ABSTRACT
Helicobacter pylori must be able to rapidly respond to fluctuating conditions within the stomach. Despite this need for constant adaptation, H. pylori encodes few regulatory proteins. Of the identified regulators, the ferric uptake regulator (Fur), the nickel response regulator (NikR), and the two-component acid response system (ArsRS) are each paramount to the success of this pathogen. While numerous studies have individually examined these regulatory proteins, little is known about their combined effect. Therefore, we constructed a series of isogenic mutant strains that contained all possible single, double, and triple regulatory mutations in Fur, NikR, and ArsS. A growth curve analysis revealed minor variation in growth kinetics across the strains; these were most pronounced in the triple mutant and in strains lacking ArsS. Visual analysis showed that strains lacking ArsS formed large aggregates and a biofilm-like matrix at the air-liquid interface. Biofilm quantification using crystal violet assays and visualization via scanning electron microscopy (SEM) showed that all strains lacking ArsS or containing a nonphosphorylatable form of ArsR (ArsR-D52N mutant) formed significantly more biofilm than the wild-type strain. Molecular characterization of biofilm formation showed that strains containing mutations in the ArsRS pathway displayed increased levels of cell aggregation and adherence, both of which are key to biofilm development. Furthermore, SEM analysis revealed prevalent coccoid cells and extracellular matrix formation in the ArsR-D52N, ΔnikR ΔarsS, and Δfur ΔnikR ΔarsS mutant strains, suggesting that these strains may have an exacerbated stress response that further contributes to biofilm formation. Thus, H. pylori ArsRS has a previously unrecognized role in biofilm formation.
IMPORTANCE Despite a paucity of regulatory proteins, adaptation is key to the survival of H. pylori within the stomach. While prior studies have focused on individual regulatory proteins, such as Fur, NikR, and ArsRS, few studies have examined the combined effect of these factors. Analysis of isogenic mutant strains that contained all possible single, double, and triple regulatory mutations in Fur, NikR, and ArsS revealed a previously unrecognized role for the acid-responsive two-component system ArsRS in biofilm formation.
INTRODUCTION
Helicobacter pylori is a Gram-negative pathogen that colonizes the gastric mucosa of humans and nonhuman primates. Approximately 50% of the world's population is infected with H. pylori, and phylogeographic evidence suggests that the bacterium may have been associated with mankind for more than 100,000 years (1). Given this long-standing association with its host and the specificity of the gastric niche, it is not surprising that H. pylori exhibits a minimalist genome indicative of reductive evolution (2, 3). As such, the H. pylori genome encodes approximately half the number of regulatory proteins seen in Haemophilus influenzae, which has a similar-size genome (3, 4); moreover, the closely related species Campylobacter jejuni also appears to encode a larger repertoire of regulatory proteins (5). Within the human stomach, H. pylori faces fluctuating stressors, such as acidity, nutrient availability, osmotic pressure, and host defenses. In order to adapt and persist within this hostile niche, H. pylori has developed a complex regulatory network that optimizes the activities of the few regulators carried in the genome (4, 6, 7). Three key nodes within this regulatory structure are represented by the following regulatory proteins: Fur, the ferric uptake regulator; NikR, a nickel response regulator; and ArsRS, an acid-responsive two-component system (7). The prominent roles of these regulators are highlighted by the fact that all three are required for wild-type colonization levels in animal models (8–11). The importance of the OmpR-like response regulator ArsR is further demonstrated by the fact that it is essential for in vitro survival. Intriguingly, the cognate histidine kinase ArsS is dispensable in vitro, and a nonphosphorylatable form of ArsR (ArsR-D52N mutant) also supports viability of H. pylori (12–14). Thus, the essentiality of ArsR must depend on the regulation of essential genes by the nonphosphorylated form of the protein (15). The nature of this regulation is not clearly understood, and despite the two states of ArsR, most transcriptional studies focus on elucidating the targets of the pH-dependent phosphorylated ArsR (ArsR∼P) regulon. Based upon this literature, current evidence indicates that ArsR∼P can act to repress or activate the expression of different sets of genes (16, 17).
In an effort to understand adaptation, numerous studies have sought to define the regulon of genes controlled by Fur, NikR, and ArsRS (10, 16–22). Examination of these transcriptome studies en masse demonstrates the vast and overlapping nature of the Fur, NikR, and ArsRS regulons. Furthermore, evidence suggests that these regulators affect expression of each other; nikR and arsR are part of the Fur regulon (23, 24), whereas NikR appears to regulate fur (20, 25). Given the overlapping nature of these regulatory cascades, it is likely not surprising that the role of each of these factors appears to be expanded compared to what is seen in other bacteria. For example, although Fur and NikR are classically thought of as metallo-regulators, in H. pylori, they also play an important role in acid acclimation (10, 26). Furthermore, Fur has been shown to be important for adaptation to oxidative, nitrosative, and osmotic stress (27–29). Thus, further study of the intricate interaction between each of these regulatory factors may provide increased information about the adaptation process of H. pylori.
One general adaptive mechanism used by many bacteria is the formation of biofilm structures that provide increased protection from a number of different stresses. Biofilm formation by H. pylori has been demonstrated both in vitro (30–32) and in vivo (33–37). While it is unclear exactly what drives biofilm formation in H. pylori, protection from antibiotics and host defenses during infection (35, 38, 39) and/or protection in putative environmental reservoir (40–43) have been proposed. Basic characterization has shown that in vitro biofilm formation is varied across strains (32) and appears to be induced under conditions, such as serum/nutrient starvation (44, 45). Initial investigations of the extracellular polymeric substances (EPS) matrix observed in in vitro biofilms revealed the presence of lipids, amino acids, and monosaccharides. Further investigation of the EPS matrix demonstrated the presence of extracellular DNA (eDNA); however, in contrast to what has been shown for other Gram-negative species, eDNA does not appear to play a structural role in H. pylori biofilms (46). An increased number of outer membrane vesicles (OMVs) are associated with H. pylori found within a biofilm compared to planktonic bacteria, suggesting a potential link between OMV production and biofilm formation (47, 48). Interestingly, OMVs have been observed in association with other Gram-negative biofilms, such as those formed by Pseudomonas aeruginosa (49).
Apart from what can be inferred from the descriptive biofilm studies, little is known about the molecular mechanisms that drive biofilm formation or the genes that are required for biofilms in H. pylori. Notable exceptions include studies that indicate that the deletion of cagE and luxS results in increased biofilm formation, whereas the deletion of additional genes (crdR, clpA, ppk, and hpA) known to contribute to biofilm development in other bacteria had no effect on biofilm formation by H. pylori (31). Proteomic analysis designed to identify proteins involved in biofilm formation identified numerous proteins that were differentially regulated between biofilm and planktonic H. pylori; intriguingly, ArsR was among the proteins upregulated (>2-fold) in the biofilm community (50). Overall, relatively little is understood about the molecular mechanisms that lead to biofilm formation in this important pathogen.
In an effort to understand the complex regulatory interaction between Fur, NikR, and ArsRS, herein, we describe the creation and basic characterization of a series of isogenic H. pylori mutant strains in which each regulatory protein was knocked out singularly or in combination. Basic characterization of these strains led to the novel observation that the ArsRS system plays a previously unappreciated role in biofilm formation in this important pathogen.
MATERIALS AND METHODS
Bacterial strains and growth.
The strains and plasmids used in the study are listed in Table 1. All isogenic mutant strains were created using H. pylori G27 as the parental strain (51, 52). Unless otherwise noted, H. pylori strains were cultured as previously described (53). Briefly, all cultures were grown at 37°C in gas evacuation jars under microaerobic conditions (5% O2, 10% CO2, and 85% N2) generated with an Anoxomat gas evacuation and replacement system (Advanced Instruments, Inc.). For plate cultures, strains were grown on horse blood agar (HBA) composed of 4% Columbia agar (Neogen Corporation), 5% defibrinated horse blood (HemoStat Laboratories, Dixon, CA), 2 mg/ml β-cyclodextrin (Sigma), and an antibiotic-antifungal cocktail (10 μg/ml vancomycin [Amresco], 5 μg/ml cefsulodin [Sigma], 2.5 U/ml polymyxin B [Sigma], 5 μg/ml trimethoprim [Sigma], and 8 μg/ml amphotericin B [Amresco]). H. pylori liquid medium consisted of brucella broth (Neogen Corporation) supplemented with 10% fetal bovine serum (FBS) (Gibco) and 10 μg/ml vancomycin. Liquid cultures were grown with shaking at 100 rpm. H. pylori strains were maintained as stock cultures at −80°C. Stock cultures were frozen in brain heart infusion broth (BD Biosciences) containing 10% FBS and 20% glycerol (EMD Chemicals, Inc.).
TABLE 1.
Primers, plasmids, and bacterial strains used in this study
| Primer, plasmid, or bacterial strain | Sequence or descriptiona | Source or reference |
|---|---|---|
| Primers | ||
| Primers for ArsR-D52N mutant construction | ||
| Up1F | GGGGATTTTTTGAGCGTTGAG | This study |
| Up2R | gttagtcacccgggtaccgagctcCACTCTGTGTTTTCTCGCTCC | This study |
| Dn1F | ctagagtcgacctgcaggcatgcaagCCATGAAAACAAAGCCTTAATTTCC | This study |
| Dn2R | CTAAATTAGGCAAAGTCAAATtCAACAACAACAAATC | This study |
| Dn3F | GATTTGTTGTTGTTGaATTTGACTTTGCCTAATTTAG | This study |
| Dn4R | CGCAAACGGCCAATGATCAC | This study |
| KanF | GTTAGTCACCCGGGTACCGAGCTC | This study |
| KanR | CTAGAGTCGACCTGCAGGCATGCAAG | This study |
| Primers for Δfur mutant confirmation | ||
| Fur-expressF1 | ATGAAAAGATTAGAAACTTTGGAA | This study |
| Fur-expressR1 | ACATTCACTCTCTTGGCATTC | This study |
| Primers for ΔnikR mutant confirmation | ||
| HP1338_Del_Conf_F | GGCCAACCATTTAAATCCAG | This study |
| HP1338_Del_Conf_R | GTTTTGATAAGCGGGCAAGA | This study |
| Primers for ΔarsS mutant confirmation | ||
| HP0165_del_ver_F | TGAAAGCATTGCGATTGAGA | This study |
| HP0165_del_ver_R | AAAACGGCTTTGATGCCTAA | This study |
| Primers for ΔarsS mutant complementation | ||
| ArsS_F | GGGGTTTTGCGTTTCTCTATC | This study |
| ArsS_compR | AATTCCGCGGTTATTGTTTTTCTTTAACCCCAC | This study |
| ArsS_compF | AATTTCTAGATTAAGATATTTTCAAGGTATCTTTAATAATAAAA | This study |
| TGCTAGAATTAGTCCCTTAAAACATTAGCAAAGTTAATCGTT | ||
| TTTAAAGTTGCAGAAATCATTGAAGGAGTTAATTGAAGTttgcgttt | ||
| ctctatcttttttaagg | ||
| ArsRProm_F | AATTTCTAGATTAAGATATTTTCAAGGTATC | This study |
| Plasmids | ||
| pDSM924 | pGEM T-easy::ΔnikR::cat | 53 |
| pDSM920 | pBluescript::ΔarsS::kan | 55 |
| pΔHP1027-K7 | pCRII-Topo::Δfur::kan | 10 |
| pDSM1525 | pTM117::arsSC | This study |
| Strains | ||
| DSM1 | WT G27 | 51 |
| DSM41 | G27 ΔflgR::cat Cmr | 14 |
| DSM145 | G27 Δfur::kan Kanr | 54 |
| DSM300 | G27 Δfur::cat Cmr | 54 |
| DSM975 | G27 ΔnikR::cat Cmr | 53 |
| DSM983 | G27 ΔarsS markerless | 53 |
| DSM986 | G27 Δfur::kan or ΔnikR::cat, Kanr Cmr | This study |
| DSM987 | G27 Δfur::cat or ΔarsS::kan, Kanr Cmr | This study |
| DSM1071 | G27 ΔarsS or nikR::cat, Cmr | 53 |
| DSM1441 | G27 ΔarsS or Δfur::kan or ΔnikR::cat, Kanr Cmr | This study |
| DSM1446 | G27 ArsR-D52N, unphosphorylatable, Kanr | This study |
| DSM1526 | G27 ΔarsS markerless/pTM117::arsSC | This study |
Lowercase letters indicate regions of overlap sequence for SOE PCR. Underlined lowercase nucleotides indicate the point mutation that encodes the D52N mutation. Bold italicized nucleotides correspond to restriction digestion sites. Cmr, chloramphenicol resistance; Kanr, kanamycin resistance.
Construction of G27 Δfur ΔnikR mutant.
The G27 Δfur ΔnikR mutant strain was constructed by naturally transforming DSM145 (Δfur::kan) (54) with pDSM924 (53), which carries a ΔnikR::cat cassette. Transformants were selected on HBA plates supplemented with 25 μg/ml kanamycin (Kan) (Gibco) and 8 μg/ml chloramphenicol (Cm) (EMD Chemicals, Inc.) to ensure colonies selected were lacking both fur (Kan) and nikR (Cm). Integration and subsequent deletion of nikR were confirmed by PCR using primers HP1338_Del_Conf_F and HP1338_Del_Conf_R. The sequences of the primers used are listed in Table 1. Colonies with amplicons of the expected size for nikR::cat were sequenced to verify the deletion of nikR. Fur deletion confirmation primers Fur-expressF1 and Fur-expressR1 are modified versions of 1027-F1 (SalI) and 1027-R6 (NotI), respectively (10), in which the restriction site has been removed. Fur-expressF1 and Fur-expressR1 were used in PCR and sequencing to verify that the Δfur mutation was unaltered, and the G27 Δfur ΔnikR mutant strain was archived as DSM986.
Construction of G27 Δfur ΔarsS mutant.
The G27 Δfur ΔarsS mutant strain was constructed by naturally transforming pDSM920 (55), carrying a ΔarsS::kan cassette, into DSM300 (Δfur::Cmr) (54). Transformants were selected on HBA plates supplemented with 25 μg/ml Kan and 8 μg/ml Cm. Kan/Cm-resistant colonies were verified by PCR using primer pairs HP0165_del_ver_F/HP0165_del_ver_R and Fur-expressF1/Fur-expressR1 for the ΔarsS and Δfur mutants, respectively. Products displaying the expected size change for the deletion of arsS and fur were sequenced using the primers described above, and the Δfur ΔarsS mutant strain was archived as DSM987.
Construction of G27 Δfur ΔnikR ΔarsS mutant.
The G27 mutant strain containing the combined deletions of fur, nikR, and arsS was constructed by transforming DSM1071 (ΔnikR ΔarsS mutant) (53) with pΔHP1027-K7 (10), which contains a Δfur::kan cassette. Transformants were selected on 25 μg/ml Kan. PCR with the primer sets Fur-expressF1/Fur-expressR1, HP1338_Del_Conf_F/HP1338_Del_Conf_R, and HP0165_del_ver_F/HP0165_del_ver_R was used to screen Kan-resistant colonies for the deletion of fur, nikR, and arsS, respectively. The Fur deletion mutant was further confirmed by sequencing with Fur-expressF1/Fur-expressR1, and the resulting Δfur ΔnikR ΔarsS mutant strain was archived as DSM1441.
Construction of G27 ArsR-D52N mutant.
Site-directed mutagenesis was used to create a nonphosphorylatable ArsR mutant strain (ArsR-D52N) (13, 15). The ArsR-D52N mutant strain was constructed in a stepwise manner using consecutive splicing by overlap extension (SOE) PCR, followed by allelic exchange. The initial SOE PCR was performed to introduce an adenine at position 154 of arsR, which results in an asparagine at amino acid position 52 in lieu of an aspartic acid. To accomplish this, arsR was amplified in two fragments: fragment one was amplified with Dn1F and Dn2R and included the arsR promoter and about 1/4 of the coding region (nucleotides 169811 to 170269, gpCP001173.1), and fragment two was amplified with Dn3F and Dn4R and included a 30-bp overlap with fragment one in addition to a majority of arsR 3′ region (nucleotides 170234 to 170584). Dn2R and Dn3F both contain the G→A transition, which is introduced into arsR during SOE PCR of the two arsR fragments. The resulting arsR construct, which carries the single-base-pair modification, was then used as the downstream section for the subsequent SOE PCRs. In addition to the arsR construct, two standard PCRs were run to amplify the region upstream of arsR (nucleotides 170645 to 171164, amplified with Up1F and Up2R) and a Kan resistance (Kanr) cassette (amplified with KanF and KanR). Next, the upstream fragment was fused to the 5′ end of the Kanr cassette through SOE PCR with Up1F and KanR. In the final step, the upstream Kanr PCR product was fused the 5′ end of the arsR construct by SOE PCR using Up1F and Dn4R. The resulting SOE PCR product was used to transform G27 (DSM1) (51). Transformants were selected on 25 μg/ml Kan. Dn1F and Dn4R primers were used to PCR amplify Kan-resistant colonies, and the resulting amplicons were screened by restriction digestion with Apo1; the G→A transition introduces an Apo1 site. Dn1F and Dn4R were used to sequence the ArsR region of colonies for which PCR products yielded the correct Apo1 digestion pattern. Twofold sequence coverage for the region was obtained to confirm the transition mutation, and the resulting strain was archived as DSM1446.
Construction of the complemented G27 ΔarsS-arsSC strain.
The arsS deletion was complemented in trans. Since arsS is the second gene in an operon with arsR, the promoter region of arsR was fused upstream of arsS using SOE PCR. Briefly, the arsR promoter region was synthesized as an oligonucleotide named ArsS_compF; this oligonucleotide was designed to contain an overlapping complementary sequence to arsS. The arsS coding region was amplified with primers ArsS_F and ArsS_compR. The promoter fragment and the arsS fragment were then mixed together and fused in a SOE PCR that employed the ArsR_promF and ArsS_compR primers; these primers were designed to incorporate restriction sites needed for downstream cloning. The resulting fragment was digested with XbaI and SacII and ligated into the similarly digested pTM117 vector (54). The ligation was then transformed into TOP10 chemically competent Escherichia coli, and transformants were selected on LB containing 25 μg/ml Kan. pTM117::arsS-positive colonies were confirmed by PCR, and the resulting pTM117::arsS complementation construct was transformed into the G27 ΔarsS mutant strain. Transformants were selected on HBA containing 25 μg/ml Kan. To prove reproducibility, five separate complementation colonies (designated ΔarsS-arsSC) were selected, were shown to express arsS by quantitative real-time PCR (qRT-PCR), and were tested in the biofilm assay. Given that all 5 isolates behaved similarly, one isolate was archived as DSM1526.
Growth curves.
Bacterial cultures were patched from freezer stocks onto HBA plates. After 48 h, sterile swabs were used to inoculate a 10-ml liquid starter culture in H. pylori liquid medium. Liquid starter cultures were grown for 20 to 24 h, and then 60-ml liquid cultures were standardized to a starting optical density at 600 nm (OD600) of 0.05 optical density units (ODU) from the overnight starter culture. Each 60-ml culture was then divided in half into two separate 125-ml flasks and grown under standard lab conditions; CFU counts and OD readings were taken from alternating culture flasks at 0, 6, 12, 20, 30, and 48 h postinoculation. The data shown represent three biologically independent experiments (see Fig. 1).
FIG 1.
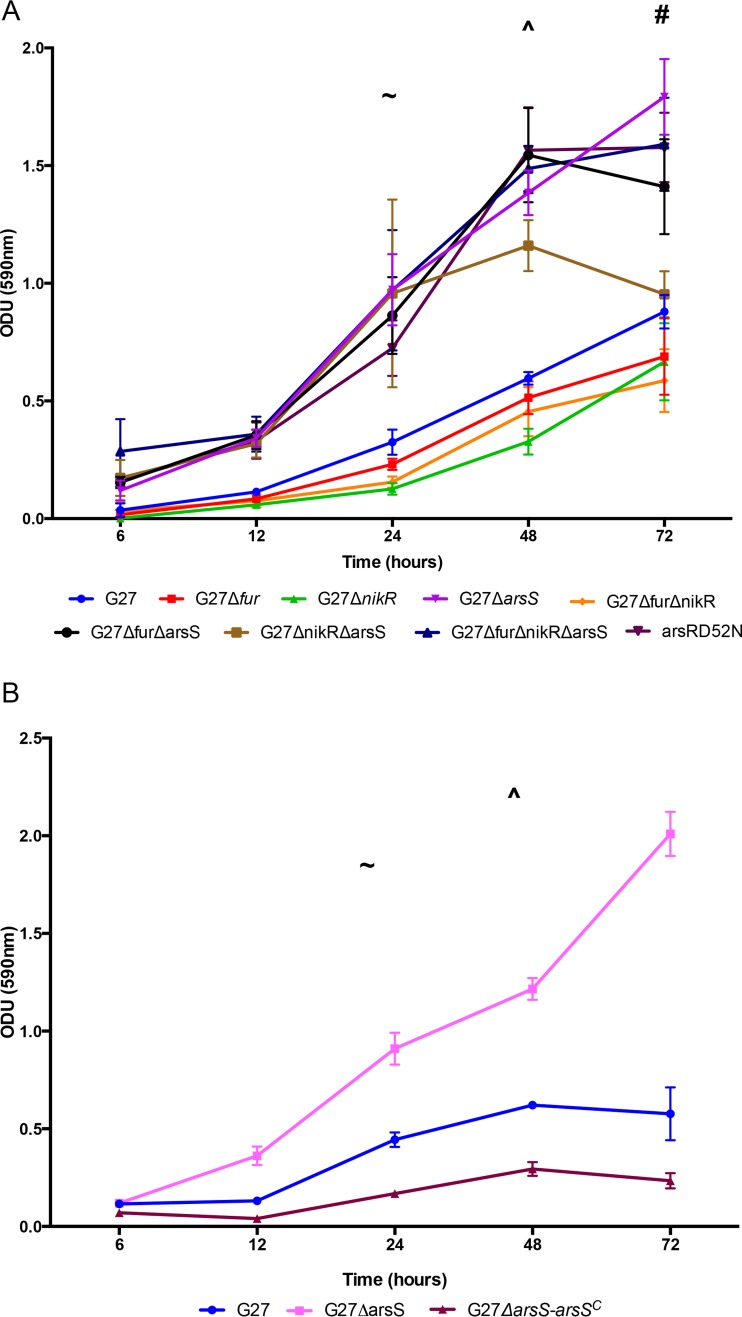
Characterization of wild-type G27 and isogenic regulatory mutant strain growth phenotypes. (A) OD600 readings were taken after 6, 12, 20, and 48 h. Mean ODU for each strain and time point is indicated by the respective symbols; error bars display standard error of the mean. The graph is representative of the results from three biologically independent experiments. (B) Enumeration of CFU was performed after 6, 12, 20, 20, and 48 h, corresponding to the OD values graphed in panel A. The mean CFU was calculated for each time point and is indicated by the respective symbols; error bars display standard error of the mean. The graph is representative of the results from three biologically independent experiments.
Biofilm quantification.
Bacterial cultures were started according to the same procedure used in the growth curves; however, biofilm assays were conducted in 24-well tissue culture plates (Corning, Inc.). Starter cultures were used to inoculate 1 ml of liquid medium per well to an OD600 of 0.1 ODU. A separate plate was used for each of the five time points, T6, T12, T24, T48, and T72. At each time point, the medium was aspirated, and each well was washed two times with phosphate-buffered saline (PBS). Plates were dried for 5 min at 37°C, and biofilms were subsequently fixed with methanol (J.T. Baker). One percent Gram's crystal violet solution (Sigma-Aldrich) was then added to each well until full (approximately 2 ml), including one empty control well to serve as a blank. The plates were incubated on the benchtop for 15 min, after which crystal violet was removed from the wells, and each well was washed three times with distilled water. The plates were again dried for 5 min at 37°C. At this point, 1.5 ml of differentiation solution (Sigma) was added to solubilize the dye contained within the biofilms. Plates were incubated with differentiation solution for 15 min, 1 ml of solubilized crystal violet solution was then transferred to a cuvette, and the absorbance (OD590) for each sample was read. The data shown represent three biologically independent experiments (see Fig. 3).
FIG 3.
Biofilm quantification. (A) Absorbance of crystal violet staining of biofilms was read at OD590 and plotted over time. ^ designates the point at which wild-type G27 displayed significant biofilm formation (P = 0.041). ∼ indicates the point at which the ΔarsS (P < 0.0001), Δfur ΔarsS (P = 0.0045), ΔnikR ΔarsS (P = 0.0012), Δfur ΔnikR ΔarsS (P = 0.0067), and ArsR-D52N (P = 0.0403) mutants displayed significant biofilm formation. # identifies the point at which the Δfur (P = 0.0082), ΔnikR (P = 0.0092), and Δfur ΔnikR (P = 0.0451) mutants displayed significant biofilm formation. The data for each strain are plotted as the mean of the results from three biologically independent experiments; error bars show the standard error of the mean. (B) Absorbance of crystal violet staining of biofilms was read at OD590 and plotted over time. Biofilm formation by the ΔarsS-arsSC mutant is shown in comparison to the ΔarsS mutant and wild-type G27 strains. ^ designates the point at which wild-type G27 displayed significant biofilm formation (P = 0.0027); ∼ indicates the point at which the ΔarsS mutant displayed significant biofilm formation (P < 0.0001). The data for wild-type G27 and the ΔarsS mutant are plotted as the mean of at least three independent experiments. The mean plotted for the ΔarsS-arsSC mutant represents 5 isolates tested in up to two biologically independent experiments. For all data sets, error bars show the standard error of the mean.
SEM.
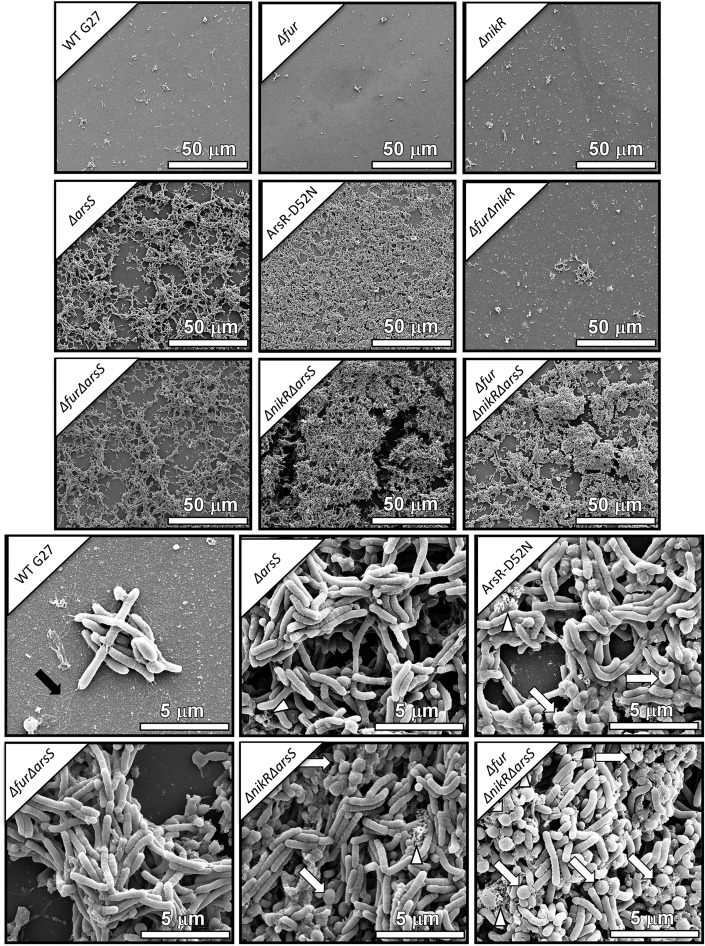
Stock cultures (OD600, 0.3 to 0.7) were inoculated 1:10 into fresh brucella broth in 12- or 24-well plates. Biofilms were captured on cell culture-treated coverslips at the bottom of the wells and analyzed by scanning electron microscopy, as previously described (56). Briefly, samples were fixed with 2.0% paraformaldehyde (Electron Microscopy Sciences) and 2.5% glutaraldehyde (Electron Microscopy Sciences) in 0.05 M sodium cacodylate (pH 7.4; Electron Microscopy Sciences) buffer for 24 h. After primary fixation, samples were washed three times with 0.05 M sodium cacodylate buffer before sequential dehydration with increasing concentrations of ethanol. After ethanol dehydration, samples were dried at the critical point using a Tousimis critical point dryer machine, mounted onto aluminum SEM sample stubs (Electron Microscopy Sciences), and sputter coated with 5 nm of gold-palladium. Afterward, samples were painted with a thin strip of colloidal silver (Electron Microscopy Sciences) at the edge to facilitate charge dissipation. Biofilms were imaged with an FEI Quanta 250 field-emission gun scanning electron microscope. The micrographs shown are representative of three biological replicates (see Fig. 4).
FIG 4.
Scanning electron micrographs of biofilm formation after 48 h. Representative micrographs from all 9 strains are shown at ×1,000 magnification. In comparison to the G27 wild-type, Δfur mutant, ΔnikR mutant, and Δfur ΔnikR mutant, which show sporadic adherent cells in small clusters, the ΔarsS, ArsR-D52N, Δfur ΔarsS, ΔnikR ΔarsS, and Δfur ΔnikR ΔarsS mutants all contain mats of cells arranged in three-dimensional (3D) architectures. The 3D structure is especially apparent in the representative image of the ΔnikR ΔarsS mutant. Biofilm-forming strains are shown in comparison to wild-type in the lower half of the figure at ×10,000 magnification. Of note, black arrows indicate flagella, white triangles show areas of extracellular matrix, and white arrows show coccoid cells. Coccoid cells were more prevalent in the ArsR-D52N, ΔnikR ΔarsS, and Δfur ΔnikR ΔarsS mutants.
Motility assay.
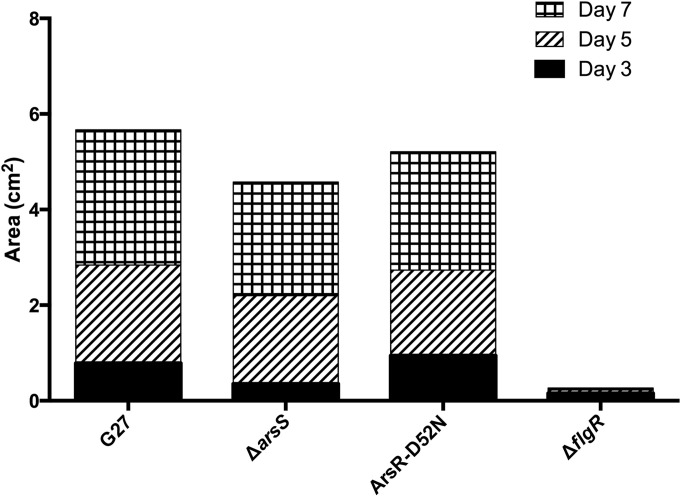
H. pylori cultures were started as described in the growth curve assay. After approximately 20 h of growth, each culture was passed through a 30-G needle to disrupt large aggregates that form in some strains, which could influence OD measurement. After passage through the needle, 0.5 ODU of each culture was pelleted and resuspended in 50 μl of H. pylori liquid medium. From this suspension, 2 μl was inoculated into motility agar plates with a 10-μl pipette tip. Motility agar consisted of 2.5% brucella broth, 0.3% agar-agar (EMD Chemicals), 10% FBS, and 10 μg/ml vancomycin. Plates were incubated at 37°C under microaerobic conditions, and the area of the halo of motility was measured 3, 5, and 7 days postinoculation. At each time point, plates were removed and imaged on an ImageQuant Las4000 (GE). Area measurements were completed in ImageJ version 1.48 (http://imagej.nih.gov/ij/) using a scale set by the known dimensions of the plate. The data presented represent three independent biological replicates (see Fig. 5).
FIG 5.
Swimming motility of wild-type G27, ΔarsS mutant, and ArsR-D52N mutant strains. The motility of wild-type G27 and the ΔarsS and ArsR-D52N mutant strains was compared using a soft agar assay. The area of the halo was measured at days 3, 5, and 7. The bar graph shows the cumulative area of motility as measured over 7 days; the solid black section corresponds to growth through day 3, the hatched segment shows additional area covered between days 3 and 5, and the checkered portion represents the increase from day 5 through 7. The bars represent the mean area of the results from three independent biological replicates.
Bacterial aggregation assay.
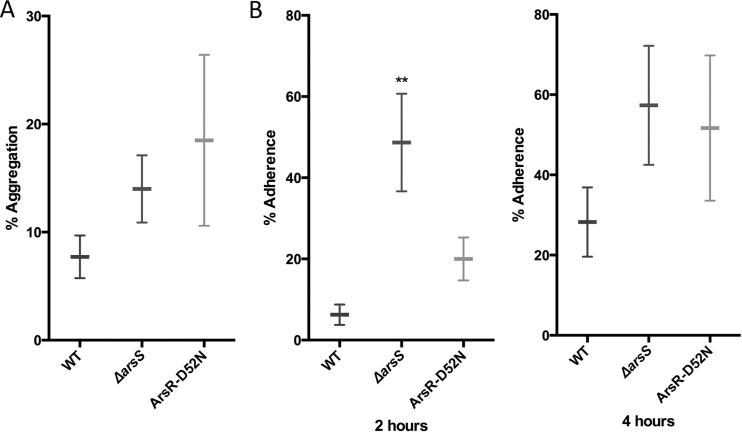
Bacterial autoaggregation was assessed as previously described (32), with the following modifications. H. pylori cultures were prepared as described in the bacterial motility assay. One optical density unit of each culture was pelleted, washed with PBS, and resuspended in 2 ml of PBS in 12 by 75-mm glass tubes, and an initial OD600 reading was taken (T0). The tubes were then placed in a shaking incubator (190 rpm, 37°C), and additional OD readings at 600 nm were taken after 2 h. Percent autoaggregation was calculated as follows: [(OD600T0 − OD600T2)/OD600T0)] × 100. The data shown represent a minimum of three biological replicates (see Fig. 6A).
FIG 6.
Autoaggregation and adherence of the ΔarsS and ArsR-D52N mutant strains. (A) Autoaggregation was assessed in wild-type G27 and the ΔarsS and ArsR-D52N mutant strains. Horizontal bars indicate the means of the results from three biological replicates, and the error bars represent the standard error of the mean. (B) Surface adherence was assessed for wild-type G27 and the ΔarsS and ArsR-D52N mutant strains after 2 and 4 h of incubation. ** indicates significantly more adherence by the ΔarsS mutant strain (P = 0.0057) than the wild type. Horizontal bars indicate the means of the results from three biological replicates, and error bars represent the standard error of the mean.
Plate adherence assay.
The plate adherence assay was conducted as previously described (57), with several modifications. Overnight liquid cultures of each strain were inoculated into 6 separate wells of a 96-well tissue culture-treated plate (Corning) at an OD600 of 0.1 ODU per well in 100 μl of H. pylori liquid medium. At various time points postinoculation, the liquid was aspirated from three wells per strain. These same wells were washed three times with PBS, after which 200 μl of 5% alamarBlue (Invitrogen) diluted in H. pylori liquid medium was added to each well. The 5% alamarBlue solution was also added to three empty wells to serve as the blank. In the three remaining wells for each strain, 100 μl of 10% alamarBlue in H. pylori liquid medium was added. These wells were used to determine the number of starting bacteria. After the addition of alamarBlue, the plates were wrapped in foil to prevent light exposure and were incubated at 37°C, under microaerobic conditions, in a static incubator for 2 h. The plate was then read using an excitation wavelength of 544 nm and emission wavelength of 590 nm, which resulted in relative fluorescence units (RFU) for each well. Triplicate wells for each experiment were averaged. From the averaged values, percent adherence was calculated as follows: (RFU of the washed wells/RFU inoculum) × 100. The data shown represent a minimum of three biologically independent experiments (see Fig. 6B).
Statistical analysis.
Statistical analyses were conducted using GraphPad Prism version 6.0g (GraphPad Software, Inc.). A two-way analysis of variance (ANOVA) with Tukey's correction for multiple comparisons was used to analyze increases in biofilm formation over time for individual strains and also for differences in biofilm formation between strains at each time point. For aggregation and adherence assays, wild-type G27 was compared to either the ΔarsS or ArsR-D52N mutant using a one-way ANOVA with Dunnett's multiple-comparison test.
RESULTS
Growth characteristics of Fur, NikR, and ArsRS mutant strains.
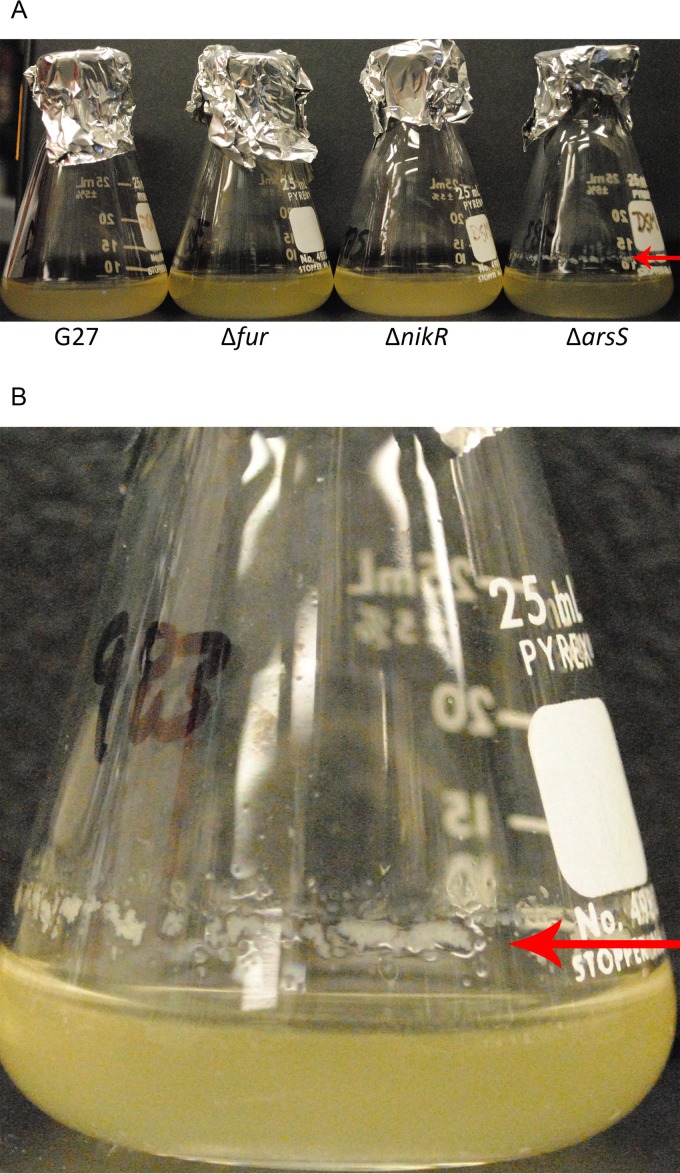
Growth curve analysis under standard laboratory conditions was conducted for each of the single, double, and triple regulatory mutants as a means to identify any substantial changes in strain health. While some minor differences were evident (Fig. 1), no overt growth defects were observed for the single or double regulatory knockout strains. However, DSM1441, the strain which lacks Fur, NikR, and ArsR, appeared to grow more slowly (Fig. 1) and took longer to recover from the frozen stock (data not shown). Moreover, visually, this strain appeared more stressed and transitioned to the coccoid form more quickly than the other strains (data not shown). For the remaining strains, while growth was fairly similar throughout exponential phase (through T24), strains lacking ArsS appeared to plateau at a lower CFU and ODU than the other strains. The difference in ODU and CFU for strains lacking ArsS was most pronounced between 24 and 30 h of growth, as cultures entered stationary phase. Around the same time point, noticeable clumping was observed in these cultures as well as a biofilm-like ring at the air-liquid interface (Fig. 2). Given this phenotype, it is possible that the slight differences in growth observed for the ΔarsS, Δfur ΔarsS, ΔnikR ΔarsS, and Δfur ΔnikR ΔarsS mutants is partially the result of bacterial clumping that skewed both optical density reading and CFU enumeration. To determine the nature of the bacterial clumping, liquid bacterial cultures were viewed using bright field microscopy; aggregates composed primarily of spiral H. pylori with some extracellular material were observed (see Fig. S1 in the supplemental material).
FIG 2.
Biofilm formation at air-liquid interface. (A) Glass flasks similar to those used for the growth curve analyses are shown. From left to right, flasks containing WT G27, Δfur mutant, ΔnikR mutant, and ΔarsS mutant strains were imaged after 72 h of growth. In comparison to the other strains, a biofilm-like rim (arrow) can be seen on the flask containing the ΔarsS strain. (B) Close-up image of the ΔarsS mutant strain shows formation of the biofilm-like ring (arrow) at the air-liquid interface.
Loss of ArsRS in G27 exacerbates biofilm formation.
Based on the observations made during the growth curve analyses, we decided to further investigate the biofilm-like phenotypes observed with the isogenic strains. First, we conducted a time course study in which biofilm formation was visually monitored over 72 h (see Fig. S2A in the supplemental material). Three distinct biofilm types were observed during this assay, and some strains displayed multiple types of biofilm formation. The major phenotypes observed were biofilm formation at the air-liquid interface, biofilm formation on the bottom of the well, and formation of a pellicle. For wild-type G27, biofilm formation was not apparent at early time points but first became visible after 24 h at the air-liquid interface (data not shown). At 48 and 72 h of growth, noticeable biofilm had developed at the air-liquid interface; additionally, by 72 h, wild-type G27 also exhibited pellicle formation (see Fig. S2A). The ΔnikR mutant strain showed similar patterns of biofilm formation. Conversely, the Δfur and Δfur ΔnikR mutants appeared to show decreased biofilm formation at the air-liquid interface and did not form a pellicle. In contrast, the ΔarsS, ArsR-D52N, Δfur ΔarsS, ΔnikR ΔarsS, and Δfur ΔnikR ΔarsS mutants formed biofilms at the air-liquid interface as well as showed various degrees of biofilm formation that extended to the bottom of the well; in some cases, the entire surface of the well was covered with biofilm (see Fig. S2B; also data not shown). In comparison, little to no biofilm was observed on the bottom of the well with wild-type G27. Strains carrying the arsS or ArsR-D52N mutation also had a granular appearance indicative of the aforementioned aggregates (see Fig. S2A).
To quantitate biofilm differences across strains, biofilm formation was monitored using crystal violet staining. This analysis was conducted temporally in order to identify any differences in rate of biofilm formation as well as the absolute quantity of biofilm formation. For the purposes of this study, the rate of biofilm formation was defined as the first time point that showed significantly increased crystal violet staining compared to that at T6, which was the earliest time point for which biofilm was quantified. Based on this definition, a strain that showed significant biofilm formation after 24 h would have an accelerated rate compared to a strain that did not show significant biofilm formation until 48 h. As shown in Fig. 3A, wild-type G27 developed a significant biofilm after 48 h; very little biofilm at the air-liquid interface was visible before 48 h (data not shown). In comparison, all strains carrying the ΔarsS or ArsR-D52N mutation showed significant biofilm development after only 24 h of growth, indicating that they formed biofilm at an accelerated rate (Fig. 3A). Conversely, the Δfur, ΔnikR, and Δfur ΔnikR mutants showed delayed biofilm development and did not show significant biofilm formation until 72 h of growth (Fig. 3A).
In addition to rate analysis, we also directly compared the total amount of biofilm formed at each time point across all of the strains (see Table S1 in the supplemental material). In light of the accelerated rate of biofilm formation observed for the ΔarsS mutant, it is perhaps not surprising that this strain also formed significantly more biofilm than the wild-type G27, Δfur mutant, ΔnikR mutant, and Δfur ΔnikR mutant strains at 24 h (see Table S1). Similarly, the Δfur ΔarsS, ΔnikR ΔarsS, and Δfur ΔnikR ΔarsS mutants all showed increased amounts of biofilm at 24 h (see Table S1). We found that the ArsR-D52N mutant did not form significantly more biofilm than wild-type G27 until after 48 h, suggesting slight phenotypic differences between the various ArsS mutant strains (see Table S1). Further support of this difference is provided by the finding that while the ΔarsS mutant no longer showed a higher level of biofilm than the wild type at 72 h, the ArsR-D52N mutant still had a significantly higher level of biofilm than G27 at this time point. Also of note, while the ΔnikR ΔarsS mutant formed significantly more biofilm than wild-type G27 at 24 h, the increase in biofilm formation plateaued by 48 h and was not significantly different than that of wild-type G27 at the later time points (Fig. 3A; see also Table S1). Even though the difference was not significant, we note that the G27 Δfur, ΔnikR and Δfur ΔnikR mutants seem to plateau at lower levels of biofilm formation than the wild type (Fig. 3A). Finally, while biofilm staining intensity for the ΔarsS, ArsR-D52N, Δfur ΔarsS, ΔnikR ΔarsS, and Δfur ΔnikR ΔarsS mutants plateau by 48 h, wild-type G27 biofilm appeared to steadily increase even at 72 h (Fig. 3A). In fact, at 72 h, only the ArsR-D52N and Δfur ΔnikR ΔarsS mutants have significantly more biofilm than wild-type G27 (see Table S1).
To confirm that a mutation of arsS was responsible for the hyperbiofilm phenotype, we constructed a strain in which the ΔarsS mutation was complemented by arsS carried on the low-copy-number pTM117 vector (54). The expression of arsS in trans completely suppressed the enhanced biofilm phenotype observed in the ΔarsS mutant strain (Fig. 3B). In fact, the complementation strain exhibited levels of biofilm formation that were less than those seen with the wild-type strain (Fig. 3B; see also Fig. S2B in the supplemental material). This decrease may indicate that higher levels of ArsS are produced from the plasmid system.
In addition to the quantitative analysis, the parental strain G27 and the isogenic mutants were also analyzed by high-resolution scanning electron microscopy (SEM) to evaluate changes in biofilm architecture on an abiotic substrate. SEM analyses revealed numerous cell-to-cell interactions occurring in biofilms and several cell morphologies, including rod, curved rod/helix, and coccoid cells (Fig. 4). Furthermore, numerous cell surface features (such as small vesicles, flagella, and branched extracellular matrix [ECM]) could be appreciated in the biofilm samples (Fig. 4). After 48 h, H. pylori G27 exhibited minimal adherence to the abiotic substrate. Conversely, the ΔarsS isogenic mutant exhibited enhanced biofilm formation and tertiary architecture compared to the parental strain (Fig. 4). The double mutants Δfur ΔarsS and ΔnikR ΔarsS, and the triple mutant Δfur ΔnikR ΔarsS all displayed enhanced biofilm formation compared to the wild-type parental strain (Fig. 4). As expected given the crystal violet results (Fig. 3A), the ArsR-D52N mutant formed a dense biofilm similar to the ΔarsS mutant strains; however, in contrast to the biofilms observed for the ΔarsS mutant, more coccoid cells were present, and there appeared to be an increase in the ECM present for the ArsR-D52N mutant strain. Likewise, an increase in coccoid cells and ECM was also seen for the Δfur ΔnikR ΔarsS and ΔnikR ΔarsS mutants, perhaps reflecting slight differences in the fitness of these strains (Fig. 4). En masse, these results indicate that the ArsRS signaling pathway may play an important role in the regulation of biofilm formation.
Loss of ArsRS does not influence H. pylori motility.
In an attempt to gain some insight into the increased biofilm formation exhibited by the ArsRS mutant strains, we next considered mechanisms known to affect biofilm formation. Previous studies suggest that flagellar genes and motility may play a significant role in adherence and/or biofilm formation in H. pylori and the closely related Campylobacter jejuni (46, 58, 59). Thus, to determine if mutations in the ArsRS system lead to alterations in H. pylori motility, swimming motility was assessed for wild-type G27 and the enhanced biofilm-forming ΔarsS and ArsR-D52N mutants. As a control, a nonmotile mutant (ΔflgR) (14), which contains a deletion in the gene encoding the master flagellar regulator FlgR, was also included. At all time points assessed (days 3, 5, and 7 after inoculation in soft agar), there were no overt difference in the area of motility observed for the wild-type and ArsRS mutant strains (Fig. 5). Thus, mutation of the ArsRS system does not appear to affect motility in H. pylori G27.
Enhanced surface adherence and autoaggregation in strains with a mutated ArsRS system.
Two additional factors that have been shown to contribute to biofilm formation are bacterial aggregation and initial attachment to a surface. Given the large bacterial aggregates that were observed during both the growth curve and biofilm assays (see Fig. S1 and S2 in the supplemental material), we reasoned that increased aggregation in strains lacking ArsS or containing the nonphosphorylatable ArsR-D52N mutation could contribute to the enhanced biofilm formation seen with these strains. Therefore, we assessed bacterial aggregation using a quantitative aggregation assay (32). Since biofilm formation occurs as early as 24 h for some strains, 20-h cultures were used for this assessment. While the observed differences were not statistically significant, we did observe increased autoaggregation for the ΔarsS and ArsR-D52N mutant strains compared to the wild-type strain (Fig. 6A). Thus, mutation of the ArsRS system appears to affect bacterial autoaggregation.
Another important step in biofilm formation is adherence to surfaces. To determine if changes in initial adherence contribute to increased biofilm formation for the ΔarsS and ArsR-D52N mutant strains, bacterial adherence to tissue culture-treated polystyrene plates were assessed. After 2 h of incubation on the plate, very few wild-type G27 were adherent (Fig. 6B). In contrast, approximately 20% of the G27 ArsR-D52N mutant inoculum had adhered to the plate by 2 h. The difference in adherence was even more pronounced with the ΔarsS mutant strain; approximately 48% of the inoculum was adherent to the plate (Fig. 6B), which was significantly more than wild-type G27 (P = 0.0057). After 4 h of incubation, the wild-type strain began adhering to the plates, with an average of 28% adherence (Fig. 6B). At the same time point, adherence of the ArsR-D52N and ΔarsS mutant strains increased to approximately 50% (Fig. 6B). These data suggest that an increase in speed and efficiency of bacterial surface adherence may contribute to the enhanced biofilm formation seen with the strains containing mutations in the ArsRS system.
DISCUSSION
Given the paucity of regulatory proteins encoded in the H. pylori genome (3, 4), numerous studies have sought to define the roles of Fur, NikR, and ArsRS in the biology of H. pylori. The vast majority of these studies have focused on resistance to various stressful environments (10, 18, 27, 60), identification of the regulon of genes controlled by each regulatory factor (16, 17, 19–21, 23, 61), or elucidation of the contribution of each regulator to colonization in an animal model (8–11). Despite the evidence supporting the fact that the Fur, NikR, and ArsRS regulons show considerable overlap, very few studies (8, 17, 25, 53) have attempted to analyze the combined effect of these regulatory factors in H. pylori biology. Herein, we constructed a series of isogenic regulatory knockout strains and evaluated H. pylori growth in vitro. We found that while differences in growth were minimal over the first 24 h, at later time points, strains containing a ΔarsS mutation began to behave differently than the other strains; both CFU counts and OD600 readings for strains lacking ArsS began to plateau and even decline after 24 h (Fig. 1A and B). Upon further examination, significant cellular aggregation (see Fig. S1 in the supplemental material) and adherence to the flask at the air-liquid interface (Fig. 2B) were observed for the ΔarsS mutant strains. Indeed, it is likely that the aggregation and adherence of these bacterial strains skewed both the CFU counts and OD readings due to loss of planktonic bacteria from the medium. SEM analysis verified increased biofilm formation in the ΔarsS mutant strains (Fig. 4), as well as in the strain carrying the ArsR-D52N mutation. In addition to the biofilm phenotype, the SEM images also showed an increase in the number of coccoid cells in strains carrying the ArsR-D52N mutation or containing a deletion of NikR and ArsS in combination (Fig. 4). In conjunction with the lower CFU counts observed for these three strains (Fig. 1B), it is likely that these strains show a quicker transition to the coccoid form. Indeed, it has been shown that in wild-type H. pylori cells, the coccoid phenotype is triggered by environmental stressors, such as nutrient deprivation (62). Therefore, one potential explanation for the increase in the coccoid state seen with the ArsR-D52N, ΔnikR ΔarsS, and Δfur ΔnikR ΔarsS mutant strains may be the induction of an aberrant stress response due to the loss of key regulatory factors required to sense the environment. Intriguingly, biofilm formation has also been proposed as a mechanism to cope with stressful conditions (63). Together, these data suggest that strains lacking a functional ArsRS system display an exacerbated stress response that results in enhanced and accelerated biofilm formation. Combining the arsS deletion with the nikR deletion further limits the ability of H. pylori to function normally and results in an increase in coccoid bacteria in addition to biofilm formation. These findings not only highlight the benefit of utilizing combined isogenic regulatory mutants to tease out the coordinated regulation of H. pylori's stress response but also suggest a role for the ArsRS system in the regulation of biofilm formation.
Of note, a similar phenomenon has been seen in C. jejuni upon deletion of the CprS histidine kinase; loss of CprS led to bacterial aggregation and biofilm formation (64). Similar to the ArsRS system in H. pylori, CprR, which is the cognate response regulator to CprS, is essential in C. jejuni (64). Further investigation of the CprRS system showed that CprS mutants display increased expression of the flagellar filament protein (FlaA), the major outer membrane protein (MOMP), and many other cellular envelope components (64, 65). Given the parallels between the ΔCprS mutant in C. jejuni and the ΔarsS mutant in H. pylori, we investigated motility and other phenotypes that may reflect changes to the outer membrane composition. We found that neither the ΔarsS nor ArsR-D52N mutant strain showed changes in motility compared to the wild-type strain (Fig. 5); this finding may not be entirely surprising, given that previous studies have suggested that flagella do not contribute directly to H. pylori's adhesive properties (44, 46, 58). In contrast to the motility phenotype, both the ΔarsS and ArsR-D52N mutant strains showed an increase in aggregation and adherence compared to wild-type G27 (Fig. 6). Both adherence and autoaggregation in H. pylori are mediated by numerous cellular components, including lipopolysaccharide (LPS) and an extensive list of OMPs (reviewed in reference 66). Early exploration of the cell surface of H. pylori revealed that this bacterium is antigenically unique (67). A major contributor to this finding may be that, compared to many other Gram-negative bacteria, H. pylori's OMP profile lacks the highly abundant nonspecific porins and is composed of numerous less-abundant species (68). Indeed, H. pylori encodes more than 60 OMPs, representing approximately 4% of the genome (68). Interestingly, transcriptomic studies of H. pylori strains lacking ArsS indicate that numerous OMPs are regulated by the ArsRS system; the majority of these OMPs appear to be repressed (16, 17). Therefore, the observed increases in adherence and autoaggregation seen in the ΔarsS mutant strains may be due to the derepression and subsequent overexpression of OMP(s) in these strains.
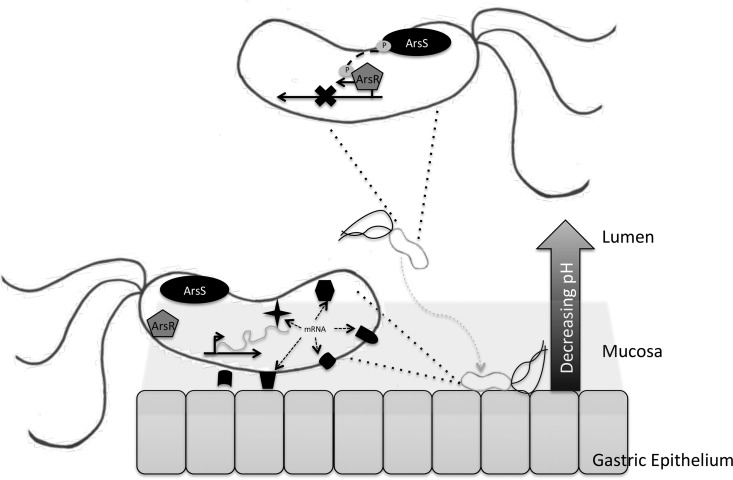
Biofilm formation in H. pylori-infected individuals is well documented (33–36), and a recent publication identified biofilm formation in an animal model of infection (37). H. pylori colonizes the gastric niche, which is highly acidic; however, H. pylori is a neutrophile and employs several mechanism to neutralize the immediate environment (69). Additionally, the pH in the stomach is not uniform; while the pH of the lumen can be as low as 1.0, the protective mucus layer covering the gastric mucosa acts as a buffer and maintains a more neutral environment (18, 70). In order to successfully colonize the stomach, H. pylori employs the use of flagella to burrow through the mucus layer and upon reaching the gastric mucosa uses several proposed adhesins to anchor itself to the epithelium (71). Thus, it seems plausible that H. pylori may utilize pH changes as an environmental indicator of proximity to the epithelial layer. Upon exposure to the highly acidic environment in the gastric lumen, ArsS phosphorylates ArsR (18), and then phosphorylated ArsR targets specific promoters for gene regulation (15, 72). Conversely, as H. pylori enters the gastric mucus layer, it experiences increasing pH. Under these conditions, ArsRS is no longer phosphorylated, which leads to the derepression of ArsR∼P-specific targets. In this model, the ArsRS system could be one mechanism by which H. pylori senses it is nearing the gastric epithelium as a way to modify the expression of components that promote colonization (Fig. 7). Deletion of ArsS or mutation of the phosphate-accepting aspartic (D) residue at position 52 nullifies the response to changing pH of the ArsRS two-component system (13). Thus, in either of the mutant strains, ArsR will remain in an unphosphorylated state, which leads to the deregulated expression of protein(s) that may contribute to enhanced biofilm formation, aggregation, and adherence observed in this investigation. This study opens several areas for future study; these include identification of the specific target(s) of ArsR∼P that leads to the observed phenotypes, determination of the effect of environmental stressors, such as acidic pH on biofilm formation, and investigation of biofilm formation in additional H. pylori strain backgrounds.
FIG 7.
Model of ArsRS regulation of colonization factors. When located in the gastric lumen (top), H. pylori is exposed to an acidic environment, which leads to autophosphorylation of ArsS and phosphotransfer to ArsR. Subsequently, ArsR∼P will bind and inhibit ArsR∼P target genes. Once H. pylori has reached the more neutral gastric epithelium (bottom), ArsRS remains unphosphorylated. This results in alleviation of repression and expression of various factors, depicted here by the various shapes, which aid in colonization and biofilm formation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Robert Shanks for helpful suggestions regarding biofilm assays and Christina Ferreira for critical comments on the manuscript.
The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the Department of Defense, the Department of Veterans Affairs, or the NIH.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00324-16.
REFERENCES
- 1.Moodley Y, Linz B, Bond RP, Nieuwoudt M, Soodyall H, Schlebusch CM, Bernhoft S, Hale J, Suerbaum S, Mugisha L, van der Merwe SW, Achtman M. 2012. Age of the association between Helicobacter pylori and man. PLoS Pathog 8:e1002693. doi: 10.1371/journal.ppat.1002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, deJonge BL, Carmel G, Tummino PJ, Caruso A, Uria-Nickelsen M, Mills DM, Ives C, Gibson R, Merberg D, Mills SD, Jiang Q, Taylor DE, Vovis GF, Trust TJ. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 3.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 4.Scarlato V, Delany I, Spohn G, Beier D. 2001. Regulation of transcription in Helicobacter pylori: simple systems or complex circuits? Int J Med Microbiol 291:107–117. doi: 10.1078/1438-4221-00107. [DOI] [PubMed] [Google Scholar]
- 5.Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, van Vliet AH, Whitehead S, Barrell BG. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 6.Danielli A, Amore G, Scarlato V. 2010. Built shallow to maintain homeostasis and persistent infection: insight into the transcriptional regulatory network of the gastric human pathogen Helicobacter pylori. PLoS Pathog 6:e1000938. doi: 10.1371/journal.ppat.1000938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danielli A, Scarlato V. 2010. Regulatory circuits in Helicobacter pylori: network motifs and regulators involved in metal-dependent responses. FEMS Microbiol Rev 34:738–752. doi: 10.1111/j.1574-6976.2010.00233.x. [DOI] [PubMed] [Google Scholar]
- 8.Bury-Moné S, Thiberge JM, Contreras M, Maitournam A, Labigne A, De Reuse H. 2004. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol Microbiol 53:623–638. doi: 10.1111/j.1365-2958.2004.04137.x. [DOI] [PubMed] [Google Scholar]
- 9.Miles S, Piazuelo MB, Semino-Mora C, Washington MK, Dubois A, Peek RM Jr, Correa P, Merrell DS. 2010. Detailed in vivo analysis of the role of Helicobacter pylori Fur in colonization and disease. Infect Immun 78:3073–3082. doi: 10.1128/IAI.00190-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gancz H, Censini S, Merrell DS. 2006. Iron and pH homeostasis intersect at the level of Fur regulation in the gastric pathogen Helicobacter pylori. Infect Immun 74:602–614. doi: 10.1128/IAI.74.1.602-614.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panthel K, Dietz P, Haas R, Beier D. 2003. Two-component systems of Helicobacter pylori contribute to virulence in a mouse infection model. Infect Immun 71:5381–5385. doi: 10.1128/IAI.71.9.5381-5385.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beier D, Frank R. 2000. Molecular characterization of two-component systems of Helicobacter pylori. J Bacteriol 182:2068–2076. doi: 10.1128/JB.182.8.2068-2076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schär J, Sickmann A, Beier D. 2005. Phosphorylation-independent activity of atypical response regulators of Helicobacter pylori. J Bacteriol 187:3100–3109. doi: 10.1128/JB.187.9.3100-3109.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDaniel TK, Dewalt KC, Salama NR, Falkow S. 2001. New approaches for validation of lethal phenotypes and genetic reversion in Helicobacter pylori. Helicobacter 6:15–23. doi: 10.1046/j.1523-5378.2001.00001.x. [DOI] [PubMed] [Google Scholar]
- 15.Marcus EA, Sachs G, Wen Y, Scott DR. 2016. Phosphorylation-dependent and phosphorylation-independent regulation of Helicobacter pylori acid acclimation by the ArsRS two-component system. Helicobacter 21:69–81. doi: 10.1111/hel.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loh JT, Gupta SS, Friedman DB, Krezel AM, Cover TL. 2010. Analysis of protein expression regulated by the Helicobacter pylori ArsRS two-component signal transduction system. J Bacteriol 192:2034–2043. doi: 10.1128/JB.01703-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pflock M, Finsterer N, Joseph B, Mollenkopf H, Meyer TF, Beier D. 2006. Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. J Bacteriol 188:3449–3462. doi: 10.1128/JB.188.10.3449-3462.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wen Y, Feng J, Scott DR, Marcus EA, Sachs G. 2006. Involvement of the HP0165-HP0166 two-component system in expression of some acidic-pH-upregulated genes of Helicobacter pylori. J Bacteriol 188:1750–1761. doi: 10.1128/JB.188.5.1750-1761.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dietz P, Gerlach G, Beier D. 2002. Identification of target genes regulated by the two-component system HP166-HP165 of Helicobacter pylori. J Bacteriol 184:350–362. doi: 10.1128/JB.184.2.350-362.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Contreras M, Thiberge JM, Mandrand-Berthelot MA, Labigne A. 2003. Characterization of the roles of NikR, a nickel-responsive pleiotropic autoregulator of Helicobacter pylori. Mol Microbiol 49:947–963. doi: 10.1046/j.1365-2958.2003.03621.x. [DOI] [PubMed] [Google Scholar]
- 21.Ernst FD, Bereswill S, Waidner B, Stoof J, Mader U, Kusters JG, Kuipers EJ, Kist M, van Vliet AH, Homuth G. 2005. Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression. Microbiology 151:533–546. doi: 10.1099/mic.0.27404-0. [DOI] [PubMed] [Google Scholar]
- 22.Choi YW, Park SA, Lee HW, Lee NG. 2009. Alteration of growth-phase-dependent protein regulation by a fur mutation in Helicobacter pylori. FEMS Microbiol Lett 294:102–110. doi: 10.1111/j.1574-6968.2009.01557.x. [DOI] [PubMed] [Google Scholar]
- 23.Lee HW, Choe YH, Kim DK, Jung SY, Lee NG. 2004. Proteomic analysis of a ferric uptake regulator mutant of Helicobacter pylori: regulation of Helicobacter pylori gene expression by ferric uptake regulator and iron. Proteomics 4:2014–2027. doi: 10.1002/pmic.200300740. [DOI] [PubMed] [Google Scholar]
- 24.Danielli A, Roncarati D, Delany I, Chiarini V, Rappuoli R, Scarlato V. 2006. In vivo dissection of the Helicobacter pylori Fur regulatory circuit by genome-wide location analysis. J Bacteriol 188:4654–4662. doi: 10.1128/JB.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delany I, Ieva R, Soragni A, Hilleringmann M, Rappuoli R, Scarlato V. 2005. In vitro analysis of protein-operator interactions of the NikR and fur metal-responsive regulators of coregulated genes in Helicobacter pylori. J Bacteriol 187:7703–7715. doi: 10.1128/JB.187.22.7703-7715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bijlsma JJ, Waidner B, Vliet AH, Hughes NJ, Hag S, Bereswill S, Kelly DJ, Vandenbroucke-Grauls CM, Kist M, Kusters JG. 2002. The Helicobacter pylori homologue of the ferric uptake regulator is involved in acid resistance. Infect Immun 70:606–611. doi: 10.1128/IAI.70.2.606-611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gancz H, Merrell DS. 2011. The Helicobacter pylori ferric uptake regulator (Fur) is essential for growth under sodium chloride stress. J Microbiol 49:294–298. doi: 10.1007/s12275-011-0396-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qu W, Zhou Y, Shao C, Sun Y, Zhang Q, Chen C, Jia J. 2009. Helicobacter pylori proteins response to nitric oxide stress. J Microbiol 47:486–493. doi: 10.1007/s12275-008-0266-0. [DOI] [PubMed] [Google Scholar]
- 29.Carpenter BM, Gancz H, Gonzalez-Nieves RP, West AL, Whitmire JM, Michel SL, Merrell DS. 2009. A single nucleotide change affects fur-dependent regulation of sodB in H. pylori. PLoS One 4:e5369. doi: 10.1371/journal.pone.0005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stark RM, Gerwig GJ, Pitman RS, Potts LF, Williams NA, Greenman J, Weinzweig IP, Hirst TR, Millar MR. 1999. Biofilm formation by Helicobacter pylori. Lett Appl Microbiol 28:121–126. doi: 10.1046/j.1365-2672.1999.00481.x. [DOI] [PubMed] [Google Scholar]
- 31.Cole SP, Harwood J, Lee R, She R, Guiney DG. 2004. Characterization of monospecies biofilm formation by Helicobacter pylori. J Bacteriol 186:3124–3132. doi: 10.1128/JB.186.10.3124-3132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yonezawa H, Osaki T, Kurata S, Zaman C, Hanawa T, Kamiya S. 2010. Assessment of in vitro biofilm formation by Helicobacter pylori. J Gastroenterol Hepatol 25(Suppl 1):S90–S94. [DOI] [PubMed] [Google Scholar]
- 33.Carron MA, Tran VR, Sugawa C, Coticchia JM. 2006. Identification of Helicobacter pylori biofilms in human gastric mucosa. J Gastrointest Surg 10:712–717. doi: 10.1016/j.gassur.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 34.Coticchia JM, Sugawa C, Tran VR, Gurrola J, Kowalski E, Carron MA. 2006. Presence and density of Helicobacter pylori biofilms in human gastric mucosa in patients with peptic ulcer disease. J Gastrointest Surg 10:883–889. doi: 10.1016/j.gassur.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 35.Cammarota G, Branca G, Ardito F, Sanguinetti M, Ianiro G, Cianci R, Torelli R, Masala G, Gasbarrini A, Fadda G, Landolfi R, Gasbarrini G. 2010. Biofilm demolition and antibiotic treatment to eradicate resistant Helicobacter pylori: a clinical trial. Clin Gastroenterol Hepatol 8:817.e3–820.e3. doi: 10.1016/j.cgh.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 36.Cellini L, Grande R, Di Campli E, Traini T, Di Giulio M, Lannutti SN, Lattanzio R. 2008. Dynamic colonization of Helicobacter pylori in human gastric mucosa. Scand J Gastroenterol 43:178–185. doi: 10.1080/00365520701675965. [DOI] [PubMed] [Google Scholar]
- 37.Attaran B, Falsafi T, Moghaddam AN. 2016. Study of biofilm formation in C57Bl/6J mice by clinical isolates of Helicobacter pylori. Saudi J Gastroenterol 22:161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaddy JA, Radin JN, Cullen TW, Chazin WJ, Skaar EP, Trent MS, Algood HM. 2015. Helicobacter pylori resists the antimicrobial activity of calprotectin via lipid A modification and associated biofilm formation. mBio 6(6):e01349-15. doi: 10.1128/mBio.01349-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yonezawa H, Osaki T, Hanawa T, Kurata S, Ochiai K, Kamiya S. 2013. Impact of Helicobacter pylori biofilm formation on clarithromycin susceptibility and generation of resistance mutations. PLoS One 8:e73301. doi: 10.1371/journal.pone.0073301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Azevedo NF, Pacheco AP, Keevil CW, Vieira MJ. 2006. Adhesion of water stressed Helicobacter pylori to abiotic surfaces. J Appl Microbiol 101:718–724. doi: 10.1111/j.1365-2672.2006.03029.x. [DOI] [PubMed] [Google Scholar]
- 41.Gião MS, Azevedo NF, Wilks SA, Vieira MJ, Keevil CW. 2008. Persistence of Helicobacter pylori in heterotrophic drinking-water biofilms. Appl Environ Microbiol 74:5898–5904. doi: 10.1128/AEM.00827-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Percival SL, Thomas JG. 2009. Transmission of Helicobacter pylori and the role of water and biofilms. J Water Health 7:469–477. doi: 10.2166/wh.2009.070. [DOI] [PubMed] [Google Scholar]
- 43.Linke S, Lenz J, Gemein S, Exner M, Gebel J. 2010. Detection of Helicobacter pylori in biofilms by real-time PCR. Int J Hyg Environ Health 213:176–182. doi: 10.1016/j.ijheh.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 44.Williams JC, McInnis KA, Testerman TL. 2008. Adherence of Helicobacter pylori to abiotic surfaces is influenced by serum. Appl Environ Microbiol 74:1255–1258. doi: 10.1128/AEM.01958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bessa LJ, Grande R, Di Iorio D, Di Giulio M, Di Campli E, Cellini L. 2013. Helicobacter pylori free-living and biofilm modes of growth: behavior in response to different culture media. APMIS 121:549–560. doi: 10.1111/apm.12020. [DOI] [PubMed] [Google Scholar]
- 46.Grande R, Di Giulio M, Di Campli E, Di Bartolomeo S, Cellini L. 2010. A model of Helicobacter pylori persistence in a case of gastric cancer. New Microbiol 33:343–349. [PubMed] [Google Scholar]
- 47.Yonezawa H, Osaki T, Kurata S, Fukuda M, Kawakami H, Ochiai K, Hanawa T, Kamiya S. 2009. Outer membrane vesicles of Helicobacter pylori TK1402 are involved in biofilm formation. BMC Microbiol 9:197. doi: 10.1186/1471-2180-9-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grande R, Di Marcantonio MC, Robuffo I, Pompilio A, Celia C, Di Marzio L, Paolino D, Codagnone M, Muraro R, Stoodley P, Hall-Stoodley L, Mincione G. 2015. Helicobacter pylori ATCC 43629/NCTC 11639 outer membrane vesicles (OMVs) from biofilm and planktonic phase associated with extracellular DNA (eDNA). Front Microbiol 6:1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schooling SR, Beveridge TJ. 2006. Membrane vesicles: an overlooked component of the matrices of biofilms. J Bacteriol 188:5945–5957. doi: 10.1128/JB.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shao C, Sun Y, Wang N, Yu H, Zhou Y, Chen C, Jia J. 2013. Changes of proteome components of Helicobacter pylori biofilms induced by serum starvation. Mol Med Rep 8:1761–1766. [DOI] [PubMed] [Google Scholar]
- 51.Baltrus DA, Amieva MR, Covacci A, Lowe TM, Merrell DS, Ottemann KM, Stein M, Salama NR, Guillemin K. 2009. The complete genome sequence of Helicobacter pylori strain G27. J Bacteriol 191:447–448. doi: 10.1128/JB.01416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N, Rappuoli R. 1993. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci U S A 90:5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carpenter BM, West AL, Gancz H, Servetas SL, Pich OQ, Gilbreath JJ, Hallinger DR, Forsyth MH, Merrell DS, Michel SL. 2015. Crosstalk between the HpArsRS two-component system and HpNikR is necessary for maximal activation of urease transcription. Front Microbiol 6:558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carpenter BM, McDaniel TK, Whitmire JM, Gancz H, Guidotti S, Censini S, Merrell DS. 2007. Expanding the Helicobacter pylori genetic toolbox: modification of an endogenous plasmid for use as a transcriptional reporter and complementation vector. Appl Environ Microbiol 73:7506–7514. doi: 10.1128/AEM.01084-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wen Y, Feng J, Scott DR, Marcus EA, Sachs G. 2007. The HP0165-HP0166 two-component system (ArsRS) regulates acid-induced expression of HP1186 alpha-carbonic anhydrase in Helicobacter pylori by activating the pH-dependent promoter. J Bacteriol 189:2426–2434. doi: 10.1128/JB.01492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gaddy JA, Tomaras AP, Actis LA. 2009. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect Immun 77:3150–3160. doi: 10.1128/IAI.00096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skindersoe ME, Rasmussen L, Andersen LP, Krogfelt KA. 2015. A novel assay for easy and rapid quantification of Helicobacter pylori adhesion. Helicobacter 20:199–205. doi: 10.1111/hel.12191. [DOI] [PubMed] [Google Scholar]
- 58.Clyne M, Ocroinin T, Suerbaum S, Josenhans C, Drumm B. 2000. Adherence of isogenic flagellum-negative mutants of Helicobacter pylori and Helicobacter mustelae to human and ferret gastric epithelial cells. Infect Immun 68:4335–4339. doi: 10.1128/IAI.68.7.4335-4339.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Svensson SL, Pryjma M, Gaynor EC. 2014. Flagella-mediated adhesion and extracellular DNA release contribute to biofilm formation and stress tolerance of Campylobacter jejuni. PLoS One 9:e106063. doi: 10.1371/journal.pone.0106063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Merrell DS, Thompson LJ, Kim CC, Mitchell H, Tompkins LS, Lee A, Falkow S. 2003. Growth phase-dependent response of Helicobacter pylori to iron starvation. Infect Immun 71:6510–6525. doi: 10.1128/IAI.71.11.6510-6525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ernst FD, Kuipers EJ, Heijens A, Sarwari R, Stoof J, Penn CW, Kusters JG, van Vliet AH. 2005. The nickel-responsive regulator NikR controls activation and repression of gene transcription in Helicobacter pylori. Infect Immun 73:7252–7258. doi: 10.1128/IAI.73.11.7252-7258.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andersen LP, Wadstrom T. 2001. Basic bacteriology and culture, p 27–38. In Mobley HLT, Mendz GL, Hazell SL (ed), Helicobacter pylori: physiology and genetics. ASM Press, Washington, DC. [PubMed] [Google Scholar]
- 63.García A, Salas-Jara MJ, Herrera C, Gonzalez C. 2014. Biofilm and Helicobacter pylori: from environment to human host. World J Gastroenterol 20:5632–5638. doi: 10.3748/wjg.v20.i19.5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Svensson SL, Davis LM, MacKichan JK, Allan BJ, Pajaniappan M, Thompson SA, Gaynor EC. 2009. The CprS sensor kinase of the zoonotic pathogen Campylobacter jejuni influences biofilm formation and is required for optimal chick colonization. Mol Microbiol 71:253–272. doi: 10.1111/j.1365-2958.2008.06534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Svensson SL, Hyunh S, Parker CT, Gaynor EC. 2015. The Campylobacter jejuni CprRS two-component regulatory system regulates aspects of the cell envelope. Mol Microbiol 96:189–209. doi: 10.1111/mmi.12927. [DOI] [PubMed] [Google Scholar]
- 66.Oleastro M, Menard A. 2013. The role of Helicobacter pylori outer membrane proteins in adherence and pathogenesis. Biology (Basel) 2:1110–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Doig P, Trust TJ. 1994. Identification of surface-exposed outer membrane antigens of Helicobacter pylori. Infect Immun 62:4526–4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alm RA, Bina J, Andrews BM, Doig P, Hancock RE, Trust TJ. 2000. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect Immun 68:4155–4168. doi: 10.1128/IAI.68.7.4155-4168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sachs G, Weeks DL, Wen Y, Marcus EA, Scott DR, Melchers K. 2005. Acid acclimation by Helicobacter pylori. Physiology (Bethesda) 20:429–438. doi: 10.1152/physiol.00032.2005. [DOI] [PubMed] [Google Scholar]
- 70.Ramsay PT, Carr A. 2011. Gastric acid and digestive physiology. Surg Clin North Am 91:977–982. doi: 10.1016/j.suc.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 71.Andersen LP. 2007. Colonization and infection by Helicobacter pylori in humans. Helicobacter 12(Suppl 2):S12–S15. [DOI] [PubMed] [Google Scholar]
- 72.Pflock M, Dietz P, Schar J, Beier D. 2004. Genetic evidence for histidine kinase HP165 being an acid sensor of Helicobacter pylori. FEMS Microbiol Lett 234:51–61. doi: 10.1111/j.1574-6968.2004.tb09512.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.