/E ratio >0.8) (Fig. 3A). In contrast, all three genera contain similar levels of CG dinucleotides (Fig. 3B). The distinct pattern of TC depletion in alpha-PVs becomes apparent when mapped on a PV phylogenetic tree (Fig. 3C). These results suggest that high basal expression of APOBEC3s at mucosal sites may have affected the evolutionary trajectory of the alpha-PV clade.
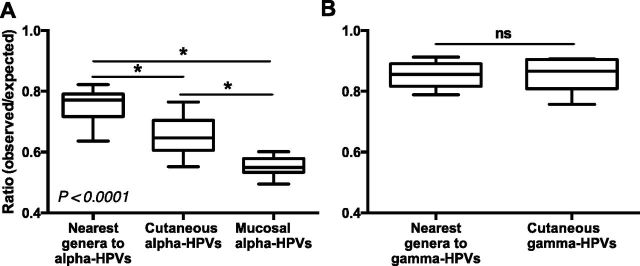
Although alpha-PVs are primarily mucosotropic, a subset is known to infect cutaneous skin (de Villiers et al. 2004). If higher APOBEC3 levels affect the genomic TC content, mucosotropic alpha-PVs should have decreased TC ratios when compared with cutaneotropic alpha-PVs. When HPVs were stratified by tissue tropism, significantly lower TC contents were observed in mucosal alpha-HPVs compared with cutaneous alpha-HPVs (Fig. 4A; Supplementary Table S2). A similar analysis of primarily cutaneous gamma-HPVs and their nearest neighbors (pi, tau, and dyoxi-PVs) revealed no differences in TC O/E ratios suggesting a specific effect of mucosal tropism on TC depletion (Fig. 4B). Interestingly, a few gamma-PVs (including HPV101, 103, and 108) that have been isolated from cervical lesions also show significant depletion of TC dinucleotides (Fig. 3C). This further supports an effect of tropism on TC dinucleotide content (Chen et al. 2007; Nobre et al. 2009). This analysis was based on available clinical data (de Villiers et al. 2004) and the tropism of many PVs is not yet clear. Taken together, our results suggest that one or more APOBEC3 family members that are highly expressed in mucosal tissues may have driven genome evolution of alpha-PVs.
Figure 4.
Extensive TC dinucleotide depletion in mucosal alpha-HPVs. The O/E ratios of TC dinucleotides in PV genomes were analyzed with genome sequences of (A) nearest genera to alpha-HPVs (one omega, seven upsilon, four omikron, two dyodelta, and one unclassified PVs; n = 15), cutaneous alpha-HPVs (n = 13), and mucosal alpha-HPVs (n = 30); and (B) nearest genera to gamma-HPVs (six pi, one dyoxi, three tau, and two unclassified PVs; n = 12) and cutaneous gamma-HPVs (n = 7). Box-and-whisker plots are shown. P-values were calculated by Student’s unpaired, two-tailed t-test. *P < 0.0001, ns, not significant.
3.4 TC depletion represents an apomorphy of alpha-PVs
Our results point towards a role for the APOBEC3 proteins in the evolution of the alpha-PV genus. However, it remains possible that low TC content was a characteristic of the most recent common ancestor (MRCA) of alpha-PVs. In this case, a founder effect may be responsible for the TC contents we observe in extant members, rather than specific APOBEC3 effects. To address this issue, thorough evolutionary trait analysis was performed to model the evolution of the TC O/E ratios and reconstruct the ancestral alpha-PV genomes.
The alpha-PV genus forms a monophyletic clade with the omega, upsilon, omicron, and dyodelta-PV genera (Fig. 3C) (Van Doorslaer 2013). Figure 5 shows the evolutionary relationship within this monophyletic clade. The computer program Continuous, as implemented within BayesTraits, was used to model the evolution of the TC O/E ratio. This approach allows for the characterization of trait evolution while automatically correcting for the influence of phylogenetic signal (Pagel 1997, 1999). These analyses indicate that the TC O/E ratio evolved according to a directional-random walk model (Bayes Factor >12). Furthermore, the Delta parameter was estimated to be significantly <1, indicating that adaptive radiation best explains the extant O/E ratios. This suggests that the initial TC depletion occurred rapidly, followed by slower rates of change among closely related species. Additionally, this approach allows for the estimation of ancestral states at important nodes of the phylogenetic tree (Fig. 5A). The ancestral state reconstruction suggests that the MRCA of the alpha, omega, upsilon, omicron, and dyodelta-PV clades had a TC O/E ratio of 0.78 (Fig. 5B). Likewise, the last common ancestor of the alpha-PV genus had a TC O/E ratio of 0.79. Therefore, the MRCA of the alpha-PVs is predicted to have a TC O/E ratio that is significantly higher than the extant members of this genus. Thus, the predicted rapid TC depletion must have occurred after the alpha-PVs began to diverge. Importantly, this provides strong evidence that TC depletion is unique to alpha-PVs and begins to illuminate the evolutionary path responsible for TC depletion.
3.5 APOBEC3 mediated editing directly affects Alphapapillomavirus evolution
It appears that the TC O/E ratios in alpha-PV genomes are dramatically lower when compared with other viral genera. Next, we determined whether APOBEC3 actively edited viral genomes. APOBEC3 deamination of TC residues can result in C-to-T transitions or C-to-G transversions that, when fixed in the viral genome, would increase the overall TT or TG content, respectively (Yu et al. 2004; Chiu and Greene 2008; Harris 2013). Although many mutations will be deleterious to the virus (Yang et al. 2007; Friedman and Stivers 2010), some of these mutations will be neutral or may even increase viral fitness (Mulder, Harari, and Simon, 2008; Jern et al. 2009; Wood et al. 2009; Kim et al. 2014). If APOBEC3 driven deamination events became fixed in the viral population, this should be reflected in a relative gain of the O/E ratios of TT or TG dinucleotides. Alternatively, if mutation followed by selection was responsible, any mutation disrupting the TC recognition motif would be selected for. Although alpha-HPV genomes are not enriched for TT dinucleotides (O/E ratio ∼1), TG dinucleotides (and CA on the opposite strand) are the most over-represented dinucleotides (Fig. 6A; increase of TG or CA dinucleotide compared with increase of any other dinucleotide, P < 0.0001 by two-way ANOVA with Tukey correction). No significant differences were noted between TG and CA using this analysis (P = 0.87). Additionally, GA dinucleotides (TC counterpart on negative strand) are also depleted in alpha-PVs thus suggesting that APOBEC3 effects are observed on both strands. These data suggest that TC depletion might in part be driven by APOBEC3-mediated deamination.
Figure 6.
Depletion of TC dinucleotides is associated with amino acid coding potential. (A) The O/E ratios of each dinucleotide in alpha-HPV (n = 64) genome sequences was calculated as described in Figure 1A. (B) E1, E2, L1, and L2 sequences from alpha-HPVs were concatenated as described in ‘Materials and Methods’ section. The O/E ratios of TC dinucleotides at various codon positions were calculated as described in ‘Materials and Methods’ section. N represents any of T, C, A, or G. (C) The TC O/E ratios at NNT CNN, TCN, and NTC in the URR were calculated as a noncoding control. Box-and-whisker plots are shown with the outliers (black triangles) identified using Tukey’s method. CA and TG dinucleotides are the most over-represented dinucleotides, while NTC cytidines are significantly depleted in alpha-PVs. P < 0.0001 by two-way ANOVA analysis with Tukey correction.
3.6 Effects of coding potential on TC depletion
Roughly 85% of the PV genome encodes for viral proteins. The previous analyses considered dinucleotide ratios for the whole viral genome, regardless of coding potential. If APOBEC3 mediated editing affected PV evolution, it is likely that TC content is correlated with the codon position of the cytidine (NNT CNN vs. TCN vs. NTC with N representing any nucleotide). Strikingly, it appears that TC reduction is most pronounced when the cytidine is in the third codon position (NTC O/E <0.5; P < 0.0001 by two-way ANOVA with Tukey correction) (Fig. 6B). Importantly, this NTC bias is not seen in the non-coding URR (Fig. 6C). Because cytidine is at the third position of these codons, substitution of cytidine in any NTC codon does not incur any significant change in the corresponding amino acid during protein synthesis (Betts and Russell 2003). These results suggest that APOBEC3 editing primarily resulted in silent and/or conserved mutations.
3.6 TC dinucleotides are dramatically depleted in the early gene regions for alpha-PVs
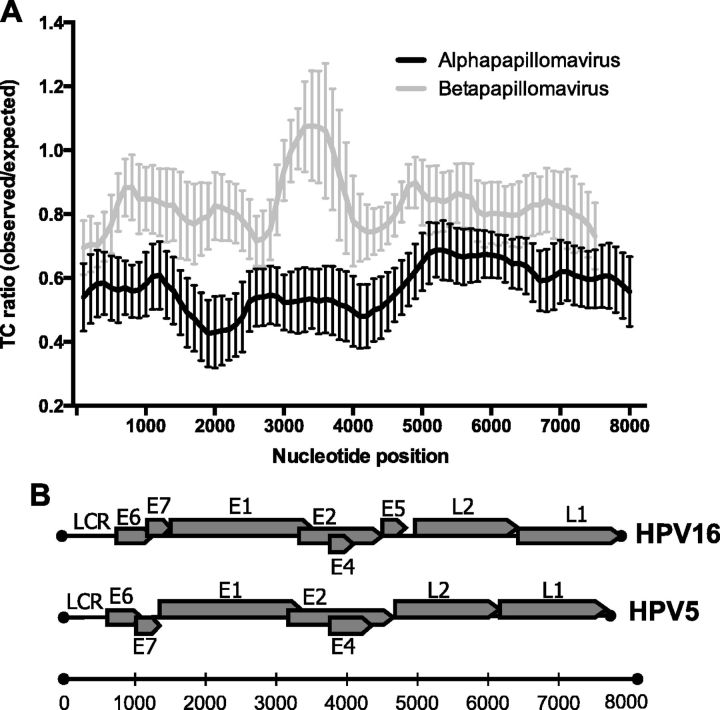
To examine if there was any bias for TC dinucleotide reduction in particular regions within alpha-PV genomes, the TC O/E ratio was calculated using a sliding window approach (a 1,000 bp window with a 100 bp overlap between two windows). Figure 7A compares genomic TC content across all alpha-and beta-PVs. The alpha-PV genomes have a lower TC ratio across the early gene regions of the viral genome (Fig. 7B).
Figure 7.
The E1 and E2 gene regions of alpha-PV genomes are the most significantly depleted sites for TC dinucleotides. (A) TC O/E ratios were calculated in 1 kb scanning windows using the CompSeq program as described in ‘Materials and Methods’ section. All alpha- (n = 77) and beta-PVs (n = 49) genomes were included. Each window represents 1,000 kb with a 100 bp step between two windows. The viral genomes were linearized in such a way that the first nucleotide corresponds to the first nucleotide following the L1 stop codon. (B) Genome maps of HPV16 and HPV5, adapted from PaVE, show the various ORFs and URR for representative alpha- and beta-PVs, respectively.
4 Discussion
PV infections have been described in most amniotic hosts. Interestingly, these infections appear to be tissue specific and show a highly restricted host range. Based on these observations, it has been suggested that PVs co-speciate with their hosts (Van Doorslaer 2013). To date, limited evidence exists supporting positive selection of PV genomes (Van Doorslaer 2013), suggesting that genetic drift is the primary driving force of PV evolution. However, most studies have investigated the role of evolution in shaping individual viral proteins, not taking into account possible evolutionary selection working at the primary nucleotide level. This study was undertaken to survey extant genomes of all known PVs and to investigate a potential role for APOBEC3-mediated restriction in the evolution of these viruses.
Viral genomes are frequently targeted by host restriction factors such as restriction nucleases, RNAi-mediated mechanisms, and DNA editing enzymes such as APOBEC3 and ADAR (Johnson 2013). In order to achieve productive infection, viruses need to evade these antiviral host mechanisms. Primate lentiviruses, such as human immunodeficiency virus type 1 (HIV-1), encode multiple viral proteins that antagonize host restriction mechanisms. For example, the HIV-1 accessory proteins Vif and Vpu counteract host restriction by hA3G and tetherin, respectively (Sheehy et al. 2002; Neil, Zang, and Bieniasz 2008). Likewise, host genomes evolve new restriction mechanisms to detect and eliminate virus infections. Well exemplified by the APOBEC3 genetic locus, host genome evolution alongside constant viral exposure may have selected for beneficial duplication events that increased the number of APOBEC3 genes. A recent study suggests that the APOBEC3 genes might have originated from one ancestral APOBEC3 gene (Münk et al. 2008). During mammalian evolution, APOBEC3 genes evolved by gene duplication, fusions, and losses, probably to adapt to various virus infections. It is likely that this ‘arms race’ has been indispensible in shaping both viral and host genomes (Duggal and Emerman 2012).
Although viral regulatory proteins may be purposed to antagonize host restriction factors, viruses may also develop more subtle ways to subvert host recognition and restriction mechanisms. We and others have previously shown that HPV infection is restricted by hA3A and that HPV genomes show signs of APOBEC3 hypermutation (Vartanian et al. 2008; Wang et al. 2014; Warren et al. 2015). Given that APOBEC3s would pose a significant barrier to HPV infection, we sought to investigate how PVs may have evolved to evade APOBEC3 restriction.
Here, our study provides evidence that PV genomes are significantly depleted in TC dinucleotides, the preferred target sites of several APOBEC3 proteins. Although all PV genomes have decreased TC content, this depletion is strongest in mucosotropic alpha-PVs (Figs. 3A and 4A). Importantly, gene expression data clearly shows significantly higher expression levels of all APOBEC3 isoforms (except A3B) in mucosal skin of cervix and vagina, compared with cutaneous skin (Fig. 2A and B). This suggests that increased APOBEC3 expression in these tissues may have affected the evolution of a subset of viral genomes.
APOBEC3-mediated TC deamination is correlated with TT and/or TG mutational signatures in tumor tissues (Nik-Zainal et al. 2012). These mutational signatures arise from distinct pathways, thus providing clues into the mechanism of deaminated cytosine processing. Although C-to-T transitions may result from error prone DNA replication across uracil lesions, C-to-T transversions largely result from errors incurred during base excision repair (BER) pathways (Nik-Zainal et al. 2012; Henderson, Chakravarthy, Fenton, 2014). TC depletion led to an increase in TG (and CA on the opposite strand), but not TT dinucleotides in alpha-PVs. This suggests that APOBEC3-mediated deamination, coupled with BER, may have partially contributed to alpha-PV genome evolution (Fig. 6A).
Although alpha-PVs appear to have minimized the TC O/E ratio across their genome (Fig. 7), further analysis of coding regions revealed that TC depletion is dependent on the position of cytidine within a codon (Fig. 6B and C). This analysis suggested that while alpha-PV genomes are able to tolerate NTC mutations, there was no decrease in TCN or NNT CNN sites. This is likely due to the degenerate nature of the genetic code (Betts and Russell 2003). Interestingly, it has been demonstrated that HPV genomes have a pronounced codon usage bias when compared with host genes (Bravo and Muller 2005; Cladel, Bertotto, and Christensen, 2010; Félez-Sánchez et al. 2015). This study may begin to shed light unto why papillomavirus genomes have evolved to use sub-optimal codons. Importantly, the observation that the third codon position is preferentially edited, suggests that these genomes are expected to still be vulnerable to APOBEC3 restriction because mutations would lead to changes in the amino acid sequence.
In addition to TC depletion, we also observed a significant reduction in CG dinucleotides in all PVs surveyed. This finding is consistent with others that have reported a significant depletion in CG residues across many small (<30 kb) DNA virus genomes (Karlin, Doerfler, Cardon 1994; Hoelzer, Shackelton, and Parrish 2008; Upadhyay et al. 2013). Previous work suggests that 5-methylcytosine modifications can negatively influence gene expression and lead to mutation via spontaneous deamination, both of which would negatively impact the course of a viral infection (Karlin, Doerfler, Cardon 1994). Additionally, TLR9 recognition of CG motifs in viral genomes can trigger antiviral immune responses such as production of type I interferons. High-risk HPV16 has previously been shown to inhibit TLR9 expression as a mechanism to avoid innate immune recognition (Hasan et al. 2007). Collectively, depletion of CG dinucleotides represents an evolutionarily conserved mechanism to evade innate immune responses.
Although further studies are necessary, our results consistently support a model by which the mucosal niche was colonized by an ancestral virus. Higher APOBEC3 levels in cervical and vaginal tissues resulted in increased editing of viral genomes and selection for those viruses with reduced TC contents. With about 85% of the genome coding for viral proteins, TC depletion appears to have been limited by coding potential. This is predicted by the evolutionary model suggesting that after the TC O/E ratio was minimized, initial rapid evolution was followed by slow drift (Fig. 5).
In conclusion, our data suggest that viral evasion strategies that select against TC dinucleotides may be an important driver of PV evolution. Furthermore, our findings reinforce that tissue tropism and host range across these, and possibly other DNA viruses, may similarly be affected by interactions with APOBEC3.
Supplementary data
Supplementary data is available at Virus Evolution online.
Acknowledgements
This work was supported by National Institutes of Health (R01 AI091968) to D.P. and (R01 CA117907-07) to J.M.E., the Intramural Research Program of the National Institute of Allergy and Infectious Diseases at the National Institutes of Health to K.V.D., and the Cancer League of Colorado to D.P. We thank Katerina Kechris for consultation on statistical analyses. We also thank Paul Lambert, Paul Ahlquist, Mario Santiago, Matt Daugherty, Rob DeSalle, and members of our lab for useful comments and suggestions.
Conflict of interest: None declared
References
- Ahasan M. M., et al. (2015). ‘APOBEC3A and C Decrease Human Papillomavirus 16 Pseudovirion Infectivity’, Biochemical and Biophysical Research Communications, 457: 295–9. [DOI] [PubMed] [Google Scholar]
- Barker D., Meade A., Pagel M. (2007). ‘Constrained Models of Evolution Lead to Improved Prediction of Functional Linkage from Correlated Gain and Loss of Genes’, Bioinformatics, 23: 14–20. [DOI] [PubMed] [Google Scholar]
- Betts M. J., Russell R. B. (2003). ‘Amino Acid Properties and Consequences of Substitutions’, Bioinformatics for Geneticists, 317: 289. [Google Scholar]
- Bogerd H. P., et al. (2006). ‘APOBEC3A and APOBEC3B are Potent Inhibitors of LTR-Retrotransposon Function in Human Cells’, Nucleic Acids Research 34: 89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo I. G., Félez-Sánchez M. (2015). ‘Papillomaviruses: Viral Evolution, Cancer and Evolutionary Medicine’, Evolution, Medicine and Public Health, 2015: 32–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo I. G., Muller M. (2005). ‘Codon Usage in Papillomavirus Genes: Practical and Functional Aspects’, Papillomavirus Report, 16: 63–72. [Google Scholar]
- Burns M. B., et al. (2013). ‘APOBEC3B is an Enzymatic Source of Mutation in Breast Cancer’, Nature 494: 366–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns M. B., Temiz N. A., Harris R. S. (2013). ‘Evidence for APOBEC3B Mutagenesis in Multiple Human Cancers’, Nature Genetics, 45: 977–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., et al. (2006). ‘APOBEC3A is a Potent Inhibitor of Adeno-Associated Virus and Retrotransposons. Current Biology, 16: 480–5. [DOI] [PubMed] [Google Scholar]
- Chen Z., et al. (2007). ‘Human Papillomavirus (HPV) Types 101 and 103 Isolated from Cervicovaginal Cells Lack an E6 Open Reading Frame (ORF) and are Related to Gamma-Papillomaviruses’, Virology, 360: 447–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu Y. -L., Greene W. C. 2008. ‘The APOBEC3 Cytidine Deaminases: An Innate Defensive Network Opposing Exogenous Retroviruses and Endogenous Retroelements’, Annual Review of Immunology, 26: 317–53. [DOI] [PubMed] [Google Scholar]
- Chiu Y. -L., et al. (2005). ‘Cellular APOBEC3G Restricts HIV-1 Infection in Resting CD4+ T Cells’, Nature, 435: 108–14. [DOI] [PubMed] [Google Scholar]
- Cladel N. M., Bertotto A., Christensen N. D. (2010). ‘Human Alpha and Beta Papillomaviruses Use Different Synonymous Codon Profiles’, Virus Genes, 40: 329–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B. R. (2006). ‘Role and Mechanism of Action of the APOBEC3 Family of Antiretroviral Resistance Factors’, Journal of Virology, 80: 1067–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D., Taboada G. L., Doallo R., Posada D. (2012). ‘jModelTest 2: More Models, New Heuristics and Parallel Computing’, Nature Methods, 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villiers E.- M., et al. 2004. ‘Classification of Papillomaviruses’, Virology, 324: 17–27. [DOI] [PubMed] [Google Scholar]
- Duggal N. K., Emerman M. (2012). ‘Evolutionary Conflicts Between Viruses and Restriction Factors Shape Immunity’, Nature Review of Immunology, 12: 687–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C. C., et al. (2005). ‘APOBEC3G Cytidine Deaminase Inhibits Retrotransposition of Endogenous Retroviruses’, Nature, 433: 430–3. [DOI] [PubMed] [Google Scholar]
- Félez-Sánchez M., et al. (2015). ‘Cancer, Warts, or Asymptomatic Infections: Clinical Presentation Matches Codon Usage Preferences in Human Papillomaviruses’, Genome Biological Evolution, 7: 2117–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J. I., Stivers J. T. (2010). ‘Detection of Damaged DNA Bases by DNA Glycosylase Enzymes’, Biochemistry, 49: 4957–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. S. (2013). ‘Cancer Mutation Signatures, DNA Damage Mechanisms, and Potential Clinical Implications’, Genome Medicine, 5: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. S., Liddament M. T. (2004). ‘Retroviral Restriction by APOBEC Proteins’, Nature Reviews Immunology, 4: 868–77. [DOI] [PubMed] [Google Scholar]
- Hasan U. A., et al. (2007). ‘TLR9 Expression and Function is Abolished by the Cervical Cancer-Associated Human Papillomavirus Type 16’, The Journal of Immunology, 178: 3186–97. [DOI] [PubMed] [Google Scholar]
- Henderson S., Chakravarthy A., Fenton T. (2014). ‘When Defense Turns into Attack’, Molecular and Cellular Oncology, 1: e29914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelzer K., Shackelton L. A., Parrish C. R. (2008). ‘Presence and Role of Cytosine Methylation in DNA Viruses of Animals’, Nucleic Acids Research, 36: 2825–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultquist J. F., et al. (2011). ‘Human and Rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H Demonstrate a Conserved Capacity to Restrict Vif-Deficient HIV-1’, Journal of Virology, 85: 11220–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmuz A., et al. (2002). ‘An Anthropoid-Specific Locus of Orphan C to U RNA-Editing Enzymes on Chromosome 22’, Genomics, 79: 285–96. [DOI] [PubMed] [Google Scholar]
- Jern P., Russell R. A., Pathak V. K., Coffin J. M. (2009). ‘Likely Role of APOBEC3G-Mediated G-to-A Mutations in HIV-1 Evolution and Drug Resistance’, PLoS Pathogens, 5: e1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W. E. (2013). ‘Rapid Adversarial Co-Evolution of Viruses and Cellular Restriction Factors’, Current Topics in Microbiology and Immunology, 371: 123–51. [DOI] [PubMed] [Google Scholar]
- Karlin S., Doerfler W., Cardon L. R. (1994). ‘Why is CpG Suppressed in the Genomes of Virtually all Small Eukaryotic Viruses but not in those of Large Eukaryotic Viruses?’ Journal of Virology, 68: 2889–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013). ‘MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability’, Molecular Biology and Evolution, 30: 772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. -Y., et al. (2014). ‘Human APOBEC3 Induced Mutation of Human Immunodeficiency Virus Type-1 Contributes to Adaptation and Evolution in Natural Infection’, PLoS Pathogens, 10: e1004281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey L., Law E. K., Brown W. L., Harris R. S. (2013). ‘Subcellular Localization of the APOBEC3 Proteins During Mitosis and Implications for Genomic DNA Deamination’, Cell Cycle, 12: 762–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRue R. S., et al. (2009). ‘Guidelines for Naming Nonprimate APOBEC3 Genes and Proteins’, Journal of Virology, 83: 494–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., et al. (2013). ‘RNA Editing of Hepatitis B Virus Transcripts by Activation-Induced Cytidine Deaminase’, Proceedings of the National Academy of Sciences, 110: 2246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddament M. T., Brown W. L., Schumacher A. J., Harris R. S. (2004). ‘APOBEC3F Properties and Hypermutation Preferences Indicate Activity against HIV-1 in vivo’, Current Biology, 14: 1385–91. [DOI] [PubMed] [Google Scholar]
- Lonsdale J., et al. (2013). ‘The Genotype-Tissue Expression (GTEx) Project’. Nature Genetics, 45: 580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). ‘Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2’, Genome Biology, 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M., Pfeiffer W., Schwartz T. (2010). ‘Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees.’ Gateway Computing Environments Workshop (GCE) 1–8. [Google Scholar]
- Minkah N., et al. (2014). ‘Host Restriction of Murine Gammaherpesvirus 68 Replication by Human APOBEC3 Cytidine Deaminases but not Murine APOBEC3’, Virology, 454–455: 215–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder L. C. F., Harari A., Simon V. (2008). ‘Cytidine Deamination Induced HIV-1 Drug Resistance’, Proceedings of the National Academy of Sciences, 105: 5501–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münk C., et al. (2008). ‘Functions, Structure, and Read-Through Alternative Splicing of Feline APOBEC3 Genes’, Genome Biology, 9: R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil S. J. D., Zang T., Bieniasz P. D. (2008). ‘Tetherin Inhibits Retrovirus Release and is Antagonized by HIV-1 Vpu’, Nature, 451: 425–30. [DOI] [PubMed] [Google Scholar]
- Nik-Zainal S., et al. ; Breast Cancer Working Group of the International Cancer Genome Consortium. (2012). ‘Mutational Processes Molding the Genomes of 21 Breast Cancers’, Cell, 149: 979–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobre R. J., et al. (2009). ‘E7 Oncoprotein of Novel Human Papillomavirus Type 108 Lacking the E6 Gene Induces Dysplasia in Organotypic Keratinocyte Cultures’, Journal of Virology’, 83: 2907–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander J. A. A., Wilgenbusch J. C., Warren D. L., Swofford D. L. (2008). ‘AWTY (Are We There Yet?): A System for Graphical Exploration of MCMC Convergence in Bayesian Phylogenetics’, Bioinformatics, 24: 581–3. [DOI] [PubMed] [Google Scholar]
- Ong C. K., et al. (1993). ‘Evolution of Human Papillomavirus Type 18: An Ancient Phylogenetic Root in Africa and Intratype Diversity Reflect Coevolution with Human Ethnic Groups’, Journal of Virology, 67: 6424–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms M., et al. (2012). ‘APOBEC3A, APOBEC3B, and APOBEC3H Haplotype 2 Restrict Human T-Lymphotropic Virus Type 1’, Journal of Virology, 86: 6097–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M. (1997). ‘Inferring Evolutionary Processes from Phylogenies’, Zoologica Scripta, 26: 331–48. [Google Scholar]
- Pagel M. (1999). ‘Inferring the Historical Patterns of Biological Evolution’, Nature, 401: 877–84. [DOI] [PubMed] [Google Scholar]
- Pham P., Bransteitter R., Petruska J., Goodman M. F. (2003). ‘Processive AID-Catalysed Cytosine Deamination on Single-Stranded DNA Simulates Somatic Hypermutation’, Nature, 424: 103–7. [DOI] [PubMed] [Google Scholar]
- Rector A., et al. (2007). ‘Ancient Papillomavirus-Host Co-Speciation in Felidae’, Genome Biology, 8: R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelton L. A., Parrish C. R., Holmes E. C. (2006). ‘Evolutionary Basis of Codon Usage and Nucleotide Composition Bias in Vertebrate DNA Viruses’, Journal of Molecular Evolution, 62: 551–63. [DOI] [PubMed] [Google Scholar]
- Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. (2002). ‘Isolation of a Human Gene that Inhibits HIV-1 Infection and is Suppressed by the Viral Vif Protein’, Nature 418: 646–50. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006). ‘RAxML-VI-HPC: Maximum Likelihood-Based Phylogenetic Analyses with Thousands of Taxa and Mixed Models’, Bioinformatics, 22: 2688–90. [DOI] [PubMed] [Google Scholar]
- Suspène R., et al. (2011). ‘Genetic Editing of Herpes Simplex Virus 1 and Epstein-Barr Herpesvirus Genomes by Human APOBEC3 Cytidine Deaminases in Culture and in vivo’ Journal of Virology, 85: 7594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suspène R., et al. (2011). ‘Genetic Editing of Herpes Simplex Virus 1 and Epstein-Barr Herpesvirus Genomes by Human APOBEC3 Cytidine Deaminases in Culture and in vivo’, Journal of Virology, 85: 7594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli P., et al. (2004). ‘Inhibition of Hepatitis B Virus Replication by APOBEC3G’, Science, 303: 1829. [DOI] [PubMed] [Google Scholar]
- Upadhyay M., et al. (2013). ‘CpG Dinucleotide Frequencies Reveal the Role of Host Methylation Capabilities in Parvovirus Evolution’, Journal of Virology, 87: 13816–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doorslaer K. (2013). ‘Evolution of the Papillomaviridae’, Virology, 445: 11–20. [DOI] [PubMed] [Google Scholar]
- Van Doorslaer K., et al. (2013). ‘The Papillomavirus Episteme: A Central Resource for Papillomavirus Sequence Data and Analysis’, Nucleic Acids Research, 41: D571–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian J. -P., Guétard D., Henry M., Wain-Hobson S. (2008). ‘Evidence for Editing of Human Papillomavirus DNA by APOBEC3 in Benign and Precancerous Lesions’, Science, 320: 230–3. [DOI] [PubMed] [Google Scholar]
- Wang Z, et al. (2014). ‘APOBEC3 Deaminases Induce Hypermutation in Human Papillomavirus 16 DNA Upon Beta Interferon Stimulation’, Journal of Virology, 88: 1308–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren C. J., et al. (2015). ‘APOBEC3A Functions as a Restriction Factor of Human Papillomavirus’, Journal of Virology, 89: 688–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood N., et al. (2009). ‘HIV Evolution in Early Infection: Selection Pressures, Patterns of Insertion and Deletion, and the Impact of APOBEC’, PLoS Pathogens, 5: e1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., et al. (2007). ‘Virion-Associated Uracil DNA Glycosylase-2 and Apurinic/Apyrimidinic Endonuclease are Involved in the Degradation of APOBEC3G-Edited Nascent HIV-1 DNA’, Journal of Biological Chemistry, 282: 11667–75. [DOI] [PubMed] [Google Scholar]
- Yu Q., et al. (2004). ‘APOBEC3B and APOBEC3C are Potent Inhibitors of Simian Immunodeficiency Virus Replication’, Journal of Biological Chemistry, 279: 53379–86. [DOI] [PubMed] [Google Scholar]
- Yu Q., et al. (2004). ‘Single-Strand Specificity of APOBEC3G Accounts for Minus-Strand Deamination of the HIV Genome’, Nature Structural and Molecular Biology, 11: 435–42. [DOI] [PubMed] [Google Scholar]
- Zhang H., et al. (2012). ‘EvolView, An Online Tool for Visualizing, Annotating and Managing Phylogenetic Trees’, Nucleic Acids Research 40: W569–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y. -H., et al. (2004). ‘Human APOBEC3F is Another Host Factor that Blocks Human Immunodeficiency Virus Type 1 Replication’, Journal of Virology, 78: 6073–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.