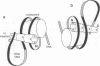Abstract
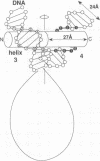
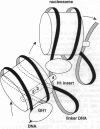
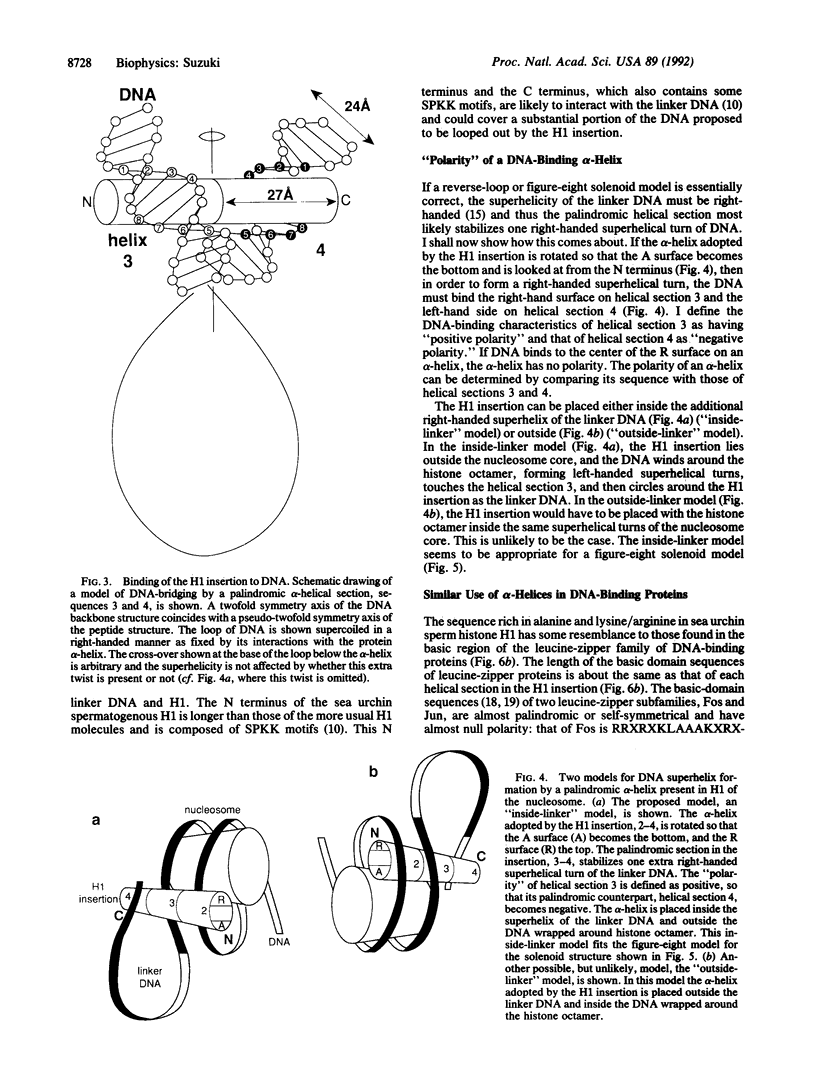
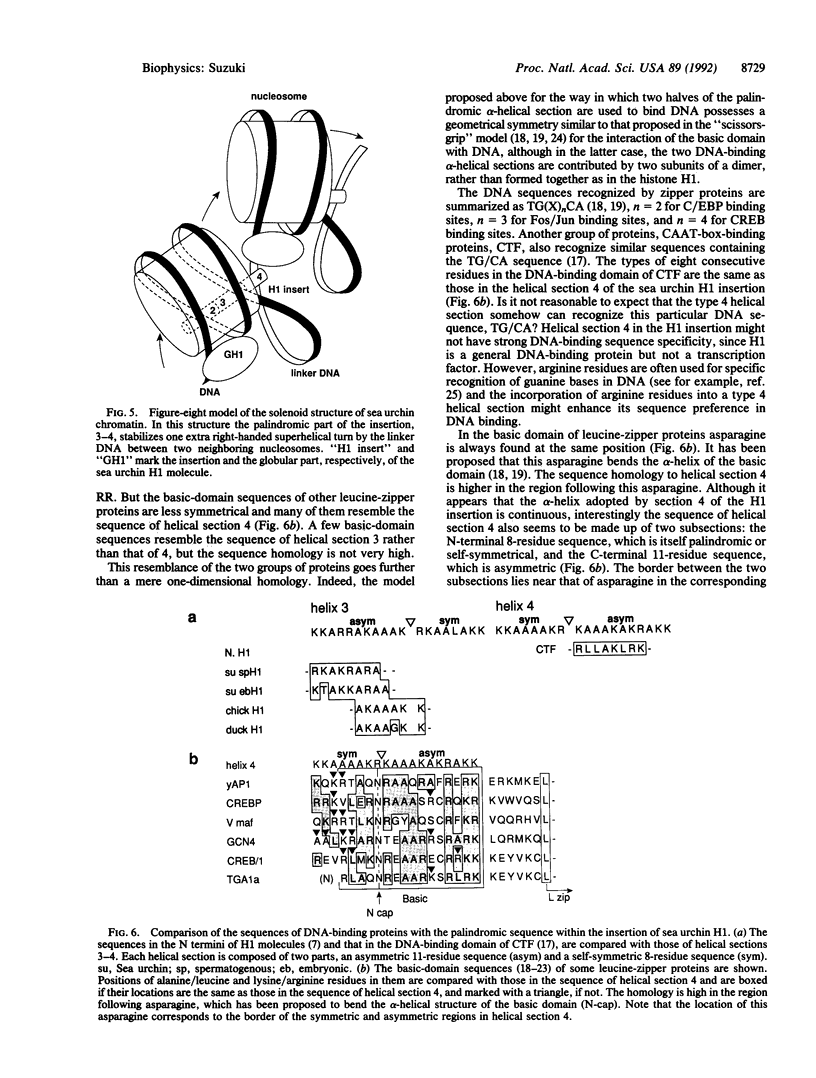
The nucleosomal DNA repeat of 240 base pairs in the chromatin structure of sea urchin sperm is exceptionally long and is accompanied by the presence of a histone H1 molecule larger than is usual in most species of chromatin. I propose how these two features are correlated and how they fit into the solenoidal model for the 300-A-diameter fiber of chromatin. Comparison of the sequence of spermatogenous H1 with other H1 sequences reveals an insert of 55 amino acid residues (residues 122-176). A 37-residue sequence in the insert (residues 140-176) has a palindromic character. I propose that each half of the palindromic sequence constitutes an alpha-helical DNA-binding unit and that the continuous alpha-helix made up of the two halves, by virtue of its palindromic nature, stabilizes the formation of an extra superhelical turn by the long linker DNA between two nucleosome cores. The N-terminal-C-terminal "polarity" of each alpha-helical section of half the palindromic sequence indicates how the arginine/lysine-rich DNA-binding surface of the alpha-helical section is used. The polarity of the H1 insertion sequence supports the so-called "reverse-loop" model or a "figure-eight" model for the path of the DNA within the solenoid structure; i.e., the linker DNA forms a right-handed superhelical turn toward the center of the solenoid structure. This use of a pair of a palindromically related alpha-helical sections has a similarity with the "scissors-grip" model for the interaction of the leucine-zipper proteins with DNA.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Butler P. J. A defined structure of the 30 nm chromatin fibre which accommodates different nucleosomal repeat lengths. EMBO J. 1984 Nov;3(11):2599–2604. doi: 10.1002/j.1460-2075.1984.tb02180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm L., Mitchell T. C. Sequence conservation in the N-terminal domain of histone H1. FEBS Lett. 1985 Nov 25;193(1):1–4. doi: 10.1016/0014-5793(85)80067-3. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giancotti V., Russo E., Cosimi S., Cary P. D., Crane-Robinson C. Secondary and tertiary structural differences between histone H1 molecules from calf thymus and sea-urchin (Sphaerechinus granularis) sperm. Biochem J. 1981 Sep 1;197(3):655–660. doi: 10.1042/bj1970655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez G. A., Yamamoto K. K., Fischer W. H., Karr D., Menzel P., Biggs W., 3rd, Vale W. W., Montminy M. R. A cluster of phosphorylation sites on the cyclic AMP-regulated nuclear factor CREB predicted by its sequence. Nature. 1989 Feb 23;337(6209):749–752. doi: 10.1038/337749a0. [DOI] [PubMed] [Google Scholar]
- Hill C. S., Martin S. R., Thomas J. O. A stable alpha-helical element in the carboxy-terminal domain of free and chromatin-bound histone H1 from sea urchin sperm. EMBO J. 1989 Sep;8(9):2591–2599. doi: 10.1002/j.1460-2075.1989.tb08398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffler J. P., Meyer T. E., Yun Y., Jameson J. L., Habener J. F. Cyclic AMP-responsive DNA-binding protein: structure based on a cloned placental cDNA. Science. 1988 Dec 9;242(4884):1430–1433. doi: 10.1126/science.2974179. [DOI] [PubMed] [Google Scholar]
- Holloway M. Neural vector. Herpes may open the way to gene therapy in neurons. Sci Am. 1991 Jan;264(1):32–32. [PubMed] [Google Scholar]
- Klug A., Rhodes D., Smith J., Finch J. T., Thomas J. O. A low resolution structure for the histone core of the nucleosome. Nature. 1980 Oct 9;287(5782):509–516. doi: 10.1038/287509a0. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Nickol J. M., Felsenfeld G., Rau D. C. Higher order structure of chromatin: orientation of nucleosomes within the 30 nm chromatin solenoid is independent of species and spacer length. Cell. 1983 Jul;33(3):831–841. doi: 10.1016/0092-8674(83)90025-9. [DOI] [PubMed] [Google Scholar]
- Mermod N., O'Neill E. A., Kelly T. J., Tjian R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell. 1989 Aug 25;58(4):741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- Morris N. R. A comparison of the structure of chicken erythrocyte and chicken liver chromatin. Cell. 1976 Dec;9(4 Pt 1):627–632. doi: 10.1016/0092-8674(76)90045-3. [DOI] [PubMed] [Google Scholar]
- Nishizawa M., Kataoka K., Goto N., Fujiwara K. T., Kawai S. v-maf, a viral oncogene that encodes a "leucine zipper" motif. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7711–7715. doi: 10.1073/pnas.86.20.7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea E. K., Klemm J. D., Kim P. S., Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991 Oct 25;254(5031):539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991 May 10;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Puigdomenech P., Romero M. C., Allan J., Sautière P., Giancotti V., Crane-Robinson C. The chromatin of sea urchin sperm. Biochim Biophys Acta. 1987 Jan 28;908(1):70–80. doi: 10.1016/0167-4781(87)90023-6. [DOI] [PubMed] [Google Scholar]
- Smith M. F., Athey B. D., Williams S. P., Langmore J. P. Radial density distribution of chromatin: evidence that chromatin fibers have solid centers. J Cell Biol. 1990 Feb;110(2):245–254. doi: 10.1083/jcb.110.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadafora C., Bellard M., Compton J. L., Chambon P. The DNA repeat lengths in chromatins from sea urchin sperm and gastrule cells are markedly different. FEBS Lett. 1976 Oct 15;69(1):281–285. doi: 10.1016/0014-5793(76)80704-1. [DOI] [PubMed] [Google Scholar]
- Suzuki M. SPKK, a new nucleic acid-binding unit of protein found in histone. EMBO J. 1989 Mar;8(3):797–804. doi: 10.1002/j.1460-2075.1989.tb03440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Sohma H., Yazawa M., Yagi K., Ebashi S. Histone H1 kinase specific to the SPKK motif. J Biochem. 1990 Sep;108(3):356–364. doi: 10.1093/oxfordjournals.jbchem.a123206. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson C. R., Sigler P. B., McKnight S. L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989 Nov 17;246(4932):911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- Widom J., Klug A. Structure of the 300A chromatin filament: X-ray diffraction from oriented samples. Cell. 1985 Nov;43(1):207–213. doi: 10.1016/0092-8674(85)90025-x. [DOI] [PubMed] [Google Scholar]
- von Holt C., Brandt W. F., Greyling H. J., Lindsey G. G., Retief J. D., Rodrigues J. D., Schwager S., Sewell B. T. Isolation and characterization of histones. Methods Enzymol. 1989;170:431–523. doi: 10.1016/0076-6879(89)70061-6. [DOI] [PubMed] [Google Scholar]