ABSTRACT
The opportunistic pathogen Candida is one of the most common causes of nosocomial bloodstream infections. Because candidemia is associated with high mortality rates and because the incidences of multidrug-resistant Candida are increasing, efforts to identify novel targets for the development of potent antifungals are warranted. Here, we describe the structure and function of the first member of a family of protein phosphatases that is specific to fungi, protein phosphatase Z1 (PPZ1) from Candida albicans. We show that PPZ1 not only is active but also is as susceptible to inhibition by the cyclic peptide inhibitor microcystin-LR as its most similar human homolog, protein phosphatase 1α (PP1α [GLC7 in the yeast Saccharomyces cerevisiae]). Unexpectedly, we also discovered that, despite its 66% sequence identity to PP1α, the catalytic domain of PPZ1 contains novel structural elements that are not present in PP1α. We then used activity and pulldown assays to show that these structural differences block a large subset of PP1/GLC7 regulatory proteins from effectively binding PPZ1, demonstrating that PPZ1 does not compete with GLC7 for its regulatory proteins. Equally important, these unique structural elements provide new pockets suitable for the development of PPZ1-specific inhibitors. Together, these studies not only reveal why PPZ1 does not negatively impact GLC7 activity in vivo but also demonstrate that the family of fungus-specific phosphatases—especially PPZ1 from C. albicans—are highly suitable targets for the development of novel drugs that specifically target C. albicans without cross-reacting with human phosphatases.
IMPORTANCE
Candida albicans is a medically important human pathogen that is the most common cause of fungal infections in humans. In particular, approximately 46,000 cases of health care-associated candidiasis occur each year in the United States. Because these infections are associated with high mortality rates and because multiple species of Candida are becoming increasingly resistant to antifungals, there are increasing efforts to identify novel targets that are essential for C. albicans virulence. Here we use structural and biochemical approaches to elucidate how a member of a fungus-specific family of enzymes, serine/threonine phosphatase PPZ1, functions in C. albicans. We discovered multiple unique features of PPZ1 that explain why it does not cross-react with, and in turn compete for, PP1-specific regulators, a long-standing question in the field. Most importantly, however, these unique features identified PPZ1 as a potential target for the development of novel antifungal therapeutics that will provide new, safe, and potent treatments for candidiasis in humans.
INTRODUCTION
Candida albicans is an opportunistic fungal pathogen that causes candidemia and is the most common cause of health care-associated Candida bloodstream infections in the United States (1). In the past, the majority of candidemia patients were immunocompromised (i.e., individuals with HIV or transplant recipients on immunosuppressant drugs, among others). However, the numbers of nonimmunocompromised patients contracting candidemia have been steadily increasing, with an estimate of 7,000 to 28,000 patients contracting nosocomial candidemia annually (2). Because the mortality rate of candidemia is 40%, these infections result in ~2,800 to 11,200 deaths per year. Unfortunately, these numbers are expected to increase as multiple species of Candida are becoming increasingly resistant to antifungal medications, including fluconazole and echinocandins (3). Although a combination of the well-established calcineurin (CN) drugs FK-506 and cyclosporine (CSA) given with the fungal inhibitor fluconazole has resulted in the very potent killing of C. albicans (4), the immunosuppressant functions of FK-506 and CSA make their use in humans problematic. Furthermore, C. albicans-specific CN inhibitors are unlikely to be achievable due to the 100% conservation of the CN active and substrate binding sites (5). Given the pressing need for new, potent antifungals, efforts to identify novel protein targets that are essential for virulence and unique to C. albicans are warranted.
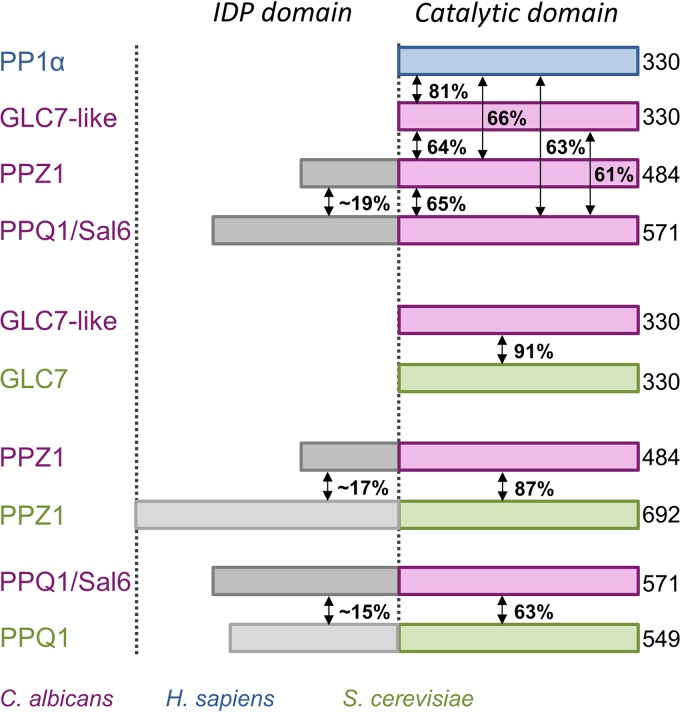
Eukaryotes contain multiple genes that encode serine/threonine protein phosphatase 1 (PP1 [in humans, PP1α, PP1β, and PP1γ]). PP1 regulates diverse and essential biological processes by dephosphorylating a variety of protein substrates. Although the intrinsic substrate specificity of PP1 is very low, by interacting with regulatory proteins to form distinct holoenzymes (~200 biochemically confirmed PP1 interactors), PP1 achieves high specificity (6–8). More than two decades ago, it was discovered that budding yeast (Saccharomyces cerevisiae), fission yeast (Schizosaccharomyces pombe), and the opportunistic fungus Candida albicans also carry a PP1 gene, coding for GLC7, Dis2, and GLC7-like, respectively (Fig. 1). There is ~80% sequence identity between C. albicans GLC7 and human PP1 isoforms (“GLC7” will be used here to refer to the PP1 homolog in fungal species) (9). Like PP1, GLC7 controls a plethora of essential biological processes, and its activity is regulated by its interaction with multiple regulatory proteins, many of which are conserved in humans (8). However, it was also discovered that fungi express a unique family of PP1-like genes, which include those coding for PPZ1 and PPQ1/Sal6 (10, 11). Unlike GLC7, these families of fungus-specific PP1-like phosphatases consist of two distinct domains: an N-terminal domain that is enriched in serines and is predicted to be unstructured (i.e., a member of the intrinsically disordered protein [IDP] family) and a C-terminal catalytic domain that has high sequence similarity to GLC7 (Fig. 1). This suggests that the fungus-specific phosphatases may also bind the GLC7-specific regulatory proteins. This was confirmed in S. cerevisiae, in which yeast two-hybrid studies demonstrated that S. cerevisiae PPZ1 (ScPPZ1) binds some, but not all, S. cerevisiae GLC7 (ScGLC7)-specific regulatory proteins (12). However, the molecular determinants that explain why fungus-specific phosphatases bind only a subset of the GLC7-specific regulators are still largely unknown.
FIG 1 .
CaPPZ1 is a PP1-like phosphatase. Domain architecture of the PP1-like phosphatases in C. albicans (GLC7-like, PPZ1, and PPQ1/Sal6 [pink]), S. cerevisiae (green), and Homo sapiens PP1α (blue). Fungus-specific PP1-like phosphatases have an N-terminal intrinsically disordered protein domain (IDP [gray]) in addition to a structured C-terminal catalytic domain (pink/green).
Furthermore, not only is PPZ1 important for cation homeostasis and cell wall biosynthesis, it is also critical for C. albicans virulence (13, 14). Namely, deletion of the Ppz1 gene reduces the ability of C. albicans to infect mice, a function that can be rescued by the reintegration of a single copy of the Ppz1 gene. In addition, PPZ1 has also been shown to play a role in the morphological changes in C. albicans associated with infectivity: that is, the transition from the yeast to the hyphal form. This is because the deletion of Ppz1 reduces the rate of hyphal growth (15). Together, these data suggest that the specific inhibition of PPZ1 will prevent this morphological transition and, as a consequence, block C. albicans infectivity without killing the commensal pathogen. The fungistatic effect of such a treatment would be more beneficial than eliminating C. albicans altogether as the latter might result in uncontrolled bacterial proliferation. Here, we used X-ray crystallography and biochemistry to elucidate the structural and functional characteristics of PPZ1 that are unique to the fungus-specific family of phosphatases. We discovered novel structural elements in PPZ1 that not only explain why PPZ1 binds less effectively to GLC7 regulators but also define new interaction surfaces that may be leveraged for the development of novel, effective antifungal therapeutics.
RESULTS
PPZ1cat is an active phosphatase with an atypical C terminus.
Like other fungus-specific phosphatases, C. albicans PPZ1 (484 amino acids [aa], 54.4 kDa) has two domains: an N-terminal intrinsically disordered protein (IDP) domain (aa 1 to 170 [PPZ1Nterm]) and a C-terminal catalytic domain (aa 171 to 484 [PPZ1cat]) (14, 16). PPZ1cat and human PP1α exhibit 66% sequence identity throughout their catalytic domains, and the 6 residues that coordinate the active site metals are 100% conserved (Fig. 1; see Fig. S1 in the supplemental material). Accordingly, full-length PPZ1 (PPZ1FL) and its individual domains (PPZ1Nterm and PPZ1cat) are readily isolated from Escherichia coli. In addition, PPZ1FL and PPZ1cat are active, as they effectively dephosphorylate a small molecule substrate mimetic (p-nitrophenyl phosphate [pNPP]) (see Fig. S2 in the supplemental material). Furthermore, their activities are identical to that of human PP1α purified from E. coli (8) or PP1c (a mixture of PP1 isoforms α, β, and γ) purified from rabbit muscle (17). However, unlike PP1α, which expresses solubly and crystallizes readily without its C-terminal disordered residues (residues 301 to 330) (8), a PPZ1 construct truncated at the corresponding residue (PPZ1catΔ466–484) is largely insoluble compared to PPZ1cat (>10-fold reduction in yield). This suggests that the PPZ1cat C-terminal region is critical for folding and/or stability. Consistent with this conclusion, the secondary structure prediction program PSIPRED (18) predicts that these residues are not disordered as they are in PP1α but instead form an α-helix (see Fig. S3 in the supplemental material).
The C-terminal residues of PPZ1cat are structured and form an α-helix.
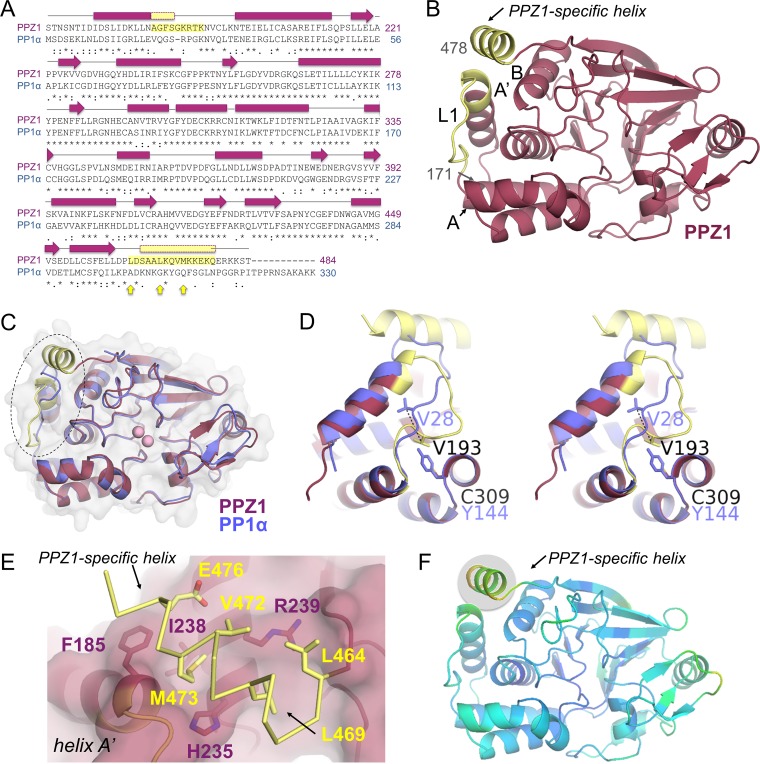
In order to understand the molecular consequences of the sequence differences between PPZ1 and PP1α and the role of the PPZ1 C-terminal residues in PPZ1 function, we determined the 3-dimensional crystal structure of PPZ1cat to 2.61 Å resolution (Table 1). The PPZ1cat structure includes residues 171 to 478. (The electron density for the last 6 residues, 479 to 484, was not observed, and thus they were not modeled.) As expected, the PPZ1cat structure adopts the canonical PP1 fold (Fig. 2A and B), comprising a mixed α/β protein, whose central loops are positioned to coordinate the active site metals. However, the structures are not identical. The root mean square deviation (RMSD) between PP1α (PDB no. 4MOV [19]) and PPZ1cat is 1.23 Å (backbone). The most significant difference is in the N-terminal region, where the PPZ1cat helix A′ extends for one more turn compared with PP1α and the loop connecting helix A′ to helix A (L1) adopts a conformation distinct from that in PP1α (Fig. 2B to D). (The RMSD of L1 between PPZ1cat and PP1α is 2.99 Å.) Although the sequences of L1 between PPZ1cat and PP1α are only 33% identical, this change of conformation is primarily due to a single amino acid change in helix C, Tyr144PP1α to Cys309PPZ1. The much smaller cysteine side chain creates a hydrophobic pocket into which the side chain of Val193PPZ1 binds (Fig. 2D). Because this pocket is not present in PP1α, the corresponding PP1α residue, Val28PP1α, interacts directly with helix A′. As a consequence, the Cβ atoms of the two side chains are separated by 5.6 Å between the two structures. The rest of the PPZ1cat L1 conformation changes to accommodate this new interaction, which in turn widens the pocket defined by helix A′, L1, and helix B (referred to here as the “Z1-helix binding pocket”).
TABLE 1 .
Data collection and refinement statistics
| Parameter | Value(s) fora: |
|
|---|---|---|
| CaPPZ1 | CaPPZ1–microcystin-LR | |
| Data collection | ||
| Space group | C2221 | P3221 |
| Cell dimensions | ||
| a, b, c (Å) | 145.0, 183.7, 69.0 | 50.7, 50.7, 201.1 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 120 |
| Resolution (Å) | 50.0–2.61 (2.66–2.61) | 50.0–2.40 (2.44–2.40) |
| Rmerge (%) | 13.3 (84.5) | 6.6 (19.7) |
| I/σ〈I〉 | 14.1 (2.1) | 30.6 (4.6) |
| Completeness (%) | 99.9 (100.0) | 98.5 (84.2) |
| Redundancy | 5.6 (5.6) | 6.1 (2.7) |
| Refinement | ||
| Resolution (Å) | 36.24–2.61 | 43.9–2.40 |
| No. of reflections | 28,311 | 12,283 |
| Rwork/Rfree | 19.4/22.2 | 17.9/23.5 |
| No. of atoms | ||
| Protein | 4,922 | 2,314 |
| Ligand/ion | 32 | 23 |
| Microcystin-LR | NAb | 71 |
| Water | 220 | 50 |
| B-factors | ||
| Protein | 33.0 | 30.8 |
| Ligand/ion | 29.9 | 46.6 |
| Microcystin | NA | 35.5 |
| Water | 30.9 | 30.1 |
| RMSDs | ||
| Bond lengths (Å) | 0.002 | 0.003 |
| Bond angles (°) | 0.72 | 0.57 |
| Ramachandran plot (%) | ||
| Favored regions | 96.1 | 96.5 |
| Allowed regions | 3.8 | 3.5 |
| Disallowed regions | 0.2 | 0.0 |
| PDB accession no. | 5JPE | 5JPF |
Values in parentheses are for the highest-resolution shell.
NA, not applicable.
FIG 2 .
The PPZ1-specific C-terminal helix. (A) Sequence alignment of CaPPZ1 (pink) and Homo sapiens PP1 (HsPP1α [blue]) with the observed secondary structural elements indicated above the sequence. Identical residues are indicated by a star, similar residues are indicated by a colon, less similar residues are indicated by a period, and dissimilar residues are indicated by a blank space. Resides from loop 1 (L1) and the PPZ1-specific helix are highlighted in yellow. Arrows (yellow) indicate the hydrophobic residues in the PPZ1-specific helix that are not present in PP1α. (B) The structure of PPZ1 is shown with the secondary structural elements discussed in the text labeled. (C) Overlay of PPZ1 (pink and yellow, as in panel B) and PP1α (blue). The change in conformation of loop L1 between the two structures is indicated by a dashed circle. (D) Stereo image of the overlay between L1 from PPZ1 and PP1α, colored as in panel C. (E) Interactions between the PPZ1-specific C-terminal helix (yellow) and the widened PPZ1-specific helix binding pocket (coral). Residues that make key interactions are shown as sticks and labeled. (F) PPZ1 colored according to residue B-factors, with yellow and green shading indicating higher B-factors.
The newly widened Z1-helix binding pocket results in the second significant structural difference between PPZ1cat and PP1α: the C-terminal residues of PPZ1cat are not disordered like they are in PP1α, but instead form an α-helix that nestles into this newly widened Z1-helix binding pocket (Fig. 2B to E). This interaction is stabilized by hydrophobic interactions between the C-terminal helix and residues from helices A′ and B, with the interaction centered on PPZ1 C-terminal helix residue Met473PPZ1 (Fig. 2E). This residue is completely buried from solvent via interactions with Leu464PPZ1, Leu469PPZ1, and Val472PPZ1 from the C-terminal helix as well as by interactions with Phe185PPZ1 from helix A′ and His235PPZ1, Ile238PPZ1, and Arg239PPZ1 from helix B. The majority of these residues are not conserved in PP1α. In particular, only a single residue (underlined) between the PPZ1 C-terminal helix and the C-terminal disordered tail of PP1α is conserved (PPZ1, 466SAALKQVMKKEKQ478; PP1α, 301KNKGKYGQFSGLN313), with the sequence of PPZ1 consisting of multiple hydrophobic residues and no helix-disrupting glycines (Leu469PPZ1, Val472PPZ1, and Met473PPZ1 versus Gly304PP1α, Gly307PP1α, and Gly311PP1α, respectively), rationalizing why the corresponding residues in PP1α are unstructured. Finally, the experimental B-factors for residues in the Z1-helix are higher than the rest of PPZ1cat (Fig. 2F), suggesting that the Z1-helix is more dynamic than the rest of the PPZ1cat.
The Z1-helix is dynamic.
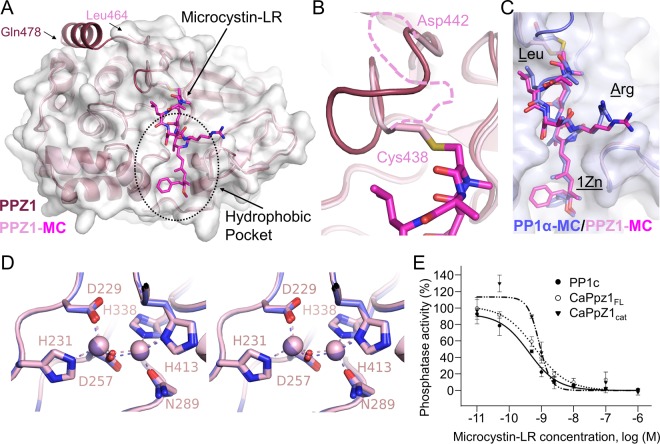
We also determined the structure of PPZ1cat bound to the marine toxin microcystin-LR (MC [PPZ1cat-MC]) (Table 1), a potent cyclic peptide inhibitor of PP1 (20). Both the MC-free and MC-bound structures of PPZ1cat are highly similar, with an RMSD of 0.54 Å (Fig. 3A). The largest difference is in the β12-β13 loop, which contains Cys438PPZ1 (Fig. 3B). This cysteine forms a covalent bond with the bound MC identical to that observed in the PP1α-MC complex (21). This causes the 438CGEFD442 loop to change conformation and become more dynamic (residues 439GEF441 were not modeled in the PPZ1cat-MC structure due to a lack of density). Unexpectedly, the Z1-helix is also no longer ordered in the PPZ1cat-MC structure (Fig. 3A). This was not due to MC binding, but instead the Z1-helix was displaced by a symmetry-related molecule in the crystal. The observation that the Z1-helix can be displaced supports the observation that the Z1-helix is more dynamic than the rest of the PPZ1cat. Importantly, the positions of L1 are identical between the PPZ1cat and PPZ1cat-MC structures, demonstrating that the conformation of L1 is intrinsic to PPZ1cat itself and not a consequence of Z1-helix binding.
FIG 3 .
The PPZ1-specific helix is dynamic. (A) Overlay of free PPZ1 (dark pink) and MC-bound PPZ1 (light pink). The C-terminal residues of both proteins are labeled. The gray surface corresponds to the PPZ1-MC complex, illustrating that the PPZ1-specific helix was not ordered and thus not modeled (see the text for details). The PPZ1 hydrophobic pocket is indicated by a dashed circle. (B) Enlarged view of the covalent bond between Cys438 and MC, resulting in a change in conformation of the β12-β13 loop. (Residues 439 to 441 were not visible in the PPZ1-MC structure and are indicated by a pink dashed line.) (C) Overlay of MC from PPZ1 (light pink) and PP1α (blue). The Leu and Arg residues in microcystin-LR are labeled, as is the 1Zn [(2S,3S,4E,6E,8S,9S)-3-amino-9-methoxy-2,6,8-trimethyl-10-phenyldeca-4,6-dienoic acid]. (D) Stereo image of the PPZ1 and PP1α metal-bound active sites. Bound Mn2+ ions are shown as spheres. (E) Dose-response curves reporting the inhibition of PP1c, PPZ1FL, and PPZ1cat activities by the MC toxin; 32P-labeled MLC20 was used as a substrate. The phosphatase activities of each enzyme without toxin were set to 100% activity. The data represent the means ± standard errors (SE) from 3 experiments.
MC binds the active site of PPZ1cat essentially identically to that observed in PP1α (Fig. 3C). Furthermore, the active sites are also identical (Fig. 3D). The bulk of the MC binds the PPZ1cat hydrophobic binding pocket (Fig. 3A), while the remainder covers and, as a consequence, blocks the active site. The largest difference between the conformations of MC in PP1α-MC and PPZ1cat-MC is a change of the rotomer conformation of the 1Zn residue phenyl group and the Arg guanidinium group (Fig. 3C). The orientation observed in PPZ1cat-MC is favored because a malonate ion from the crystallization mother liquor is bound in the rotomer position populated in the PP1α structure; thus both positions are expected to be equally likely in the absence of malonate. Furthermore, because the residues that mediate MC binding are highly conserved between PPZ1cat and PP1α (79% identical and 90% similar [see Fig. S1 in the supplemental material]), MC is predicted to inhibit their activities with similar 50% inhibitory concentration (IC50) values. We tested this in vitro using both small molecule (pNPP)- and peptide (myosin light chain [pMLC])-based substrates, which show that the IC50 values of MC for all constructs are essentially identical (IC50s for pMLC of 0.96 nM for PPZ1FL, 0.72 nM for PPZ1cat, and 0.43 nM for PP1c, and IC50s for pNPP of 11.8 nM for PPZ1cat and 4.4 nM for PP1α) (Fig. 3E; see Fig. S2 in the supplemental material).
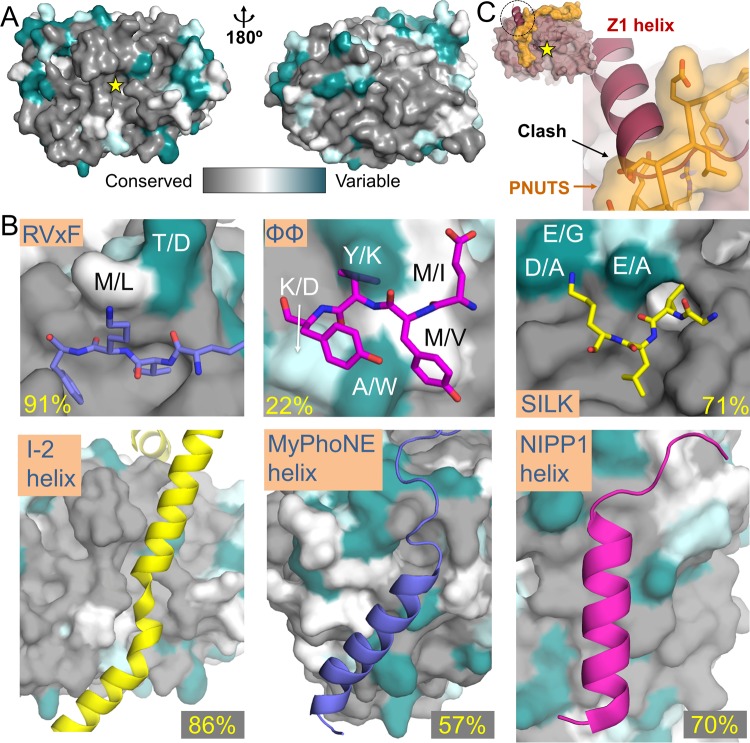
The sequence and structural conservation of regulatory protein binding pockets in PPZ1cat is highly variable.
While PP1 exhibits broad specificity, it acts in a highly specific manner by forming stable complexes (holoenzymes) with a host of regulatory proteins that direct PP1 activity toward specific substrates and localize PP1 to specific regions of the cell (8, 22). Recent structural studies have revealed that PP1 binds these regulators using small linear interaction motifs (SLiMs) (7, 19, 23, 24). The most well-known is the RVxF SLiM, which is found in ~70% of all known regulators (22). Others include the MyPhoNE, SILK, ΦΦ, and Arg SLiMs (6, 7, 19, 23, 25). Mapping of the sequence differences onto the structure of PPZ1cat shows that the bulk of the changes are distally located from the active site (Fig. 4A). This comparison also reveals that the RVxF interaction residues are 91% conserved between PPZ1 and PP1α (the only difference being a conservative Met290PP1α-to-Leu455PPZ1 substitution [Fig. 4B; see Fig. S1 in the supplemental material]), suggesting that all PP1 regulators that contain an RVxF sequence should bind PPZ1. Other sites are similarly conserved. For example, inhibitor-2 (I-2) is a specific protein inhibitor of PP1. In addition to an RVxF and SILK motif, it binds PP1 using a long helix, which binds across the PP1 active site (25). Like the RVxF motif, the I-2 helix binding pocket is highly conserved in PPZ1 (86% identical, 93% conserved Fig. 4B; see Fig. S1) suggesting that I-2 can bind productively to PPZ1. Likewise, the conservation of the NIPP1-helix interaction pocket (70% identical and 80% conserved [Fig. 4B; see Fig. S1]), also suggesting that this PP1 regulatory protein can bind productively to PPZ1cat.
FIG 4 .
PP1 regulatory protein binding pockets are differentially conserved in PPZ1. (A) Sequence conservation between PPZ1 and PP1α mapped onto the surface of PPZ1. The yellow star indicates the location of the PP1 active site. (B) Close-up views of the RVxF, ΦΦ, SILK, I-2 helix, MyPhoNE helix, and NIPP1 helix binding pockets on PPZ1, with regulators that bind these pockets shown as sticks (RVxF, ΦΦ, and SILK) or a cartoon (I-2 helix, MyPhoNE helix, and NIPP1 helix). The sequence changes between PP1α/PPZ1 are indicated as in panel A. The sequence identity of the binding pockets is reported (see Fig. S1 in the supplemental material). (C) Overlay of a PP1 regulator (PNUTS [shown here in orange]) that bridges the ΦΦ and Arg motif binding pockets onto PPZ1cat (rose). The clash between the regulator and Z1-specific helix is shown.
In contrast, other SLiM binding pockets are much less conserved in PPZ1. For example, the SILK binding sites are only 71% identical and 79% similar between PPZ1 and PP1α (Fig. 4B; see Fig. S1 in the supplemental material). Three of the 4 amino acid differences result in a change from an acidic to uncharged residue (Glu56PP1α/Ala221PPZ1, Asp166PP1α/Ala331PPZ1, and Glu167PP1α/Gly332PPZ1). As these acidic residues coordinate the basic “K” of the SILK motif, the “K” may not be necessary for PPZ1 to bind SILK motif-containing proteins. The same is true for the MyPhoNE binding pocket (23) (Fig. 4B; see Fig. S1). While the majority of residues that define the pocket are largely similar—57% identical and 75% similar—multiple residues differ between the two proteins (Asp179PP1α/Val344PPZ1, Gln198PP1α/Phe363PPZ1, Gly215PP1α/Glu380PPZ1, and His237PP1α/Ser402PPZ1). The Gly215PP1α/Glu380PPZ1 substitution results in the largest clash in the superimposed structures, with the Glu380PPZ1 side chain clashing with that of Trp17MYPT1, suggesting that these changes impact MYPT1 binding.
The SLiM binding pocket that is most different between PP1α and PPZ1 is the ΦΦ interaction site, with an identity of 22% and a similarity of 67% (7, 19) (Fig. 4B; see Fig. S1 in the supplemental material). The most significant differences are the replacement of Tyr78PP1α, which defines the ΦΦ interaction pocket, with Lys243PPZ1 and the replacement of Ala279PP1α with Trp444PPZ1. Both Lys243PPZ1 and Trp444PPZ1 are large, bulky residues that hinder access to the pocket. This suggests that PP1 regulators that contain a ΦΦ motif will bind PPZ1 with lower affinity than PP1α. Finally, although the Arg interaction pocket is perfectly conserved between both PP1α and PPZ1 (see Fig. S1), the presence of the Z1-helix in PPZ1 is expected to negatively impact the binding of regulators that use this site for binding, especially those that also use the ΦΦ binding pocket. Because the Z1-helix binds between these two sites, its presence will block access to the Arg site (Fig. 4C), reducing the ability of PPZ1 to bind regulators that contain these sequential SLiMs.
PPZ1 binds only a subset of PP1 regulatory proteins.
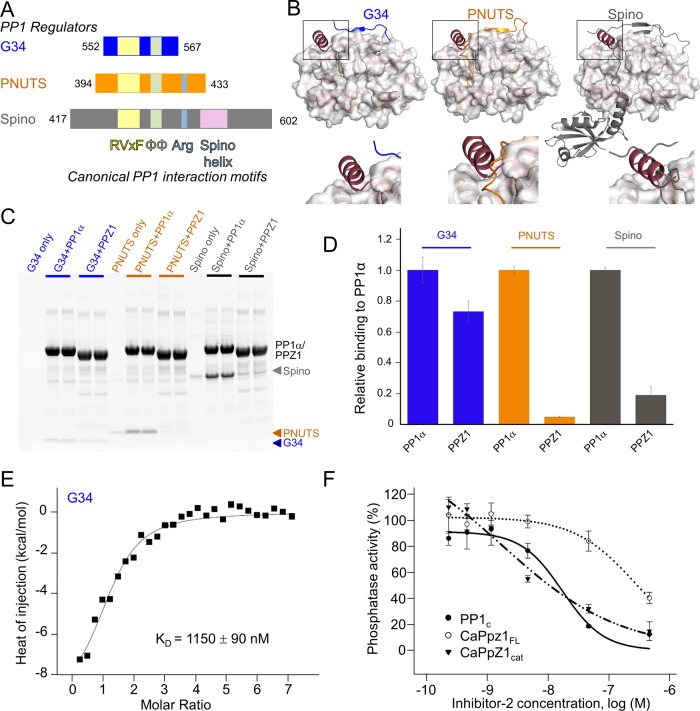
To test if PPZ1 binds differentially to known PP1 regulators, we used pulldown assays with three well-characterized regulators—GADD34, PNUTS, and spinophilin (Fig. 5A and B). The minimal PP1-binding domains of these regulators bind PP1α with strong affinities (Equilibrium dissociation constant [KD] values are 62 nM for GADD34552–567 [26], 9.3 nM for PNUTS394–433 [19], and 8.7 nM for spinophilin417–602 [27]). As predicted, the pulldown assays show that PPZ1cat binds GADD34 less effectively than PP1α (Fig. 5C and D). Because the RVxF site is nearly perfectly conserved between PP1 and PPZ1cat, this suggests that the weakened affinity is largely due to an inability of GADD34552–567 to productively bind the ΦΦ motif binding pocket. To quantify the reduction in binding, we used isothermal titration calorimetry (ITC). The affinity of GADD34 for PPZ1cat decreases more than 19-fold, resulting in a KD of only 1,150 ± 90 nM (Fig. 5E). Consistent with this, peptides that contain only RVxF motifs bind PP1 with KDs in the micromolar range (28), similar to those observed for GADD34552–567 and PPZ1cat.
FIG 5 .
Most PP1 regulatory proteins bind to PPZ1 less effectively. (A) Domain structure of the regulators tested (G34, GADD34552–567; PNUTS, PNUTS394–433; Spino, spinophilin417–602) with their canonical PP1 interaction motifs highlighted in different colors. (B) Structures of the GADD34 (blue; PDB no. 4XPN [26]), PNUTS (orange; PDB no. 4MOY [19]), and spinophilin (gray; PDB no. 3EGG [27]) PP1 holoenzymes, with close-ups of the Z1 helix interaction pocket. (C) The relative binding of PP1α and PPZ1 with the regulators described in panel A was determined using a pulldown assay. (D) Densitometry analysis of the gel in panel B. (E) Binding isotherm of GADD34552–567 with PPZ1cat (KD, 1,150 ± 90 nM). (F) Dose-response curves of phosphatase activity with increasing concentrations of I-2 with PP1c, PPZ1cat, and PPZ1FL phosphatases. The data represent the means ± SE from 3 experiments.
While the PP1 binding domain of GADD34 contains only an RVxF motif and ΦΦ motif, other regulators, such as PNUTS394–433 and spinophilin417–602 contain additional motifs that facilitate PP1 binding (PNUTS, RVxF-ΦΦ-Arg; spinophilin, RVxF–ΦΦ–Arg–spinophilin417–602–helix [Fig. 5A and B]). We used pulldown assays to determine if the altered ΦΦ binding pocket and the presence of the Z1-helix negatively impact the binding of these regulators to PPZ1 (Fig. 5C). The results show that PPZ1, compared to PP1α, does not effectively pull down either PNUTS or spinophilin (~85% less binding [Fig. 5C and D]). Furthermore, mutation of either the ΦΦ or Arg motif in spinophilin results in a similar reduction in PP1 binding, to levels similar to those observed for PPZ1 and wild-type spinophilin (see Fig. S4 in the supplemental material). Together, these data demonstrate that the altered ΦΦ-binding pocket coupled with the presence of the Z1-helix (Fig. 5B) negatively impacts the ability of PPZ1 to interact with regulators that require these sites for binding.
The presence of the N-terminal IDP domain reduces the ability of I-2 to inhibit PPZ1.
Unlike GADD34, PNUTS, and spinophilin, I-2 does not bind PP1 at the ΦΦ- or PPZ1-specific binding pockets. Instead, it binds PP1 using the SILK–RVxF–I-2–helix interaction pockets (25). With the exception of the SILK motif binding pocket, these interaction sites in PPZ1 are largely conserved with those in PP1α. Furthermore, the change in the SILK binding pocket suggests that PPZ1 no longer has a strict requirement for the “K” residue (Fig. 4B). This suggests that I-2 will inhibit both PPZ1 and PP1α with equal potencies. To test this, we measured the ability of I-2 to bind and inhibit PPZ1 activity and compared it to that determined for PP1c. The data show that both PPZ1cat and PP1c are inhibited by I-2 with nearly equivalent IC50s, measured to be 7 and 12 nM, respectively (Fig. 5F). However, unexpectedly, we also discovered that I-2 is a much less potent inhibitor against PPZ1FL, which includes the ~160-amino-acid N-terminal IDP domain (see Fig. S3 in the supplemental material). With PPZ1FL, the IC50 of I-2 increases 39-fold to 278 nM; this increase was observed for both recombinant I-2 and I-2 partially purified from rabbit muscle (not shown). (The latter result is consistent with previous studies [29] that showed that S. cerevisiae PPZ1FL is also poorly inhibited by I-2.) Furthermore, the specific activities of the two PPZ1 constructs also differ significantly, with 4.6 mU/mg for PPZ1FL and 1,200 mU/mg for PPZ1cat. These data suggest that the N-terminal domain likely masks one or more binding sites for I-2 (but not the active site, as the PPZ1FL protein is fully susceptible to MC inhibition [Fig. 3E]) and prevents, via an as yet undetermined mechanism, PPZ1 inhibition by this protein inhibitor.
DISCUSSION
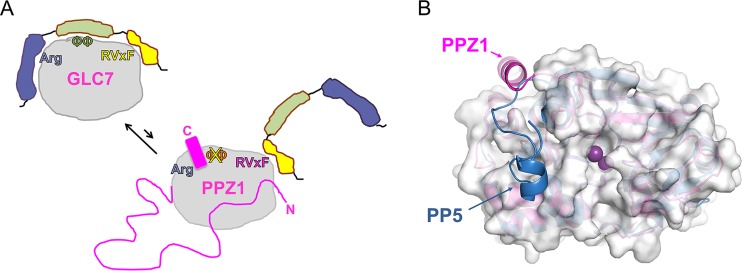
Our structural and biochemical studies reveal why, in spite of their high sequence similarity, fungus-specific phosphatases like PPZ1 bind only a subset of the GLC7-specific regulators (Fig. 6A). Specifically, we discovered that there are three distinct mechanisms by which PPZ1 inhibits the binding of many GLC7-specific regulators. First, sequence differences between PPZ1cat and PP1α in canonical SLIM interaction pockets, especially the ΦΦ binding pocket, inhibit the subset of regulators that requires these sites for binding (see Fig. S1 in the supplemental material). Second, the presence of the N-terminal IDP domain, which is not found in GLC7, also negatively impacts the binding of at least some regulators (e.g., I-2), likely through an as yet undefined steric mechanism (Fig. 5 and 6A). Third, structural differences between PPZ1cat and PP1α can also negatively impact regulator binding. In particular, our data show that the presence of the Z1-specific helix hinders access to the C-terminal groove by regulators that bind the Arg site via the ΦΦ site (Fig. 5). In addition, the dynamic nature of this helix raises the intriguing possibility that it may also serve a regulatory role in PPZ1 function. Finally, the binding pocket of the Z1-specific helix is also unique, as that of PP5, the only other known serine/threonine protein phosphatase with a C-terminal helix, binds at the front, versus the top, of the core catalytic domain (Fig. 6B) (30). Together, these results show that instead of PPZ1 competing with GLC7 for its regulators, the regulators preferentially bind and control the activity of GLC7 (Fig. 6A). This resolves a long-standing question about why the presence of the fungus-specific phosphatases does not globally disrupt GLC7 function (12).
FIG 6 .
Features of PPZ1 that limit its interaction with GLC7 regulators. (A) Cartoon illustrating that the presence of the disordered N-terminal domain, the lack of conservation of key PP1/GLC7 interaction domains (such as the ΦΦ binding pocket), and the presence of the PPZ1-specific helix negatively impact the binding of PPZ1 with many GLC7-specific regulators, leaving the regulators to preferentially bind and regulate the activity of GLC7. (B) Overlay of PPZ1 (pink) and PP5 (blue; PDB no. 1S95 [30]), illustrating that the C-terminal helices of the two proteins bind different pockets in the core PSP catalytic domain.
Our studies also revealed that multiple pockets in PPZ1cat are unique to this phosphatase and thus can be exploited for drug development. While the PPZ1cat and PP1α active sites are perfectly conserved, the conformation of loop L1 is unique in PPZ1cat, as is its folded Z1-specific helix. Both of these new structural elements create novel binding surfaces that are not present in human isoforms of PP1. Thus, they are useful for the development of specific, potent antifungals. Because their active sites are perfectly conserved, one strategy for developing a fungus-specific inhibitor of PPZ1 would be to use “fragment linking” (31): i.e., linking a small molecule that targets one of the unique PPZ1-specific binding pockets with a small molecule that, for example, targets the PP1 active site. This would result in an inhibitor that selectively targets PPZ1, as it would bind PPZ1 with higher affinity than human PP1 isoforms. Because PPZ1 is critical for C. albicans hypha formation, a morphological change associated with infectivity, this type of PPZ1-specific inhibitor would stop C. albicans infections without killing the commensal pathogen. This would prevent uncontrolled bacterial proliferation that can accompany C. albicans elimination and thus will result in novel, potent drugs with minimal side effects for the treatment of candidemia.
MATERIALS AND METHODS
Protein expression, purification for pNPP-based assays, and crystallography.
The gene encoding PPZ1cat (aa 171 to 484) was synthesized (GeneArt; Invitrogen), subcloned into the RP1B bacterial expression plasmid (32), and expressed largely as previously described (8). Specifically, the plasmid was cotransformed with the pGRO7 plasmid, which encodes the GroEL/GroES chaperone (TaKaRa), into E. coli BL21(DE3) cells (Invitrogen). Cells were grown in LB medium supplemented with 1 mM MnCl2 at 30°C to an optical density at 600 nm (OD600) of ~0.5, at which point arabinose was added (2 g/liter) to induce the expression of the GroEL/GroES chaperone. At an OD600 of ~1, the temperature was lowered to 10°C, and the expression of PPZ1 was induced using 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside): the protein was allowed to express for ~20 h at 10°C. The cells were harvested by centrifugation, suspended in fresh LB medium (again supplemented with 1 mM MnCl2 and 200 µg/ml of chloramphenicol to inhibit the ribosome), and agitated for ~5 h at 10°C. Harvested cells were frozen and stored at −80°C.
All purifications were performed at 4°C. Cells expressing His6-tagged tobacco etch virus (TEV) protease sequence-PPZ1cat were lysed in lysis buffer (25 mM Tris [pH 8.0], 700 mM NaCl, 5 mM imidazole, 1 mM MnCl2, 0.1% Triton X-100) using high-pressure homogenization (Avestin C3 EmulsiFlex) in the presence of an EDTA-free protease inhibitor cocktail (Roche). The lysate was clarified by centrifugation at 45,500 × g, filtered through a 0.22-µm-pore polyethersulfone (PES) membrane filter (Millipore), and then loaded onto Ni2+-nitrilotriacetic acid (NTA) resin (GE Healthcare) preequilibrated in buffer A (25 mM Tris [pH 8.0], 700 mM NaCl, 5 mM imidazole, 1 mM MnCl2). Bound His6-TEV-PPZ1cat was washed with 100 ml of buffer A, followed by more stringent wash with 100 ml “stringent wash buffer” consisting of 94% buffer A and 6% buffer B (25 mM Tris [pH 8.0], 700 mM NaCl, 250 mM imidazole, 1 mM MnCl2). The bound His6-TEV-PPZ1cat was eluted using 100% buffer B and immediately purified by size exclusion chromatography (SEC) using Superdex 75 26/60 preequilibrated in 20 mM Tris (pH 8.0), 500 mM NaCl, 0.5 mM Tris(2-carboxyethyl)phosphine (TCEP), and 1 mM MnCl2. Fractions containing the His6-TEV-PPZ1cat protein were pooled and incubated overnight with TEV protease at 4°C. A second Ni2+-NTA “subtraction” purification was used to separate cleaved PPZ1cat from the cleaved His6 tag and TEV; the flowthrough, which contained the cleaved PPZ1cat, was pooled and purified in a final step using SEC. PPZ1cat was pooled, concentrated, and used immediately for crystallization experiments.
Protein expression and/or purification for dephosphorylation assays.
The catalytic subunit of rabbit skeletal muscle protein phosphatase 1 (PP1c) was isolated as described (33). The 20-kDa myosin light chain (MLC20) and the myosin light chain kinase (MLCK) were obtained from turkey gizzard, and MLC20 was phosphorylated by MLCK in the presence of [γ-32P]ATP and Mg2+ as described previously (34). Recombinant 6×His-tagged inhibitor-2 (I-2) was prepared as described previously (28); I-2 was also isolated from rabbit skeletal muscle (35) for some control experiments.
The coding sequences for full-length Candida PPZ1FL and its conserved C-terminal catalytic domain, termed PPZ1cat, were generated by PCR from the pET28-CaPPZ1 plasmid (GenBank accession no. GQ357913 [14, 36]) and cloned into the E. coli expression vector pGEX6p-1 (Amersham Biosciences); both constructs were sequence verified (UD Genomed, Ltd.). Both constructs were then transformed into E. coli BL21(DE3) RIL cells (Agilent), and expression of N-terminal glutathione S-transferase (GST)-tagged proteins was induced with 0.6 mM IPTG (Sigma) at 18°C. Overnight incubation was used to express GST-PPZ1cat, while a shorter, 3-h incubation period was required to reduce inclusion body formation and ensure optimal production of soluble GST-PPZ1FL. In both cases, 0.5 mM MnCl2 was included in the culture medium, and 1 mM MnCl2 was present during the subsequent purification steps. The fusion proteins were purified using glutathione-Sepharose 4B (GE Healthcare) resin, and the GST tag was removed using the PreScission protease (GE Healthcare) during elution (following the instructions of the manufacturer). The protein concentration was determined using the Bradford assay, and protein purity was verified by SDS-PAGE.
Protein phosphatase assays.
The activities of the recombinant C. albicans phosphatases were measured with 32P-labeled MLC20 substrate as previously described (34), with the exception that the substrate concentration was 1 µM and 2 mM MnCl2 was included in the assay mixtures. The activity of rabbit PP1 was also determined under identical conditions as a control. The 50% inhibitory concentrations (IC50s) of microcystin-LR (obtained from Enzo Life Sciences or purified as previously described [37]) were determined by using either the protein substrate or a small molecule substrate as described previously (38), except that pNPP (Sigma-Aldrich) was used instead of 3-O-methylfluorescein phosphate (OMFP). Reactions were carried out in 96-well plates (Costar). MC concentrations were prepared by serial dilution and added to 150 µl of buffered enzyme (18 nM protein in 40 mM HEPES [pH 7.0], 1.33 mM dithiothreitol [DTT], 1.33% [vol/vol] Triton X-100, 0.133 mg/ml bovine serum albumin [BSA], 1.33 mM sodium ascorbate, 1.33 mM MnCl2) in wells containing the reaction mixtures. The high-signal controls were prepared by adding dimethyl sulfoxide (DMSO), matching the amount present in the reaction mixture with the highest concentration of MC. The low-signal controls were the reaction mixtures containing the highest concentration of microcystin-LR. The reactions were initiated by the addition of pNPP substrate and incubated at room temperature for 30 min. The reactions were stopped using 100 µl of 300 mM potassium phosphate (pH 10). The absorbance was measured at 405 nm using an Epoch spectrophotometer (BioTek). The percentage of activity of each protein was calculated using the equation [(absorbance − low-signal control)/(high-signal control − low signal control)] × 100%. The IC50 was then calculated using SigmaPlot 12.5.
Crystallization and structure determination.
Crystals of apo-PPZ1cat were obtained using hanging drop vapor diffusion in 1.8 M ammonium citrate tribasic (pH 7.0) in a 2:1 protein/crystallization condition ratio. To generate PPZ1cat-MC crystals, PPZ1cat was first incubated with MC at a 1:1 molar ratio for 15 min before crystallization using hanging drop vapor diffusion in 0.06 M citric acid (pH 4.1) plus 16% (wt/vol) polyethylene glycol (PEG) 3350. For data collection, crystals were first cryo-protected with 30% glycerol in mother liquor (PPZ1cat) or 3.4 M Na-malonate (pH 4.0) (PPZ1cat-MC) and then flash-frozen in liquid nitrogen. X-ray data were collected in house at 100 K using a Rigaku FR-E+ Superbright rotating copper anode X-ray generator with a Saturn 944+ HG charge-coupled device (CCD) detector (Brown University Structural Biology Facility). The data were phased using molecular replacement (Phaser as implemented in PHENIX [39]) using PP1 (PDB no. 4MOV [19]) as the search model. Clear electron density of MC could be observed bound to the active site. The initial models were built using Phenix.AutoBuild (40) followed by iterative rounds of refinement in PHENIX and manual building using Coot (41).
Isothermal titration calorimetry.
GADD34552–567 peptide was purchased from Biosynthesis, Lewisville, TX. The peptide was dissolved directly in ITC buffer (20 mM Tris [pH 8.0], 500 mM NaCl, 0.5 mM TCEP, 1 mM MnCl2). GADD34552–567 was titrated into PPZ1cat using a VP-ITC microcalorimeter at 25°C (Microcal, Inc.). Data from the ITC runs were analyzed using Origin 7.0.
Pulldown assay.
GADD34552–567 was purchased. PNUTS394–433 and spinophilin417–602 were purified according to a method described previously (19, 27). Spinophilin417–602 ΦΦ→AA, R469D, and R469E mutants were generated using site-directed mutagenesis. PP1α7–330 and PPZ1cat were purified by the method described above but retained the His tag. After SEC (20 mM Tris [pH 8.0], 500 mM NaCl, 0.5 mM TCEP, 1 mM MnCl2), 50% (vol/vol) glycerol was added to both PP1α7–330 and PPZ1cat. The final concentrations of the proteins were adjusted to 5 µM. Ten micromoles of each PP1 regulator was added into 500 µl of PP1α7–330 or PPZ1cat. The mixture was incubated under rocking (4°C) for 60 min to allow for complex formation. A 15-μl bed volume of Ni-NTA resin (GE Healthcare) was added into tubes, and the mixture was incubated at 4°C for 60 min. The beads were pelleted by centrifugation (2000 × g) and washed three times with 500 µl of SEC buffer. Forty microliters of SDS loading buffer was added to the beads, and the samples were boiled at 80°C for 5 min and analyzed on NuPAGE 4 to 12% bis-Tris gels. Gels were stained overnight with SYPRO ruby protein gel stain (Life Technologies) according to the manufacturer’s protocols and scanned using a Typhoon 9410 laser scanner (GE Healthcare) with an excitation wavelength of 457 nm and emission filter of 610 nm following destaining. Densitometry was performed using ImageQuant TL 7.0 software for quantification of the band intensity.
Accession number(s).
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession no. 5JPE for PPZ1 and 5JPF for PPZ1-MC.
SUPPLEMENTAL MATERIAL
Conservation of PP1 interaction pockets between HsPP1α (1α) and CaPPZ1 (Z1). The residues that define the metal, toxin, and regulatory motif binding pockets in HsPP1α and the corresponding residues in CaPPZ1 are shown. Residues that are different between the two proteins are in boldface and shaded in a darker color. The sequence identity (ID) and similarity (Sim) of the distinct binding pockets are calculated above each list. Download
MC potently inhibits recombinant HsPP1α and CaPPZ1 activity measured with pNPP. The data represent the mean ± standard deviation (SD) from 4 experiments. Download
Secondary structure prediction for CaPPZ1. The N-terminal domain (gray box) is predicted to lack secondary structural elements (pink cylinders, α-helices; yellow arrows, β-strands), while the catalytic domain (pink box) is predicted to be structured. The C-terminal residues in PPZ1 are predicted to be helical (green box). “Conf” represents the confidence of the prediction, with higher bars indicating greater confidence scores. Download
Mutation of the spinophilin ΦΦ and Arg motifs negatively impacts its ability to bind PP1α. Pull-down assays with (A) PP1α or (B) PPZ1 and either wild-type (wt) spinophilin or spinophilin variants in which the ΦΦ or Arg PP1 interaction motifs are mutated (SpinoΦΦ→AA, SpinoR469D, and SpinoR469E). (C) Densitometry and quantification of pulldown results shown in panels A and B. PP1α is in blue and PPZ1 in red. Mutation of either the ΦΦ or Arg motif in spinophilin reduces the amount of spinophilin pulled down by PP1α to levels nearly identical to those obtained with PPZ1. Download
ACKNOWLEDGMENTS
This research is based in part on data obtained at the Brown University Structural Biology Core Facility, which is supported by the Division of Biology and Medicine, Brown University and W.P.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Citation Chen E, Choy MS, Petrényi K, Kónya Z, Erdődi F, Dombrádi V, Peti W, Page R. 2016. Molecular insights into the fungus-specific serine/threonine protein phosphatase Z1 in Candida albicans. mBio 7(4):e00872-16. doi:10.1128/mBio.00872-16.
REFERENCES
- 1.Pappas PG, Rex JH, Lee J, Hamill RJ, Larsen RA, Powderly W, Kauffman CA, Hyslop N, Mangino JE, Chapman S, Horowitz HW, Edwards JE, Dismukes WE, NIAD Mycoses Study Group . 2003. A prospective observational study of candidemia: epidemiology, therapy, and influences on mortality in hospitalized adult and pediatric patients. Clin Infect Dis 37:634–643. [DOI] [PubMed] [Google Scholar]
- 2.Wenzel RP, Gennings C. 2005. Bloodstream infections due to Candida species in the intensive care unit: identifying especially high-risk patients to determine prevention strategies. Clin Infect Dis 41(Suppl 6):S389–S393. doi: 10.1086/430923. [DOI] [PubMed] [Google Scholar]
- 3.Sanglard D. 2002. Resistance of human fungal pathogens to antifungal drugs. Curr Opin Microbiol 5:379–385. doi: 10.1016/S1369-5274(02)00344-2. [DOI] [PubMed] [Google Scholar]
- 4.Uppuluri P, Nett J, Heitman J, Andes D. 2008. Synergistic effect of calcineurin inhibitors and fluconazole against Candida albicans biofilms. Antimicrob Agents Chemother 52:1127–1132. doi: 10.1128/AAC.01397-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grigoriu S, Bond R, Cossio P, Chen JA, Ly N, Hummer G, Page R, Cyert MS, Peti W. 2013. The molecular mechanism of substrate engagement and immunosuppressant inhibition of calcineurin. PLoS Biol 11:e1001492. doi: 10.1371/journal.pbio.1001492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hendrickx A, Beullens M, Ceulemans H, Den Abt T, Van Eynde A, Nicolaescu E, Lesage B, Bollen M. 2009. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem Biol 16:365–371. doi: 10.1016/j.chembiol.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 7.O’Connell N, Nichols SR, Heroes E, Beullens M, Bollen M, Peti W, Page R. 2012. The molecular basis for substrate specificity of the nuclear NIPP1:PP1 holoenzyme. Structure 20:1746–1756. doi: 10.1016/j.str.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peti W, Nairn AC, Page R. 2013. Structural basis for protein phosphatase 1 regulation and specificity. FEBS J 280:596–611. doi: 10.1111/j.1742-4658.2012.08509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng ZH, Wilson SE, Peng ZY, Schlender KK, Reimann EM, Trumbly RJ. 1991. The yeast GLC7 gene required for glycogen accumulation encodes a type 1 protein phosphatase. J Biol Chem 266:23796–23801. [PubMed] [Google Scholar]
- 10.Da Cruz e Silva EF, Hughes V, McDonald P, Stark MJ, Cohen PT. 1991. Protein phosphatase 2Bw and protein phosphatase Z are Saccharomyces cerevisiae enzymes. Biochim Biophys Acta 1089:269–272. doi: 10.1016/0167-4781(91)90023-F. [DOI] [PubMed] [Google Scholar]
- 11.Chen MX, Chen YH, Cohen PT. 1993. PPQ, a novel protein phosphatase containing a Ser + Asn-rich amino-terminal domain, is involved in the regulation of protein synthesis. Eur J Biochem 218:689–699. doi: 10.1111/j.1432-1033.1993.tb18423.x. [DOI] [PubMed] [Google Scholar]
- 12.Venturi GM, Bloecher A, Williams-Hart T, Tatchell K. 2000. Genetic interactions between GLC7, PPZ1 and PPZ2 in Saccharomyces cerevisiae. Genetics 155:69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noble SM, French S, Kohn LA, Chen V, Johnson AD. 2010. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet 42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adám C, Erdei E, Casado C, Kovács L, González A, Majoros L, Petrényi K, Bagossi P, Farkas I, Molnar M, Pócsi I, Ariño J, Dombrádi V. 2012. Protein phosphatase CaPpz1 is involved in cation homeostasis, cell wall integrity and virulence of Candida albicans. Microbiology 158:1258–1267. doi: 10.1099/mic.0.057075-0. [DOI] [PubMed] [Google Scholar]
- 15.Nagy G, Hennig GW, Petrenyi K, Kovacs L, Pocsi I, Dombradi V, Banfalvi G. 2014. Time-lapse video microscopy and image analysis of adherence and growth patterns of Candida albicans strains. Appl Microbiol Biotechnol 98:5185–5194. doi: 10.1007/s00253-014-5696-5. [DOI] [PubMed] [Google Scholar]
- 16.Posas F, Casamayor A, Morral N, Ariño J. 1992. Molecular cloning and analysis of a yeast protein phosphatase with an unusual amino-terminal region. J Biol Chem 267:11734–11740. [PubMed] [Google Scholar]
- 17.Cohen PT, Schelling DL, da Cruz e Silva OB, Barker HM, Cohen P. 1989. The major type-1 protein phosphatase catalytic subunits are the same gene products in rabbit skeletal muscle and rabbit liver. Biochim Biophys Acta 1008:125–128. doi: 10.1016/0167-4781(89)90181-4. [DOI] [PubMed] [Google Scholar]
- 18.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. 2013. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res 41:W349–W357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choy MS, Hieke M, Kumar GS, Lewis GR, Gonzalez-DeWhitt KR, Kessler RP, Stein BJ, Hessenberger M, Nairn AC, Peti W, Page R. 2014. Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code. Proc Natl Acad Sci U S A 111:4097–4102. doi: 10.1073/pnas.1317395111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacKintosh C, Beattie KA, Klumpp S, Cohen P, Codd GA. 1990. Cyanobacterial microcystin-LR is a potent and specific inhibitor of protein phosphatases 1 and 2A from both mammals and higher plants. FEBS Lett 264:187–192. doi: 10.1016/0014-5793(90)80245-E. [DOI] [PubMed] [Google Scholar]
- 21.Goldberg J, Huang HB, Kwon YG, Greengard P, Nairn AC, Kuriyan J. 1995. Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1. Nature 376:745–753. doi: 10.1038/376745a0. [DOI] [PubMed] [Google Scholar]
- 22.Bollen M, Peti W, Ragusa MJ, Beullens M. 2010. The extended PP1 toolkit: designed to create specificity. Trends Biochem Sci 35:450–458. doi: 10.1016/j.tibs.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terrak M, Kerff F, Langsetmo K, Tao T, Dominguez R. 2004. Structural basis of protein phosphatase 1 regulation. Nature 429:780–784. doi: 10.1038/nature02582. [DOI] [PubMed] [Google Scholar]
- 24.Egloff MP, Johnson DF, Moorhead G, Cohen PT, Cohen P, Barford D. 1997. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J 16:1876–1887. doi: 10.1093/emboj/16.8.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurley TD, Yang J, Zhang L, Goodwin KD, Zou Q, Cortese M, Dunker AK, DePaoli-Roach AA. 2007. Structural basis for regulation of protein phosphatase 1 by inhibitor-2. J Biol Chem 282:28874–28883. doi: 10.1074/jbc.M703472200. [DOI] [PubMed] [Google Scholar]
- 26.Choy MS, Yusoff P, Lee IC, Newton JC, Goh CW, Page R, Shenolikar S, Peti W. 2015. Structural and functional analysis of the GADD34:PP1 eIF2alpha phosphatase. Cell Rep 11:1885–1891. doi: 10.1016/j.celrep.2015.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ragusa MJ, Dancheck B, Critton DA, Nairn AC, Page R, Peti W. 2010. Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites. Nat Struct Mol Biol 17:459–464. doi: 10.1038/nsmb.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirschi A, Cecchini M, Steinhardt RC, Schamber MR, Dick FA, Rubin SM. 2010. An overlapping kinase and phosphatase docking site regulates activity of the retinoblastoma protein. Nat Struct Mol Biol 17:1051–1057. doi: 10.1038/nsmb.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Posas F, Bollen M, Stalmans W, Ariño J. 1995. Biochemical characterization of recombinant yeast PPZ1, a protein phosphatase involved in salt tolerance. FEBS Lett 368:39–44. doi: 10.1016/0014-5793(95)00593-X. [DOI] [PubMed] [Google Scholar]
- 30.Swingle MR, Honkanen RE, Ciszak EM. 2004. Structural basis for the catalytic activity of human serine/threonine protein phosphatase-5. J Biol Chem 279:33992–33999. doi: 10.1074/jbc.M402855200. [DOI] [PubMed] [Google Scholar]
- 31.Fattori D, Squarcia A, Bartoli S. 2008. Fragment-based approach to drug lead discovery: overview and advances in various techniques. Drugs R D 9:217–227. doi: 10.2165/00126839-200809040-00002. [DOI] [PubMed] [Google Scholar]
- 32.Peti W, Page R. 2007. Strategies to maximize heterologous protein expression in Escherichia coli with minimal cost. Protein Expr Purif 51:1–10. doi: 10.1016/j.pep.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 33.Tóth A, Kiss E, Herberg FW, Gergely P, Hartshorne DJ, Erdödi F. 2000. Study of the subunit interactions in myosin phosphatase by surface plasmon resonance. Eur J Biochem 267:1687–1697. doi: 10.1046/j.1432-1327.2000.01158.x. [DOI] [PubMed] [Google Scholar]
- 34.Erdödi F, Tóth B, Hirano K, Hirano M, Hartshorne DJ, Gergely P. 1995. Endothall thioanhydride inhibits protein phosphatases-1 and -2A in vivo. Am J Physiol 269:C1176–C1184. [DOI] [PubMed] [Google Scholar]
- 35.Shenolikar S, Ingebritsen TS. 1984. Protein (serine and threonine) phosphate phosphatases. Methods Enzymol 107:102–129. doi: 10.1016/0076-6879(84)07007-5. [DOI] [PubMed] [Google Scholar]
- 36.Kovács L, Farkas I, Majoros L, Miskei M, Pócsi I, Dombrádi V. 2010. The polymorphism of protein phosphatase Z1 gene in Candida albicans. J Basic Microbiol 50(Suppl 1):S74–S82. doi: 10.1002/jobm.200900434. [DOI] [PubMed] [Google Scholar]
- 37.Máthé C, Beyer D, Erdodi F, Serfozo Z, Székvölgyi L, Vasas G, M-Hamvas M, Jámbrik K, Gonda S, Kiss A, Szigeti ZM, Surányi G. 2009. Microcystin-LR induces abnormal root development by altering microtubule organization in tissue-cultured common reed (Phragmites australis) plantlets. Aquat Toxicol 92:122–130. doi: 10.1016/j.aquatox.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 38.Tierno MB, Johnston PA, Foster C, Skoko JJ, Shinde SN, Shun TY, Lazo JS. 2007. Development and optimization of high-throughput in vitro protein phosphatase screening assays. Nat Protoc 2:1134–1144. doi: 10.1038/nprot.2007.155. [DOI] [PubMed] [Google Scholar]
- 39.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC, Adams PD. 2008. Automated structure solution with the PHENIX suite. Methods Mol Biol 426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Conservation of PP1 interaction pockets between HsPP1α (1α) and CaPPZ1 (Z1). The residues that define the metal, toxin, and regulatory motif binding pockets in HsPP1α and the corresponding residues in CaPPZ1 are shown. Residues that are different between the two proteins are in boldface and shaded in a darker color. The sequence identity (ID) and similarity (Sim) of the distinct binding pockets are calculated above each list. Download
MC potently inhibits recombinant HsPP1α and CaPPZ1 activity measured with pNPP. The data represent the mean ± standard deviation (SD) from 4 experiments. Download
Secondary structure prediction for CaPPZ1. The N-terminal domain (gray box) is predicted to lack secondary structural elements (pink cylinders, α-helices; yellow arrows, β-strands), while the catalytic domain (pink box) is predicted to be structured. The C-terminal residues in PPZ1 are predicted to be helical (green box). “Conf” represents the confidence of the prediction, with higher bars indicating greater confidence scores. Download
Mutation of the spinophilin ΦΦ and Arg motifs negatively impacts its ability to bind PP1α. Pull-down assays with (A) PP1α or (B) PPZ1 and either wild-type (wt) spinophilin or spinophilin variants in which the ΦΦ or Arg PP1 interaction motifs are mutated (SpinoΦΦ→AA, SpinoR469D, and SpinoR469E). (C) Densitometry and quantification of pulldown results shown in panels A and B. PP1α is in blue and PPZ1 in red. Mutation of either the ΦΦ or Arg motif in spinophilin reduces the amount of spinophilin pulled down by PP1α to levels nearly identical to those obtained with PPZ1. Download