SUMMARY
During embryonic development, cells must establish fates, morphologies and behaviors in coordination with one another to form a functional body. A prevalent hypothesis for how this coordination is achieved is that each cell’s fate and behavior is determined by a defined mixture of RNAs. Only recently has it become possible to measure the full suite of transcripts in a single cell. Here we quantify genome-wide mRNA abundance in each cell of the C. elegans embryo up to the 16-cell stage. We describe spatially dynamic expression, quantify cell-specific differential activation of the zygotic genome, and identify genes that were previously unappreciated as being critical for development. We present an interactive data visualization tool that allows broad access to our dataset. This genome-wide single-cell map of mRNA abundance, alongside the well-studied life history and fate of each cell, describes at a cellular resolution the mRNA landscape that guides development.
eTOC BLURB
Tintori et al. performed single-cell RNA-seq on each cell of the C. elegans embryo through the 16-cell stage. Cells were identified by their transcriptomes post hoc. These data provide a Resource that reveals cell-specific genome activation and genes critical for development. An interactive data visualization allow exploration of the resulting dataset.
INTRODUCTION
An outstanding challenge of developmental biology is to explain how differential gene expression promotes the fundamental processes of embryonic development. Such processes include determining the fate of each cell, moving cells relative to each other to produce structures such as organs, and changing the composition and shape of each cell to perform metabolic or structural functions. Genomic approaches developed over the past decade have made it possible to generate comprehensive rosters of every transcript’s abundance in an organism or tissue during key developmental events. In this study, we have measured the mRNA abundances, genome-wide, in each cell of the early C. elegans embryo. In doing so, we have quantified the divergence of the genetic expression of these cells as they begin to perform diverse functions in the embryo.
The C. elegans embryo is a powerful and well-established system for studying cell biology and development (Figure 1A), and was chosen as a model organism in part because the entirety of development can be tracked with single-cell resolution (Sulston et al. 1983). The timing and orientation of every cell division, apoptotic event, and cell migration has been documented, and the exact lineal relationship of any cell to any other is known. Yet performing genomic studies with a matching resolution has been a challenge. Until recently, genomic protocols required collection of embryos in bulk, but C. elegans fertilization is staggered, rendering embryos asynchronous with each other. There is no practical system in place for culturing single cell types, leaving the only source of bulk biological material imprecisely staged samples that are usually composed of mixed cell types. Low-input RNA-sequencing (RNA-seq) methods developed within the last five years offer a solution to the genomics problem; a single C. elegans cell can be precisely identified and defined both in space and time.
Figure 1. Single-cell mRNA-seq libraries for complete sets of cells from C. elegans embryos of the 1-, 2-, 4-, 8- and 16-cell stages.
(A) Terminal cell fates of descendants of each cell of the 16-cell embryo. Terminal fates were calculated from Sulston et al. 1983, and refer to cell fates at the time of the first larval hatching.
(B) Schematic of samples that were hand-dissected and prepared for scRNA-seq. The 4-cell stage is diagrammed below for illustration.
(C) The total mass of mRNA detected from each embryo (diamonds). Embryos whose total mass of mRNA differed from the average by more than one standard deviation (plotted outside of gray band) were excluded from subsequent analyses.
(D) The number of genes whose transcripts were detected in each whole embryo (diamonds).
(E) The number of genes whose transcripts were detected in each individual cell (circle).
(F) Key of the names of each cell from the zygote to the 16-cell stage.
See also Table S1
Understanding the full suite of mRNAs expressed in the C. elegans embryo has long been of interest. Whole-embryo mRNA timecourses revealed that thousands of genes are dynamically regulated at these early stages (Baugh et al. 2003; Baugh et al. 2005). Aided by advances in low-input RNA-seq technology of the last few years, researchers have interrogated the transcriptomes of the embryo by manually dissecting cells and performing RNA-seq. Due to the difficulty of identifying cells once they are dissected, only the 2-cell stage embryo has been sequenced at an entirely single-cell resolution (Hashimshony et al. 2012; Hashimshony et al. 2015; Osborne Nishimura et al. 2015). One study has performed transcript profiling of some single cells and some clusters of cells from later stages (Hashimshony et al. 2015). In this study we have sequenced each cell of an individual embryo in replicate for embryos up to the 16-cell stage. We hand-dissected complete sets of single cells from each embryo, and developed a unique strategy for identifying the dissected cells.
Many of the interesting phenomena of early development are transcriptionally regulated in C. elegans, including morphogenesis and cell fate specification (Edgar et al. 1994; Sommermann et al. 2010; Broitman-Maduro et al. 2006). Much of what we know about the genetics of these events has been gleaned from traditional genetic screens, which have a blind spot for pleiotropic genes and genes with partially redundant functions (Wieschaus 1997; Sawyer et al. 2011). With high-throughput sequencing, we can identify the genes whose transcript abundances correlate with morphogenesis, differentiation, or other phenomena, regardless of challenges such as pleiotropy or redundancy.
Here we present a transcriptional lineage of early C. elegans development – a map of genome-wide transcript abundance in each cell through the first stages of development. We generated this map by performing single cell RNA-seq (scRNA-seq) on each cell from the zygote to the 16-cell stage. We address previously unanswered questions about the differential activation of the zygotic genome in each cell, describe spatially dynamic gene expression, and identify previously unknown genes that are critical for development. Finally, we introduce a publicly available interactive data visualization tool that we developed to maximize the usefulness of our dataset to the scientific community.
RESULTS
Transcriptome Diversity Among Cells of the Embryo Increases Over Time
Each cell at each stage in the early C. elegans embryo has a name, a known life history and fate, and is identifiable by its position relative to other cells (Sulston et al. 1983). We performed scRNA-seq on manually-dissected, individual cells from 1-, 2-, 4-, 8- and 16-cell stage embryos, with a minimum of 5 replicates for each sample (Figure 1B). We note that due to asynchronous cell divisions there is no true 16-cell stage, but we use this term for convenience (details in Experimental Methods). We sequenced the mRNA of each cell separately, knowing which embryo the cell came from but not knowing its identity. We then used its transcript profile to identify its cell type post hoc. Cell size and cell division timing gave us some clues of the identities of 19 of the 31 cell types. For example, all the anterior (AB descendant) cells at the 8-and 16-cell stages divide in synchrony with each other, and the germ cell precursors at the 2- to 8-cell stages are considerably smaller than the rest of the cells (purple in Figure S1, and Extended Methods). These visual clues provided independent support for the results of our post hoc cell identity assignments (Figure S1 and described below).
In total, we generated 219 transcriptomes, describing quantitative expression levels for 8,575 detected genes (>25 RPKM). We aggregated data from cells of the same embryo to calculate whole-embryo statistics. To calculate the mass of mRNA in each cell, and thereby each whole embryo, we used spike-in controls from the External RNA Control Consortium (Baker et al. 2005). The mass of mRNA detected was relatively constant between stages, though embryos of later timepoints showed higher variability (Figure 1C). Among 31 whole embryos, five embryos had an mRNA mass more than one standard deviation above or below the average and were excluded from further analysis (details in Table S1). To evaluate changes in transcriptome complexity over time in individual cells and in whole-embryos, we calculated the number of mRNA species detected in each single-cell transcriptome and whole-embryo aggregation (Figure 1D,E). We noticed an increase in transcriptome complexity in whole embryos over time (>25 RPKM in any contributing cell), but a decrease in complexity in individual cells. The increase in whole-embryo complexity could be due to either cell-specific activation of the zygotic genome, or to the fact that a larger number of single-cell libraries constitute the whole embryo total at later stages, potentially allowing for fewer false negatives when compared to the small number of transcriptomes that make up whole-embryo values at earlier stages.
Before we could test the validity of the transcriptomes generated, we first needed to identify the cell type of origin for each transcriptome.
Posterior Cells of the Embryo Have Distinct Signatures Involving Hundreds of Cell-Specific Transcripts
Many of the cell types we sampled are enriched for transcripts of one or a few known marker genes, which we were able to use to assign identities to our transcriptomes. A multi-gene clustering approach has been shown to be more effective at grouping replicates of a cell type than a single- or few-gene approach (Björklund et al. 2016; Jaitin et al. 2014; Grün et al. 2015; Satija et al. 2015). We used an iterative Principle Component Analysis (PCA) strategy (described below) to group transcriptomes by cell type, thereby collapsing our 219 transcriptomes down to 18 groups of identical or related cell types. We then used known marker genes to assign identities to each of these 18 groups (Figure 2). The 18 groups were P0, AB, P1, ABa, ABp, EMS, P2, ABxx (granddaughters of AB), MS, E, C, P3, ABxxx (great granddaughters of AB), MSx (daughters of MS), Ex (daughters of E), Cx (daughters of C), D and P4. Some of the groups that contained multiple cell types were later sorted into more specific groups (Figure 3).
Figure 2. Replicates of each cell type were grouped by transcript signatures and identified by candidate gene expression.
(A–C) Transcriptomes of cells from the 2-cell stage (A) were subjected to Principle Component Analysis (PCA) (B) using only data from reproducibly differentially enriched genes, as selected by our algorithm (details in Experimental Procedures). (C) Genome browser tracks of the last exon of erm-1 (AB-enriched) and cpg-2 (P1-enriched). Colors correspond to embryo of origin. Heights of tracks indicate read count density. All y-axes of genome browser tracks are scaled consistently within each panel.
(D–G) Transcriptomes of cells from the 4-cell stage (D) were subjected to PCA (E). (F) PCA of the 10 transcriptomes that were not resolved in (E). (G) Genome browser tracks of the last exon of erm-1 (AB-enriched), med-2 (EMS-enriched) and cpg-2 (P2-enriched).
(H–L) Transcriptomes of cells from the 8-cell stage (H) were subjected to PCA using iteratively generated sets of informative genes (I–K). (L) Genome browser tracks of the last exon of W02F12.3 (ABxx-enriched), tbx-35 (MS-enriched), end-3 (E-enriched), mex-5 (C- and P3-enriched), and cpg-2 (P3-enriched).
(M–Q) Transcriptomes of cells from the 16-cell stage (M) were subjected to PCA using iteratively generated sets of informative genes (N–P). (Q) Genome browser tracks of the last exon of T09B4.1 (ABxxx-specific), ceh-51 (MSx-specific), end-1 (Ex-specific), pal-1 (Cx- and P4-specific), and cpg-2 (P4-specific). See Figures S1 and S2 for further identification of D and P4 transcriptomes. See also Figure S1, S2
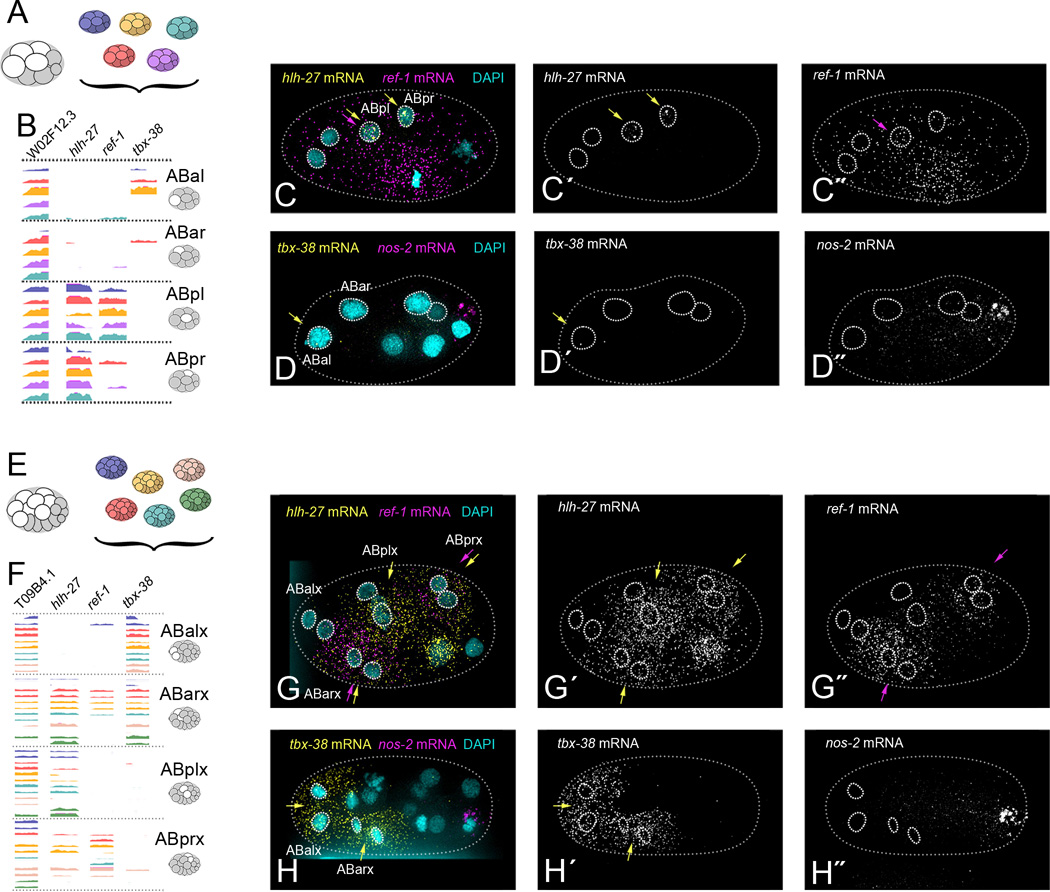
Figure 3. Differential transcript enrichment of notch target genes in cells that could not be distinguished by global transcript signatures.
(A) AB descendants from five replicates of the 8-cell stage embryo.
(B) Genome browser tracks of ABxx transcriptomes, sorted into groups based on expression of notch target genes hlh-27, ref-1 and tbx-38 (Extended Methods). Last exons only are shown.
(C) Example of smFISH targeting hlh-27 (C´, yellow arrows) and ref-1 (C′, purple arrows) transcripts in intact 6- or 8-cell stage embryos (hlh-27 pattern seen in 100% of embryos, n=4. ref-1 pattern seen in 75% of embryos, n=4. Remaining embryo showed ubiquitous ref-1 staining).
(D) Example of smFISH targeting tbx-38 (D´, yellow arrows) in intact 8-cell stage embryos (pattern seen in 33% of embryos, n=3. 67% of embryos showed equal tbx-38 expression in ABal and ABar). (D′) nos-2 (P3-specific) marks the posterior of the embryo.
(E) AB descendants from six replicates of the 16-cell stage embryo.
(F) Genome browser tracks of ABxxx transcriptomes, sorted into four groups based on a PCA using only notch target gene expression (shown in Figure S2D). Last exons only are shown.
(G) Example of smFISH targeting hlh-27 (G´, yellow arrows) and ref-1 (G′, purple arrows) transcripts in intact 15-cell stage embryos (both patterns seen in 100% of embryos, hlh-27 n=5, ref-1 n=2).
(H) Example of smFISH targeting tbx-38 (H´, yellow arrows) in intact 15-cell stage embryos (pattern seen in 100% of embryos, n=14). nos-2 (P4-specific) marks the posterior of the embryo. See Figures S1 and S2 for further identification of ABx, ABxx, and ABxxx transcriptomes.
See also Figure S1, S2
To filter for informative genes to use in our PCAs, we designed an algorithm to select genes that are reproducibly differentially enriched between cells of the embryo (details in Experimental Procedures). To group replicates of each cell type together, we performed a PCA on all transcriptomes of a given stage using just those filtered genes. We inspected plots of the first and second principle components for distinct groups consisting of one transcriptome from each embryo, which suggest grouping by shared cell-specific features (Figure 2B,E,I,N). We interpreted a group with exactly one cell from each embryo as comprising the replicates of a single (albeit unknown) cell type.
Each PCA tended to isolate only the most dramatically distinct cell types (Figure 2E,I,N). To then identify cell types with more subtle distinguishing features, we removed the transcriptomes that had already clustered out into independent groups, and re-ran the gene selection algorithm and PCA with just the remaining cells. In this way, we continuously enhanced our resolution and split groups of cells off based on increasingly subtle differences (Figure 2F,J,K,O,P; arrows show the cluster of remaining transcriptomes that were put through the next PCA iteration). We chose this iterative PCA approach because it allowed us to take advantage of a unique feature of the C. elegans embryo: Each embryo sampled from a given stage generated an identical number of transcriptomes, representing exactly the same set of cell types. Many transcriptome clustering methods define clusters of unspecified size (Yan et al. 2013; Jaitin et al. 2014; Grün et al. 2015; Zeisel et al. 2015), but for this experiment it was most informative to identify groups consisting of exactly one transcriptome from each embryo (see Discussion). The simplest way to achieve this was to inspect the results of a PCA plot for isolated groups of transcriptomes that consisted of one transcriptome from each replicate (Figure 2).
The cells of the 2-cell stage embryo (AB and P1) have noticeably different sizes, which allowed us to identify these cells during sample collection. We were able to use this previous knowledge to test the accuracy of our gene selection algorithm and PCA approach. We found that our strategy did in fact allow us to independently and accurately distinguish between these two cell types; all AB cells fell on one side of the first principle component, while all P1 cells fell on the other side of the principle component (Figure 2B). The germ cell precursors in subsequent stages (P2 at the 4-cell stage and P3 at the 8-cell stage) were noticeably smaller than the others and so were also identified upon collection. These cells successfully segregated from the other cells types by our algorithm and PCA (Figure 2E,I). The independent identification of these cells as replicates of each other further validated our algorithm as an effective unsupervised method for selecting informative genes.
To assign identities to groups of cells distinguished by PCA, we examined genes that are known to be expressed in specific cell types. For example, med-2 is known to be expressed in EMS at the four-cell stage (Maduro et al. 2001). Our transcriptome data shows high med-2 levels exclusively and robustly in one distinct group of replicates at the four-cell stage (Figure 2G). Based on these observations, we concluded that this cluster consists of the EMS transcriptomes. Similarly, using known markers of cell identity, we verified AB and P1 cells at the 2-cell stage, AB daughter cells (ABa and ABp, referred to collectively here as ABx) and P2 cells at the 4-cell stage, MS, E, C, P3 and AB granddaughter cells (ABxx) at the 8-cell stage, and MS daughters (MSx), E daughters (Ex), C daughters (Cx), D, P4, and AB great-granddaughters (ABxxx) at the 16-cell stage (Figure 2, Extended Methods).
Anterior Cells of the Embryo were Indistinguishable from Each Other by an Unsupervised Multi-Gene Approach, but Show Differential Enrichment of Notch Target Gene mRNAs
For both the 8-cell and 16-cell stages, our PCA approach did not visibly distinguish the descendants of AB from each other (Figure 2K). These results indicate that the transcriptomes of AB descendants at these stages were very similar to each other. This is consistent with the fact that very few genes are known to be differentially expressed between these cells (Priess 2005). To distinguish between these transcriptomes, we examined them for transcripts of a few genes whose proteins are known to be differentially expressed between these cells, namely members of the notch signaling pathway, hlh-27, ref-1 and tbx-38 (Neves & Priess 2005). We queried all transcriptomes of the AB descendants at the 8- or 16-cell stages for transcripts of these three genes, and found that they offered enough information to partition these transcriptomes into four cell types at the 8-cell stage and four pairs at the 16-cell stage (Figure 3B,F, Extended Methods).
To match each hand-sorted group of transcriptomes to a specific cell identity, we performed single molecule FISH (smFISH) on these notch targets in intact 8- and 16-cell embryos. We analyzed micrographs to determine which cell of the embryo expressed each of the distinct combinations of notch targets seen in our data. At the 8-cell stage hlh-27 transcripts were the most highly enriched in the ABpl and ABpr cells, ref-1 transcripts were enriched in ABpl cells, and tbx-38 transcripts were detected at very low levels primarily in ABal (Figure 3C,D). At the 16-cell stage hlh-27 was enriched in all AB descendants except the ABalx (ABala and ABalp) cells, ref-1 was detected in ABarx and ABprx cells, and tbx-38 was detected in ABalx and ABarx cells (Figure 3G,H). This smFISH data in combination with the scRNA-seq data for these notch targets allowed us to sort and identify transcriptomes into the four cell types at the 8-cell stage (ABal, ABar, ABpl and ABpr), and into four pairs of cell types at the 16-cell stage (ABalx, ABarx, ABplx and ABprx; Extended Methods).
The notch targets mentioned above are critical for cell fate specification of these anterior cells, and are activated via signaling from neighboring cells (Priess 2005). A previous study that sequenced all AB descendants together after allowing them to grow outside of their native embryonic environment showed no hlh-27 expression in these cells, suggesting that key fate-determining signaling events may have been prevented (Hashimshony et al. 2015). This indicates that processing cells around 10 minutes after dissection, as we did, produces results that more accurately reflect the biology of intact embryos.
Together, our data reveal that the transcriptomes of AB descendants are almost indistinguishable from one another except for transcripts of a few genes, whereas P1 descendants show hundreds of differences from one another. Some pairs of cells were ultimately indistinguishable from each other by our method, but because each cell was sequenced independently, this decreased resolution reflects the biology of the cells.
The Transcriptional Lineage Expands Upon Known Gene Expression Patterns During Development and Increases Their Resolution
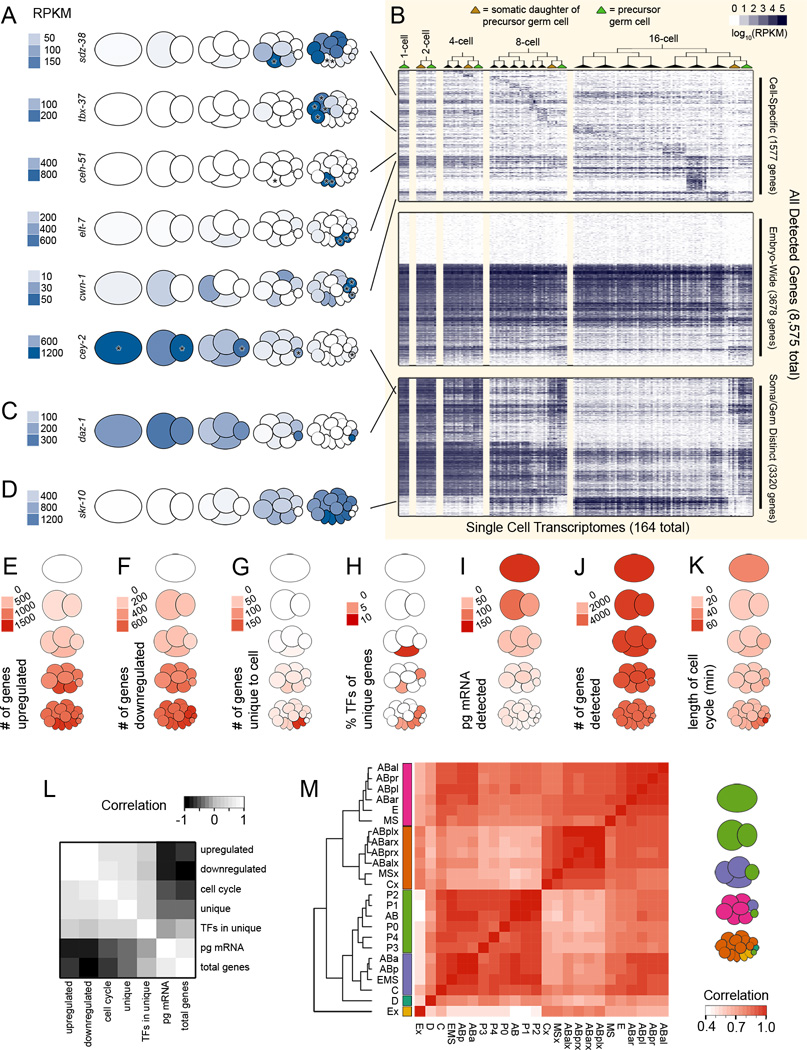
Having assigned cell identities to each transcriptome in our dataset, we first confirmed that the data and our identity assignments reflected certain known expression patterns. We queried our dataset for expression patterns of sdz-38 (which encodes a putative zinc finger protein that is expressed in the MS cell; Robertson et al. 2004), tbx-37 (a T-box transcription factor found in ABa descendants; Neves & Priess 2005), ceh-51 (a homeodomain protein expressed in the MS lineage; Broitman-Maduro et al. 2009), elt-7 (a GATA-type transcription factor that induces gut specification in the E descendants; Sommermann et al. 2010), cwn-1 (a wnt ligand expressed in the C and D cells; Gleason et al. 2006), and cey-2 (a putative RNA binding factor restricted to the germ line; Seydoux & Fire 1994). None of these genes were used to previously identify each cell type (Figures 2 and 3), so were able to use their expression patterns to independently test the validity of our data and our cell assignments. Our scRNA-seq data reflect the expected patterns for all six of these genes (Figure 4A, key in Figure 1F), and additionally quantify their expression in each cell, as well as that of the other 8,569 detected genes (Figure 4B).
Figure 4. Differential activation of the zygotic genome in each cell lineage.
(A) Transcript abundances of six genes with previously known expression patterns, heat-mapped on to pictograms of the embryo (key in Figure 1F). Asterisks indicate the cells in which we expected expression, based on the literature; sdz-38 expected in E, Ex (Ea and Ep); tbx-37 expected in ABalx (ABala and ABalp), ABarx (ABara and ABarp); ceh-51 expected in MS, MSx (MSa and MSp); elt-7 expected in Ex (Ea and Ep); cwn-1 expected in Cx (Ca and Cp), D; cey-2 expected in P0, P1, P2, P3, P4 (references in Main Text).
(B) Heatmap of transcript abundances of all 8,575 detected genes (y-axis) in each cell throughout time and space (x-axis). Only transcriptomes that passed quality filtration were plotted (164 out of 219). The y-axis along the top third of the heatmap is scaled twice as large as the bottom two thirds, to show detail. See Figure S3 for comparisons to related previously published datasets.
(C) Transcript abundance data for daz-1 (a maternally inherited gene required for meiosis; Karashima et al. 2000), an example of a transcript we detected in only the germ cells and their sister cells.
(D) Transcript abundance data for skr-10 (a member of the ubiquitin ligase complex; Yamanaka et al. 2002), an example of a transcript we detected in only somatic cells.
(E) The number of upregulated genes for each cell type. Genes were scored as upregulated in a cell if their transcripts were at least twice as abundant as in any ancestors of that cell.
(F) The number of downregulated genes for each cell type. Genes were scores as downregulated in a cell if their transcript abundances were half or less that of an ancestor.
(G) The number of cell-specific, or unique, genes. Genes were scored as unique to a cell type if their transcript abundance was at least 10 times higher than in any other cell type in the dataset.
(H) Percentage of each cell type’s unique genes, as defined in (G), that are transcription factors.
(I) Mass of mRNA per cell as calculated using concentrations of control mRNA spike-ins.
(J) Number of genes detected above 25 RPKM in each cell.
(K) Length of cell cycle for each cell.
(L) Pearson correlation of E-K across all cell types (excluding germ cell precursors, which are transcriptionally distinct; Schaner & Kelly 2006).
(M) Matrix of the correlation coefficients of all cell types’ transcriptomes. Six branches of highly correlated cell types are color coded in the cartoon to the right.
See also Figure S3
Low-input transcriptomes for some of these cells, including AB and P1, have previously been generated (Hashimshony et al. 2012; Osborne Nishimura et al. 2015; Hashimshony et al. 2015), as have whole-embryo microarray timecourses of C. elegans development (Baugh et al. 2003; Baugh et al. 2005). We compared our scRNA-seq data to data from two previous studies (Hashimshony et al. 2015, Osborne Nishimura et al. 2015) that each used different methods to sequence mRNA from AB and P1 cells (2-cell stage). We calculated enrichment index values for each gene (a product of the gene’s AB/P1 fold change and the gene’s average expression, Experimental Procedures). To measure the agreement between each study, we compared the enrichment index values calculated from each study’s data. All studies were positively correlated with one another to similar extents, and the correlation increased when only significantly differentially enriched genes were compared (Figure S3A).
We analyzed data from the 2005 whole-embryo microarray timecourse (Baugh et al. 2005) to test if our low-input transcriptomes reflect the patterns identified by a higher-input but lower-sensitivity experiment. We searched for genes whose detected transcript levels either increased or decreased by twofold over time in the microarray data and identified 1,935 and 2,164 genes respectively. Transcripts whose detected levels increased or decreased in our dataset included 91% and 97% of those showing this pattern in the earlier dataset. In addition, we identified 7,763 other transcripts whose detected levels increased or decreased over time, many of which had very low expression levels, presumably undetectable by microarray, and 1,053 for which there was no microarray probe in the previous experiment (Figure S3B). This result suggests that even though the transcriptomes we present here were generated from just picograms of mRNA, they capture the patterns described by a higher-input method, but with much greater sensitivity and resolution.
Because samples are not pooled during scRNA-seq, false negatives are likely common. It is difficult to distinguish false negatives (due to uncaptured RNA molecules) from true negatives (due to stochastic gene expression) in scRNA-seq data, but the fact that our dataset, when averaged by cell type, identified 91% and 97% of the genes Baugh et al. 2005 identified as increasing or decreasing in abundance suggests that our false negative rate is well below 10%.
Transcriptional Dichotomy Between Germ Cells and the Soma
Visualization of transcript levels for all 8,575 detected genes across all cell types revealed three broad trends of gene expression (Fig. 4B): First were transcripts only detected in subsets of cell types, suggesting cell-specific transcription (Fig 4B top). Second were transcripts detected at a relatively high abundance in the zygote, and at lower levels over time in an embryo-wide fashion, suggesting global mRNA degradation (Fig. 4B center). Third were transcripts that were detected as differentially abundant between somatic cells and germ cell precursors (Fig 4B bottom). Within this third group, in some cases transcripts became undetectable over time in the somatic cells but remained detectable in the germ cell and the immediate sister of the germ cell (as in daz-1, a gene required for oogenesis; Karashima et al. 2000, Figure 4C). In other cases, genes became detectable over time in the somatic cells, while remaining undetectable in germ cells and their sisters (as in skr-10, a core component of the ubiquitin-ligase complex; Yamanaka et al. 2002; Figure 4D). Transcriptional quiescence in the germline is a feature that many organisms share (Deshpande et al. 2004; Cheung & Rando 2013). Our dataset identifies thousands of transcripts affected by this phenomenon, and quantifies their abundances.
Differential Activation of the Zygotic Genome Among Cell Lineages
Fundamental events of embryonic development start earlier in the C. elegans embryo than in many other model organisms. Cell-fate determining steps begin as early as the 2-cell stage, and gastrulation begins at the 26-cell stage. Within the embryo, certain cells engage in these events earlier than others. For example, gastrulation begins earliest in the E descendants and follows later in other cells (Nance et al. 2005). By further example, at the 16-cell stage the P1 descendants (which we will refer to as posterior cells) include 4 cells that are already restricted to a single fate, while none of the AB descendants (which we will refer to as anterior cells) are as fully fate-restricted (Figure 1A). Based on this, we hypothesized that transcriptomes change more dramatically in the more fate-restricted posterior cells than the anterior cells. To quantify the extent to which transcriptomes of each lineage change over time, we asked how many genes were detected as having increased or decreased transcript levels in each cell when compared to the cell’s parent. We found a higher number of both increasing and decreasing detected transcript levels in the non-germ descendants of the P1 cell (Figure 4E,F) than in AB descendants, supporting our hypothesis that there is more dynamic gene regulation in these cells than in the AB descendants. We wondered whether this apparent increased dynamism of gene regulation (number of transcripts increasing or decreasing in abundance) in the non-germ posterior cells could be related to other features of these cells, such as greater mass of mRNA (Figure 4I), greater transcriptome complexity (Figure 4J), or longer cell cycle (Figure 4K; Wormbase 2007). By the 16-cell stage, the total mass of mRNA and the number of detected transcripts in each cell negatively correlated with the dynamism of gene regulation in the posterior cells (average R = −0.52 and −0.19) while the length of the cell cycle positively correlated with the dynamism of gene regulation (average R = 0.51, Figure 4L). This suggests to us that there are cellular features broadly associated with a cell lineage’s progression through the maternal to zygotic transition, including fewer total transcripts and a longer cell cycle.
To quantify the extent to which each cell’s transcriptome is unique, we evaluated the number of genes with transcripts detected exclusively in that cell and no others. Again we saw higher numbers of unique transcripts in the non-germ descendants of the P1 cell. The cell type with the highest number of uniquely expressed genes (176) was the Ex cells (Ea and Ep; Figure 4G). These cells have already established an endoderm-specific transcription program (Maduro 2010), and are minutes away from initiating gastrulation by moving from the outside of the embryo to the inside (Nance et al. 2005). These are also the first cells that have a gap phase in their cell cycles, taking 40 minutes to divide compared to ~20 minutes in the other cells of this stage (Edgar & McGhee 1988). Because many of the posterior cells become restricted to a single fate before the anterior cells do, we hypothesized that the posterior cells might express a greater number of cell-specific transcription factors. For each cell type, we calculated the percentage of that cell’s unique genes that were transcription factors. We found a larger proportion of mRNAs encoding transcription factors uniquely in the posterior cells (Figure 4H), suggesting that these cells are initiating lineage-specific transcriptional programs.
As a cell’s transcriptome becomes distinct from that of its neighbors, there are likely several processes involved, including differential transcription, degradation, and segregation of transcripts during cell division. While all of these processes contribute to the development of an embryo, it is likely that our dataset is most informative regarding transcriptional events. Because of the high false negative rate in scRNA-seq data, cell-specific detection of a transcript is more reliable than cell-specific absence. For this reason we focused most of our following analyses on increases in transcript abundance in specific cells, rather than decreases. Instances where transcripts of a gene are twice as enriched in a daughter cell compared to its parent could be due to transcription or differential enrichment, and our dataset cannot currently distinguish between the two.
The well-documented cell lineage of C. elegans tells us the exact lineal relationship between any pair of cells, uniquely allowing us to compare transcriptomes in both space and time. To analyze whether cell-specific features of the transcriptome were maintained over time, we generated a correlation matrix comparing the transcriptomes of all cell types to one another (Figure 4M). Cells of the 1- and 2-cell stage and all germ cell precursors clustered together with high correlation, indicating that germ cell specific features were common across stages. Otherwise, the strongest correlations were between cells of different lineages but a common stage, suggesting prominent stage-specific expression.
Genes with Spatially Dynamic Expression
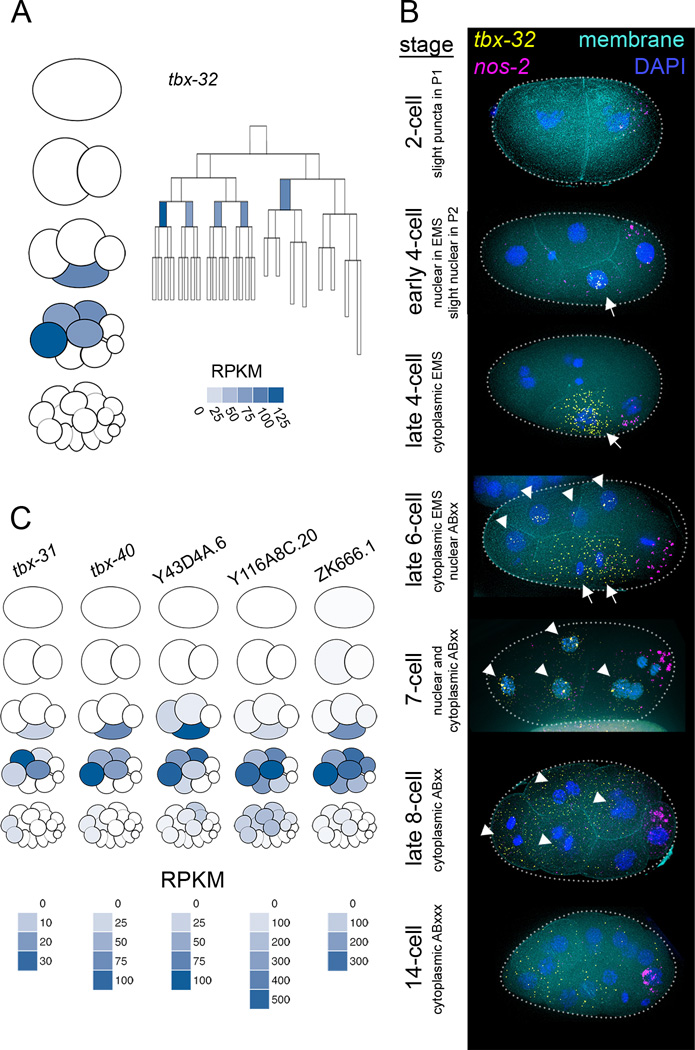
When a given transcript is detected across multiple temporal stages in an embryo, the most parsimonious explanation is that the transcript is inherited from parent cells to daughter cells lineally. While we expect some genes to contradict this assumption and be uniquely expressed in cells that are not related by lineage, such a scenario cannot be detected in a whole-embryo timecourse. The present dataset has a high enough resolution both temporally and spatially that we were able to identify transcripts whose overall expression is continuous throughout consecutive stages, but that are detected in different cell lineages throughout those stages. One such example is tbx-32 (Figure 5A), which was robustly detected in EMS at the 4-cell stage but absent in the daughters of EMS (E and MS) at the following stage. Instead tbx-32 transcripts appeared in anterior cells ABal, ABar, ABpl and ABpr (also referred to here as ABxx), that are not directly related to EMS by lineage.
Figure 5. Spatially dynamic gene expression is revealed by high resolution data.
(A) A cell lineage map and a pictogram of the 1- through 16-cell stages. Color corresponds to transcript abundance data for tbx-32 in each cell type.
(B) smFISH of tbx-32. 100% of 2-cell stage embryos (n=2), 83% of 4-cell stage embryos (n=6, one embryo showed ubiquitous staining), 100% of 6- to 8-cell stage embryos (n=6), and 100% of 12- to 15-cell stage embryos (n=3) showed this pattern.
(C) Pictograms for 5 genes showing transcript enrichment patterns similar to that of tbx-32.
To test the validity of this cross-lineage expression pattern, we performed smFISH on intact embryos. We detected tbx-32 transcripts in EMS at the 4-cell stage and in AB descendants at the 8-cell stage, as our RNA-seq data predicted (Figure 5B). tbx-32 transcripts were more abundant in the 16-cell stage by smFISH than we anticipated from our RNA-seq dataset, but partially degraded transcripts may be more detectable by smFISH (which recognizes many regions of the transcript) than by the RNA-seq method we used (which requires the presence of a polyadenylated tail for detection). This smFISH data allowed us to describe the tbx-32 expression pattern with an even higher temporal resolution than in the transcriptional lineage. The smFISH data revealed nuclear localization of tbx-32 transcripts early in the EMS and ABxx cell cycles, and cytoplasmic localization later in these cell cycles. This sequence of localizations suggests that the dynamic pattern of tbx-32 expression is due to zygotic transcription in these cells. We found five more genes (tbx-31, tbx-40, Y43D4A.6, Y116A8C.20, ZK666.1; Figure 5C) that have patterns similar to tbx-32, suggesting that a common mechanism may be regulating all of these genes.
scRNA-seq Data Reveals Synexpressed Sets of Paralogous Genes
The C. elegans genome is a snapshot of an evolving document. Continuous duplication and mutation events have produced a genome with many sets of paralogous genes in varying states of divergence. An estimated 32% of C. elegans genes have one or more paralogs (Woollard 2005), and we hypothesized that these sets of paralogs are more likely than a random pair of genes to be synexpressed (having transcripts whose expression patterns are highly correlated; Niehrs & Pollet 1999). To test this hypothesis we searched our data for groups of genes that were both synexpressed and similar to one another in sequence.
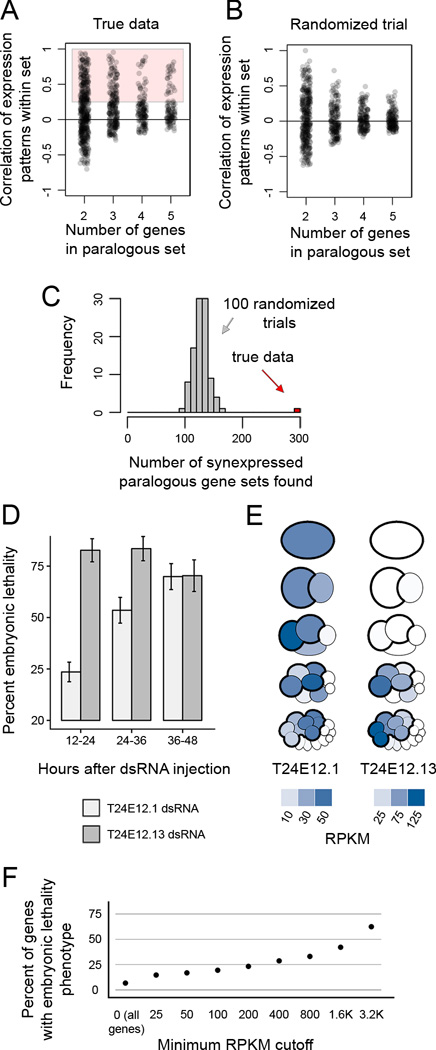
We found 295 sets of 2–5 genes that were synexpressed and paralogous (Extended Experimental Procedures; Figure 6A, Table S3). As a control, we scrambled the gene names in our dataset 100 times and repeated the analysis, finding on average 128 synexpressed paralogous gene sets in these permutations (Figure 6B,C). The 295 sets identified using unscrambled data consisted of 640 genes, of which only 126 have a known phenotype (19.7%; WormMine 2016).
Figure 6. Previously unappreciated paralogous, synexpressed genes are critical for development.
(A) Correlations of expression patterns for sets of 2–5 genes that are similar to each other in sequence. 295 sets of genes had a correlation coefficient greater than 0.25, and were considered paralogous and synexpressed.
(B) Gene set correlations in a scrambled dataset.
(C) Histogram of the number of synexpressed paralogous gene sets detected in our dataset (red bar) and in 100 datasets randomized by scrambling gene names without replacement (gray bars).
(D) Lethality phenotype observed in embryos in which T24E12.1 and T24E12.13 were targeted by co-injection of dsRNA. Error bars represent 95% confidence interval. See Figure S4 for embryonic lethality in single injections and other pairs of genes co-injected.
(E) Pictograms showing quantitative transcript abundance data for the genes highlighted in (D,E).
(F) Percent of genes targeted by RNAi in Kamath et al. 2003 that show embryonic lethality. Genes are filtered by their transcript abundance as detected in present study.
scRNA-seq Reveals Genes that are Required for Embryonic Development
To test whether our dataset could lead us to genes that are critical for development but have not yet been appreciated as such, we selected a small group of genes to target by dsRNA injection and test for embryonic lethality. dsRNA injection is more labor-intensive than feeding methods but generally results in more penetrant phenotypes (Ahringer 2006). We selected nine pairs of synexpressed paralogous genes out of the 295 sets identified in Figure 6C, and prepared dsRNAs to target each gene. We co-injected each pair into RNAi-hypersensitive rrf-3 mutant worms. Worms injected with dsRNA targeting one of the nine pairs (T24E12.1 and T24E12.13) produced offspring with 94% embryonic lethality (Figure S4). To test which of these genes was critical for development we injected dsRNA targeting each of them separately into N2 worms. High levels of embryonic lethality were observed in both conditions (82.7% for T24E12.1 and 70.3% for T24E12.13; Figure 6D), suggesting that both these genes are critical for development. Transcripts of these two genes are enriched in AB descendants at the 8- and 16-cell stages (Figure 6E). Their mRNA expression patterns were somewhat staggered, with T24E12.1 being more highly detected at earlier stages and in the posterior AB descendants, while T24E12.13 was detected at higher levels in the anterior AB descendants. Knocking down T24E12.13 by RNAi in previous studies resulted in no detectable phenotype (Kamath et al. 2003; Sönnichsen et al. 2005), while its paralog, T24E12.1, had never previously been tested (Wormbase 2015).
We hypothesized that genes detected in our dataset are more likely than a random set to be relevant to development. To test this, we compared our data to that from a genome-wide RNAi screen (Kamath et al. 2003). 6.7% of the total genes tested in the screen had an embryonic phenotype (Figure 6F). Among the genes tested in the screen which were also detected in our data above 25 RPKM, 14.7% had an embryonic phenotype. As the detected transcript abundance increased, the likelihood that the gene has an embryonic lethality phenotype increased. This suggests that our dataset may be informative in guiding researchers toward untested genes that are functionally relevant to development.
An Interactive Data Visualization Tool to Explore Our Gene Expression Data
To maximize the accessibility of our data, we developed an interactive data visualization tool (available in Chrome and Firefox browsers at http://tintori.bio.unc.edu). With this tool, the user can select which two cells or embryos they wish to compare, and generate a differential gene expression plot that highlights all of the transcripts enriched specifically in either sample (Figure 7). All detected transcripts are plotted by their fold change between any two selected samples, and their average expression level. These metrics were chosen because they are less abstracted than p-value and therefore more intuitive, but the user can also filter the data by adjusted p-value using the slider next to the plot.
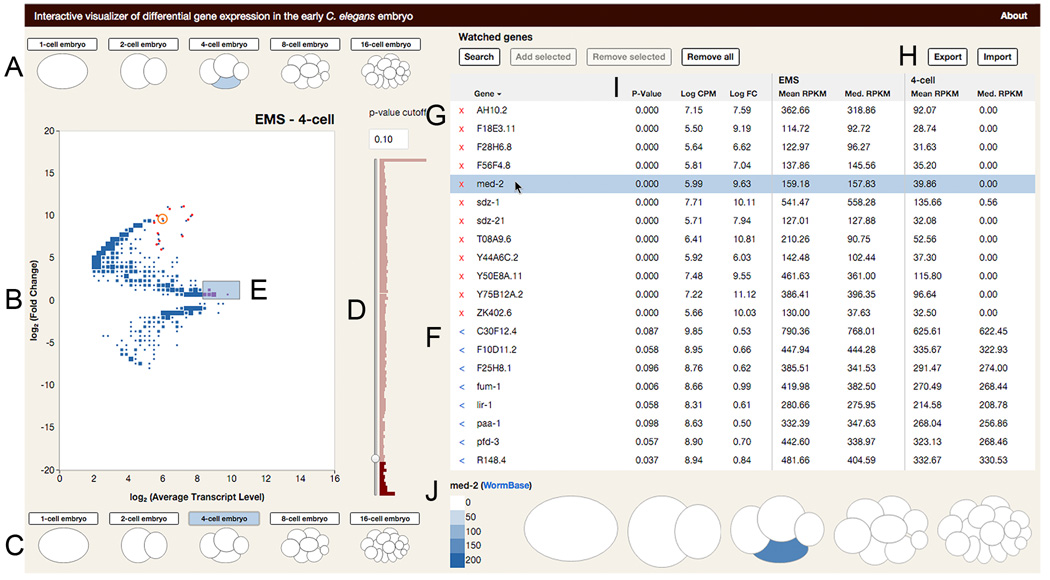
Figure 7. An interactive data visualization tool for querying the transcriptional lineage.
Still image of data visualization tool. Full version available in Chrome and Firefox browsers at http://tintori.bio.unc.edu.
(A–C) Sample selection. The user clicks on the cells or whole embryos they wish to compare on the top (A) and bottom (C) of the plot. When a new sample is selected, the plot (B) is redrawn to reflect the selected comparison. Size of points in B scales to the number of genes represented by each dot.
(D–H) Gene selection. (D) The user can filter genes by adjusted P-value of differential enrichment between samples. (E) Clicking on a point or selecting a swath of points on the plot adds genes and their data to the Selected Genes table (F). Known genes can be added directly, by typing their names into the search bar. (G) The Watched Genes table is curated by adding Selected Genes individually or in bulk. (H) The Watched Genes table can be exported, and lists of genes can be imported to the Watched Genes table in bulk.
(I–J) Gene expression metrics. (I) The gene tables are sortable by name, average expression level, fold change, significance of differential enrichment, and expression levels in either sample being compared. (J) Clicking on a gene in the table reveals a cartoon of the embryo over all five stages. Each cell is colored corresponding to the transcript level of the highlighted gene.
The interactive tool allows hypothesis-driven analyses (in which the user can query known genes of interest) as well as exploratory analyses (in which the user can discover new genes of interest). Our scRNA-seq data may be used to explore many fundamental aspects of development, such as specification of distinct cell types such as muscle or intestine, and cell behaviors such as cell cycle control or morphogenesis. We hope our visualization tool will invite researchers working on these and other topics to explore our dataset.
DISCUSSION
A transcriptional lineage to complement the completely defined cell lineage of the C. elegans embryo
For decades the C. elegans embryo has been a powerful tool for studying cell biology and development, largely because of its invariant cell lineage (Sulston et al. 1983). Here we present a transcriptional lineage that, when paired with the cell lineage, describes a genome-wide suite of transcripts present in early embryonic cells. Because all cells sampled have a precisely known relationships to each other, this dataset allows a comparison of transcriptomes in space and time, as these cells progressively diverge in fate, morphology, and behavior. As technology improves, scRNA-seq of cells beyond the 16-cell stage will become feasible, ideally allowing the possibility of a transcriptional map for every cell at every stage of development. The challenge of post hoc cell identification, explored in a previous study (Hashimshony et al. 2012) and in this manuscript, will continue to be relevant at these later stages of development.
Several research groups have previously performed scRNA-seq on human and mouse cells, and identified their cell types post hoc by the transcriptomes (Grün et al. 2015; Yan et al. 2013; Biase et al. 2014; Xue et al. 2013; Zeisel et al. 2015; Jaitin et al. 2014; Trapnell et al. 2014; Satija et al. 2015; Achim et al. 2015; Pollen et al. 2014). The invariant development of C. elegans provides a constraint on the possible identities of each transcriptome. This advantage is not present in other systems and can help guide cell-type identification. For example, because each 4-cell embryo yields exactly 1 transcriptome each of exactly 4 cell types, we know that out of our total of 20 unidentified transcriptomes from this stage (4 cells x 5 replicates), exactly 5 of them are from P2 cells, 5 are from EMS cells, 5 are from ABa cells, and 5 are from ABp cells. Because we know that a cell from one embryo will have exactly one counterpart in each other embryo, we were able to plot Principle Component Analyses of all transcriptomes, as in Figure 2E,F, and look for clusters containing one cell from each embryo. Given the unique constraints of this system, such clustering suggests that all replicates of a single cell-type are grouping together. The fact that known cell-type markers show transcript enrichment patterns that are consistent with our replicate grouping indicates the accuracy of our gene filtration and iterative PCA approach (Figure 2C,G,L,Q).
Another difference between our study and previous scRNA-seq studies that identified cell types post hoc is that the C. elegans cells used in the present study divide about every 20 minutes, whereas the human and mouse cells of previous studies divide approximately every 24 hours. Given this comparatively short cell cycle, it is remarkable that the posterior C. elegans cells have such distinct transcript signatures, and by the same token perhaps not surprising that the anterior cells are difficult to distinguish.
Previous studies have described transcriptomes at these stages of development at a lower spatial resolution (Baugh et al. 2003; Baugh et al. 2005; Hashimshony et al. 2012; Hashimshony et al. 2015). By leaving embryos intact until immediately before sample collection, sequencing every individual cell at each stage, and using technology that captures full-length mRNAs, we have expanded upon these previous datasets. Our method preserves fate-determining cell signaling events, allows for comparisons between groups of cells that were never sequenced separately before (such as AB descendants) and allows for inquiry into cell-specific variation in transcript splicing.
Cells of the Early Embryo Can Be Identified by Their Transcriptomes Alone
We have assigned a cell identity to each transcriptome based on its transcript abundance data, cross-referenced to known expression patterns and in situ RNA hybridization. The transcriptomes of some cell types (particularly the P1 descendants) grouped together tightly, were clearly distinct from other cell types, and had identities confirmed by well-studied genes, making us confident in our assessments. For the anterior cell types whose transcriptomes were less distinct from one another, we have a lower confidence in our assignments (as in Figure 3D´). We consider this paucity of distinguishing features to be an interesting biological result, suggesting that it would make little difference if transcriptomes identities were mis-assigned between these cell-types. The current understanding of these cells’ developmental potential supports the notion that they should be difficult to distinguish from each other. For example, the sister cells ABa and ABp of the 4-cell stage are initially developmentally equivalent, and the differences between them are not established until after cytokinesis separates them (Priess & Thomson 1987). In the future, if features are identified that more clearly differentiate these cell types, our existing single-cell transcriptomes can be revisited with those features in mind.
Previous studies that have measured transcript abundance in cells of the early embryo have either measured whole-embryo transcript levels (Baugh et al. 2003; Baugh et al. 2005; Levin et al. 2012), or measured only parts of the embryo at a single-cell resolution and the rest of the embryo in clusters of related cells (Hashimshony et al. 2015). These clusters of cells were sampled by dissecting embryos starting at the 2-cell stage and allowing the isolated cells to divide in culture, then sequencing the group of descendants. This allowed descendants of founder cells to be harvested at later time points than in our study, but kept the cells naïve to critical signaling events that take place in intact embryos. With our dataset, by leaving all cells intact in the embryo until minutes before sampling, we captured single-cell transcriptomes while allowing the cell-cell signaling necessary for proper development to occur, and we detected the transcriptional results of this signaling (Figure 3B,F).
A Stark Contrast in mRNA Composition Between Germ Cell Precursors and Somatic Cells
One pattern that is apparent when comparing gene expression across all cell types (Figure 4B) is that there is a prevalent distinction between the mRNA composition of the somatic cells and the germ cells (including the somatic sister of each germ cell precursor). Previous studies, such as Seydoux & Fire 1994, have observed this contrast in transcript composition between the germ and soma. Their reliance on in situ hybridization necessarily restricted the number of such genes they were able to study (10 genes), whereas the present genome-wide study expands their findings to thousands of genes. Differences between “immortal” germ cells and “mortal” somatic cells have fascinated researchers for over a century (Weismann 1893; Boveri 1910; Schierenberg & Strome 1992; Lai & King 2013; Lehmann & Ephrussi 2007; Yamanaka 2007). The present dataset quantitatively identifies thousands of genes with differential transcript abundances between the germ and soma. Furthermore, the dataset includes before, during, and after snapshots of somatic descendants of germ cell precursors, in their transition from the germ-like profiles of their parent cell to the somatic profiles of their descendants. This provides a rich view of how a cell’s transcriptome changes as it transitions from a germ state to a somatic state over time.
Cross-lineage Expression Patterns Highlight Genes that May Share Mechanisms of Gene Regulation
tbx-32 and the five other genes with similar expression patterns are examples of genes whose expression is not continuous from parent to daughter cell, but rather appears in one cell type (EMS) at one stage, then in a different lineage of cells (ABxx) at the next stage. The EMS cell at the 4-cell stage and one of these ABxx cells (ABar) at the 8-cell stage have another feature in common, which is that both orient their mitotic spindles in response to Wnt signaling (Walston et al. 2004).The fact that this specific expression pattern is shared by several genes suggests that a common mechanism may be regulating all of these genes, possibly the previously characterized Wnt signaling. Alternatively, these six genes may play a role in establishing which cells are capable of responding to Wnt signaling.
Identifying Critical Regulators of Development
Testing a small subset of genes, we identified two that are critical for embryonic development (Figure 6D). This indicates that our dataset may be well-suited to highlight previously unappreciated key regulators. These two genes are similar in sequence, and have similar but slightly staggered transcript enrichment patterns (Figure 6E). The staggering of these two patterns may represent subfunctionalization after a gene duplication event. This observation suggests that by considering both homology and spatiotemporal transcript abundance, our dataset may reveal patterns about divergence in sequence and function after a gene duplication event.
Although we saw embryonic lethal phenotypes in only 2 of the 18 genes we tested by dsRNA injection, we expect that a higher proportion of the genes highlighted by our dataset are likely to be important, for example for embryonic functions not required for hatching, for postembryonic development, or for stress tolerance.
EXPERIMENTAL PROCEDURES
Worm husbandry and embryo dissections
All worms were grown at 20°C and dissected at room temperature (21–24°C). Single embryos were selected at 10–20 minutes before the desired stage and dissected based on Edgar & Goldstein 2012 (details in Extended Experimental Procedures).
RNA preparation, sequencing, and analysis
cDNA was generated using the SMARTer Ultra Low RNA Input for Illumina Sequencing Kit, and sequencing libraries were prepared using the Nextera XT kit, both according to manufacturers instructions. Sequences from this study are available at NCBI GEO GSE77944. Identical reads were collapsed before analysis (details in Extended Experimental Procedures). RPKM values for all genes in each sample available in Table S2. Differential expression analyses were conducted using edgeR (Robinson 2010).
Assigning Cell Identities to Each Transcriptome
Transcriptomes for P0, AB, P1, ABx, EMS, P2, ABax, ABpl, ABpr, MS, E, C, P3, ABxxx, MSx, Ex, Cx, D/P4 were identified as such as described in Figures 2 and 3. ABx transcriptomes were resolved into ABa and ABp based on similarities to the transcriptomes of their daughter cells. ABax transcriptomes were resolved into ABal and ABar based on transcript abundance of genes differentially expressed between tbx-38 positive and tbx-38 negative cells. ABxxx transcriptomes were resolved into ABalx, ABarx, ABplx and ABprx based on PCA using transcript abundance data for notch target genes. D/P4 transcriptomes were resolved to D and P4 as in Figure 2. Details in Extended Experimental Procedures.
Defining synexpressed, paralogous sets of genes
Genes were BLASTed against the C. elegans EST collection with an e-value threshold of 10−15. This cut-off was chosen based on end-1 and end-3, a known example of paralogous genes that overlap in function. e=10−15 was the most conservative cut off that resulted in end-1 and end-3 appearing in each others’ list of BLAST hits. We considered sets of genes to be synexpressed if their average correlation coefficient exceeded 0.25 (Figure 6A).
RNAi
RNAs were combined and diluted to a total of 1 ug/uL for each condition. 15–22 young adult worms were injected for each condition. rrf-3 mutant worms (PK1429) were used for experiments in Figure S4, and N2 worms were used for experiments in Figure 6D. Embryonic lethality was calculated as the percent of unhatched embryos remaining 24 hours after mothers were removed from the plate, out of the total unhatched and hatched progeny.
Single molecule fluorescent in situ hybridization
N2 (Figure 3) or LP306 worms (containing a GFP membrane marker, Figure 5) were grown at 20°C and embryos were prepared as in Shaffer et al. 2013 and Ji & van Oudenaarden 2012 (details in Extended Experimental Procedures).
Supplementary Material
HIGHLIGHTS.
-
-
RNA-seq on each cell of the early C. elegans embryo complements the known lineage
-
-
Cell-specific activation of the zygotic genome was defined for each cell
-
-
High resolution scRNA-seq data reveals complex gene expression patterns
-
-
An interactive online data visualization tool allows easy exploration of the dataset
Acknowledgments
The authors would like to thank Corbin Jones and the UNC High Throughput Sequencing Facility for their generosity during scRNA-seq protocol optimization, and Jay Zhang for sharing his expertise in preparing scRNA-seq libraries. BG and SCT were supported by NIH grant R01 GM083071. JDL, EON and SCT were supported by NIH grant R01 GM104050. SCT was supported by NIH grant F31 HD088128 and by a NSF GRFP award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Experiments were conceived of by SCT, JDL and BG.
smFISH was performed by EON, all other experiments and analyses were performed by SCT.
Data visualization tool conceived of by SCT and PG, and coded by PG.
Manuscript written by SCT.
The authors declare no conflicts of interest.
REFERENCES
- Achim K, Pettit J-B, Saraiva LR, Gavriouchkina D, Larsson T, Arendt D, Marioni JC. High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin. Nature Biotechnology. 2015;33(5):503–509. doi: 10.1038/nbt.3209. http://doi.org/10.1038/nbt.3209. [DOI] [PubMed] [Google Scholar]
- Ahringer J. Reverse Genetics. WormBook : the Online Review of C. Elegans Biology. 2006:1–6. http://doi.org/10.1895/wormbook.1.47.1. [Google Scholar]
- Baker SC, Bauer SR, Beyer RP, Brenton JD, Bromley B, Burrill J, et al. The External RNA Controls Consortium: a progress report. Nature Methods. 2005;2(10):731–734. doi: 10.1038/nmeth1005-731. http://doi.org/10.1038/nmeth1005-731. [DOI] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Claggett JM, Hill-Harfe K, Wen JC, Slonim DK, et al. The homeodomain protein PAL-1 specifies a lineage-specific regulatory network in the C. elegans embryo. Development (Cambridge, England) 2005;132(8):1843–1854. doi: 10.1242/dev.01782. http://doi.org/10.1242/dev.01782. [DOI] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Slonim DK, Brown EL, Hunter CP. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development (Cambridge, England) 2003;130(5):889–900. doi: 10.1242/dev.00302. [DOI] [PubMed] [Google Scholar]
- Biase FH, Cao X, Zhong S. Cell fate inclination within 2-cell and 4-cell mouse embryos revealed by single-cell RNA sequencing. Genome Research. 2014;24(11):1787–1796. doi: 10.1101/gr.177725.114. http://doi.org/10.1101/gr.177725.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björklund ÅK, Forkel M, Picelli S, Konya V, Theorell J, Friberg D, et al. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nature Immunology. 2016 doi: 10.1038/ni.3368. http://doi.org/10.1038/ni.3368. [DOI] [PubMed] [Google Scholar]
- Boveri T. Über die Teilung centrifugierter Eier von Ascaris megalocephala. Archiv Für Entwicklungsmechanik Der Organismen. 1910;30(2):101–125. http://doi.org/10.1007/BF02263806. [Google Scholar]
- Broitman-Maduro G, Lin KT-H, Hung WWK, Maduro MF. Specification of the C. elegans MS blastomere by the T-box factor TBX-35. Development (Cambridge, England) 2006;133(16):3097–3106. doi: 10.1242/dev.02475. http://doi.org/10.1242/dev.02475. [DOI] [PubMed] [Google Scholar]
- Broitman-Maduro G, Owraghi M, Hung WWK, Kuntz S, Sternberg PW, Maduro MF. The NK-2 class homeodomain factor CEH-51 and the T-box factor TBX-35 have overlapping function in C. elegans mesoderm development. Development (Cambridge, England) 2009;136(16):2735–2746. doi: 10.1242/dev.038307. http://doi.org/10.1242/dev.038307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung TH, Rando TA. Molecular regulation of stem cell quiescence. Nature Reviews. Molecular Cell Biology. 2013;14(6):329–340. doi: 10.1038/nrm3591. http://doi.org/10.1038/nrm3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G, Calhoun G, Schedl P. Overlapping mechanisms function to establish transcriptional quiescence in the embryonic Drosophila germline. Development (Cambridge, England) 2004;131(6):1247–1257. doi: 10.1242/dev.01004. http://doi.org/10.1242/dev.01004. [DOI] [PubMed] [Google Scholar]
- Edgar LG, Goldstein B. Culture and Manipulation of Embryonic Cells. Methods in Cell Biology. 2012;107:151–175. doi: 10.1016/B978-0-12-394620-1.00005-9. http://doi.org/10.1016/B978-0-12-394620-1.00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar LG, McGhee JD. DNA synthesis and the control of embryonic gene expression in C. elegans. Cell. 1988;53(4):589–599. doi: 10.1016/0092-8674(88)90575-2. [DOI] [PubMed] [Google Scholar]
- Edgar LG, Wolf N, Wood WB. Early transcription in Caenorhabditis elegans embryos. Development (Cambridge, England) 1994;120(2):443–451. doi: 10.1242/dev.120.2.443. [DOI] [PubMed] [Google Scholar]
- Gleason JE, Szyleyko EA, Eisenmann DM. Multiple redundant Wnt signaling components function in two processes during C. elegans vulval development. Developmental Biology. 2006;298(2):442–457. doi: 10.1016/j.ydbio.2006.06.050. http://doi.org/10.1016/j.ydbio.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Grün D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525(7568):251–255. doi: 10.1038/nature14966. http://doi.org/10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- Hashimshony T, Feder M, Levin M, Hall BK, Yanai I. Spatiotemporal transcriptomics reveals the evolutionary history of the endoderm germ layer. Nature. 2015;519(7542):219–222. doi: 10.1038/nature13996. http://doi.org/10.1038/nature13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Reports. 2012;2(3):666–673. doi: 10.1016/j.celrep.2012.08.003. http://doi.org/10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, et al. Massively Parallel Single-Cell RNA-Seq for Marker-Free Decomposition of Tissues into Cell Types. Science (New York, N.Y.) 2014;343(6172):776–779. doi: 10.1126/science.1247651. http://doi.org/10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji N, van Oudenaarden A. Single molecule fluorescent in situ hybridization (smFISH) of C. elegans worms and embryos. WormBook : the Online Review of C. Elegans Biology. 2012:1–16. doi: 10.1895/wormbook.1.153.1. http://doi.org/10.1895/wormbook.1.153.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421(6920):231–237. doi: 10.1038/nature01278. http://doi.org/10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Karashima T, Sugimoto A, Yamamoto M. Caenorhabditis elegans homologue of the human azoospermia factor DAZ is required for oogenesis but not for spermatogenesis. Development (Cambridge, England) 2000;127(5):1069–1079. doi: 10.1242/dev.127.5.1069. [DOI] [PubMed] [Google Scholar]
- Lai F, King ML. Repressive translational control in germ cells. Molecular Reproduction and Development. 2013;80(8):665–676. doi: 10.1002/mrd.22161. http://doi.org/10.1002/mrd.22161. [DOI] [PubMed] [Google Scholar]
- Lehmann R, Ephrussi A. Ciba Foundation Symposium 182 - Germline Development. Chichester, UK: John Wiley & Sons, Ltd; 2007. Germ Plasm Formation and Germ Cell Determination in Drosophila; pp. 282–304. http://doi.org/10.1002/9780470514573.ch16. [DOI] [PubMed] [Google Scholar]
- Levin M, Hashimshony T, Wagner F, Yanai I. Developmental Milestones Punctuate Gene Expression in the Caenorhabditis Embryo. Developmental Cell. 2012;22(5):1101–1108. doi: 10.1016/j.devcel.2012.04.004. http://doi.org/10.1016/j.devcel.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Maduro MF. Cell fate specification in the C. elegans embryo. Developmental Dynamics : an Official Publication of the American Association of Anatomists. 2010;239(5):1315–1329. doi: 10.1002/dvdy.22233. http://doi.org/10.1002/dvdy.22233. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Meneghini MD, Bowerman B, Broitman-Maduro G, Rothman JH. Restriction of mesendoderm to a single blastomere by the combined action of SKN-1 and a GSK-3beta homolog is mediated by MED-1 and −2 in C. elegans. Molecular Cell. 2001;7(3):475–485. doi: 10.1016/s1097-2765(01)00195-2. [DOI] [PubMed] [Google Scholar]
- Nance J, Lee J-Y, Goldstein B. Gastrulation in C. elegans. WormBook : the Online Review of C. Elegans Biology. 2005:1–13. doi: 10.1895/wormbook.1.23.1. http://doi.org/10.1895/wormbook.1.23.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves A, Priess JR. The REF-1 family of bHLH transcription factors pattern C. elegans embryos through Notch-dependent and Notch-independent pathways. Developmental Cell. 2005;8(6):867–879. doi: 10.1016/j.devcel.2005.03.012. http://doi.org/10.1016/j.devcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Niehrs C, Pollet N. Synexpression groups in eukaryotes. Nature. 1999;402(6761):483–487. doi: 10.1038/990025. http://doi.org/10.1038/990025. [DOI] [PubMed] [Google Scholar]
- Osborne Nishimura E, Zhang JC, Werts AD, Goldstein B, Lieb JD. Asymmetric transcript discovery by RNA-seq in C. elegans blastomeres identifies neg-1, a gene important for anterior morphogenesis. PLoS Genetics. 2015;11(4):e1005117. doi: 10.1371/journal.pgen.1005117. http://doi.org/10.1371/journal.pgen.1005117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Shuga J, Wang X, Leyrat AA, Lui JH, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nature Biotechnology. 2014;32(10):1053–1058. doi: 10.1038/nbt.2967. http://doi.org/10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess JR. Notch signaling in the C. elegans embryo. WormBook : the Online Review of C. Elegans Biology. 2005:1–16. doi: 10.1895/wormbook.1.4.1. http://doi.org/10.1895/wormbook.1.4.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess JR, Thomson JN. Cellular interactions in early C. elegans embryos. Cell. 1987;48(2):241–250. doi: 10.1016/0092-8674(87)90427-2. [DOI] [PubMed] [Google Scholar]
- Robertson SM, Shetty P, Lin R. Identification of lineage-specific zygotic transcripts in early Caenorhabditis elegans embryos. Developmental Biology. 2004;276(2):493–507. doi: 10.1016/j.ydbio.2004.09.015. http://doi.org/10.1016/j.ydbio.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nature Biotechnology. 2015;33(5):495–502. doi: 10.1038/nbt.3192. http://doi.org/10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer JM, Glass S, Li T, Shemer G, White ND, Starostina NG, et al. Overcoming redundancy: an RNAi enhancer screen for morphogenesis genes in Caenorhabditis elegans. Genetics. 2011;188(3):549–564. doi: 10.1534/genetics.111.129486. http://doi.org/10.1534/genetics.111.129486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaner CE, Kelly WG. Germline chromatin. WormBook : the Online Review of C. Elegans Biology. 2006:1–14. doi: 10.1895/wormbook.1.73.1. http://doi.org/10.1895/wormbook.1.73.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierenberg E, Strome S. The establishment of embryonic axes and determination of cell fates in embryos of the nematode Caenorhabditis elegans. Seminars in Developmental Biology. 1992;3:25–33. http://doi.org/10.1234/12345678. [Google Scholar]
- Seydoux G, Fire A. Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development (Cambridge, England) 1994;120(10):2823–2834. doi: 10.1242/dev.120.10.2823. [DOI] [PubMed] [Google Scholar]
- Shaffer SM, Wu M-T, Levesque MJ, Raj A. Turbo FISH: a method for rapid single molecule RNA FISH. PloS One. 2013;8(9):e75120. doi: 10.1371/journal.pone.0075120. http://doi.org/10.1371/journal.pone.0075120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommermann EM, Strohmaier KR, Maduro MF, Rothman JH. Endoderm development in Caenorhabditis elegans: the synergistic action of ELT-2 and −7 mediates the specification→differentiation transition. Developmental Biology. 2010;347(1):154–166. doi: 10.1016/j.ydbio.2010.08.020. http://doi.org/10.1016/j.ydbio.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sönnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434(7032):462–469. doi: 10.1038/nature03353. http://doi.org/10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Developmental Biology. 1983;100(1):64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature Biotechnology. 2014;32(4):381–386. doi: 10.1038/nbt.2859. http://doi.org/10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walston T, Tuskey C, Edgar L, Hawkins N, Ellis G, Bowerman B, et al. Multiple Wnt signaling pathways converge to orient the mitotic spindle in early C. elegans embryos. Developmental Cell. 2004;7(6):831–841. doi: 10.1016/j.devcel.2004.10.008. http://doi.org/10.1016/j.devcel.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Weismann A. The germ-plasm. London: W. Scott; 1893. [Google Scholar]
- Wieschaus E. Nobel Lecture. In: Ringertz N, editor. Nobel Lectures. Singapore: 1997. [Google Scholar]
- Woollard A. Gene duplications and genetic redundancy in C. elegans. WormBook : the Online Review of C. Elegans Biology. 2005:1–6. doi: 10.1895/wormbook.1.2.1. http://doi.org/10.1895/wormbook.1.2.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormbase. WS170. 2007 Feb 09; http://ws170.wormbase.org/
- Wormbase. WS252. 2015 Dec 04; http://ws252.wormbase.org/
- WormMine. WS250. 2015 Oct 29; http://wormbase.org/tools/wormmine/
- Xue Z, Huang K, Cai C, Cai L, Jiang C-Y, Feng Y, et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500(7464):593–597. doi: 10.1038/nature12364. http://doi.org/10.1038/nature12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka A, Yada M, Imaki H, Koga M, Ohshima Y, Nakayama K-I. Multiple Skp1-related proteins in Caenorhabditis elegans: diverse patterns of interaction with Cullins and F-box proteins. Current Biology : CB. 2002;12(4):267–275. doi: 10.1016/s0960-9822(02)00657-7. [DOI] [PubMed] [Google Scholar]
- Yamanaka S. Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell. 2007;1(1):39–49. doi: 10.1016/j.stem.2007.05.012. http://doi.org/10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Yan L, Yang M, Guo H, Yang L, Wu J, Li R, et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nature Structural & Molecular Biology. 2013;20(9):1131–1139. doi: 10.1038/nsmb.2660. http://doi.org/10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science (New York, N.Y.) 2015;347(6226):1138–1142. doi: 10.1126/science.aaa1934. http://doi.org/10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.