Abstract
Although normally regarded harmless commensals, enterococci may cause a range of different infections in humans, including urinary tract infections, sepsis, and endocarditis. The acquisition of vancomycin resistance by enterococci (VRE) has seriously affected the treatment and infection control of these organisms. VRE are frequently resistant to all antibiotics that are effective treatment for vancomycin-susceptible enterococci, which leaves clinicians treating VRE infections with limited therapeutic options. With VRE emerging as a global threat to public health, we aimed to isolate, identify enterococci species from tilapia and their resistance to van-mediated glycopeptide (vanA and vanC) as well as the presence of enterococcal surface protein (esp) using conventional and molecular methods. The cultural, biochemical (Vitek 2 system) and polymerase chain reaction results revealed eight Enterococcus isolates from the 80 fish samples (10%) to be further identified as E. faecalis (6/8, 75%) and E gallinarum (2/8, 25%). Intraperitoneal injection of healthy Nile tilapia with the eight Enterococcus isolates caused significant morbidity (70%) within 3 days and 100% mortality at 6 days post-injection with general signs of septicemia. All of the eight Enterococcus isolates were found to be resistant to tetracycline. The 6/6 E. faecalis isolates were susceptible for penicillin, nitrofurantoin, gentamicin, and streptomycin. On the other hand 5/6 were susceptible for ampicillin, vancomycin, chloramphenicol, and ciprofloxacin. The two isolates of E. gallinarum were sensitive to rifampicin and ciprofloxacin and resistant to vancomycin, chloramphenicol, and erythromycin. Molecular characterization proved that they all presented the prototypic vanC element. On the whole, one of the two vancomycin resistance gene was present in 3/8 of the enterococci isolates, while the esp virulence gene was present in 1/8 of the enterococci isolates. The results in this study emphasize the potential role that aquatic environments are correlated to proximity to anthropogenic activities in determining the antimicrobial resistance patterns of Enterococcus spp. recovered from fish in the river Nile in Giza, Elmounib, Egypt as a continuation of our larger study on the reservoirs of antibiotic resistance in the environment.
Keywords: nile tilapia, antibiotic resistance phenotype, esp, vanA and vanC genes, E. faecalis, E. gallinarum
Introduction
Some of the features that have made tilapia an advantageous fish species for culture are: rapid growth rate, firm and white muscle, the ability to survive in poor water conditions, and high reproductive success with limited requirements during incubation (Nandlal and Pickering, 2004). They belong to the base of the food chain, which makes them suitable to consume low cost and wide range of food sources with reduced ecological impact (Beveridge and Baird, 1998). In Egypt, the inland waters capture reached 250,196 in 2013 (FAO, 2015-2016). Egypt Aquaculture Production in 2010 was 900,000 tons of which Nile tilapia constituted more than 55% of the total production (FAO, 2015-2016) and in 2008, Egypt produced 13.8% of the world’s cultured tilapias (FAO, 2015-2016).
The global increase in intensive fish farming has been accompanied by bacterial infections that are usually treated with antibiotics added to fish foodstuffs as therapeutic agents, prophylactics and growth promoters (Muñoz-Atienza et al., 2013) resulting in the emergence and spread of antibiotic-resistant bacteria in the aquaculture environment and increasing the risk of antibiotic resistance bacterial transfer to humans and in the alterations of the bacterial microbiota of the aquatic environment. The main problem with the use of antibiotics in aquaculture is that many of the antibiotics that are used are from antibiotic classes that are also used in the treatment of human bacterial infections. The problem of increasing resistance is an ongoing and evolving issue and to be recognized at the global level by several international organizations with the so-called Tripartite comprising the WHO, FAO and the OIE leading the discussions on critical needs for human and animal health, and on the relation between the usage of antimicrobials in animal production and the consequences for human health which led to draft the third revision of the “WHO List of Critically Important Antimicrobials for Human Medicine” (WHO, 2012b).
Enterococci are widely distributed in nature (Klare et al., 2003), also known to be present in the intestine of healthy fish, and are potential probiotic candidates in aquaculture. Enterococci have been known to be resistant to most antibiotics used in clinical practice (Lukášoá and Šustáčková, 2003) and have been implicated in severe human infections as a consequence of associated determinants of virulence and antimicrobial resistance with the vancomycin-resistant prevalence to the harbored vanA gene (Palazzo et al., 2011). Clinically relevant enterococci (Enterococcus faecalis, and E. gallinarum, the vanC-type resistance group) are frequent causes of nosocomial infections, like urinary tract infections and sepsis (Menichetti, 2005). Antibiotic resistance genes exist not only in nature and in humans but also in fish as an enormous reservoir of genes that can be transferred to human pathogens either by direct contact or indirectly through the consumption of contaminated fish. Examples of antibiotic resistance genes detected in aquaculture over the last 7 years are summarized in Rolain (Rolain, 2013). Since development of the second revision in 2009 (WHO, 2012a), the committee faced several new issues of which vancomycin-resistant enterococci was one of them. In 1993 the first non-human reservoir of vanA vancomycin resistant E. faecium were identified (Pruden and Shore, 2011).
As vancomycin resistant by enterococci (VRE) are emerging as a global threat to public health, therefore, the objective of the study was to improve the understanding of antibiotic resistance through characterization of antibiotic-resistant VRE isolates associated with the fish retail market. Assessment and identification of the Enterococcus species was implanted by conventional and molecular (16S rRNA and ddlE.faecalis) assays isolated from tilapia and to evaluate the virulence impression esp which encodes for the Enterococcus surface protein (ESP), which participates in biofilm formation and is associated with colonization and persistence in the urinary tract promotes adhesion, colonization and evasion of the immune system, and to play some role in antibiotic resistance (Foulquie Moreno et al., 2006). The two vancomycin antimicrobial resistance genes vanA (associated with a high level of inducible resistance to vancomycin and cross resistance to teicoplanin) and vanC (mediated by the chromosomal VANC1 gene, which is constitutively present in E. gallinarum conferring relatively low resistance levels to vancomycin and is not transferable) were also molecularly assayed.
Materials and Methods
Fish Collection and Sampling
Eighty random fish were sampled from five different market types: supermarkets, fish mongers and fish vendors, landing sites, wholesale markets. Fish were placed in a cooler containing ice and transported back to lab. The liver, kidney, brain, spleen and ascitic fluid were removed by dissecting the fish. Twenty-five grams of each sample was homogenized in 225 mL of bile esculin azide broth (BBL) in Stomacher Lab Blender 400 for 30 s from which 0.1 and 0.01 mL were spread onto the surface of bile esculin azide agar plates (BBL) and incubated at 45°C for 24 and 48 h. The initial dilutions were treated similarly. In order to guarantee the absence of contamination, typical enterococci colonies (black) on bile esculine azide agar were streaked onto blood agar (Oxoid) plates prepared with 5% of defibrinated sheep blood and incubated at 37°C for 24 and 48 h. The phenotypic characterization of bacterial isolates was studied to determine their colony morphology, cell morphology, motility, Gram stain and to be finally characterized using the Vitek 2 system (bioMerieux SA, Marcy I’Etoile, France).
Molecular Identification of Enterococcus Species
Identification of presumptive enterococci was confirmed using the polymerase chain reaction (PCR): one mL of culture was aseptically spreaded on MRS agar plates and was incubated at 25°C for 3–5 days. Eventually, the forming colonies were transferred onto another new MRS agar plates until discrete colonies were obtained. Each discrete colony was further subcultured for several times to ensure that a pure culture was obtained. The virulent strain was identified by biochemical characterization based on the ability of utilization of different carbon sources by Vitek 2 (bioMerieux, Lyon, France). The genomic DNA was extracted using Wizard Genomic DNA Purification Kit (Promega, USA) and used as a template for PCR using Enterococcus genus-specific primers to amplify the conserved sequence of the 16S rRNA gene (Karsidani et al. (2010) that yield a 112 bp PCR product as outlined in Table 1. PCR products were analyzed by electrophoresis on 1–2% (w/v) agarose (Pronadisa, Madrid, Spain) gels stained with Gel red (Biotium, Fremont, CA, USA), and visualized with the Gel Doc 1000 documentation system (Bio-Rad, Madrid, Spain).
Table 1.
Oligonucleotide primers sequences and size of the PCR-targeted products.
| Microorganism | Target gene | bp fragment | Primer sequence (5′–3′) | Annealing temp (°C) | Reference |
|---|---|---|---|---|---|
| E. faecalis/E. gallinarum | 16S rRNA (Genus -specific primers) | 112 bp | F TAC TGA CAA ACC ATT CAT GAT G | 59 (E. faecalis) | Karsidani et al., 2010 |
| R AAC TTC GTC ACC AAC GCG AAC | 50 (E. gallinarum) | ||||
| vanA (Glycopeptide resistance genotype Vancomycin resistance) | 732 bp | F GGG AAA ACG ACA ATT GC | 59 (E. faecalis) | Biavasco et al., 2007; Karsidani et al., 2010 | |
| R GTA CAA TGCG GCC GTTA | 50 (E. gallinarum) | ||||
| vanC1 (Glycopeptide resistance genotype Vancomycin resistance) | 822 bp | F GGT ATC AAG GAA ACC TC | 59 (E. faecalis) | ||
| R CTT CCG CCA TCA TAG CT | 50 (E. gallinarum) | ||||
| E. faecalis |
ddlE.faecalis (Species-specific primers) |
941 bp | F ATC AAG TAC AGT TAG TCT | 55 | |
| R ACG ATT CAA AGC TAA CTG | |||||
|
esp (virulence factor) |
932 bp | F TTG CTA ATG CTA GTC CAC GACC | 55 | ||
| R GCG TCA ACA CTT GCA TTG CCG AA |
Detection of In vivo Potential Virulence
Experimental Infection
The study was approved by the Cairo University ethical committee for animal research, Egyptian animal welfare agency, Cairo, Egypt. The procedures used for animal care and housing were in accordance with the U.S. Department of Agriculture through the Animal Welfare Act (7USC 2131) 1985 and Animal Welfare Standards incorporated in 9 CFR Part 3, 1991.
In order to determine the virulent capacities of the eight isolates, infectivity trials were conducted through the intraperitoneal injection of each of the eight Enterococcus isolates using Nile tilapia as a model. Prior to challenge, the fish were randomly sampled to be subjected to microbiological analysis which indicated that they were free of enterococci. As a control, E. faecalis strains V583 and OG1RF, as well as E. faecium 64/3 were used. The procedure carried out by Martins et al. (2008) was adopted. Eighty fish weighing 55.0 ± 5.0 g were distributed in 12 aerated aquaria of 100 cm × 40 cm × 25 cm and acclimatized for 10 days before assay. During the course of the experiments (21 days) a daily 50% water renovation was provided and feeding the fish with commercial diet. During this period, the water temperature was maintained at 25.0 ± 0.2°C, pH 7.0 ± 0.2 and ammonia 0.5 ± 0.1 mg L-1. The colonies confirmed as Enterococcus sp. by PCR (E. faecalis, E. gallinarum) were prepared as an inoculum of the bacteria diluted in tubes containing infusion of heart and brain (BHI; Difco, Detroit, MI, USA) to reach the concentration of 1 × 107 CFU/mL estimated by serial-dilution method (1:10). The experiment was done by challenging the fish with 107 CFU/ml of each of the live Enterococcus species (E. faecalis, E. gallinarum). The 80 fish were divided into eight groups, each group comprised of 10 fish was intraperitoneally challenged with each individual Enterococcus isolate. The control fish (unchallenged group) received 0.1 μl of saline solution (0.85% NaCl) through i.p. injection. The clinical signs of the fish were observed continuously within 24 h duration of the experiment and fish mortality was daily checked during this period. The liver, brain and kidney from the dead fish were collected and microbiologically analyzed for re-isolation and identification of the inoculated strain by bacterial culture and PCR. The degree of virulence was expressed as lethal dose 50% (LD50) calculated as described by Reed and Müench (1938).
Bacteria Culture
The swab from the organs that were collected four hourly were immediately streaked onto the blood agar plates and incubated at 30°C for 24 h. Gram staining were performed to identify Gram-positive cocci in chain or paired and catalase test negative organisms. Finally, the colonies were further characterized using the Vitek 2 system (bioMerieux).
Polymerase Chain Reaction
For confirmation of E. faecalis and E. gallinarum, total cellular DNA was extracted using Wizard Genomic DNA Purification Kit (Promega, USA) according to manufacturer’s protocol. The extracted DNA was then further evaluated by PCR for E. faecalis and E. gallinarum -specific section of 16S rRNA. PCR products were analyzed by electrophoresis as previously indicated.
PCR Detection of the Potential Virulence Factor esp in E. faecalis and E. gallinarum
Detection of the esp (enterococcal surface protein) gene in the eight enterococci was performed by PCR (Table 1). The positive control strain for detection of potential virulence factor was E. faecalis P36 for esp. PCR-amplifications was performed from total bacterial DNA obtained using the Wizard DNA Purification Kit (Promega, Madrid, Spain) in 25 μl reaction mixtures with 1 μl of purified DNA, 0.7 μM of each primer, 0.2 mM of each dNTP, buffer 1×, 1.5 mM MgCl2 and 0.75 U of Platinum Taq DNA polymerase (Invitrogen, Madrid, Spain). PCR products were analyzed by electrophoresis as previously indicated.
Determination of Antibiotic Resistance Phenotype
Antibiotic susceptibility of the eight enterococci was determined by overlaying antibiotic-containing disks (Oxoid) on Diagnostic Sensitivity Test Agar (Oxoid) previously seeded with approximately 1 × 105 CFU/ml of each enterococcal isolate. The antibiotics tested and their concentrations are listed in Table 2. Inhibition zone diameters were measured after overnight incubation of the plates at 37°C. Resistance phenotypes were recorded as recommended by the CLSI (2011). All isolates were subjected to the minimal inhibitory concentration (MIC) to vancomycin and the assessments were conducted using the microbroth dilution method as previously recommended (CLSI, 2011). E. faecalis ATCC 29212 and Staphylococcus aureus ATCC29213 were used as quality control strains.
Table 2.
Antibiotic interpretation for Enterococcus species.
| On the WHO’s critically important antimicrobials list (WHO, 2012b) | Antibiotic disk | Concentration | Antibiotic interpretation |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
E. faecalis (n = 6) |
E. gallinarum (n = 2) |
|||||||||
| Sensitive |
Resistant |
Sensitive |
Resistant |
|||||||
| n | % | n | % | n | % | n | % | |||
| Penicillins | ||||||||||
| Critically important | Penicillin | 10 μg | 6 | 100 | 0 | 0 | 1 | 50 | 1 | 50 |
| Critically important | Ampicillin | 10 μg | 5 | 83.3 | 1 | 16.7 | 1 | 50 | 1 | 50 |
| Glycopeptides | ||||||||||
| Critically important | Vancomycin | 30 μg | 5 | 83.3 | 1 | 16.7 | 0 | 0 | 2 | 100 |
| Macrolides | ||||||||||
| Critically important | Erythromycin | 15 μg | 2 | 33.3 | 4 | 66.7 | 0 | 0 | 2 | 100 |
| Tetracyclines | ||||||||||
| Highly important | Tetracycline | 30 μg | 0 | 0 | 6 | 100 | 0 | 0 | 2 | 100 |
| Fluoroquinolones | ||||||||||
| Critically important | Ciprofloxacin | 5 μg | 5 | 83.3 | 1 | 16.7 | 2 | 100 | 0 | 0 |
| Nitrofurantoins | ||||||||||
| Important | Nitrofurantoin | 300 μg | 6 | 100 | 0 | 0 | 1 | 50 | 1 | 50 |
| Phenicols | ||||||||||
| Highly important | Chloramphenicol | 30 μg | 5 | 83.3 | 1 | 16.7 | 0 | 0 | 2 | 100 |
| Ansamycins | ||||||||||
| Critically important | Rifampicin | 5 μg | 2 | 33.3 | 4 | 66.7 | 2 | 100 | 0 | 0 |
| Aminoglycosides | ||||||||||
| Critically important | Gentamicin | 10 μg | 6 | 100 | 0 | 0 | 1 | 50 | 1 | 50 |
| Critically important | Streptomycin | 10 μg | 6 | 100 | 0 | 0 | 1 | 50 | 1 | 50 |
Detection of Genetic Determinants of Antimicrobial Resistance
Total bacterial DNA was extracted by the GenEluteTM Bacterial Genomic DNA Kit (Sigma-Aldrich, St. Louis, MO, USA). Vancomycin-resistant isolates were studied for the presence of the glycopeptides resistance genes VanA and VanC1 by PCR (Table 1).
Results
The clinical picture and postmortem lesions of the experimentally infected tilapia were seen as: external lesions (Figure 1): eye lesions: in the form of unilateral or bilateral eye redness/opacity; Skin lesions: detached scales, extensive skin congestion, ulcers, hemorrhage, or dark discoloration in the form of strips; Fins: congestion at the base of the fins, or even hemorrhagic; Abdomen: slightly distended in some cases; Anal opening: congested with protruded anal opening.
FIGURE 1.

Tilapia fish showing externally eye opacity, dark skin discoloration in the form of strips, congested base at the pelvic fin, hemorrhagic pelvic fin, and distended abdomen.
Postmortem lesions (Figure 2): general signs of septicemia; Muscle: redness; Kidney: renal congestion; Liver: friable, pale and congested, dark gall bladder; Spleen: enlarged and congested; Heart: congested; Intestine: intestinal congestion and devoid of food; Gill: congested; Abdominal cavity: hemorrhagic ascites.
FIGURE 2.
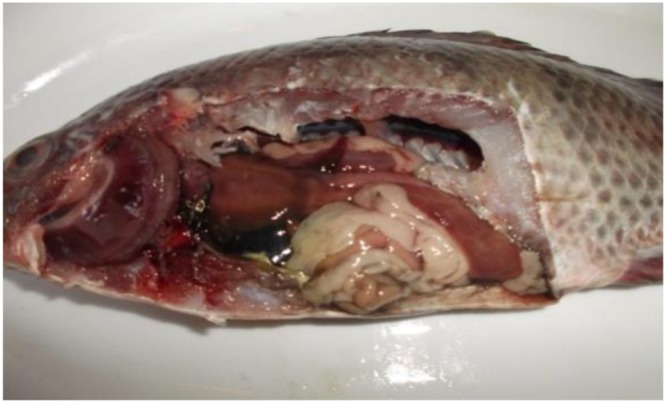
Tilapia fish postmortem internal findings revealed typical signs of septicemia in which severely congested gill, kidney, spleen, intestine and heart, dark gall bladder, in addition to hemorrhagic ascites, friable mottled liver, muscle redness, and parts of the intestine were devoid of food.
Biochemical identification of the Enterococcus species was successfully implemented through the Vitek 2 system. The cultural, biochemical (Vitek 2 system) and PCR results revealed eight Enterococcus strains from the 80 fish samples (10%) to be further identified as E. faecalis (6/8, 75%) and E gallinarum (2/8, 25%). Although the eight isolates were non-hemolytic, they were then proven to be pathogenic through the in vivo experimental infection. Intraperitoneal injection of healthy Nile tilapia with the eight Enterococcus isolates caused significant morbidity (70%) within 3 days and 100% mortality at 6 days post-injection with general signs of septicemia.
The antibiotic sensitivity test for the Enterococcus species showed that 6/6 of the E. faecalis were susceptible for penicillin, nitrofurantoin, gentamicin, and streptomycin, while all isolates (8/8) were resistant to tetracycline (Table 2). Only 1/6 E. faecalis isolates was resistant to vancomycin. On the other hand, 5/6 of the E. faecalis were susceptible for ampicillin, vancomycin, chloramphenicol, and ciprofloxacin. The lowest rate of sensitivity was observed for rifampicin and erythromycin (2/6). The 2/2 isolates of E. gallinarum were sensitive to rifampicin and ciprofloxacin, but resistant to vancomycin, chloramphenicol, and erythromycin. Among the eight isolates, 1/2 E. gallinarum and 4/6 E. faecalis were resistant to at least three antibiotics. High level of multidrug resistance to critically important antibiotics was detected in E. gallinarum strains (6/11 of E. gallinarum versus 5/11 of E. faecalis). Vancomycin resistant isolates exhibited high and consistent MIC values (≤230 μg/ml).
As indicated in Table 3, the vanC and vanA resistance genes were present in combination in one of the two E. gallinarum isolates (50%), while the second isolate carried the vanC gene only (50%). The E. faecalis (six isolates), exhibited the following pattern: the vanA gene was found in 1/6 isolates, while none of the vanA and vanC were indicated in 5/6 isolates.
Table 3.
Genetic profile of isolated bacteria.
| Bacterial isolates | Total n = isolates | Genetic profile | n = of +ive isolates | % |
|---|---|---|---|---|
| E. gallinarum | 2 | 16S rRNA+ VanC | 1 | 50 |
| 16S rRNA+ VanA+ VanC | 2 | 100 | ||
| E. faecalis | 6 | 16S rRNA + VanC+ ddlE.faecalis | 0 | 0 |
| 16S rRNA + ddlE.faecalis + esp | 1 | 16.6 | ||
| 16S rRNA + ddlE.faecalis + VanA | 1 | 16.6 | ||
| 16S rRNA + ddlE.faecalis | 4 | 50 |
Detection of in vivo potential virulence: experimental infection.
The virulence gene esp was found in 1/6 of the E. faecalis isolates and absent from the 2/2 E. gallinarum isolates.
On the whole, one of the two vancomycin resistance gene was present in 37.5% of the enterococci isolates (3/8), while the esp virulence gene was present in one out of the eight enterococci isolates (12.5%).
Discussion
Public health impacts from antibiotic use in agriculture and aquaculture have already drawn much attention in the last decade (Heuer et al., 2009; Marshall and Levy, 2011; Kuehn, 2012; WHO, 2012a). Therefore, it is important to monitor the occurrence of antibiotic resistant bacteria in surface waters used for drinking and irrigation purposes since these bacteria are able to spread through food to humans.
For species differentiation, the motility and pigment tests were performed to phenotypically distinguish among species where E. faecalis was non-motile whereas E. gallinarum was motile (CDC, 2010). In vivo experimentation, using local strains of Nile tilapia, was utilized to elucidate and assess the in vivo survival and pathogenic potential of the eight enterococci (E. faecalis, n = 6 and E. gallinarum, n = 2) strains. Growth in vitro did not differ between these two strains. In the Nile tilapia model, the eight isolates were rapidly lethal and pathogenic. Koch’s postulates confirmed E. faecalis and E. gallinarum as the causative agents of enterococcosis among the captured Nile tilapia in Egypt.
Various lists of critically important antibiotics, such as those published by the WHO (2012b) are also given for prophylaxis in industrial fish aquaculture (Cabello, 2006) to fight disease or/and growth promoters to end up on our table as seafood. This has resulted in an increase of both antibiotic resistance bacteria in fish pathogens and in antibiotic resistance in the environment (Cabello, 2006) which must consequently in an act to prioritize the importance of strict regulatory use of antibiotics. Researchers at the Arizona State University’s Biodesign institute examined antibiotic use in shrimp, salmon, catfish, trout, tilapia and swai, originating from 11 countries (Bangladesh, Canada, Chile, China, Indonesia, Mexico, Panama, Scotland, Thailand, USA, and Vietnam). Of the 47 antibiotics evaluated, they discovered traces of just five (Done and Halden, 2015) of which, oxytetracycline was the most prevalent in the study samples. They hypothesized that the very low number of antibiotics (5/47) recovered was because antibiotics have a period of time between when they are administered and when the fish is processed (Done and Halden, 2015). However, regulations in this area vary between countries (Luening, 2015). As in Vietnam (Pham et al., 2015), the Egyptian Farmers’ decision-making processes about antimicrobial use, are influenced by biased sources of information, such as drug manufacturers, sellers and by financial incentives.
Muñoz-Atienza et al. (2013) found that, the percentage of enterococcal strains showing acquired antibiotic resistance was 68%. Interestingly, our results found E. faecalis with a high percentage of resistance to ciprofloxacin and/or norfloxacin, rifampicin, and glycopeptides with a very low resistance (5%) to erythromycin and tetracycline. In spite of the high prevalence of acquired antibiotic resistance found in their enterococci of aquatic origin, they showed low incidence or absence of resistance to the clinically relevant antibiotics vancomycin, ampicillin, penicillin and gentamicin, which is in agreement with previous studies but in contrast to our findings. Fortunately, our observations indicated a low level of antibiotic resistance in the E. faecalis isolates making it a non-feared infectious agent in intensive care ward (Higuita and Huycke, 2014).
From the five recognized genes, vanA, vanB, vanC, vanD, and vanE contributing to vancomycin resistance in enterococci. (CDC, 2010), we focused on two genes vanA and vanC. These genes were previously studied in Egypt in large animals and poultry so it was our opinion to extend them to fish. In addition to motility and pigment tests, an organism’s susceptibility profile also helps differentiate vanA isolates from vanC isolates (CDC, 2010). Identification of VRE to species level aids in confirming whether an isolate has intrinsic (vanC) or acquired resistance (vanA). The vanA is mediated by newly acquired gene clusters not previously found in enterococci and described primarily in E. faecalis and vanA-resistant strains possess inducible, high-level resistance to vancomycin. On the other hand, the vanC resistance phenotype was described in E. gallinarum, which demonstrate intrinsic, low-level resistance to vancomycin. Knowledge of the type of resistance is critical for infection control purposes as vanA is transferable and can spread from organism to organism while in contrast, vanC genes is not transferable and has been associated less commonly with serious infections or associated with outbreaks (CDC, 2010).
The vanA resistance gene seems to be widespread in enterococci of natural ecosystems, in addition to clinical settings, and it would be interesting to track the routes of dissemination of these resistant microorganisms that have great importance in public health (Barros et al., 2012). Vancomycin resistance in enterococci isolated from food has a variable global pattern and the vancomycin resistant strains appears to be disparate according to the geographic region where they emerge (Franz et al., 2001; Giraffa, 2002; Gomes et al., 2008; Van Tyne and Gilmore, 2014). In Europe, this divergence is attributed to the use of antimicrobial agents as growth promoter whereas in the United States, it is attributed to the wide hospital usage of vancomycin (Van Tyne and Gilmore, 2014).
Enterococci should be considered a significant zoonotic pathogen and a possible reservoir of genes encoding resistance potentially transferred to other bacterial species (Kuhn et al., 2000) as there is ample evidence that many antibiotic resistance determinants found in pathogenic human bacteria have a fish origin (Cabello, 2006; Penders et al., 2013; Rolain, 2013). Spreading of resistance genes is a high risk as they are highly persistent and do not disappear from aquaculture sites, even after several years after eliminating the use of antibiotics (Tamminen et al., 2011). Thus, resistant bacteria from the fish that are consumed by humans, as well as direct contact between humans and fish, can be a source of antibiotic resistance gene transfer (Schjorring and Krogfelt, 2011). It must be emphasized that, 5/6 of the E. faecalis isolates were recovered from the river Nile in Giza ElMounib while the remaining one isolate was from an aquaculture site located in ElTal Elkebir in the Delta. This opens the discussion as to the significance and seriousness of the ElMounib location. In Egypt, poverty levels, overcrowdness, informal settlements in such areas as in ElMounib are issues that complicate the problem of antibiotic resistance where the presence of pathogenic enterococci in sewage-contaminated river water where bacterial contamination of surface water and particularly contamination with fecal-derived bacteria, has long been a water quality issue owing to the potential for disease transmission (Kinge et al., 2010; Romanis, 2013). It has been reported in the literature that surface waters have become a major reservoir of multi-antibiotic resistant pathogenic bacteria due to contamination by agricultural waste, animal excreta, the effluent water used for irrigation purposes and sewage disposal (Lupo et al., 2012). Consequently, the enterococci carrying ARG disseminated in the River Nile via waste water or run-off from livestock facilities and agriculture in the area under investigation contaminates the Nile tilapia.
The elevated levels of ARGs in aquatic environments are correlated to proximity to anthropogenic activities with special reference to environmental impact of agricultural effluent, irrigation water and fishing (Berglund, 2015), large-scale industrial agricultural facilities, which raise food animals at high-density and using antibiotics for treatment or as growth promoters (Burkholder et al., 2007; Gilchrist et al., 2007; Greger and Koneswaran, 2010). Similarly, impacts from large scale and widespread antibiotic use in aquaculture need to be re-assessed (Knapp et al., 2010; Weir et al., 2012). In aquaculture, fish infections are treated through the administration of antibiotics directly into the water, avoiding any kind of purification processes (Kümmerer, 2009). The presence of these organisms in the fish samples is disquieting as this could lead to major human health problems. This emphasizes the need for constant evaluation of the wastewater treatment effluents to ensure compliance to set guidelines (Olaniran et al., 2015).
The fact that the E. faecalis and E. gallinarum strains eval- uated in this work that were lacking the esp gene, might be related with their non-clinical origin and absence of ampicillin resistance. This was observed in previous studies which reported that esp are more common in ampicillin-resistant/vancomycin-resistant E. faecium (VREF) than in ampicillin-susceptible/VREF strains (Vankerckhoven et al., 2004; Muñoz-Atienza et al., 2013). In this context, the increase in the incidence of VREF at hospital settings has been attributed mainly to the spread of ampicillin-resistant VREF exhibiting esp (Klare et al., 2005; Werner et al., 2008; Muñoz-Atienza et al., 2013). The discrepancies found when comparing our data with those obtained in other studies could be due to several factors: (i) differences in the antibiotics used and sources isolated, (ii) to the resistance mechanism of Enterococcus spp. under antibiotic selection pressure induced by antibiotic use differing with the aquaculture environment, (iii) It could be that various global factors are relevant to the antibiotic resistance of bacteria, (iv) the differences among fish groups might be due to various non-antibiotic factors (e.g., cadmium) driving the co-selection of antibiotic resistance according to different methods of fish management.
Our PCR results showed that E. faecalis and E. gallinarum strains evaluated in this work showed that, the esp which encode a cell wall-associated protein involved in immune evasion, was absent in the two E. gallinarum strains and found in 1/6 of the E. faecalis strains isolated. The same gene was not found in any of the tested Lactic Acid Bacteria by Muñoz-Atienza et al. (2013). As screening for the presence of virulence genes to evaluate the potential virulence of enterococci could be speculative, empirical testing with a disease challenge was conducted. Experimental pathogenicity test in vivo proved that, the eight isolates of the Enterococcus species were pathogenic. The infection or pathogenicity process of Enterococcus spp. is very complex and is said to involve different virulent and pathogenicity factors which either act together or separately at different stages of infection (Al-Bahry et al., 2014). Several authors have analyzed a number of virulence factors in E. faecalis, including a structurally novel toxin, a surface protein, esp (Shankar et al., 1999), which contributes to colonization of the bladder in a model of urinary tract infection (Shankar et al., 2001) and confers to the biofilm production capability of enterococci (Toledo-Arana et al., 2001).
Conclusion
The present study supports the view that there is a risk of transfer of resistant bacteria to humans from consumption of fish and that follow-up studies are required to: (a) investigate the extent of antibiotic use in Egyptian aquaculture farms, (b) to determine the molecular basis of antimicrobial resistance to the different antibiotics, (c) the potential for transfer of resistance genes from aquaculture isolates to human pathogens, (d) some assessment of the risk of transfer of resistant organisms (or genes) to humans via food chain, and (e) the threats imposed by environmental contamination with antibiotic resistant bacteria. In addition to antibiotic resistance dissemination into the surrounding environment, the use of antibiotics in aquaculture facilities without proper sanitation techniques, the antibiotics can accumulate in the sediment at the bottom of aquaculture ponds for extended periods of time, creating constant pressure to bacterial populations that increase the risk of antibiotic resistance in bacterial pathogens found in the fish water environment (Baquero et al., 2008; Pruden and Shore, 2011) and to the personnel working in the facility. Therefore, subsequent action should be taken to enforce the restricted use of certain antibiotics that are critically important for the treatment of human illnesses.
Author Contributions
KO developed the concept, designed experiments, collected, and analyzed data and prepared the manuscript; MA, IR, FH, and AA gave technical support and conceptual advice; AO and NF performed PCR assays. All authors discussed the results and implications and commented on the manuscript at all stages.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Al-Bahry S. N., Mahmoud I. Y., Paulson J. R., Al-Musharafi S. K. (2014). Survival and growth of antibiotic resistant bacteria in treated wastewater and water distribution system and their implication in human health: a review. Int. Arabic J. Antimicrob. Agents 4 1–11. [Google Scholar]
- Baquero F., Martinez J. L., Canton R. (2008). Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 19 260–265. 10.1016/j.copbio.2008.05.006 [DOI] [PubMed] [Google Scholar]
- Barros J., Andrade M., Radhouani H., Lopez M., Igrejas G., Poeta P., et al. (2012). Detection of vawA-Containing Enterococcus species in faecal microbiota of gilthead seabream (Spams aurata). Microbes. Environ. 27 509–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund B. (2015). Environmental dissemination of antibiotic resistance genes and correlation to anthropogenic contamination with antibiotics. Infect. Ecol. Epidemiol. 5:28564 10.3402/iee.v5.28564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge M. C. M., Baird D. J. (1998). “Feeding mechanism and feeding ecology,” in Tilapias: Their Biology and Exploitation eds Beverigde M. C. M., McAndrew B. J. (London: Chapman and Hall; ). [Google Scholar]
- Biavasco F., Foglia G., Paoletti C., Zandri G., Magi G., Guaglianone E., et al. (2007). VanA-type enterococci from humans, animals, and food: species distribution, population structure, tn1546 typing and location, and virulence determinants. Appl. Environ. Microbiol. 73 3307–3319. 10.1128/AEM.02239-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkholder J., Libra B., Weyer P., Heathcote S., Kolpin D., Thorne P. S., et al. (2007). Impacts of waste from concentrated animal feeding operations on water quality. Environ. Health Perspect. 115 308–312. 10.1289/ehp.8839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello F. C. (2006). Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ. Microbiol. 8 1137–1144. 10.1111/j.1462-2920.2006.01054.x [DOI] [PubMed] [Google Scholar]
- CDC (2010). Vancomycin-resistant Enterococci (VRE) and the Clinical Laboratory Centers for Disease Control and Prevention National. (Center) for Emerging and Zoonotic Infectious Diseases (NCEZID) Division. (of) Healthcare Quality Promotion (DHQP). Atlanta, GA: Centers for Disease Control and Prevention National. [Google Scholar]
- CLSI (2011). Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; 21st Informational Supplement. M100-S21. Wayne, PA: CLSI. [Google Scholar]
- Done H. Y., Halden R. U. (2015). Reconnaissance of 47 antibiotics and associated microbial risks in seafood sold in the United States. J. Hazard. Mat. 282 10–17. 10.1016/j.jhazmat.2014.08.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2015-2016). Aquaculture topics and activities. The State of the World’s Aquatic Genetic Resources for Food and Agriculture. In: FAO Fisheries and Aquaculture Department [online]. Rome. Updated 22 June 2016. [Cited 10 July 2016]. Available at: http://www.fao.org/fishery/AquaticGeneticResources/en [Google Scholar]
- Foulquie Moreno M. R., Sarantinopoulos P., Tsakalidou E., De Vuyst L. (2006). The role and application of enterococci in food and health. Int. J. Food Microbiol. 106 1–24. 10.1016/j.ijfoodmicro.2005.06.026 [DOI] [PubMed] [Google Scholar]
- Franz C., Muscholl-Silberhorn A., Yousif N., Vancanneyt M., Swings J., Holzapfel W. (2001). Incidence of virulence factors and antibiotic resistance among Enterococci isolated from food. Appl. Environ. Microbiol. 67 4385–4389. 10.1128/AEM.67.9.4385-4389.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist M. J., Greko C., Wallinga D. B., Beran G. W., Riley D. G., Thorne P. S. (2007). The potential role of concentrated animal feeding operations in infectious disease epidemics and antibiotic resistance. Environ. Health Perspect. 115 313–316. 10.1289/ehp.8837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraffa G. (2002). Enterococci from foods. FEMS Microbiol. Rev. 26 163–171. 10.1111/j.1574-6976.2002.tb00608.x [DOI] [PubMed] [Google Scholar]
- Gomes B. C., Esteves C. T., Palazzo I. C. V., Darini A. L., Felis G. E., Sechi L. A.et al. (2008). Prevalence and characterization of Enterococcus spp. isolated from Brazilian foods. Food Microbiol. 25 668–675. 10.1016/j.fm.2008.03.008 [DOI] [PubMed] [Google Scholar]
- Greger M., Koneswaran G. (2010). The public health impacts of concentrated animal feeding operations on local communities. Fam. Community Health 33 11–20. 10.1097/FCH.0b013e3181c4e22a [DOI] [PubMed] [Google Scholar]
- Heuer O. E., Kruse H., Grave K., Collignon P., Karunasagar I., Angulo F. J. (2009). Human health consequences of use of antimicrobial agents in aquaculture. Clin. Infect. Dis. 49 1248–1253. 10.1086/605667 [DOI] [PubMed] [Google Scholar]
- Higuita N. I. A., Huycke M. M. (2014). “Enterococcal Disease, epidemiology, and implications for treatment,” in Enterococci: from Commensals to Leading Causes of Drug Resistant Infection eds Gilmore M. S., Clewell D. B., Ike Y., Shankar N. (Boston, MA: Eye and Ear Infirmary; ). [Google Scholar]
- Karsidani S. H., Soltani M., Nikbakhat-Brojeni G., Ghasemi M., Skall H. (2010). Molecular epidemiology of zoonotic streptococcosis/lactococcosis in rainbow trout (Oncorhynchus mykiss) aquaculture in Iran. Iran. J. Microbiol. 2 198–209. [PMC free article] [PubMed] [Google Scholar]
- Kinge C. N. W., Ateba C. N., Kawadza D. T. (2010). Antibiotic resistance profiles of Escherichia coli isolated from different water sources in the Mmabatho locality, North-West province, South Africa. South Afr. J. Sci. 106 44–49. 10.4102/sajs.v106i1/2.14 [DOI] [Google Scholar]
- Klare I., Konstabel C., Badstübner D., Werner G., Witte W. (2003). Occurrence and spread of antibiotic resistances in Enterococcus faecium. Int. J. Food Microbiol. 88 269–290. 10.1016/S0168-1605(03)00190-9 [DOI] [PubMed] [Google Scholar]
- Klare I., Konstabel C., Mueller-Bertling S., Werner G., Strommenger B., Kettlitz C., et al. (2005). Spread of ampicillin/vancomycin-resistant Enterococcus faecium of the epidemic-virulent clonal complex-17 carrying the genes esp and hyl in German hospitals. Eur. J. Clin. Microbiol. Infect. Dis. 24 815–825. 10.1007/s10096-005-0056-0 [DOI] [PubMed] [Google Scholar]
- Knapp C. W., Dolfing J., Ehlert P. A. I., Graham D. W. (2010). Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ. Sci. Technol. 44 580–587. 10.1021/es901221x [DOI] [PubMed] [Google Scholar]
- Kuehn B. M. (2012). FDA claims to curb farm use of antibiotics. JAMA 307 2244–2245. 10.1001/jama.2012.4560 [DOI] [PubMed] [Google Scholar]
- Kuhn I., Ivesen A., Burman L. G., Olsson-Liljequist B., Franklin A., Finn M., et al. (2000). Epidemiology and ecology of enterococci, with special reference to antibiotic resistant strains, in animals, humans and the environment. Int. J. Antimicrob. Agents 14 337–342. 10.1016/S0924-8579(00)00146-1 [DOI] [PubMed] [Google Scholar]
- Kümmerer K. (2009). Antibiotics in the aquatic environment—a review – Part I. Chemosphere 75 435–441. 10.1016/j.chemosphere.2008.12.006 [DOI] [PubMed] [Google Scholar]
- Luening (2015). Available at: http://aquaculturenorthamerica.com/research/survey-identifies-antibiotics-in-farm-raised-seafood/#. [Google Scholar]
- Lukášoá J., Šustáčková A. (2003). Enterococci and Antibiotic Resistance. Acta Vet. Brno 72 315–323. 10.2754/avb200372020315 [DOI] [Google Scholar]
- Lupo A., Coyne S., Berendonk T. U. (2012). Origin and evolution of antibiotic resistance: the common mechanisms of emergence and spread in water bodies. Front. Microbiol. 3:18 10.3389/fmicb.2012.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall B. M., Levy S. B. (2011). Food animals and antimicrobials: impacts on human health. Clin. Microbiol. Rev. 24 718–733. 10.1128/CMR.00002-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins M. L., Mouriño J. L. P., Amaral G. V., Vieira F. N., Dotta G., Jatobá A. M. B., et al. (2008). Haematological changes in Nile tilapia experimentally infected with Enterococcus sp. Braz. J. Biol. 68 657–661. 10.1590/S1519-69842008000300025 [DOI] [PubMed] [Google Scholar]
- Menichetti F. (2005). Current and emerging serious gram-positive infections. Clin. Microbiol. Infect. 11(Suppl. 3) 22–28. 10.1111/j.1469-0691.2005.01138.x [DOI] [PubMed] [Google Scholar]
- Muñoz-Atienza E., Gómez-Sala B., Araújo C., Campanero C., del Campo R., Hernández P. E., et al. (2013). Antimicrobial activity, antibiotic susceptibility and virulence factors of lactic acid bacteria of aquatic origin intended for use as probiotics in aquaculture. BMC Microbiol. 13:15 10.1186/1471-2180-13-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandlal S., Pickering T. (2004). Tilapia Fish Farming in Pacific Island Countries. Vol 1. Tilapia Hatchery Operation. Noumea: Secretariat of the Pacific Community. [Google Scholar]
- Olaniran A. O., Nzimande S. B. T., Mkize N. G. (2015). Antimicrobial resistance and virulence signatures of Listeria and Aeromonas species recovered from treated wastewater effluent and receiving surface water in Durban, South Africa. BMC Microbiol. 15:234 10.1186/s12866-015-0570-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo I. C. V., Pitondo-Silva A., Levy C. E., da Costa Darini A. L. (2011). Changes in vancomycin-resistant Enterococcus faecium causing outbreaks in Brazil. J. Hosp. Infect. 79 70–74. 10.1016/j.jhin.2011.04.016 [DOI] [PubMed] [Google Scholar]
- Penders J., Stobberingh E. E., Savelkoul P. H., Wolffs P. F. (2013). The human microbiome as a reservoir of antimicrobial resistance. Front. Microbiol. 4:87 10.3389/fmicb.2013.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham D. K., Chu J., Do N. T., Brose F., Degand G., Delahaut P., et al. (2015). Monitoring antibiotic use and residue in freshwater aquaculture for domestic use in vietnam. EcoHealth 12 480–489. 10.1007/s10393-014-1006-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruden A., Shore L. S. (2011). Hormones and Pharmaceuticals Generated by Concentrated Animal Feeding Operations Transport in Water and Soil. New York, NY: Springer. [Google Scholar]
- Reed L. J., Müench H. (1938). A simple method of estimating fifty percent end points. Amer. J. Hyg. 27 493–497. [Google Scholar]
- Rolain J. M. (2013). Food and human gut as reservoirs of transferable antibiotic resistance encoding genes. Front. Microbiol. 4:173 10.3389/fmicb.2013.00173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanis M. (2013). Evaluation of Agricultural Effluent and Irrigation Water As Sources of Antibiotic Resistant Escherichia coli. M.Sc. thesis, University of Stellenbosch; Stellenbosch. [Google Scholar]
- Schjorring S., Krogfelt K. A. (2011). Assessment of bacterial antibiotic resistance transfer in the gut. Int. J. Microbiol. 2011 1–10. 10.1155/2011/312956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar N., Lockatell C. V., Baghdayan A. S., Drachenberg C., Gilmore M. S., Johnson D. E. (2001). Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect. Immun. 69 4366–4372. 10.1128/IAI.69.7.4366-4372.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar V., Baghdayan A. S., Huycke M. M., Lindahl G., Gilmore M. S. (1999). Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect. Immun. 67 193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamminen M., Karkman A., Lohmus O., Muziasari W. I., Takasu H., Wada S., et al. (2011). Tetracycline resistance genes persist at aquaculture farms in the absence of selection pressure. Environ. Sci. Technol. 45 386–391. 10.1021/es102725n [DOI] [PubMed] [Google Scholar]
- Toledo-Arana A., Valle J., Solano C., Arrizubieta M. J., Cucarella C., Lamata M., et al. (2001). The enterococcal surface protein, esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 67 4538–4545. 10.1128/AEM.67.10.4538-4545.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tyne D., Gilmore M. S. (2014). Friend turned foe: evolution of enterococcal virulence and antibiotic resistance. Ann. Rev. Microbiol. 68 337–356. 10.1146/annurev-micro-091213-113003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vankerckhoven V., Van Autgaerden T., Vael C., Lammens C., Chapelle S., Rossi R., et al. (2004). Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J. Clin. Microbiol. 42 4473–4479. 10.1128/JCM.42.10.4473-4479.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir M., Rajić A., Dutil L., Uhland C., Bruneau N. (2012). Zoonotic bacteria and antimicrobial resistance in aquaculture: opportunities for surveillance in Canada. Can. Vet. J. 53 619–622. [PMC free article] [PubMed] [Google Scholar]
- Werner G., Coque T. M., Hammerum A. M., Hope R., Hryniewicz W., Johnson A., et al. (2008). Emergence and spread of vancomycin resistance among enterococci in Europe. Euro. Surveill. 13 1–11. [PubMed] [Google Scholar]
- WHO (2012a). Report of the 3rd Meeting of the WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance, 14–17 June 2011 Oslo, Norway. Geneva: World Health Organization; 70. [Google Scholar]
- WHO (2012b). World Health Organization. Critically Important Antimicrobials for Human Medicine 3rd Edn Geneva: World Health Organization. [Google Scholar]


