Abstract
Thiopurine therapy, commonly used in autoimmune conditions, can be complicated by life-threatening leukopenia. This leukopenia is associated with genetic variation in TPMT (encoding thiopurine S-methyltransferase). Despite a lower frequency of TPMT mutations in Asians, the incidence of thiopurine-induced leukopenia is higher in Asians than in individuals of European descent. Here we performed an Immunochip-based 2-stage association study in 978 Korean subjects with Crohn’s disease treated with thiopurines. We identified a nonsynonymous SNP in NUDT15 (encoding p.Arg139Cys) that was strongly associated with thiopurine-induced early leukopenia (odds ratio (OR) = 35.6; Pcombined = 4.88 × 10−94). In Koreans, this variant demonstrated sensitivity and specificity of 89.4% and 93.2%, respectively, for thiopurine-induced early leukopenia (in comparison to 12.1% and 97.6% for TPMT variants). Although rare, this SNP was also strongly associated with thiopurine-induced leukopenia in subjects with inflammatory bowel disease of European descent (OR = 9.50; P = 4.64 × 10−4). Thus, NUDT15 is a pharmacogenetic determinant for thiopurine-induced leukopenia in diverse populations.
Azathioprine (AZA) and its close analog 6-mercaptopurine (6-MP) are thiopurines widely used in the treatment of patients with cancers, organ transplantation, and autoimmune or inflammatory diseases, including inflammatory bowel diseases (IBD)1,2. However, thiopurine-associated leukopenia is a substantial hazard that occurs in up to 5% of individuals with IBD of European descent3–5. The association between thiopurine-induced leukopenia and TPMT mutations is well established, and TPMT testing before thiopurine exposure is recommended by the US Food and Drug Administration (FDA). However, only about a quarter of individuals with IBD who have thiopurine-associated leukopenia carry a TPMT mutation, suggesting the existence of additional factors and also questioning the usefulness of pretesting for TPMT status in this situation6–8. Interestingly, although the frequency of TPMT mutations is lower in Asians than in individuals of European descent (~3% versus ~10%)9–13, the frequency at which thiopurine-induced leukopenia occurs in Asians is considerably higher11,12,14. Thiopurine-associated leukopenia (defined by a white blood cell (WBC) count of <3,000 cells/mm3) was observed in 31.2% and 39.6% of Korean individuals with Crohn’s disease at median AZA doses of 1.34 and 1.80 mg/kg/d, respectively11,14. We therefore conducted an association analysis in a single-center Korean population with Crohn’s disease to identify additional genetic variation associated with thiopurine-induced leukopenia. The workflow of the study is shown in Supplementary Figure 1. In total, 66 of 978 eligible subjects developed leukopenia within 8 weeks of commencing thiopurine therapy (defined as early leukopenia cases15). A further 280 experienced leukopenia after the first 8 weeks (defined as late leukopenia cases15), and 632 did not experience leukopenia, despite exposure to ≥1 mg/kg/d AZA for ≥8 weeks (defined as controls) (Supplementary Table 1).
Using the Immunochip (Illumina), we conducted a discovery association analysis of 95,405 genotyped SNPs in 33 cases with early leukopenia and 307 controls without leukopenia (Supplementary Fig. 1) that were randomly selected from the whole cohort of 978 participating subjects. An excess of low P values was observed at the tail of the quantile-quantile distribution (Supplementary Fig. 2), but there was no evidence of inflation due to population stratification (λGC = 1.039). We observed four associations with leukopenia at P < 1 × 10−5 at rs9843344 (3p25.1, FBLN2), rs1986731 (6p22.3, the CMAHP pseudogene), rs2945770 (8q24.22, ST3GAL1) and rs17109616 (14q31.1, NRXN3) (Supplementary Fig. 3). In addition, we observed suggestive association at TPMT (rs1142345, TPMT*3C) (OR = 7.11; P = 1.61 × 10−4). Aiming to improve the coverage of genetic variants, we also performed the discovery analysis for 436,011 imputed SNPs and found 4 additional loci with association P < 1 × 10−5 (1q32, 8q22, 13q14 and 14q31) (Supplementary Fig. 3). Only the SUCLA2-NUDT15-MED4 region at 13q14 showed multiple signals achieving genome-wide significance (P < 5 × 10−8) (rs79076357, rs1168552132, rs142829497 and rs73481212), with the most significant association seen at rs142829497 (OR = 24.2; P = 2.36 × 10−23).
One SNP from each of the eight loci showing association at P < 1 × 10−5, as well as rs116855232 in NUDT15, which leads to an amino acid substitution (basic arginine to polar cysteine) at position 139 of the protein, and rs1142345 within TPMT, were selected for replication in an additional 33 independent cases with early leukopenia and 325 controls. The assay for SNP rs142829497 failed, and, consequently, we instead analyzed rs73481212 (in complete linkage disequilibrium (LD) with rs142829497; r2 = 1.00). Two SNPs within the SUCLA2-NUDT15-MED4 region at 13q14 were replicated, with associations achieving genome-wide significance (Supplementary Table 2). A nonsynonymous SNP, rs116855232 (encoding p.Arg139Cys), showed the most significant and strongest association with early leukopenia in the combined analysis of the discovery (Immunochip) and replication samples (OR = 35.6; Pcombined = 4.88 × 10−94) (Table 1). The other SNP, rs73481212, was in strong LD with rs116855232 (encoding p.Arg139Cys) (r2 = 0.93). The TPMT*3C allele (rs1142345) also showed consistent association with early leukopenia in the replication sample (P < 0.05) but not at a genome-wide significant level in the combined analysis (Pcombined = 2.95 × 10−5) (Supplementary Table 2).
Table 1.
Association of NUDT15 rs116855232 (p.Arg139Cys) with thiopurine-induced early leukopenia in the discovery, replication and combined samples
| Study group | Cases
|
Controls
|
||||||
|---|---|---|---|---|---|---|---|---|
| n | RAF | OR (95% CI) | P valuea | PBDb | n | RAF | OR (95% CI) | |
| Discovery | 33 | 0.530 | 39.65 (20.03–78.47) | 2.79 × 10−48 | 307 | 0.028 | 1.00 | |
| Replication | 33 | 0.576 | 32.57 (17.41–60.93) | 7.18 × 10−48 | 325 | 0.040 | 1.00 | |
| Combined | 66 | 0.553 | 35.63 (22.47–56.51) | 4.88 × 10−94 | 0.677 | 632 | 0.034 | 1.00 |
RAF, risk allele frequency; OR, odds ratio; CI, confidence interval.
P values were calculated using allelic association tests. Combined P values were calculated using the Cochran-Mantel-Haenszel test.
Asymptotic P value of the Breslow-Day test for heterogeneity in the OR.
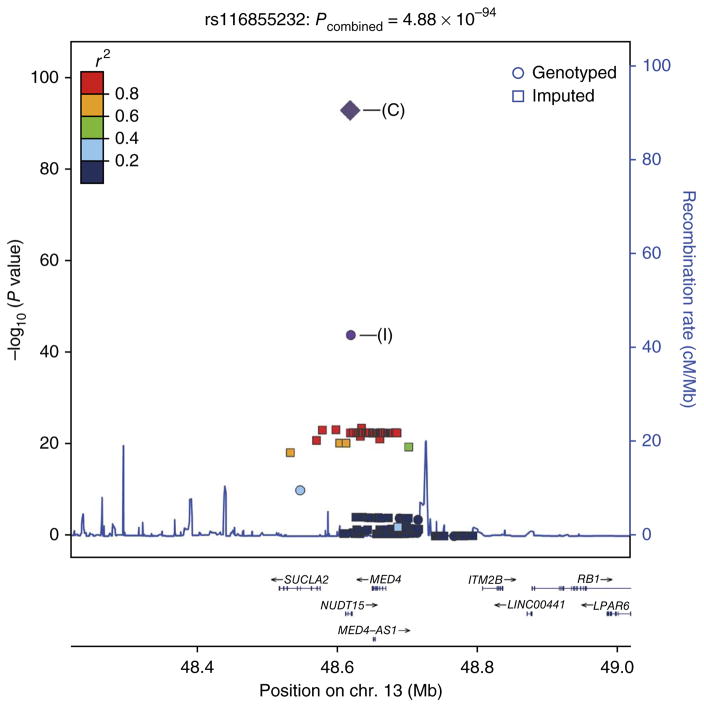
As rs116855232 is within an LD region of ~185 kb in length that includes SUCLA2, NUDT15 and MED4, we searched for additional nonsynonymous SNPs using exome sequencing data from 59 Koreans (S.H. Oh, J.B., K.M. Kim, H.C. & Y.J. Lee et al., unpublished data). We identified two additional nonsynonymous SNPs, rs7320366 in SUCLA2 and rs186364861 in NUDT15, that were in moderate or low LD with rs116855232 (r2 = 0.33 and 0, respectively). Only rs7320366 showed significant albeit more modest association with leukopenia (OR = 5.03; Pcombined = 3.55 × 10−20) (Supplementary Table 3). Subsequent conditional logistic regression analysis indicated that the association of rs7320366 in SUCLA2 was completely abolished by conditioning on the association effect at rs116855232 in NUDT15, but the association at rs116855232 remained highly significant after conditioning on rs7320366 (Fig. 1 and Supplementary Fig. 4).
Figure 1.
Regional association plot of the SUCLA2-NUDT15-MED4 locus at 13q14. The plot shows the results for rs116855232, which has the strongest association with early leukopenia. SNPs are plotted according to their chromosomal positions (NCBI Build 37) with −log10 P values from the Immunochip analysis in the region spanning ~185 kb. The −log10 P values of the lead SNP are shown for the Immunochip study (I) and the combined study (C). Strength of LD (r2 values) between the lead SNP and the other SNPs is indicated by color. Genes within the region of interest are annotated at the bottom, with arrows indicating the direction of transcription. The estimated recombination rates from the 1000 Genomes Project (March 2012) CHB (Han Chinese in Beijing, China) and JPT (Japanese in Tokyo, Japan) samples are plotted to reflect local LD structure. Plots were generated using LocusZoom.
Next, we tested for association with cases of late leukopenia and confirmed the effect of the NUDT15 variant encoding p.Arg139Cys in this population as well (OR = 5.29; P = 5.77 × 10−21) (Supplementary Table 4). Finally, using the same criteria for leukopenia as used in the Korean population, we found that the NUDT15 allele encoding p.Arg139Cys (overall allele frequency in individuals of European descent of ~0.004) was significantly associated with thiopurine-associated leukopenia in a US IBD cohort of 1,188 subjects with wild-type TPMT (allele frequency of 2.74% in leukopenia cases versus 0.31% in thiopurine-treated IBD cases without leukopenia; OR = 9.50; P = 4.64 × 10−4).
Combining the discovery and replication Korean samples, we found that the NUDT15 allele encoding p.Arg139Cys was present in 89.4% (59/66) of the early leukopenia cases but was present in only 6.8% (43/632) of the controls (Supplementary Table 5), suggesting that the presence of this NUDT15 allele had a sensitivity of 89.4% (59/66) and specificity of 93.2% (589/632) for early leukopenia, with an area under the curve (AUC) value of 0.92 (Supplementary Fig. 5 and Supplementary Table 5). Assuming a prevalence of 7% for early leukopenia in the Korean population (66/978), 16 cases would need to be tested to avoid 1 case of early leukopenia. Subjects carrying one copy of the NUDT15 risk allele had 88 times higher risk (heterozygous OR) for developing thiopurine-induced early leukopenia than non-carriers (Supplementary Table 5), and no controls were homozygous for the NUDT15 risk allele. We also estimated the absolute risk of leukopenia for the different NUDT15 genotypes (Supplementary Table 6). In addition, as the number of copies of the NUDT15 risk allele increased, the dose of thiopurines at which leukopenia occurred decreased, the interval from the onset of thiopurine therapy to the development of leukopenia decreased and the grade of the observed leukopenia increased (Table 2). Together, these data suggest a gene-dose effect of the NUDT15 risk allele in the development of thiopurine-associated leukopenia. In contrast, TPMT variants showed only moderate association with early leukopenia (Supplementary Table 7). In this study, we found TPMT mutations in only 3.8% (13/346) of Korean Crohn’s disease cases with thiopurine-associated leukopenia, in keeping with published data10–12,16 and at much lower frequency than the ~25% reported in IBD cases of European descent6,7. Moreover, if we excluded 9 cases with both the TPMT*3C and NUDT15 risk alleles from the analysis, only 1.2% (4/346) of Korean Crohn’s disease cases who experienced leukopenia during thiopurine therapy had TPMT mutations. These results suggest that TPMT genotyping before initiating thiopurine therapy is of limited use in Asians. However, the current clinical practice in Korea is to commence treatment with thiopurines at a low dose and slowly titrate the dose according to response to minimize adverse events. This practice may have led to an underestimation of the effects for both TPMT and NUDT15 variants. The NUDT15 risk allele encoding p.Arg139Cys is much more common in Asians than in individuals of European descent, with reported allele frequencies of 10.4% in Koreans, 7% in Japanese, 13% in Chinese and 2% in an admixed American population. This risk allele is present but at a very low frequency in individuals of European descent; nevertheless, individuals of European descent carrying the NUDT15 risk allele are also at significant risk of leukopenia during thiopurine therapy.
Table 2.
Comparison of dose of azathioprine, interval from onset of therapy to leukopenia and grade of leukopenia in relation to the NUDT15 rs116855232 genotype (p.Arg139Cys) of cases with leukopenia
| Genotype
|
P valueb | |||
|---|---|---|---|---|
| Homozygote (TT) (n = 14) | Heterozygote (CT) (n = 133) | Non-carrier (CC) (n = 199) | ||
| Azathioprine dose (mg/kg/d)a | 0.86 (0.50–1.09) | 1.06 (0.26–2.84) | 1.53 (0.14–3.12) | 4.93 × 10−11 |
| Interval from onset of therapy to leukopenia (d)a | 19 (9–28) | 135 (12–3,300) | 465 (21–3,705) | 1.03 × 10−17 |
| Leukopeniac | ||||
| Grade 3 or 4 | 14 (100.0) | 10 (7.5) | 4 (2.0) | 4.85 × 10−19 |
| Grade 4 | 12 (85.7) | 3 (2.3) | 0 (0.0) | 5.20 × 10−19 |
Grade 3 leukopenia is defined by a WBC count between 1,000 and 2,000 cells/mm3. Grade 4 leukopenia is defined by a WBC count of less than 1,000 cells/mm3.
Values are medians (range).
P values for all categorical variables were calculated using the Fisher’s exact test. P values for continuous variables were calculated using the Kruskal-Wallis test.
Values shown are the number of cases and percentage of cases.
NUDT15 (nucleoside diphosphate–linked moiety X-type motif 15) is a 164-amino-acid protein that belongs to the nudix hydrolase enzyme family, whose members are capable of hydrolyzing compounds with the general structure of a nucleoside diphosphate linked to another moiety, X. Although its substrate in vivo remains unclear, NUDT15 can degrade both 8-oxo-dGTP and 8-oxo-dGDP in vitro, suggesting that it might remove an oxidatively damaged form of guanine (7,8-dihydro-8-oxoguanine) from DNA and the nucleotide pool, thereby preventing misincorporation of 8-oxo-dGTP into DNA and preventing A:T to C:G transversions17. To investigate the functional role of the coding variant of NUDT15 (p.Arg139Cys) in the thiopurine-induced cytotoxicity effect, we measured the sensitivity of Jurkat cells to 6-MP after they were transfected with wild-type or mutant NUDT15 construct. After treatment with 7.5 μM 6-MP for 24 h, the number of viable Jurkat cells transfected with mutant NUDT15 construct was lower in comparison to the number of control cells transfected with wild-type construct (P = 0.00012; Supplementary Fig. 6). The levels of the apoptosis marker surface Annexin V were also significantly increased in mutant-transfected cells compared to those transfected with wild-type construct (P = 0.018; Supplementary Fig. 6), supporting a role for mutant NUDT15 in thiopurine-induced cytotoxicity. As Jurkat cells carry wild-type TPMT and NUDT15, it appears that the mutant NUDT15 has a dominant role in sensitivity to 6-MP.
In summary, we have identified and replicated a variant associated with substantially elevated risk of thiopurine-associated leukopenia in diverse populations. Our findings explain, in part, the higher prevalence of this phenomenon in Asians despite a lower prevalence of TPMT mutations. Our unbiased approach identified a nonsynonymous and potentially damaging polymorphism in a gene involved in purine metabolism that demonstrates clinical usefulness for anticipating patients at risk of this potentially life-threatening condition. Our findings have implications beyond IBD, as thiopurines are used in a number of oncological and immune-based conditions, as well as beyond the Korean population, given the allele frequency of this polymorphism in other Asian populations as well as the increased diversity of populations in regions such as North America and Europe. Future studies to explore the precise mechanism by which the NUDT15 p.Arg139Cys variant influences thiopurine-induced leukopenia and to identify additional genetic susceptibility loci for thiopurine-induced leukopenia within this pathway are warranted.
ONLINE METHODS
Study population
Of a total of 1,191 Korean Crohn’s disease cases who used thiopurines, 978 were included in the present study (for details, see Supplementary Fig. 1 and the Supplementary Note). Of the 978 subjects, 66 experienced leukopenia within the first 8 weeks of thiopurine therapy (defined as cases with early leukopenia15), 280 experienced leukopenia after the first 8 weeks (defined as cases with late leukopenia15), and 632 did not experience leukopenia and had been treated with ≥1 mg/kg/d AZA for ≥8 weeks (defined as controls). To investigate whether the association was present in individuals of European descent, we used a US cohort recruited from the IBD Center at Cedars-Sinai Medical Center. Further description of the characteristics of the study population is provided in the Supplementary Note. All participants provided written informed consent. The study was approved by the institutional review board of the Asan Medical Center.
Use of thiopurines and assessment of leukopenia
Our clinical approach to prescribing thiopurines in Crohn’s disease is to start at a low dose of either AZA or 6-MP and to increase the medication dosage slowly over several months until the target weight-based dose is achieved. It is not our clinical practice to assess TPMT status before initiating thiopurine therapy. In this cohort, decisions regarding dose adjustment or discontinuation of thiopurine drugs due to adverse events were made by the physician responsible on a case-by-case basis. Details on the dosing schedule, monitoring of complete blood counts and assessment of leukopenia are provided in the Supplementary Note.
SNP genotyping and association analysis
We used Immunochip data from 504 Crohn’s disease cases from a study that included Immunochip genotyping of 726 Crohn’s disease cases and 469 healthy controls (S.-K.Y., M.H., H.C., W.Z. & Y.J. et al., unpublished data). The remaining 222 Crohn’s disease cases were not included in the current study because they received either insufficient (n = 112) or no (n = 110) thiopurine therapy. Our discovery analysis was focused on early leukopenia, using the 33 cases with early leukopenia and 307 controls.
Genotyping with the Immunochip was performed at the Cedars-Sinai Medical Center. The Immunochip includes 196,524 genetic variants in genetic susceptibility loci previously shown to confer risk for autoimmune or inflammatory diseases and other regions with suggestive evidence of association. SNP and sample quality control analyses were performed with the use of established procedures. Briefly, all SNPs on the X, Y and mitochondrial chromosomes, as well as copy number variation–related SNPs, were excluded. As a part of quality control, SNPs were excluded if they had a call rate lower than 90%, a minor allele frequency (MAF) of <0.01 and significant deviation from Hardy-Weinberg equilibrium in controls (P < 2.5 × 10−7). Similarly, we removed all the samples with a genotyping rate less than 96% from further analysis. Only 95,405 SNPs were left for further quality control. We then examined the potential genetic relatedness of the 340 samples on the basis of pairwise identity by state using PLINK version 1.07 software (see URLs). Sample duplications and/or genetic relatedness were not found. Subsequently, we used PCA (principal-component analysis)-based methods to detect population outliers and stratification in the software package EIGENSTRAT 3.0 (see URLs)18. For PCA, all 340 samples (33 early leukopenia cases and 307 controls) were analyzed together with the 194 reference samples from the International HapMap Project, and no outliers were identified (Supplementary Fig. 7). After quality control measures were implemented, a total of 95,405 SNPs in 340 samples were used in the association analysis.
For the test of replication, 10 selected SNPs were successfully genotyped using TaqMan SNP genotyping assays in an additional 33 early leukopenia cases and 325 clinically confirmed controls. The most significant signal was replicated in 280 late leukopenia cases and 632 controls from the discovery and replication samples. With a two-stage design, the current study had good power (78%) to detect a genetic risk variant with a population frequency of 5% and risk effect (OR) of 15 and great power (90%) to detect a risk variant with a population frequency of 5% and an OR of 20 or a population frequency of 10% and an OR of 15. We also determined TPMT genotypes (rs1142345) in all participants. Genotype data for the SNPs selected on the basis of imputation data were confirmed in the discovery samples. Genotype data for rs116855232 in the US cohort were generated using the Human Exome BeadChip (Illumina). Logistic regression was performed with adjustment for the top four principal components to control for potential confounding. Seventy-three cases and 1,115 controls with both Exome BeadChip and phenotype data were included in the analysis.
Imputation
To improve the coverage of genetic variants, we imputed untyped genotypes in the discovery samples using the Asian reference panel (JPT and CHB) from the 1000 Genomes Project databases (February 2012 release) and IMPUTE version 2.0 (see URLs)19. Imputed genotypes with a genotype probability of <90%, as well as SNPs with an imputation certainty of <80% (based on information scores from imputation results), a MAF of <1% and a missing rate of >10% of genotypes, were eliminated from further analysis. A total of 436,011 imputed SNPs passed quality control and were combined with 95,405 genotyped SNPs for association analysis.
Transfection with NUDT15 and apoptosis analysis
To investigate the functional role of the coding variant (p.Arg139Cys) of NUDT15 in the thiopurine-induced cytotoxicity effect, we measured the sensitivity of cells to 6-MP after they were transfected with wild-type or mutant NUDT15. Expression plasmid encoding Myc-DDK–tagged NUDT15 (Origene) was modified to generate mutant NUDT15 (Cys139) using site-directed mutagenesis (Cosmogenetech). An immortalized line of human T lymphocyte cells, Jurkat cells (American Type Culture Collection; mycoplasma free), were cultured in RPMI 1640 (Welgene) supplemented with 10% FBS and 1% antibiotics. Cells were transfected with either wild-type (Arg139) or mutant (Cys139) NUDT15 construct using Lipofectamine 2000 (Invitrogen). After transfection, cells were allowed to recover at 37 °C in the presence of 5% CO2 for 24 h before treatment with 7.5 μM 6-MP (Sigma-Aldrich). The sensitivity of the cells to 7.5 μM 6-MP was measured by cell survival and apoptosis assays. To evaluate cell survival, cells were diluted with medium for a 4× dilution and with trypan blue viability stain for a 2× dilution. Viable cells were counted on a hemocytometer using a microscope at 100× magnification. A FITC Annexin V/Dead Cell Apoptosis kit (Invitrogen) was used for apoptosis assays. Cells were washed in PBS, resuspended (~1 × 106 cells/ml) in 100 μl of 1× Annexin binding buffer (10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl2, pH 7.4) and then incubated with 1 μl of propidium iodide (100 μg/ml; diluted in 1× Annexin binding buffer) and 5 μl of FITC Annexin V at room temperature for 15 min. We then added 400 μl of 1× Annexin binding buffer. Apoptosis after drug treatment was assessed by BD FACS Canto II (BD Biosciences) using Annexin V and propidium iodide staining. The data were analyzed by Student’s t test, and P < 0.05 was considered statistically significant.
Statistical analysis
All association analyses were performed via logistic regression with an additive model of inheritance using PLINK version 1.07 (see URLs). The quantile-quantile plot was generated using R version 2.13.1 (see URLs) to evaluate the overall significance of the discovery association analysis and the potential impact of population stratification. The impact of population stratification was also evaluated by calculating the genomic control inflation factor. The Manhattan plot of −log10 P values was generated using Haploview version 4.2 (see URLs). Replication analysis was performed by analyzing the follow-up samples separately and then performing a combined analysis of all cases and controls. Association analysis of the combined samples was performed using Cochran-Mantel-Haenszel stratification analysis. The Breslow-Day test was used to test for heterogeneity of the OR estimates between the discovery and validation samples. We tested the independence of the multiple associations observed within the SUCLA2-NUDT15-MED4 region at 13q14 using conditional logistic regression analysis.
Categorical variables were expressed as proportions and compared using the χ2 test or Fisher’s test if the number of subjects in any cell of 2 × 2 table was five or less. Continuous variables were summarized as medians with ranges and compared using the Mann-Whitney U test or Kruskal-Wallis test. Statistical analysis was performed using SPSS for Windows, version 18.0.
Supplementary Material
Acknowledgments
We would like to thank all participants who provided the DNA and clinical information necessary for this study. J.L. and W.Z. are supported by the Biomedical Research Council, the Agency for Science, Technology and Research (A*STAR; BMRC SPF2014/001), Singapore. D.P.B.M., T.H. and M.D. are supported by the US National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases (P01DK046763, U01DK062413, U01AI067068, R01HS021747), the European Union (IBD-BIOM 305479), the Leona M. and Harry B. Helmsley Charitable Trust (2011PG-MED004, 2014PG-IBD014) and internal funding from the F. Widjaja Foundation Inflammatory Bowel and Immunobiology Research Institute. This work was supported by a Mid-Career Researcher Program grant through NRF (National Research Foundation of Korea) to K.S. (2010-0015648, 2014R1A2A1A09005824) funded by the Ministry of Science, Information and Communication Technology (ICT) and Future Planning and by a Korean Health Technology R&D Project grant to S.-K.Y. (A120176) from the Ministry of Health and Welfare, the Republic of Korea.
Footnotes
URLs. 1000 Genomes Project, http://www.1000genomes.org/; dbSNP, http://www.ncbi.nlm.nih.gov/SNP/; EIGENSTRAT, http://genepath.med.harvard.edu/~reich/EIGENSTRAT.htm; Haploview, http://www.broad.mit.edu/mpg/haploview/; IMPUTE2, https://mathgen.stats.ox.ac.uk/impute/impute_v2.html; LocusZoom, http://csg.sph.umich.edu/locuszoom/; PLINK, http://pngu.mgh.harvard.edu/~purcell/plink/; R, http://www.r-project.org/.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
K.S. and S.-K.Y. obtained financial support and conceived and designed the study. K.S. supervised genotyping, data analysis and interpretation, and S.-K.Y. supervised all the sample collection, data analysis and interpretation. J.L. supervised data analysis and interpretation. S.-K.Y., B.D.Y., K.-J.K., S.H.P., S.-K.P., D.-H.Y. and I.L. recruited subjects and participated in diagnostic evaluation. S.-K.Y., S.H.P. and S.-K.P. assessed clinical features. T.H., M.D. and D.P.B.M. supervised the Immunochip genotyping and provided their data. M.H., W.Z., Y.J. and H.C. performed data analyses. M.H., J.B. and H.C. prepared DNA samples and performed genotyping. J.B. performed in vitro transfection experiments and data analysis. K.S. and S.-K.Y. drafted the manuscript. K.S., S.-K.Y., D.P.B.M. and J.L. revised the manuscript.
References
- 1.Timmer A, McDonald JW, Tsoulis DJ, Macdonald JK. Azathioprine and 6-mercaptopurine for maintenance of remission in ulcerative colitis. Cochrane Database Syst Rev. 2012;9:CD000478. doi: 10.1002/14651858.CD000478.pub3. [DOI] [PubMed] [Google Scholar]
- 2.Prefontaine E, Sutherland LR, Macdonald JK, Cepoiu M. Azathioprine or 6-mercaptopurine for maintenance of remission in Crohn’s disease. Cochrane Database Syst Rev. 2009;1:CD000067. doi: 10.1002/14651858.CD000067.pub2. [DOI] [PubMed] [Google Scholar]
- 3.Present DH, Meltzer SJ, Krumholz MP, Wolke A, Korelitz BI. 6-Mercaptopurine in the management of inflammatory bowel disease: short- and long-term toxicity. Ann Intern Med. 1989;111:641–649. doi: 10.7326/0003-4819-111-8-641. [DOI] [PubMed] [Google Scholar]
- 4.Connell WR, Kamm MA, Ritchie JK, Lennard-Jones JE. Bone marrow toxicity caused by azathioprine in inflammatory bowel disease: 27 years of experience. Gut. 1993;34:1081–1085. doi: 10.1136/gut.34.8.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraser AG, Orchard TR, Jewell DP. The efficacy of azathioprine for the treatment of inflammatory bowel disease: a 30 year review. Gut. 2002;50:485–489. doi: 10.1136/gut.50.4.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colombel JF, et al. Genotypic analysis of thiopurine S-methyltransferase in patients with Crohn’s disease and severe myelosuppression during azathioprine therapy. Gastroenterology. 2000;118:1025–1030. doi: 10.1016/s0016-5085(00)70354-4. [DOI] [PubMed] [Google Scholar]
- 7.Dewit O, et al. Limitations of extensive TPMT genotyping in the management of azathioprine-induced myelosuppression in IBD patients. Clin Biochem. 2011;44:1062–1066. doi: 10.1016/j.clinbiochem.2011.06.079. [DOI] [PubMed] [Google Scholar]
- 8.Booth RA, et al. Assessment of thiopurine S-methyltransferase activity in patients prescribed thiopurines: a systematic review. Ann Intern Med. 2011;154:814–823. doi: 10.7326/0003-4819-154-12-201106210-00009. [DOI] [PubMed] [Google Scholar]
- 9.Kumagai K, et al. Allelotype frequency of the thiopurine methyltransferase (TPMT) gene in Japanese. Pharmacogenetics. 2001;11:275–278. doi: 10.1097/00008571-200104000-00012. [DOI] [PubMed] [Google Scholar]
- 10.Cao Q, Zhu Q, Shang Y, Gao M, Si J. Thiopurine methyltransferase gene polymorphisms in Chinese patients with inflammatory bowel disease. Digestion. 2009;79:58–63. doi: 10.1159/000205268. [DOI] [PubMed] [Google Scholar]
- 11.Kim JH, et al. Influences of thiopurine methyltransferase genotype and activity on thiopurine-induced leukopenia in Korean patients with inflammatory bowel disease: a retrospective cohort study. J Clin Gastroenterol. 2010;44:e242–e248. doi: 10.1097/MCG.0b013e3181d6baf5. [DOI] [PubMed] [Google Scholar]
- 12.Takatsu N, et al. Adverse reactions to azathioprine cannot be predicted by thiopurine S-methyltransferase genotype in Japanese patients with inflammatory bowel disease. J Gastroenterol Hepatol. 2009;24:1258–1264. doi: 10.1111/j.1440-1746.2009.05917.x. [DOI] [PubMed] [Google Scholar]
- 13.Collie-Duguid ES, et al. The frequency and distribution of thiopurine methyltransferase alleles in Caucasian and Asian populations. Pharmacogenetics. 1999;9:37–42. doi: 10.1097/00008571-199902000-00006. [DOI] [PubMed] [Google Scholar]
- 14.Lee HJ, et al. The safety and efficacy of azathioprine and 6-mercaptopurine in the treatment of Korean patients with Crohn’s disease. Intest Res. 2009;7:22–31. [Google Scholar]
- 15.Lewis JD, et al. Timing of myelosuppression during thiopurine therapy for inflammatory bowel disease: implications for monitoring recommendations. Clin Gastroenterol Hepatol. 2009;7:1195–1201. doi: 10.1016/j.cgh.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fangbin Z, et al. Should thiopurine methyltransferase genotypes and phenotypes be measured before thiopurine therapy in patients with inflammatory bowel disease? Ther Drug Monit. 2012;34:695–701. doi: 10.1097/FTD.0b013e3182731925. [DOI] [PubMed] [Google Scholar]
- 17.Takagi Y, et al. Human MTH3 (NUDT18) protein hydrolyzes oxidized forms of guanosine and deoxyguanosine diphosphates: comparison with MTH1 and MTH2. J Biol Chem. 2012;287:21541–21549. doi: 10.1074/jbc.M112.363010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 19.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.