Abstract
Campylobacter jejuni is one of the leading foodborne pathogens worldwide. C. jejuni is isolated from a wide range of foods, domestic animals, wildlife, and environmental sources. The currently available culture-based isolation methods are not highly effective for wastewater samples due to the low number of C. jejuni in the midst of competing bacteria. To detect and isolate C. jejuni from wastewater samples, in this study, we evaluated a few different enrichment conditions using five different antibiotics (i.e., cefoperazone, vancomycin, trimethoprim, polymyxin B, and rifampicin), to which C. jejuni is intrinsically resistant. The selectivity of each enrichment condition was measured with Ct value using quantitative real-time PCR, and multiplex PCR to determine Campylobacter species. In addition, the efficacy of Campylobacter isolation on different culture media after selective enrichment was examined by growing on Bolton and Preston agar plates. The addition of polymyxin B, rifampicin, or both to the Bolton selective supplements enhanced the selective isolation of C. jejuni. The results of 16S rDNA sequencing also revealed that Enterococcus spp. and Pseudomonas aeruginosa are major competing bacteria in the enrichment conditions. Although it is known to be difficult to isolate Campylobacter from samples with heavy contamination, this study well exhibited that the manipulation of antibiotic selective pressure improves the isolation efficiency of fastidious Campylobacter from wastewater.
Keywords: Campylobacter jejuni, wastewater, isolation, antibiotics, qRT-PCR
Introduction
Campylobacter is the major bacterial cause of foodborne infection, annually accounting for approximately 166 million foodborne illnesses around the world (Kirk et al., 2015). In addition to the clinical symptoms of gastroenteritis, Campylobacter is the major risk factor of Guillain–Barré syndrome (GBS), a neurological disorder causing muscular paralysis, as a post-infection complication (Hughes and Cornblath, 2005). Among pathogenic Campylobacter species, C. jejuni and C. coli are most frequently associated with human infection (Kaakoush et al., 2015). Thus far, the consumption of contaminated poultry is the primary cause of developing human campylobacteriosis (Whiley et al., 2013).
Despite the well-known fastidious nature of Campylobacter (Silva et al., 2011), Campylobacter is isolated from environmental sources, such as lake, river, sea, and sewage, suggesting that environmental water is a possible vehicle that transmits Campylobacter to humans (Jones, 2001). C. jejuni is the pathogenic species that is mainly related to water-borne campylobacteriosis worldwide (Pitkanen, 2013). In Canada, Campylobacter outbreaks caused by cross contamination related with meltwater and heavy rainfall are problematic to public health (Millson et al., 1991; Clark et al., 2003). However, the isolation of Campylobacter implicated in water-borne outbreak appear to be challenging, not only due to rapid loss in culturability of isolates from the environment (Wingender and Flemming, 2011) and from stool samples (Bullman et al., 2012), but also due to the time gap between the initial infection and outbreak investigation (Hanninen et al., 2003; Jakopanec et al., 2008). Therefore, regular monitoring system of water resources by using culture-based methods is likely to underestimate the prevalence of Campylobacter spp. in the environment. This might mislead our understanding of the role played by the environmental sources in human infection and possibly the contamination of food chain by Campylobacter, even though Campylobacter is most frequently detected in animal fecal samples (29.7%), untreated human sewage (25.6%), and surface water (26.6%), according to a study in Alberta, Canada, among the three major foodborne pathogens, including Campylobacter, Salmonella, and Escherichia coli O157:H7 (Jokinen et al., 2011).
Various culture supplements have been examined to improve selective isolation of Campylobacter spp. (Corry et al., 1995). For example, ISO method 2005 has been applied for the detection of thermo-tolerant campylobacters from water, and alternative culture-based methods in combination with molecular end-point confirmation (Hokajarvi et al., 2013; Pitkanen, 2013). Sample volume, incubation time, enrichment volume, passage of enrichment, and PCR-primer specificity all play an important role (Levesque et al., 2011) and enrichment procedures as well (Rosef et al., 2001). Khan et al. (2009, 2013) compared two methods (i.e., centrifugation vs. membrane filtration) for the isolation and detection of Campylobacter from agriculture watersheds, and reported the effect of incubation temperature on the detection rates and the type of dominant Campylobacter species detected from water samples. However, wastewater samples are even more challenging than samples from agricultural watersheds due to the relatively low number of Campylobacter in comparison with the high levels of microbial competitors and PCR inhibitors in wastewater (Koenraad et al., 1997; Abulreesh et al., 2005; Schrader et al., 2012).
To overcome the limitations in traditional culture-based methods for the detection of foodborne pathogens, molecular methods, such as PCR, have become practical and widely used due to the speed and reproducibility (Law et al., 2015). Recently, direct quantitative PCR was applied to the detection of Campylobacter in river water and showed the possibility of an alternative method for Campylobacter detection (Van Dyke et al., 2010). Nevertheless, the detection validation of Campylobacter with culture-based methods would still be necessary to reveal the direct correlation between a clinical illness and its etiological agents.
In this study, we present amendments to existing culture methods to improve the enrichment and isolation of Campylobacter spp. from wastewater. We used raw sewage influent samples since they are contaminated more heavily than effluent samples. By adopting different incubation temperatures and several antibiotics, to which C. jejuni is intrinsically resistant, we developed an improved enrichment method to recover culturable C. jejuni from wastewater samples. The efficiency of Campylobacter isolation was evaluated using quantitative real-time PCR (qRT-PCR) targeting genus-specific 16S rDNA primers, and a second end-point multiplexed PCR with species-specific primers. By using 16S rDNA amplicon sequencing, in addition, we identified the major bacteria in wastewater that compete with Campylobacter under the selective enrichment conditions.
Materials and Methods
Bacterial Strains, Culture Conditions, and Primers
Campylobacter jejuni ATCC 33560 and NCTC 11168 were routinely cultured in Mueller Hinton (MH) media at 42°C under microaerobic conditions (5% O2, 10% CO2, and 85% N2). The primers used in the study are described in Table 1.
Table 1.
Primers used in this study.
| Primer | Sequence (5′→ 3′) | Reference |
|---|---|---|
| *CampyLvl-16S-F | CCT GAM GCA GCA ACG CC | de Boer et al., 2013 |
| *CampyLvl-16S-R | CGG AGT TAG CCG GTG CTT ATT | |
| *CampyLvl-16S-P | CTC CGA AAA GTG TCA TCC T | |
| **CampyYM-16S-F | GGA TGA CAC TTT TCG GAG C | Yamazaki-Matsune et al., 2007 |
| **CampyYM-16S-R | CAT TGT AGC ACG TGT GTC | |
| **C. hyointest-23S-F | ATA ATC TAG GTG AGA ATC CTA G | |
| **C. hyointest-23S-R | GCT TCG CAT AGC TAA CAT | |
| **C. coli-ask-F | GGT ATG ATT TCT ACA AAG CGA G | |
| **C. coli-ask-R | ATA AAA GAC TAT CGT CGC GTG | |
| **C. fetus-cstA-F | GGT AGC CGC AGC TGC TAA GAT | |
| **C. fetus-cstA-R | AGC CAG TAA CGC ATA TTA TAG TAG | |
| **C. lari-glyA-F | TAG AGA GAT AGC AAA AGA GA | |
| **C. lari-glyA-R | TAC ACA TAA TAA TCC CAC CC | |
| **C. jejuni-cj0414-F | CAA ATA AAG TTA GAG GTA GAA TGT | |
| **C. jejuni-cj0414-R | CCA TAA GCA CTA GCT AGC TGA T | |
| **C. upsal-lpxA-F | CGA TGA TGT GCA AAT TGA AGC | |
| **C. upsal-lpxA-R | TTC TAG CCC CTT GCT TGA TG | |
| ***IAC-F | CTA ACC TTC GTG ATG AGC AAT CG | Deer et al., 2010 |
| ***IAC- R | GAT CAG CTA CGT GAG GTC CTA C | |
| ***IAC-P | AGC TAG TCG ATG CAC TCC AGT CCT CCT | |
| 27F | AGA GTT TGA TCM TGG CTC AG | Weisburg et al., 1991 |
| 1492R | TAC GGY TAC CTT GTT ACG ACT T |
*Primers used in qRT- PCR to detect Campylobacter genus, **Primers for multiplex PCR to detect Campylobacter species, ***Primer used as Internal Control template (IAC).
Enrichment Conditions for Post Grit Samples from Wastewater Treatment Facilities
Raw sewage samples (post grit influent; PG) were collected from two different wastewater treatment facilities (Pine Creek and Bonnybrook) in Calgary, Alberta, in November and December, 2014. The samples were stored at 4–8°C and processed within 12 h after arrival. The wastewater samples (100 ml) were concentrated by centrifugation at 9000 rpm for 20 min at 4°C (Sorvall RC-5B), and pellets were resuspended in 4 ml of Bolton broth (Oxoid) for further enrichment process as described by Chenu et al. (2013) with minor modifications. Briefly, four different kinds of Bolton Broth (Oxoid) enrichment broth were prepared: (1) Bolton’s with Campylobacter-selective supplements [BN; cefoperazone 20 μg/ml, vancomycin 20 μg/ml, trimethoprim 20 μg/ml, and cycloheximide 50 μg/ml, Dalynn], (2) BN plus 10 μg/ml rifampicin [BNR], (3) BN plus 5 IU/ml polymyxin B [BNP], and (4) BN with both rifampicin and polymyxin B [BNRP]. Independently, 1 ml of pellet suspension was transferred to three wells in a 96-well plate and serially diluted to determine most probable number (MPN). For the 1st enrichment procedure, the plates were incubated at 37 or 42°C for 40–48 h under microaerobic conditions. Then, the culture broths were transferred to a 2nd enrichment medium consisting of the same antimicrobial supplements with 150 μg/ml 2,3,5-triphenyl-tetrazolium chloride (TTC, Sigma) and incubated for 24 h. TTC is a color indicator to show metabolic activity, and the inclusion of the dye in the assay aids in detection of levels of growth (Gabrielson et al., 2002). The cultures were subject to qRT-PCR and multiplex PCR.
Validation of C. jejuni Growth with Antibiotic Supplements
C. jejuni ATCC 33560, which is a quality control (QC) strain for antimicrobial susceptibility testing of C. jejuni (Clinical and Laboratory Standards Institute [CLSI], 2010), and NCTC 11168 were employed to evaluate the growth capability of C. jejuni under different enrichment conditions. Four different kinds of enrichment broth were prepared as described above. C. jejuni ATCC 33560 and NCTC 11168 were cultured on MH agar plates at 42°C for 24 h and harvested with fresh MH broth. The bacterial suspension was adjusted to an OD600 of 0.07 and incubated at 42°C with shaking at 200 rpm under microaerobic conditions. To determine the growth of C. jejuni strains, the samples were taken at 0, 3, 6, 12, and 24 h, and CFU and OD600 values were measured.
Confirmation of Campylobacter Growth using qRT-PCR
To confirm if Campylobacter was successfully enriched, 50 μl of culture broth was transferred to 96 well PCR plates and heated to 95°C for 10 min to extract DNA. Quantitative PCR was performed using an ABI 7500 (Applied Biosystems) system with Campylobacter genus-specific 16S rDNA primers (de Boer et al., 2013). The internal control template (IAC) and primers were included in reaction mixtures to measure inhibitory effects in enrichment samples (Deer et al., 2010). Amplification was carried out with following conditions: 50°C for 2 min and 95°C for 30 s; 40 cycles at 95°C for 3 s and 60°C for 30 s. Ct values were evaluated to determine the growth of Campylobacter and 3-tube MPN estimates.
Confirmation of Campylobacter spp. using Multiplex PCR
To identify Campylobacter spp. in the enrichment broths, multiplex PCR was performed for 42°C enrichment broths as described elsewhere with primer sets for 16S rDNA and six species-specific primers (Yamazaki-Matsune et al., 2007). Same templates used in qRT-PCR were also employed for multiplex PCR. The amplification reaction was performed following conditions: 95°C for 15 min; 40 cycles at 95°C for 30 s, 58°C for 1 min and 30 s, 72°C for 1 min; 72°C for 7 min.
Isolation of Campylobacter spp. and Identification of Non-Campylobacter Isolates by 16S rRNA Sequencing from Wastewater
To isolate Campylobacter spp. from enrichment cultures, wells showing the lowest Ct value in qRT-PCR results in the 2nd enrichment plate were selected. The cultures were prepared with 10-fold serial dilutions and sub-cultured on Bolton agar plates with Bolton supplement (BB, Dalynn) or Bolton agar plates with Preston supplement (BP, Oxoid). Following 2–3 days incubation at 42°C under microaerobic conditions, several colonies showing different shape, color, and transparency were randomly picked and transferred to the same fresh broth. After 2 days incubation, 50 μl of the cultures was harvested and boiled at 95°C for 10 min. Genus-specific 16S rDNA PCR amplification was carried out to distinguish between Campylobacter and non-Campylobacter (Linton et al., 1996). Amplification was performed following conditions: 94°C for 1 min; 35 cycles at 94°C for 30 s, 52°C for 30 s, 72°C for 1 min; 72°C for 5 min. PCR amplicons were visualized using 2% agarose gel with SYBR safe DNA gel stain solution (Invitrogen). To identify non-Campylobacter competitors growing in the selective enrichment conditions, 16S rDNA was amplified with universal bacterial domain primers (27F and 1492R) for 100 Campylobacter genus-specific 16S rDNA negative isolates (Weisburg et al., 1991). Amplification was conducted following conditions: 94°C for 1 min; 35 cycles at 94°C for 30 s, 50°C for 30 s, 72 °C for 1 min and 30 s; 72°C for 5 min. The amplified PCR products were purified and commercially sequenced by Sanger sequencing method (Macrogen, Inc., South Korea), and the results were analyzed by using Blastn1.
Results
Campylobacter jejuni Growth in the Presence of Additional Antibiotic Supplements
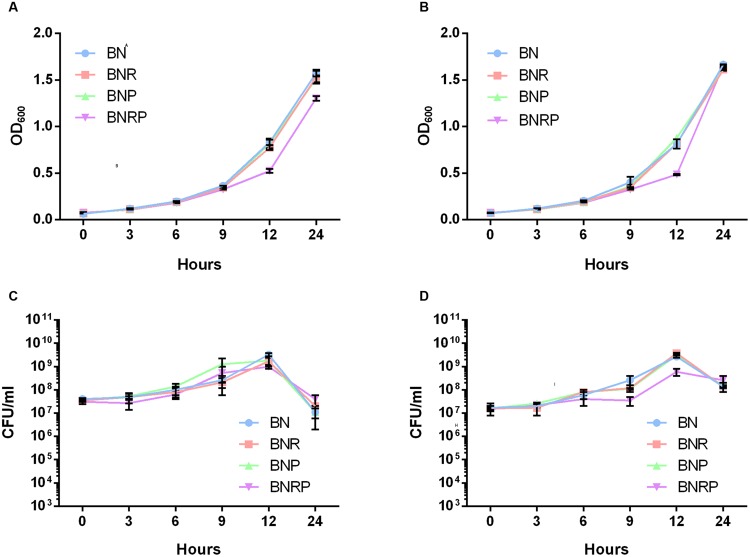
To improve the frequency of C. jejuni isolation from wastewater samples that are heavily contaminated with various microorganisms, we decided to increase antibiotic selective pressure by using different combinations of multiple antibiotics to which C. jejuni is intrinsically resistant (Taylor and Courvalin, 1988; Corry et al., 1995). For the growth testing, we used C. jejuni ATCC 33560, a QC strain for antibiotic susceptibility testing (Clinical and Laboratory Standards Institute [CLSI], 2010), and C. jejuni NCTC 11168, the first genome-sequenced Campylobacter strain (Parkhill et al., 2000). Whereas the Bolton selective supplement (BN) consists of three antibiotics, including cefoperazone, vancomycin, and trimethoprim, the Preston Campylobacter-selective supplement contains polymyxin B, rifampicin, and trimethoprim. The two selective supplements for Campylobacter isolation commonly contain trimethoprim. In the experiment, BN was used as basic antimicrobial supplements, and polymyxin B and/or rifampicin were added to BN to increase antibiotic selective pressure. The addition of either polymyxin B or rifampicin to BN did not affect the growth. The supplementation with both rifampicin and polymyxin B slightly reduced the OD600 at 12 h; however, there was no significant difference in growth in the four different enrichment conditions (Figure 1). The results indicate that C. jejuni can grow in the presence of combinations of the multiple antibiotics to which C. jejuni is naturally resistant.
FIGURE 1.
Growth of Campylobacter jejuni ATCC 33560 and NCTC 11168 in four different antimicrobial enrichment conditions at 42°C. Measurement of OD600 and CFU counting in C. jejuni ATCC 33560 (A,C) and C. jejuni NCTC 11168 (B,D). BN, Bolton broth with Bolton Campylobacter-selective supplement; BNR, BN supplemented with rifampicin; BNP, BN supplemented with polymyxin B; BNRP, BN supplemented with and rifampicin and polymyxin B.
Ct Values of qRT-PCR in Campylobacter Detection under different Enrichment Conditions
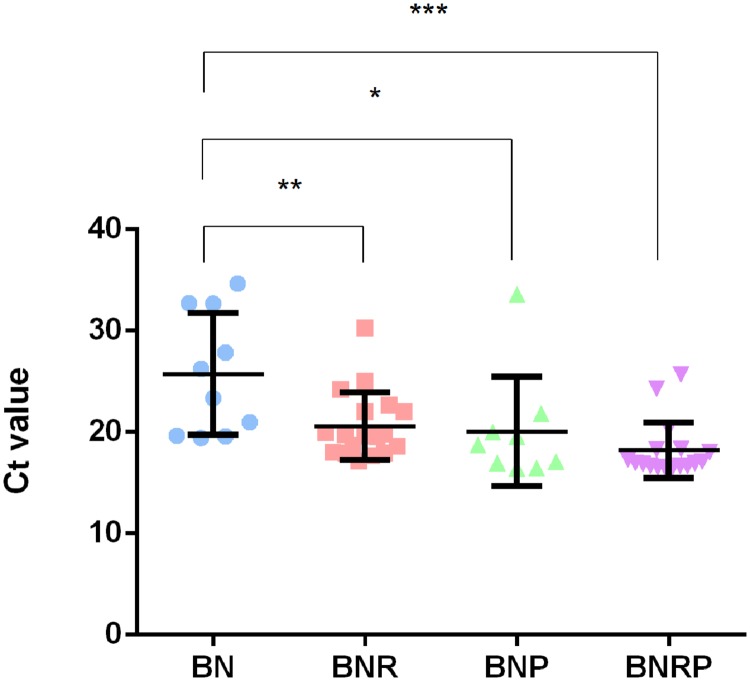
The Ct values of qRT-PCR for the detection of Campylobacter varied depending on the antimicrobial enrichment. The addition of one of the antibiotics (i.e., either rifampicin or polymyxin B) significantly decreased the Ct value, meaning that Campylobacter population was increased by the selective enrichment. Furthermore, supplementation of both antibiotics showed the lowest Ct value compared to the other enrichment conditions (Figure 2), indicating that the increased antibiotic selective pressure enhanced the enrichment of Campylobacter in raw sewage samples. Positive samples were more frequently detected at 42°C than 37°C, and non-interpretable results, where Ct values could not be determined, were sometimes observed at 37°C (data not shown). This suggests that contaminating bacteria cannot be effectively inhibited at 37°C.
FIGURE 2.
Distribution of Ct values in four different enrichment conditions at 42°C. Bolton broth with Bolton supplement (BN), BN with rifampicin (BNR), BN with polymyxin B (BNP), and BN with rifampicin and polymyxin B (BNRP). Statistical analysis was conducted with one-way analysis of variance (ANOVA) using GraphPad Prism 6 (GraphPad Software Inc., USA). *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
Multiplex PCR Detection of Campylobacter spp. under different Enrichment Conditions
In addition to qRT-PCR detection, multiplex PCR was performed to determine the species of Campylobacter isolates. The results of multiplex PCR demonstrated that the primary Campylobacter spp. were C. jejuni and C. coli (Table 2). C. jejuni and C. coli were more frequently detected by the addition of rifampicin compared to polymyxin B. In many cases, positive results were discrepant between qRT-PCR and multiplex PCR (54% in qRT-PCR in comparison with multiplex PCR). For example, the same sample that was Campylobacter-negative based on qRT-PCR was shown to be positive by multiplex PCR (data not shown).
Table 2.
The number of positive detection of Campylobacter 16S rDNA, C. jejuni, C. coli, and C. lari with multiplex PCR in four different enrichment conditions at 42°C.
| Detection with multiplex PCR | BN | BNR | BNP | BNRP |
|---|---|---|---|---|
| 16S rDNA only* | 10 (40%) | 3 (12%) | 9 (33%) | 2 (9%) |
| 16S rDNA + C. jejuni | 7 (28%) | 9 (36%) | 11 (41%) | 9 (39%) |
| 16S rDNA + C. coli | 4 (16%) | 6 (24%) | 4 (15%) | 7 (30%) |
| 16S rDNA + C. jejuni + C. coli | 3 (12%) | 7 (28%) | 2 (7%) | 5 (22%) |
| 16S rDNA + C. lari | 1 (4%) | 0 | 1 (4%) | 0 |
| Total 16s rDNA* positive | 25 (100%) | 25 (100%) | 27 (100%) | 23 (100%) |
*Positive in multiplex PCR detection with Campylobacter 16S rDNA primers but negative for species-specific detection.
Enhanced Campylobacter Isolation from Raw Sewage by Increased Antibiotic Selective Pressure
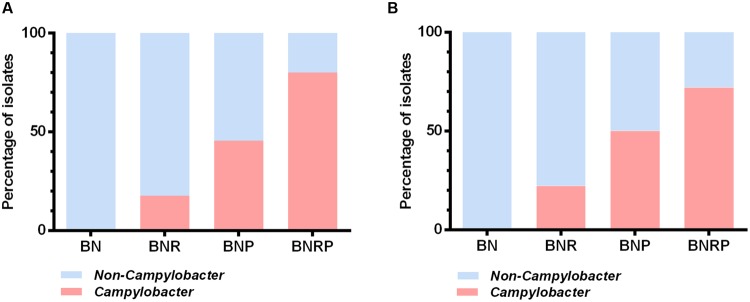
The frequency of Campylobacter isolation from raw sewage was determined under the four different antibiotic enrichment conditions. To examine the effect of agar media on the Campylobacter isolation, we plated the enrichment cultures on Bolton and Preston agars, common culture media for Campylobacter. Consistent with the qRT-PCR results, the addition of rifampicin, polymyxin B, and both antibiotics significantly increased the isolation frequency for Campylobacter and decreased the isolation frequency of non-Campylobacter (Figure 3). In particular, BNRP showed the highest isolation rate of Campylobacter, whereas BN did not recover any Campylobacter spp. (Figure 3). Whereas the antibiotic enrichment significantly affected the isolation frequency, Bolton and Preston agar media did not make any differences in the isolation frequency (Figure 3). Morphologically, small pinkish or transparent colonies usually turned out to be Campylobacter (data not shown). To identify the major non-Campylobacter populations growing on the selective enrichment media, we randomly selected 100 colonies based on colony morphologies and performed 16S rDNA amplicon sequencing. The major non-Campylobacter spp. included Enterococcus, E. coli, Klebsiella, Proteus, and Pseudomonas (Table 3). The supplementation of additional antibiotics, either single (i.e., BNR and BNP) or both (i.e., BNPR), suppressed the growth of other bacterial populations. However, Enterococcus spp., such as Enterococcus durans and Enterococcus faecium, were still isolated in BNPR (Table 3). Importantly, increased antibiotic selective pressure improved the frequencies of isolating Campylobacter from wastewater (Table 3).
FIGURE 3.
Percentage distribution of Campylobacter and non-Campylobacter isolates in four different enrichment conditions at 42°C. After antimicrobial enrichment, strains were isolated by growing on Bolton agar plates supplemented with Bolton selective supplement (BB; A) and Bolton agar plates supplemented with Preston selective supplement (BP; B). The results are based on PCR detection with primers for Campylobacter 16S rDNA. The number of isolates in BB is as follows; BN 18, BNR 17, BNP 22, and BNRP 25. The number of isolates in BP is as follows; BN 18 BNR 18, BNP 24, and BNRP 25.
Table 3.
Distribution of Campylobacter and non-Campylobacter strains in four different enrichment conditions at 42°C.
| Species | BN | BNR | BNP | BNRP |
|---|---|---|---|---|
| E. coli | 9 (25%) | 10 (28.6%) | 1 (2.2%) | 0 |
| E. fergusonii | 3 (8.3%) | 4 (11.4%) | 0 | 0 |
| E. durans | 0 | 5 (14.3%) | 7 (15.2%) | 8 (16%) |
| E. faecium | 4 (11.1%) | 6 (17.1%) | 6 (13%) | 4 (8%) |
| P. aeruginosa | 5 (13.9%) | 3 (8.6%) | 5 (10.9%) | 0 |
| P. penneri | 0 | 0 | 1 (2.2%) | 0 |
| P. mirabilis | 0 | 0 | 4 (8.7%) | 0 |
| K. pneumoniae | 15 (41.7%) | 0 | 0 | 0 |
| Campylobacter | 0 | 7 (20%) | 22 (47.8%) | 38 (76%) |
| Total number of isolates | 36 (100%) | 35 (100%) | 46 (100%) | 50 (100%) |
Discussion
In this study, we improved the efficacy of C. jejuni isolation from wastewater by increasing antibiotic selective pressure in the enrichment step. The addition of rifampicin, polymyxin B, or both to the enrichment media affected the Ct values of qRT-PCR results (Figure 2). According to the distribution of Ct values, the addition of the antibiotic(s) decreased Ct values, meaning that antibiotic supplements improved the growth of Campylobacter. In particular, rifampicin significantly reduced Ct values (Figure 2). A few studies have thus far reported that increased selective pressure enhances Campylobacter isolation from food. Yoo et al. (2014) reported that the addition of rifampicin (10 μg/ml) or polymyxin B (5 IU/ml) to Bolton agar (Bolton agar with Bolton supplement) restrained the growth of non-Campylobacter without any inhibition of C. jejuni and C. coli in fresh produce foods. Chon et al. (2013) demonstrated that the addition of high concentrations of polymyxin B to the mBolton supplement in enrichment procedure improved the efficiency of C. jejuni and C. coli recovery and suppressed background competing bacteria. Consistently, our results showed that the supplementation with additional antibiotics improved the efficacy of C. jejuni isolation even from heavily contaminated wastewater samples. In addition, we also identified bacterial populations that compete with Campylobacter under the four different selective enrichment conditions. The inputs of Campylobacter entering the influent of wastewater treatment facilities in this study would be primarily from sewage effluent in Calgary and also possibly from wildlife, such as migrating birds (Cody et al., 2015). Depending on the treatment procedure, the incidence rate of Campylobacter in sewage effluent can be altered, and cross contamination between water resources and sewage is associated with water-borne Campylobacter outbreaks (Jones, 2001; Pitkanen, 2013).
The 16S rDNA amplicon sequencing analysis of individual colonies from the enrichment plates revealed that Escherichia, Pseudomonas, Klebsiella, and Enterococcus were the major competing bacteria in C. jejuni isolation from wastewater (Table 3). Baylis et al. (2000) identified competitor organisms in foods by using Preston and Bolton selective supplement media, showing that Yersinia, Enterobacter, Escherichia, Enterococcus, Pseudomonas, and Klebsiella are representative competitors. This is quite similar to our results from the BN enrichment conditions. Escherichia were frequently isolated in BN (Table 3), presumably because extended-spectrum beta-lactamase (ESBL)-producing E. coli may reduce the selectivity of Bolton supplement and consequently E. coli growth would suppress Campylobacter (Moran et al., 2011). Although the supplementation of additional antibiotic(s) suppressed the overgrowth of competing bacteria and enriched Campylobacter, Enterococcus survived well in the presence of five different antibiotics as it was frequently isolated with Campylobacter (Table 3). The survival of Enterococcus in the presence of vancomycin (20 μg/mL) in Bolton supplement indicated that Enterococcus isolated from the enrichment broth is vancomycin-resistant enterococci (VRE), a drug-resistant strain of serious public health concern (Cetinkaya et al., 2000). This study aimed at developing an improved culture method to isolate Campylobacter from wastewater, and we used the influent samples, not the effluent, since the influent is more contaminated than the effluent. Therefore, the results do not provide the information about the level of Campylobacter contamination in the effluent that may have a direct impact on public health compared to the influent data.
In this study, we demonstrated that antibiotic selective pressure and culture temperature are the critical factors for C. jejuni isolation from raw sewage. The BN, BNR, BNP, and BNRP conditions showed similar MPN values at 42°C; however, only BNRP showed reasonable MPN values and BNR and BNP showed relatively lower MPN numbers at 37°C compared to those at 42°C (data not shown). The results exhibited that culture temperature also plays an important role in the selective enrichment of C. jejuni. Humphrey et al. showed the effect of antibiotics and temperature on the recovery rate in cold-damaged C. jejuni. The sub-lethally injured cells are more sensitive to antibiotics in 43°C than 37°C, affecting the restoration of C. jejuni (Humphrey, 1986). In previous studies, Humphrey et al. also suggested that pre-incubation at 37°C for 4–18 h followed 42 or 37°C incubation for 48 h would be beneficial to the recovery of Campylobacter in comparison with 42°C (Humphrey, 1989; Humphrey and Muscat, 1989). Whereas Khan et al. (2013) demonstrated that the detection frequency of Campylobacter spp. was higher at 37°C in BN than 42°C, C. jejuni was detected more frequently at 42°C than at 37°C. Consistently, our results suggested that 42°C seems to enhance C. jejuni growth in raw sewage samples.
The additional antibiotic(s) plus an increased incubation temperature (i.e., 42°C) improved the isolation rates of C. jejuni and C. coli from heavily contaminated raw sewage samples. The addition of rifampicin and polymyxin B specifies the selective enrichment of thermo-tolerant Campylobacter spp., such as C. jejuni and C. coli, the major human pathogenic species (Kaakoush et al., 2015). Based on our findings, increased antibiotic selective pressure and culture temperature are the key parameters impacting the success in C. jejuni isolation from heavily contaminated wastewater samples. Additionally, rifampicin appears to be effective in improving the selectivity of Campylobacter enrichment for PCR-based quantitative methods, whereas both rifampicin and polymyxin B are required to suppress competing bacterial growth and improve the selectivity of C. jejuni isolation with culture-based methods.
Author Contributions
Design of the project: JK, NA, NN, and BJ; Performance of the experiments: JK, EO, GB, and SB; Data analysis: JK, EO, GB, SB, LC, NA, NN, and BJ; Writing of the manuscript: JK, NA, and BJ.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was supported by Alberta Innovates-Energy and Environment Solutions (AI-EES). The laboratory facilities were supported by the Canada Foundation for Innovation (CFI).
References
- Abulreesh H. H., Paget T. A., Goulder R. (2005). Recovery of thermophilic Campylobacters from pond water and sediment and the problem of interference by background bacteria in enrichment culture. Water Res. 39 2877–2882. 10.1016/j.watres.2005.05.004 [DOI] [PubMed] [Google Scholar]
- Baylis C. L., Macphee S., Martin K. W., Humphrey T. J., Betts R. P. (2000). Comparison of three enrichment media for the isolation of Campylobacter spp. from foods. J. Appl. Microbiol. 89 884–891. 10.1046/j.1365-2672.2000.01203.x [DOI] [PubMed] [Google Scholar]
- Bullman S., O’leary J., Corcoran D., Sleator R. D., Lucey B. (2012). Molecular-based detection of non-culturable and emerging campylobacteria in patients presenting with gastroenteritis. Epidemiol. Infect. 140 684–688. 10.1017/S0950268811000859 [DOI] [PubMed] [Google Scholar]
- Cetinkaya Y., Falk P., Mayhall C. G. (2000). Vancomycin-resistant enterococci. Clin. Microbiol. Rev. 13 686–700. 10.1128/CMR.13.4.686-707.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenu J. W., Pavic A., Cox J. M. (2013). A novel miniaturized most probable number method for the enumeration of Campylobacter spp. from poultry-associated matrices. J. Microbiol. Methods 93 12–19. 10.1016/j.mimet.2013.01.013 [DOI] [PubMed] [Google Scholar]
- Chon J. W., Kim H., Yim J. H., Park J. H., Kim M. S., Seo K. H. (2013). Development of a selective enrichment broth supplemented with bacteriological charcoal and a high concentration of polymyxin B for the detection of Campylobacter jejuni and Campylobacter coli in chicken carcass rinses. Int. J. Food. Microbiol. 162 308–310. 10.1016/j.ijfoodmicro.2013.01.018 [DOI] [PubMed] [Google Scholar]
- Clark C. G., Price L., Ahmed R., Woodward D. L., Melito P. L., Rodgers F. G., et al. (2003). Characterization of waterborne outbreak-associated Campylobacter jejuni, Walkerton, Ontario. Emerg. Infect. Dis. 9 1232–1241. 10.3201/eid0910.020584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute [CLSI] (2010). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria; Approved Guideline. M45-A2 2nd Edn Wayne, PA: CLSI. [Google Scholar]
- Cody A. J., Mccarthy N. D., Bray J. E., Wimalarathna H. M., Colles F. M., Jansen Van Rensburg M. J., et al. (2015). Wild bird-associated Campylobacter jejuni isolates are a consistent source of human disease, in Oxfordshire, United Kingdom. Environ. Microbiol. Rep. 7 782–788. 10.1111/1758-2229.12314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry J. E. L., Post D. E., Colin P., Laisney M. J. (1995). Culture media for the isolation of Campylobacters. Int. J. Food. Microbiol. 26 43–76. 10.1016/0168-1605(95)00044-K [DOI] [PubMed] [Google Scholar]
- de Boer R. F., Ott A., Guren P., Van Zanten E., Van Belkum A., Kooistra-Smid A. M. D. (2013). Detection of Campylobacter species and Arcobacter butzleri in stool samples by use of real-time multiplex PCR. J. Clin. Microbiol. 51 253–259. 10.1128/JCM.01716-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deer D. M., Lampel K. A., Gonzalez-Escalona N. (2010). A versatile internal control for use as DNA in real-time PCR and as RNA in real-time reverse transcription PCR assays. Lett. Appl. Microbiol. 50 366–372. 10.1111/j.1472-765X.2010.02804.x [DOI] [PubMed] [Google Scholar]
- Gabrielson J., Hart M., Jarelöv A., Kühn I., Mckenzie D., Möllby R. (2002). Evaluation of redox indicators and the use of digital scanners and spectrophotometer for quantification of microbial growth in microplates. J. Microbiol. Methods 50 63–73. 10.1016/S0167-7012(02)00011-8 [DOI] [PubMed] [Google Scholar]
- Hanninen M. L., Haajanen H., Pummi T., Wermundsen K., Katila M. L., Sarkkinen H., et al. (2003). Detection and typing of Campylobacter jejuni and Campylobacter coli and analysis of indicator organisms in three waterborne outbreaks in Finland. Appl. Environ. Microbiol. 69 1391–1396. 10.1128/AEM.69.3.1391-1396.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokajarvi A. M., Pitkanen T., Siljanen H. M., Nakari U. M., Torvinen E., Siitonen A., et al. (2013). Occurrence of thermotolerant Campylobacter spp. and adenoviruses in Finnish bathing waters and purified sewage effluents. J. Water Health. 11 120–134. 10.2166/wh.2012.192 [DOI] [PubMed] [Google Scholar]
- Hughes R. A., Cornblath D. R. (2005). Guillain-Barré syndrome. Lancet 366 1653–1666. 10.1016/S0140-6736(05)67665-9 [DOI] [PubMed] [Google Scholar]
- Humphrey T. J. (1986). Techniques for the optimum recovery of cold injured Campylobacter jejuni from milk or water. J. Appl. Bacteriol. 61 125–132. 10.1111/j.1365-2672.1986.tb04265.x [DOI] [PubMed] [Google Scholar]
- Humphrey T. J. (1989). An appraisal of the efficacy of pre-enrichment for the isolation of Campylobacter jejuni from water and food. J. Appl. Bacteriol. 66 119–126. 10.1111/j.1365-2672.1989.tb02461.x [DOI] [PubMed] [Google Scholar]
- Humphrey T. J., Muscat I. (1989). Incubation-temperature and the isolation of Campylobacter jejuni from food, milk or water. Lett. Appl. Microbiol. 9 137–139. 10.1111/j.1472-765X.1989.tb00308.x [DOI] [Google Scholar]
- Jakopanec I., Borgen K., Vold L., Lund H., Forseth T., Hannula R., et al. (2008). A large waterborne outbreak of campylobacteriosis in Norway: the need to focus on distribution system safety. BMC. Infect. Dis. 8:128 10.1186/1471-2334-8-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokinen C., Edge T. A., Ho S., Koning W., Laing C., Mauro W., et al. (2011). Molecular subtypes of Campylobacter spp. Salmonella enterica, and Escherichia coli O157:H7 isolated from faecal and surface water samples in the Oldman River watershed, Alberta, Canada. Water Res. 45 1247–1257. 10.1016/j.watres.2010.10.001 [DOI] [PubMed] [Google Scholar]
- Jones K. (2001). Campylobacters in water, sewage and the environment. J. Appl. Microbiol. 90 68S–79S. 10.1046/j.1365-2672.2001.01355.x [DOI] [PubMed] [Google Scholar]
- Kaakoush N. O., Castano-Rodriguez N., Mitchell H. M., Man S. I. M. (2015). Global epidemiology of Campylobacter Infection. Clin. Microbiol. Rev. 28 687–720. 10.1128/CMR.00006-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I. U., Hill S., Nowak E., Edge T. A. (2013). Effect of incubation temperature on the detection of thermophilic Campylobacter species from freshwater beaches, nearby wastewater effluents, and bird fecal droppings. Appl. Environ. Microbiol. 79 7639–7645. 10.1128/AEM.02324-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I. U. H., Gannon V., Loughborough A., Jokinen C., Kent R., Koning W., et al. (2009). A methods comparison for the isolation and detection of thermophilic Campylobacter in agricultural watersheds. J. Microbiol. Methods 79 307–313. 10.1016/j.mimet.2009.09.024 [DOI] [PubMed] [Google Scholar]
- Kirk M. D., Pires S. M., Black R. E., Caipo M., Crump J. A., Devleesschauwer B., et al. (2015). World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med. 12:e1001921 10.1371/journal.pmed.1001921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenraad P. M. F. J., Rombouts F. M., Notermans S. H. W. (1997). Epidemiological aspects of thermophilic Campylobacter in water-related environments: a review. Water Environ. Res. 69 52–63. 10.2175/106143097X125182 [DOI] [Google Scholar]
- Law J. W.-F., Ab Mutalib N.-S., Chan K.-G., Lee L.-H. (2015). Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front. Microbiol. 5:770 10.3389/fmicb.2014.00770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque S., St-Pierre K., Frost E., Arbeit R. D., Michaud S. (2011). Determination of the optimal culture conditions for detecting thermophilic Campylobacters in environmental water. J. Microbiol. Methods 86 82–88. 10.1016/j.mimet.2011.04.001 [DOI] [PubMed] [Google Scholar]
- Linton D., Owen R. J., Stanley J. (1996). Rapid identification by PCR of the genus Campylobacter and of five Campylobacter species enteropathogenic for man and animals. Res. Microbiol. 147 707–718. 10.1016/S0923-2508(97)85118-2 [DOI] [PubMed] [Google Scholar]
- Millson M., Bokhout M., Carlson J., Spielberg L., Aldis R., Borczyk A., et al. (1991). An outbreak of Campylobacter jejuni gastroenteritis linked to meltwater contamination of a municipal well. Can. J. Public Health 82 27–31. [PubMed] [Google Scholar]
- Moran L., Kelly C., Cormican M., Mcgettrick S., Madden R. H. (2011). Restoring the selectivity of bolton broth during enrichment for Campylobacter spp. from raw chicken. Lett. Appl. Microbiol. 52 614–618. 10.1111/j.1472-765X.2011.03046.x [DOI] [PubMed] [Google Scholar]
- Parkhill J., Wren B. W., Mungall K., Ketley J. M., Churcher C., Basham D., et al. (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403 665–668. 10.1038/35001088 [DOI] [PubMed] [Google Scholar]
- Pitkanen T. (2013). Review of Campylobacter spp. in drinking and environmental waters. J. Microbiol. Methods 95 39–47. 10.1016/j.mimet.2013.06.008 [DOI] [PubMed] [Google Scholar]
- Rosef O., Rettedal G., Lageide L. (2001). Thermophilic campylobacters in surface water: a potential risk of campylobacteriosis. Int. J. Environ. Health Res. 11 321–327. 10.1080/09603120120081791 [DOI] [PubMed] [Google Scholar]
- Schrader C., Schielke A., Ellerbroek L., Johne R. (2012). PCR inhibitors - occurrence, properties and removal. J. Appl. Microbiol. 113 1014–1026. 10.1111/j.1365-2672.2012.05384.x [DOI] [PubMed] [Google Scholar]
- Silva J., Leite D., Fernandes M., Mena C., Gibbs P. A., Teixeira P. (2011). Campylobacter spp. as a foodborne pathogen: a review. Front. Microbiol. 2:200 10.3389/fmicb.2011.00200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. E., Courvalin P. (1988). Mechanisms of antibiotic-resistance in Campylobacter species. Antimicrob. Agents Chemother. 32 1107–1112. 10.1128/AAC.32.8.1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke M. I., Morton V. K., Mclellan N. L., Huck P. M. (2010). The occurrence of Campylobacter in river water and waterfowl within a watershed in southern Ontario, Canada. J. Appl. Microbiol. 109 1053–1066. 10.1111/j.1365-2672.2010.04730.x [DOI] [PubMed] [Google Scholar]
- Weisburg W. G., Barns S. M., Pelletier D. A., Lane D. J. (1991). 16s ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiley H., Van Den Akker B., Giglio S., Bentham R. (2013). The role of environmental reservoirs in human campylobacteriosis. Int. J. Environ. Res. Public Health 10 5886–5907. 10.3390/ijerph10115886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender J., Flemming H. C. (2011). Biofilms in drinking water and their role as reservoir for pathogens. Int. J. Hyg. Environ. Health 214 417–423. 10.1016/j.ijheh.2011.05.009 [DOI] [PubMed] [Google Scholar]
- Yamazaki-Matsune W., Taguchi M., Seto K., Kawahara R., Kawatsu K., Kumeda Y., et al. (2007). Development of a multiplex PCR assay for identification of Campylobacter coli Campylobacter fetus, Campylobacter hyointestinalis subsp hyointestinalis, Campylobacter jejuni, Campylobacter lari and Campylobacter upsaliensis. J. Med. Microbiol. 56 1467–1473. [DOI] [PubMed] [Google Scholar]
- Yoo J. H., Choi N. Y., Bae Y. M., Lee J. S., Lee S. Y. (2014). Development of a selective agar plate for the detection of Campylobacter spp. in fresh produce. Int. J. Food Microbiol. 189 67–74. 10.1016/j.ijfoodmicro.2014.07.032 [DOI] [PubMed] [Google Scholar]