ABSTRACT
Neisseria gonorrhoeae is naturally competent for transformation. The first step of the transformation process is the uptake of DNA from the environment into the cell. This transport step is driven by a powerful molecular machine. Here, we addressed the question whether this machine imports single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA) at similar rates. The fluorescence signal associated with the uptake of short DNA fragments labeled with a single fluorescent marker molecule was quantified. We found that ssDNA with a double-stranded DNA uptake sequence (DUS) was taken up with a similar efficiency as dsDNA. Imported ssDNA was degraded rapidly, and the thermonuclease Nuc was required for degradation. In a nuc deletion background, dsDNA and ssDNA with a double-stranded DUS were imported and used as the substrates for transformation, whereas the import and transformation efficiencies of ssDNA with single-stranded DUS were below the detection limits. We conclude that the DNA uptake machine requires a double-stranded DUS for efficient DNA recognition and transports ssDNA and dsDNA with comparable efficiencies.
IMPORTANCE Bacterial transformation enables bacteria to exchange genetic information. It can speed up adaptive evolution and enhances the potential of DNA repair. The transport of DNA through the outer membrane is the first step of transformation in Gram-negative species. It is driven by a powerful molecular machine whose mechanism remains elusive. Here, we show for Neisseria gonorrhoeae that the machine transports single- and double-stranded DNA at comparable rates, provided that the species-specific DNA uptake sequence is double stranded. Moreover, we found that single-stranded DNA taken up into the periplasm is rapidly degraded by the thermonuclease Nuc. We conclude that the secondary structure of transforming DNA is important for the recognition of self DNA but not for the process of transport through the outer membrane.
INTRODUCTION
Transformation describes the import and inheritable integration of DNA from the environment. The putative benefits of transformation are genome repair by external DNA as well as the acquisition of novel genes. Various factors determine whether extracellular DNA is efficiently used as a substrate for transformation, including its sequence, methylation, and secondary structure (1–3).
Since the integration of newly acquired DNA into the genome is based on homologous recombination, sequence diversity limits interspecies gene transfer. Moreover, the genomes of the Pasteurellaceae and Neisseriaceae are enriched with specific DNA uptake sequences (DUSs) conveying the species-specific recognition of DNA via various dialects (4) (5). For Neisseria gonorrhoeae, this sequence consists of 12 nucleotides and reads 5′-ATGCCGTCTGAA-3′. Within this sequence, the DUS core, 5′-CTG-3′, is the most important for transformation, and transversions in this core reduce DNA import by 2 orders of magnitude (4). The minor pilin ComP is important for the DUS specificity of DNA import (6). ComP was shown to display a preference for binding to the DNA uptake sequence, most likely through an electropositive stripe exposed by the pilus (5). In particular, the inner core of the DUS was shown to be important for binding to ComP and for transformation (7).
Another mechanism for limiting interspecies gene transfer is provided by DNA-degrading enzymes (2). For N. gonorrhoeae, the thermonuclease Nuc is an interesting candidate, but its influence on transformation efficiency is unknown. Gonococcal Nuc can degrade double-stranded DNA (dsDNA) and single-stranded DNA (ssDNA) of various origins (8). Methylation of gonococcal chromosomal DNA limits Nuc's activity. Moreover, Nuc can degrade neutrophil extracellular traps (9). While Nuc carries a signal peptide for secretion, active secretion into the supernatant has not been observed, and therefore, a periplasmic location was proposed (8).
The secondary structure of the transforming DNA affects the efficiency of transformation. Early work indicated that transformation with ssDNA was inefficient (10). In particular, the transformation activity of ssDNA in Haemophilus influenzae has been attributed to the inefficient denaturation of dsDNA (11). Various recent reports indicate, however, that ssDNA is used as a substrate for transformation. In particular, N. gonorrhoeae strains containing the gonococcal genetic island (GGI) secrete ssDNA using a type IV secretion system (12). Coculture experiments provided evidence that the secreted DNA was used for transformation (13). Moreover, the secretin PilQ binds ssDNA more strongly than dsDNA (14). Duffin and Seifert introduced a Watson DUS and a Crick DUS into single-stranded DNA from M13 phage for measuring the efficiency of ssDNA transformation (15). They found that the Crick DUS enhanced the transformation efficiency 180- to 470-fold compared to that for ssDNA without DUS, while the Watson DUS modestly enhanced the transformation efficiency. The effect was strongly dependent on the gonococcal strain. While the transformation rate of ssDNA with the Crick DUS was indistinguishable from that of dsDNA in strain MS11, it was an order of magnitude lower in strain FA1090 (15). It is reasonable to assume that the transformation rate is affected by the rates of DNA uptake through the outer and/or inner membrane, by the stability of ssDNA in the cytoplasm, and by the recombination rate. It is unclear which of these steps reduces the transformation rate of ssDNA for strain FA1090.
Mechanistically, the process of transformation proceeds in separable steps. They include binding of DNA at the extracellular side, transport through the cell envelope, protection from nuclease attack within the cytoplasm, and recombination into the chromosome (16–19). For Gram-negative species, an additional transport step is required to take up the transforming DNA from the environment into the periplasm of the cell. Transport through the outer membrane and through the inner membrane has been shown to be separable for Neisseria gonorrhoeae (20, 21), Helicobacter pylori (22), and Vibrio cholerae (23). For H. pylori and V. cholerae, the two steps of DNA import have been shown to be temporally uncoupled, but they are spatially coupled in the case of V. cholerae (22, 23). With the exception of H. pylori, a conserved set of genes is essential for transformation of all known naturally competent species. Here, we use the nomenclature for N. gonorrhoeae. Proteins belonging to the type IV pilus system mediate initial binding to the cell surface (6, 17). PilQ proteins form a pore within the outer membrane that supports export of the pilus fiber, DNA binding, and DNA uptake (14, 24, 25). Within the periplasm, ComE rapidly colocalizes with the imported DNA (21, 26). ComE binds to both double-stranded and single-stranded DNA (27). Single-stranded DNA enters the cytoplasm through a channel formed by ComA (20). For N. gonorrhoeae, there is evidence that ssDNA forms transiently in the periplasm (28), but it is unclear whether it forms directly after entry into the periplasm or during transport through the inner membrane. The molecular machines that drive DNA uptake during transformation are powerful molecular motors (29, 30), but the physical mechanism that drives the directed transport of DNA through the outer and inner membrane remains elusive.
In this work, we addressed the question whether ssDNA is taken up by the DNA import motor at a rate and with an efficiency comparable to those for dsDNA. To this end, we compared the fluorescence intensity resulting from the import of ssDNA with dsDNA, each carrying a single fluorescent dye molecule. We found that the stationary-state signal generated by the DNA taken up was severely affected by the thermonuclease Nuc, since Nuc degraded ssDNA faster than it degraded dsDNA. In a nuc deletion background, ssDNA uptake was undetectable. Hybridization of a short oligonucleotide covering the DUS was sufficient for supporting DNA uptake. The rate of DNA uptake was similar for dsDNA and ssDNA with double-stranded DUS (dsDUS), and the total amount of DNA at steady state was lower for dsDNA. The transformation probabilities using dsDNA, dsDUS DNA, and ssDNA reflected the DNA uptake probabilities. We conclude that double-stranded DUS is required for the efficient uptake of DNA through the outer membrane. Our data provide evidence that the DNA uptake machine imports ssDNA and dsDNA at similar speeds once the DUS is recognized.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
We used two derivatives of MS11 (see Table S1 in the supplemental material). Strain GV1 carried an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible recAind copy and a frameshift mutation in the minor pilin pilV (31–33). RecA is essential for homologous recombination. It does not influence the rate of DNA uptake but inhibits pilin antigenic variation. Deletion of pilV increases the amount of imported DNA. This strain was used for the experiments unless otherwise noted. Furthermore, a recAind pilQ strain was used (25).
The nuc deletion strain was generated by an in-frame replacement of the nuc open reading frame (ORF) by the short Kanr ORF. A 504-bp fragment upstream of the nuc ORF was generated using primers HG3 and HG4. The Kanr ORF from pUP6 was amplified using primers HG1 and HG2. A 564-bp fragment downstream of the nuc ORF was generated using primers HG5a and HG6. The three fragments were joined by fusion PCR using primers HG3 and HG6. The PCR product was purified and subsequently amplified using primers HG11 and HG12. After purification, GV1 was transformed with the PCR product, resulting in replacement of the nuc ORF by the Kanr ORF (Δnuc ΔpilV). The transformants were selected on GC agar containing kanamycin. Insertion was verified by PCR using primers HG3 and HG6 and sequencing of the purified PCR product.
N. gonorrhoeae was grown overnight at 37°C in 5% CO2 on agar plates containing gonococcal base agar (10 g/liter Bacto agar [BD Biosciences, Bedford, MA], 5 g/liter NaCl [Roth, Darmstadt, Germany], 4 g/liter K2HPO4 [Roth], 1 g/liter KH2PO4 [Roth], 15 g/liter Bacto proteose peptone no. 3 [BD], 0.5 g/liter soluble starch [Sigma-Aldrich, St. Louis, MO]) and the following supplements: 1 g/liter d-glucose (Roth), 0.1 g/liter l-glutamine (Roth), 0.289 g/liter l-cysteine-HCl·H2O (Roth), 1 mg/liter thiamine pyrophosphate (Sigma-Aldrich), 0.2 mg/liter Fe(NO3)3 (Sigma-Aldrich), 0.03 mg/liter thiamine HCl (Roth), 0.13 mg/liter 4-aminobenzoic acid (Sigma-Aldrich), 2.5 mg/liter β-NAD (Roth), and 0.1 mg/liter vitamin B12 (Sigma-Aldrich). Gonococcal colonies were resuspended in GC medium before each experiment.
DNA uptake assay.
Several bacterial colonies of 16- to 20-h-old cultures grown on GC agar were resuspended with a 10-μl inoculation syringe in 100 μl DNA uptake medium (GC medium supplemented with IsoVitaleX and 7 mM MgCl2) to an optical density at 600 nm (OD600) of 0.1. Labeled DNA fragments were added to the cell suspension to a final concentration of 1 ng/μl. The cells were incubated with DNA at 37°C in 5% CO2 and subsequently treated with 10 U DNase I (recombinant; Fermentas) for another 15 min at 37°C. After washing with phosphate-buffered saline, 50 μl of this dilution was applied to clean coverslips for microscopic analysis. For DNA stability experiments, the cells were washed with DNA uptake medium. Fragments labeled at the end and in the middle were purchased, with modification, from Sigma-Aldrich. Each condition was characterized independently on at least three different days using the same stock of labeled DNA. Hybridization of the complementary strand to an ssDNA fragment with a single dye molecule attached did not significantly affect the fluorescence intensity. Spectroscopic analysis of the fluorescence intensity showed a difference of less than 8% between ssDNA and dsDNA when they each carried a single dye molecule.
Microscopy and quantitative analysis of single-cell fluorescence.
For fluorescence quantification experiments, an inverted microscope (TI-E; Nikon) was used at room temperature. A 120-W metal halogenide fluorescent lamp (Intensilight; Nikon) served as the illumination source. Images were taken with an electron-multiplying charge-coupled-device (EMCCD) camera (IXON X3897; Andor). A 100× oil immersion CFI apochromat TIRF objective (numerical aperture [NA], 1.49; Nikon) was used.
For time-lapse experiments, an inverted microscope (TI-2000; Nikon) was used at 37°C. A 120-W metal halogenide fluorescent lamp (Intensilight; Nikon) served as the illumination source. Images were taken with an EMCCD camera (Cascade II:512; Photometrics). A 100× oil immersion CFI Plan Fluor objective (NA, 1.3; Nikon) was used.
Day-to-day variations in the brightness of the fluorescence lamp were detected by using test beads on a number 3 focal check fluorescence microscope test slide (Invitrogen). The intensities stayed relatively stable, with the day-to-day variance being less than ±10%. A correction factor corresponding to this reference intensity was calculated every day and applied to the respective data sets. The image analysis procedures have been described previously (21). In this study, the algorithm for detecting cellular fluorescence as a function of time was slightly modified for reducing the noise: the fluorescence intensity of all individual cells within one field of view was determined and averaged, yielding a single data point (see Fig. 4).
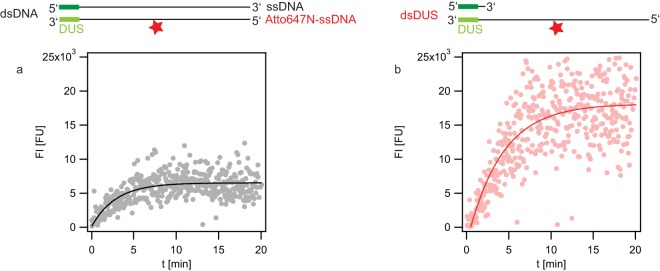
FIG 4.
Import kinetics of dsDNA and dsDUS DNA. Gonococci (Ng058 Δnuc ΔpilV) were incubated with DNA fragments containing a single dye molecule at the center (top, stars). The fluorescence intensity of single cells was recorded as a function of time. Each data point depicts the average fluorescence intensity of all cells within one microscopic field of view, averaging ∼20 individual cells. (a) dsDNA; (b) dsDUS DNA. Solid lines, fits to exponential functions.
Transformation assay.
Cell cultures of the MS11 wild-type (wt) strain grown for 16 h on GC agar were harvested in transformation medium (GC medium with IsoVitaleX and 7 mM MgCl2) and diluted to OD600 of 0.2. MgCl2 is a cofactor in DNA uptake and strongly increases the transformation efficiency. Five hundred microliters of the cell suspension was mixed with 2.5 μg of the transformation plasmid pSY6 isolated from Escherichia coli DH5α/pSY6 by use of a plasmid maxipreparation or 45 pmol of a synthetic DNA fragment, and the mixture was incubated for 30 min at 37°C and 250 rpm. Afterwards, the cell and DNA mixture was diluted by adding it to 2 ml transformation medium in cell culture flasks with ventilation caps. The cell culture flasks were incubated for 3 h at 37°C and 250 rpm in 5% CO2. The above-mentioned pSY6 is a transformation plasmid containing the DNA uptake sequence and a modified gyrase B subunit, leading to nalidixic acid resistance only when the subunit is incorporated into the genome. The synthetic fragments are either double or single stranded and consist of the 300-bp stretch flanking the mutation in pSY6 which confers nalidixic acid resistance, as well as a terminal DUS to support DNA uptake. After incubation, the cells were resuspended in 200 μl of transformation medium, which corresponds to a dilution factor of 100. Finally, 50 μl of several serial dilutions was evenly spread on GC agar plates (dilutions, 10−5, 10−6, 10−7, and 10−8) and on GC agar plates with 2 μg/ml nalidixic acid (dilutions, 10−4, 10−5, 10−6, and 10−7 for pSY6 and 100, 10−1, 10−2, and 10−3 for synthetic DNA fragments). The cells were grown for a further 45 h at 37°C in 5% CO2, before the number of colonies on each plate was counted separately.
RESULTS
Double-stranded DUS supports uptake of otherwise single-stranded DNA.
We set out to compare the DNA uptake efficiency of single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA). To minimize the probability of hairpin formation, short complementary oligonucleotides with a length of 100 bases were synthesized. The sequence was randomly chosen from λ phage DNA and contained the 12-bp DNA uptake sequence (DUS) (see Table S2 in the supplemental material [Cy5-ssDNA-end]). The Crick strand with the DUS at the 3′ end carried a single Cy5 dye molecule at the 5′ end. The Crick strand was chosen because it showed the highest transformation rate in previous experiments (15). The DNA fragments were labeled with a single dye molecule to minimize the effect of labeling on the interaction of DNA with components of the DNA uptake machine. dsDNA was generated by hybridization between the unlabeled and the labeled strand. All experiments were performed in a ΔpilV recAind strain background. Deletion of the gene for the minor pilin, pilV, significantly enhances the amount of imported DNA (34). Therefore, the fluorescence signal resulting from uptake of fluorescently labeled DNA was large compared to the signal of the wt (21). Since we were interested only in the difference between the efficiencies of dsDNA and ssDNA uptake, we chose to work in this genomic background to obtain a strong signal. RecA is required for pilin antigenic variation (35). To avoid variation of the pilin subunit during the course of our experiments, we chose to work in a recAind background without induction. While recA is essential for recombination during transformation, it is dispensable for DNA uptake into the periplasm.
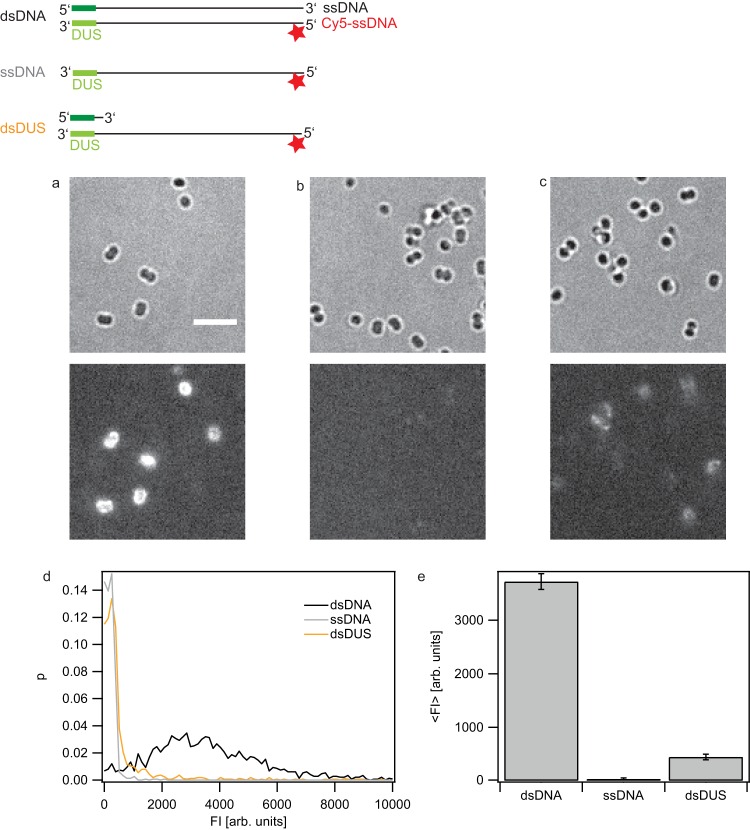
Gonococci were incubated with DNA for 1 h in a gently shaking tube and subsequently treated with DNase. The extended incubation time was chosen to compare our data to previous results (21). Gonococci were imaged (Fig. 1a and b), and the fluorescence intensities of individual cells were measured using a previously described procedure (21). From this analysis the distribution of fluorescence intensities of individual cells was obtained (Fig. 1c). The distribution of fluorescence intensities after Cy5-dsDNA uptake was broad (Fig. 1c) and reminiscent of the distribution obtained by the uptake of longer (0.3- to 10-kbp) fragments labeled with multiple dyes per fragment (21). The amount of imported Cy5-ssDNA was considerably smaller than the amount of Cy5-dsDNA (Fig. 1d and e).
FIG 1.
dsDUS supports the import of ssDNA. Gonococci (Ng005 ΔpilV) were incubated for 1 h with DNA fragments containing a single dye molecule at the 5′ end (top, stars). Subsequently, they were treated with DNase I. The fragments consisted of dsDNA (a), ssDNA (b), and ssDNA with a 16-base complementary oligonucleotide containing the DUS (c). (Top) Bright-field images; (bottom) fluorescence images. (d) Probability distribution of the total fluorescence intensity of individual cells incubated with dsDNA, ssDNA, or 100 bases of ssDNA with a 16-base complementary oligonucleotide containing the DUS. (e) Average fluorescence intensity of individual cells. Error bars, standard deviations from three independent experiments; gray bars, fragments with a sequence from λ phage DNA with a single Cy5 dye molecule at the 5′ end. Data are for >1,500 cells for each condition. arb. units, arbitrary units.
The DUS strongly enhances the probability of transformation and DNA uptake. It was conceivable that only the double-stranded DUS (dsDUS) was recognized by gonococci. We addressed the question whether a DNA fragment with the dsDUS but otherwise ssDNA was taken up. When the complementary strand was added and dsDNA was formed, uptake of dsDNA was detected (Fig. 1c). To ensure that end labeling did not introduce an artifact, we repeated the experiment with an Atto 647N dye attached at position 37 of the DNA fragment. Labeling in the middle of the fragment produced the same result, indicating that the position of the dye did not affect the outcome of the experiment (see Fig. S1 in the supplemental material). Compared to the level of the background in experiments using continuously labeled DNA, the level of the background was strongly reduced with individual labels, allowing a better quantitative comparison of the efficiency of dsDNA, ssDNA, and dsDUS uptake. Previous work characterizing the binding affinity of the minor pilin ComP to the DUS exclusively used dsDNA (5). The structure of the DUS-binding protein ComP shows that an electropositive stripe and a DNA-docking platform are consistent with the binding of dsDNA.
In conclusion, the uptake of single-stranded DNA by gonococci is detectable, provided that the DUS is double stranded.
Nuc rapidly degrades imported single-stranded DNA.
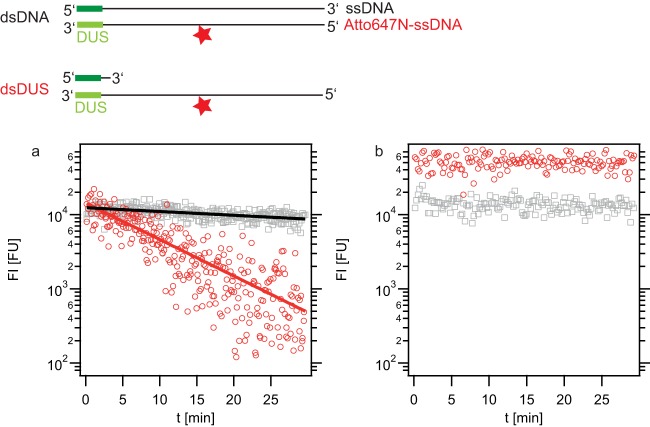
In the next step, the stability of ssDNA within the periplasm was addressed. So far, the amount of time that elapsed between the start of DNase treatment and the time of imaging was variable. In the next step, the onset of imaging was controlled. Again, bacteria were incubated with dsDNA or dsDUS DNA (an ssDNA fragment with double-stranded DUS). Subsequently, extracellular DNA was removed, DNase was added, and the fluorescence intensity was monitored starting directly after addition of DNase. The fluorescence signal associated with dsDNA decayed slowly (Fig. 2a). The decay kinetics were in agreement with an exponential decay, with a characteristic time of decay of dsDNA (τdecds) of 85 ± 7 min. The decay of dsDUS DNA was considerably faster and occurred at a characteristic time of decay of ssDNA (τdecss) of 9 ± 1 min. We verified that the decay kinetics did not depend on DNase addition. To this end, the DNase treatment was omitted and imaging started directly after the washing step where DNA was removed. The decay kinetics did not show a significant difference (see Fig. S2 in the supplemental material), with τdecds of 110 ± 30 min and τdecss of 8 ± 1 min. Moreover, the initial fluorescence levels at time zero [FI(t = 0)] were comparable between Fig. 2a and Fig. S2 in the supplemental material, indicating that the fluorescence intensity was mainly caused by internalized DNA and that DNA binding to the outside of the cell does not contribute significantly to the fluorescence signal.
FIG 2.
Decay of imported dsDNA and dsDUS DNA in a nuc deletion strain. Strains Ng005 ΔpilV (a) and Ng058 Δnuc ΔpilV (b) were incubated for 1 h with DNA fragments containing a single dye molecule at the center of the fragment (top, stars). Single-cell fluorescence intensity was monitored starting immediately after DNA removal and addition of DNase I. Gray symbols, dsDNA; red symbols, dsDUS; t, time; solid lines, exponential fits to the data.
Recently, the thermonuclease Nuc was speculated to reside within the periplasm (8). Therefore, we tested whether Nuc was responsible for the decay of imported DNA. To this end, we performed the experiment explained above in a Δnuc ΔpilV strain. Both dsDNA and dsDUS were stable for an extended period of time (Fig. 2b), indicating that Nuc was responsible for the fast decay observed in Fig. 2a. Fitting of an exponential decay function was not possible. The fluorescence intensity associated with the uptake of dsDUS DNA was significantly higher (∼3.5 times) than the signal associated with the uptake of dsDNA. The observation that in the presence of Nuc the initial fluorescence intensities were comparable (Fig. 2a) can be explained by the fact that several minutes elapsed from the time of removal of extracellular DNA to the start of imaging. Since Nuc causes the faster decay of dsDUS than of dsDNA, they were present at similar amounts when imaging started.
Since the fluorescence signal associated with imported DNA was stable for 30 min in the nuc deletion strain, the fluorescence signal could be used as a measure for the amount of DNA residing within the cell after 1 h of exposure to extracellular DNA. First, we verified that DNA uptake was dependent on PilQ, the protein that forms the outer membrane channel required for DNA uptake (24). pilQ deletion caused an ∼200-fold reduction of the average fluorescence intensity in the presence of dsDNA (Fig. 3b). This signal could be considered the basal level, because DNA uptake was inhibited.
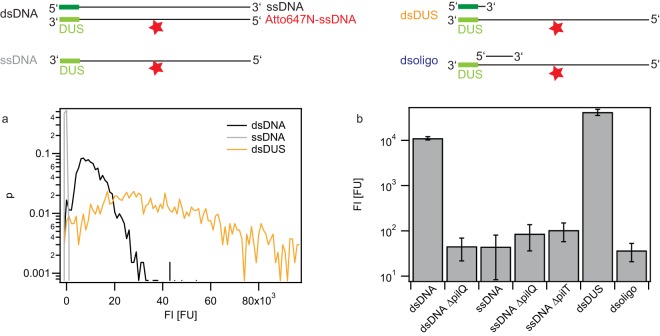
FIG 3.
Carrying capacities of various DNA fragments in a nuc deletion strain. Gonococci were incubated for 1 h with DNA fragments containing a single dye molecule at the center (top, stars). Subsequently, they were treated with DNase I. (a) Probability distribution of the total fluorescence intensity of individual cells. Black, strain Ng058 Δnuc ΔpilV with dsDNA; gray, strain Ng058 Δnuc ΔpilV with ssDNA; orange, strain Ng058 Δnuc ΔpilV with dsDUS. (b) Average fluorescence intensity of individual cells. dsDNA, Ng058 Δnuc ΔpilV incubated with dsDNA; dsDNA ΔpilQ, Ng163 ΔpilQ Δnuc ΔpilV incubated with dsDNA; ssDNA, Ng058 Δnuc ΔpilV incubated with ssDNA; ssDNA ΔpilQ, Ng163 ΔpilQ Δnuc ΔpilV incubated with ssDNA; ssDNA ΔpilT, Ng162 ΔpilT Δnuc ΔpilV incubated with ssDNA; dsDUS, Ng058 Δnuc ΔpilV incubated with ssDNA with double-stranded DUS; dsoligo, Ng058 Δnuc ΔpilV incubated with ssDNA with a 16-bp double-stranded region adjacent to DUS. Error bars, standard deviations from three independent experiments. Data are for >1,000 cells for each condition.
The fluorescence associated with ssDNA uptake did not exceed the basal level, indicating that the level of ssDNA uptake was below the detection limit of our setup (Fig. 3). Deletion of pilQ or of the type IV pilus retraction ATPase pilT (36) did not change the fluorescence level.
The signal generated by dsDUS DNA exceeded the signal generated by dsDNA, in agreement with the findings presented in Fig. 2b (Fig. 3a and b). We verified that short stretches of dsDNA other than the DUS did not support the uptake of otherwise ssDNA by hybridizing a 16-base fragment adjacent to the DUS motif. The average fluorescence signal was comparable to the signal generated by purely ssDNA (Fig. 3b), demonstrating that the double-stranded DUS is required for generating a significant DNA uptake signal.
We conclude that the thermonuclease Nuc degrades imported ssDNA and dsDNA. In a nuc deletion background, the signal associated with the uptake of ssDNA was at least 200-fold lower than the signal associated with the uptake of dsDNA. Double-stranded DUS on otherwise ssDNA was sufficient to achieve a signal exceeding the dsDNA signal.
Uptake kinetics of ssDNA containing double-stranded DUS.
We found that gonococci take up single-stranded DNA but Nuc rapidly degrades ssDNA. Therefore, we used a nuc deletion strain for monitoring the kinetics of dsDUS (ssDNA with double-stranded DUS) uptake. Cells were incubated with dsDNA or dsDUS, and images were acquired continuously (Fig. 4). The fluorescence intensities associated with DNA uptake were determined by quantitative image analysis. Both dsDNA and dsDUS uptake revealed saturation kinetics and were well described by an exponential fit. The characteristic time of uptake of dsDNA (τupds) was 2.9 ± 0.3 min, and the characteristic time of uptake of dsDUS DNA (τupss) was 4.1 ± 0.4 min. The carrying capacities, given as the maximum fluorescence intensity (FImax), were 6,300 ± 400 fluorescent units (FU) for dsDNA and 20,000 ± 800 FU for dsDUS.
In summary, dsDNA and dsDUS DNA uptake showed saturation kinetics with comparable characteristic times.
Transformation efficiencies of dsDNA, ssDNA, and dsDUS.
Finally, the question whether the substrate dependence of the transformation efficiency reflected the substrate dependence of DNA uptake was addressed. Unlabeled ssDNA fragments with a length of 300 bp were synthesized (see Table S2 in the supplemental material). The fragments contained part of the sequence of the gyrB gene with the point mutation that confers resistance against nalidixic acid and the DUS (ngch_L01). dsDNA was generated by PCR using primers kh033 and kh034. When these fragments were used as transforming DNA in the MS11 wt, they conferred antibiotic resistance and enabled us to measure the transformation frequency as described in Materials and Methods. In the absence of transforming DNA, the probability was lower than 3 × 10−8 (Fig. 5). We attribute the resistance in these clones to spontaneous mutations. Transformation with the pSY6 plasmid containing the modified gyrB gene yielded a transformation probability (ptrafo) of 0.13 ± 0.03. The transformation probability of the double-stranded 300-bp fragment was significantly higher than the background (ptrafo, 3 × 10−6 ± 2 × 10−6). Thus, although the probability of transformation is orders of magnitude lower for the fragment than for the plasmid, transformation was clearly quantifiable. The transformation probability of single-stranded DNA with double-stranded DUS (dsDUS) was 8 × 10−8 ± 2 × 10−8; i.e., the level of transformation was significantly higher than the background. When ssDNA fragments were used, no transformation was detectable. Furthermore, we assessed whether deletion of nuc affected the transformation probabilities by repeating the transformation assay in a Δnuc strain. The transformation probability of dsDUS DNA was ∼3 times higher than that with the wt background. nuc deletion did not significantly affect the transformation probability when dsDNA or ssDNA was used as the substrate (Fig. 5).
FIG 5.
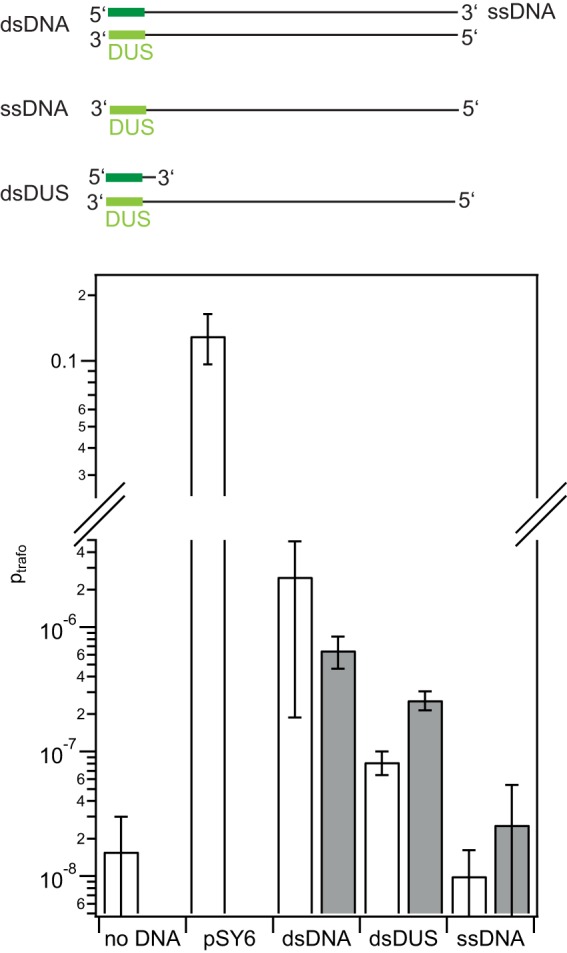
Transformation probability with dsDNA, ssDNA, and dsDUS. Transformation assays (with wt strain MS11) were performed with unlabeled DNA containing part of the gene encoding the modified gyrase B that confers resistance against nalidixic acid. pSY6 is a plasmid containing the DUS and the sequence of the modified gyrase B. The fragments consisted of dsDNA, ssDNA, and ssDNA with a 16-base complementary oligonucleotide containing double-stranded DUS. White bars, wt; gray bars, a Δnuc strain; error bars, standard errors of the means obtained from three or more independent experiments.
Taken together, the transformation assays show that short fragments of dsDNA and ssDNA with dsDUS are used as the substrates for transformation in our system, but the probability of ssDNA transformation does not exceed the background level.
DISCUSSION
Physical constraints on the molecular mechanisms of DNA uptake.
The DNA import machines are powerful translocation motors (22, 30), but the physical mechanism that generates the directed translocation of DNA is unclear. Here, we show that in the nuc deletion background, the rates of import of ssDNA with dsDUS and of dsDNA are comparable. This finding puts constraints on possible mechanisms for DNA uptake through the outer membrane. One potential translocation mechanism would be a cyclic molecular motor (37). In this scenario, the motor would bind to the DNA, translocate the DNA through a conformational change, and subsequently release the DNA prior to undergoing a second conformational change back to its initial position. Usually, energy provided by ATP hydrolysis is essential for generating directional bias. Such a cyclic molecular motor would require a well-defined secondary DNA structure for specific binding. For example, the translocation motor required for packaging of DNA into bacteriophage ϕ29 requires electrostatic contacts with adjacent pairs of backbone phosphates spaced every 10 bp on one DNA strand (38). Our results strongly suggest that such a scenario requiring tight coupling is not the underlying mechanism.
The fact that ssDNA is imported at a similar rate as dsDNA, provided that the DUS is double stranded, is consistent with the following picture. First, dsDUS binds to ComP, which is part of a complex formed by proteins belonging to the type IV pilus system. Once the DNA has entered the periplasm through the secretin pore formed by PilQ, ComE binds to the DNA and inhibits backwards diffusion, imposing a directional bias. Importantly, ComE binds both dsDNA and ssDNA independently of its sequence (27). ComE likely forms a translocation ratchet, where Brownian motion or pilus activity drives the movement of the DNA through the uptake pore and binding of ComE within the periplasm inhibits backwards diffusion (16). In contrast to a cyclic molecular motor, this mechanism can function independently of spatial constraints like the DNA secondary structure.
ComE governs the carrying capacity of the periplasm for DNA in a gene dosage-dependent fashion (21). We found that the carrying capacity of ssDNA with dsDUS was significantly (∼3 times) higher than that of dsDNA. One potential reason is the fact that the persistence length of ssDNA is considerably shorter. As a consequence, the radius of the random coil formed by ssDNA is shorter than that of the random coil formed by dsDNA, allowing a higher packaging density. Another reason could be the different affinities of ComE binding to ssDNA and dsDNA.
Nuc degrades imported DNA.
We showed that imported single-stranded DNA was rapidly degraded and that degradation was dependent on the thermonuclease Nuc. Deletion of nuc increased the probability of transformation when dsDUS was used as a substrate. This observation supports the idea that transforming DNA can be stored in the periplasm prior to its import into the cytoplasm (21).
The fluorescence decay of double-stranded DNA occurred at a considerably lower rate. This observation is consistent with our earlier observation that 10 kbp of dsDNA was detectable in the periplasm at a time scale of hours but that the amount of periplasmic DNA decreased, allowing turnover (21). Recombinant gonococcal Nuc was shown to degrade both ssDNA and dsDNA (8), in agreement with our observations. The failure to detect Nuc or its DNA-degrading activity in the supernatant led to the suggestion that Nuc may be localized in the periplasm (8). In a recent study, Nuc was found to have extracellular activity, but it remained unclear whether extracellular Nuc arose through secretion or through lysis (9). The fact that Nuc contains an N-terminal signal peptide supports the idea that Nuc resides within the periplasm at least transiently.
Using the pilQ control, we showed that the cell-associated fluorescence signal was caused by fluorescent DNA residing within the cell. Thus, Fig. 2 shows the kinetics of DNA degradation within the cell. Since the fluorescent label is not likely to enter the cytoplasm (22) (21), we conclude that we observed the degradation of DNA within the periplasm. Nuc was shown to degrade both ssDNA and dsDNA, although the degradation kinetics were not compared quantitatively (8). Here, we observed that the fluorescence signal associated with imported ssDNA was lost considerably faster than the signal associated with dsDNA. The underlying reasons may be (i) the faster degradation of ssDNA by Nuc or (ii) the faster transport of short pieces of ssDNA through the outer membrane because ssDNA has a shorter persistence length than dsDNA and, thus, its mean coil size per nucleotide is considerably smaller. We presume that the degradation of fluorescently labeled DNA within the periplasm does not cause a loss of the fluorescence signal, as long as the dye molecules reside within the periplasm. Instead, the loss of fluorescence becomes detectable when fluorescent labels attached to degraded DNA are rapidly transported from the periplasm to the environment. Therefore, the characteristic times of fluorescence decay measured in this study are considered to be the upper limit and the degradation process (excluding the transport of the dye out of the cell) may occur even faster. For transforming chromosomal DNA of gonococcal origin, the degradation rate is likely to be lower due to its methylation (8).
Single-stranded DNA enters the cytoplasm through a channel formed by ComA (20). For N. gonorrhoeae, it is currently not clear at what time point transforming dsDNA becomes single stranded. There is evidence that ssDNA forms transiently in the periplasm (28). Rapid degradation of ssDNA by Nuc in the periplasm can explain why ssDNA was detected only transiently.
Efficiencies of transformation and DNA uptake.
N. gonorrhoeae is constitutively competent for transformation. It preferentially takes up extracellular chromosomal DNA of its own species because it contains multiple repeats of the DNA uptake sequence. However, the major source of transforming DNA is still unclear. Single-stranded DNA is detectable within early gonococcal biofilms, when gene transfer is higher than that in later stages of biofilm development (39, 40). This raises the question about the role of secreted ssDNA in gene transfer. Indeed, previous studies showed that ssDNA can serve as a substrate for transformation by N. gonorrhoeae. The reported transformation efficiencies were similar for ssDNA with the Crick DUS and dsDNA in strain MS11, but the transformation efficiency of ssDNA was reduced 24 times in strain FA1090 (15). Earlier reports showed similar efficiencies of ssDNA and dsDNA transformation (41). Here we found that uptake of short ssDNA fragments is highly inefficient compared to that of dsDNA. Moreover, and in contrast to previous reports, the probability of ssDNA transformation did not exceed the background probability of spontaneous mutation, whereas ssDNA containing dsDUS DNA was used as a substrate for transformation.
We propose that different lengths and preparations of the DNA substrates most likely explain the discrepancy. In fact, for the same reason, Duffin and Seifert (15) point out the difficulty of comparing their results with the results of Stein (41). In our study, synthetic DNA fragments were used to avoid the formation of undesired double strands. First, short fragments minimize the probability of hairpin formation. Second, synthetic DNA prohibits contamination with cDNA that may form residual amounts of double-stranded DNA. The transformation probability increases with substrate length (37). Although the probability is low for the 300-base fragments used here, it is clearly quantifiable (Fig. 5). In previous studies, phage DNA with a length of ∼7 kbp was used (15). Southern blotting was used to verify that contamination with dsDNA was absent. While dsDNA was undetectable, the formation of secondary structures is possible. For example, if a hairpin were generated in the DUS region, the structural similarities to dsDUS would enhance the DNA uptake efficiency via interaction with ComP. Hairpin formation, particularly in long single-stranded DNA molecules, is likely to generate a double-stranded or partially double-stranded DUS, increasing the interaction with ComP and enhancing the DNA uptake probability. Partial hairpin formation is also likely to promote ssDNA uptake in the natural situation (12). On the gonococcal chromosome, DUSs are frequently arranged as inverted repeats, but they do not affect the transformation efficiency of double-stranded plasmids (42). In this way, secreted ssDNA could be recognized and taken up regardless of the requirement for dsDUS.
Conclusion.
We found that gonococci efficiently take up both single-stranded and double-stranded DNA, provided that they contain a double-stranded DNA uptake sequence. The fact that the secondary structure of the transforming DNA is not important for the kinetics of DNA import strongly suggests that the motor biasing DNA translocation toward the periplasm is loosely coupled to the DNA, in agreement with a translocation ratchet mechanism.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mike Koomey for the donation of bacterial strains, Enno Oldewurtel for support with image analysis, and Hank Seifert, Paul Duffin, and the Berenike Maier lab for helpful discussions.
This work was supported by the Deutsche Forschungsgemeinschaft through grant MA3898.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00464-16.
REFERENCES
- 1.Claverys JP, Martin B, Polard P. 2009. The genetic transformation machinery: composition, localization, and mechanism. FEMS Microbiol Rev 33:643–656. doi: 10.1111/j.1574-6976.2009.00164.x. [DOI] [PubMed] [Google Scholar]
- 2.Matic I, Taddei F, Radman M. 1996. Genetic barriers among bacteria. Trends Microbiol 4:69–72. doi: 10.1016/0966-842X(96)81514-9. [DOI] [PubMed] [Google Scholar]
- 3.Rotman E, Seifert HS. 2014. The genetics of Neisseria species. Annu Rev Genet 48:405–431. doi: 10.1146/annurev-genet-120213-092007. [DOI] [PubMed] [Google Scholar]
- 4.Frye SA, Nilsen M, Tonjum T, Ambur OH. 2013. Dialects of the DNA uptake sequence in Neisseriaceae. PLoS Genet 9:e1003458. doi: 10.1371/journal.pgen.1003458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cehovin A, Simpson PJ, McDowell MA, Brown DR, Noschese R, Pallett M, Brady J, Baldwin GS, Lea SM, Matthews SJ, Pelicic V. 2013. Specific DNA recognition mediated by a type IV pilin. Proc Natl Acad Sci U S A 110:3065–3070. doi: 10.1073/pnas.1218832110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aas FE, Wolfgang M, Frye S, Dunham S, Lovold C, Koomey M. 2002. Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type IV pilus expression. Mol Microbiol 46:749–760. doi: 10.1046/j.1365-2958.2002.03193.x. [DOI] [PubMed] [Google Scholar]
- 7.Berry JL, Cehovin A, McDowell MA, Lea SM, Pelicic V. 2013. Functional analysis of the interdependence between DNA uptake sequence and its cognate ComP receptor during natural transformation in Neisseria species. PLoS Genet 9:e1004014. doi: 10.1371/journal.pgen.1004014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steichen CT, Cho C, Shao JQ, Apicella MA. 2011. The Neisseria gonorrhoeae biofilm matrix contains DNA, and an endogenous nuclease controls its incorporation. Infect Immun 79:1504–1511. doi: 10.1128/IAI.01162-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juneau RA, Stevens JS, Apicella MA, Criss AK. 2015. A thermonuclease of Neisseria gonorrhoeae enhances bacterial escape from killing by neutrophil extracellular traps. J Infect Dis 212:316–324. doi: 10.1093/infdis/jiv031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biswas GD, Sparling PF. 1981. Entry of double-stranded deoxyribonucleic acid during transformation of Neisseria gonorrhoeae. J Bacteriol 145:638–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mulder C, Doty P. 1968. Residual activity of denatured transforming DNA of Haemophilus influenzae: a naturally occurring cross-linked DNA. J Mol Biol 32:423–435. doi: 10.1016/0022-2836(68)90019-3. [DOI] [PubMed] [Google Scholar]
- 12.Salgado-Pabon W, Jain S, Turner N, van der Does C, Dillard JP. 2007. A novel relaxase homologue is involved in chromosomal DNA processing for type IV secretion in Neisseria gonorrhoeae. Mol Microbiol 66:930–947. doi: 10.1111/j.1365-2958.2007.05966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dillard JP, Seifert HS. 2001. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol Microbiol 41:263–277. doi: 10.1046/j.1365-2958.2001.02520.x. [DOI] [PubMed] [Google Scholar]
- 14.Assalkhou R, Balasingham S, Collins RF, Frye SA, Davidsen T, Benam AV, Bjoras M, Derrick JP, Tonjum T. 2007. The outer membrane secretin PilQ from Neisseria meningitidis binds DNA. Microbiology 153:1593–1603. doi: 10.1099/mic.0.2006/004200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duffin PM, Seifert HS. 2012. Genetic transformation of Neisseria gonorrhoeae shows a strand preference. FEMS Microbiol Lett 334:44–48. doi: 10.1111/j.1574-6968.2012.02612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allemand JF, Maier B. 2009. Bacterial translocation motors investigated by single molecule techniques. FEMS Microbiol Rev 33:593–610. doi: 10.1111/j.1574-6976.2009.00166.x. [DOI] [PubMed] [Google Scholar]
- 17.Chen I, Dubnau D. 2004. DNA uptake during bacterial transformation. Nat Rev Microbiol 2:241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 18.Kidane D, Ayora S, Sweasy JB, Graumann PL, Alonso JC. 2012. The cell pole: the site of cross talk between the DNA uptake and genetic recombination machinery. Crit Rev Biochem Mol Biol 47:531–555. doi: 10.3109/10409238.2012.729562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maier B. 2012. Competence and transformation. In Graumann PL. (ed), Bacillus, cellular and molecular biology. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 20.Facius D, Meyer TF. 1993. A novel determinant (comA) essential for natural transformation competence in Neisseria gonorrhoeae and the effect of a comA defect on pilin variation. Mol Microbiol 10:699–712. doi: 10.1111/j.1365-2958.1993.tb00942.x. [DOI] [PubMed] [Google Scholar]
- 21.Gangel H, Hepp C, Muller S, Oldewurtel ER, Aas FE, Koomey M, Maier B. 2014. Concerted spatio-temporal dynamics of imported DNA and ComE DNA uptake protein during gonococcal transformation. PLoS Pathog 10:e1004043. doi: 10.1371/journal.ppat.1004043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stingl K, Muller S, Scheidgen-Kleyboldt G, Clausen M, Maier B. 2010. Composite system mediates two-step DNA uptake into Helicobacter pylori. Proc Natl Acad Sci U S A 107:1184–1189. doi: 10.1073/pnas.0909955107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seitz P, Blokesch M. 2014. DNA transport across the outer and inner membranes of naturally transformable Vibrio cholerae is spatially but not temporally coupled. mBio 5:e01409-14. doi: 10.1128/mBio.01409-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drake SL, Koomey M. 1995. The product of the pilQ gene is essential for the biogenesis of type IV pili in Neisseria gonorrhoeae. Mol Microbiol 18:975–986. doi: 10.1111/j.1365-2958.1995.18050975.x. [DOI] [PubMed] [Google Scholar]
- 25.Wolfgang M, van Putten JP, Hayes SF, Dorward D, Koomey M. 2000. Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J 19:6408–6418. doi: 10.1093/emboj/19.23.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seitz P, Pezeshgi Modarres H, Borgeaud S, Bulushev RD, Steinbock LJ, Radenovic A, Dal Peraro M, Blokesch M. 2014. ComEA is essential for the transfer of external DNA into the periplasm in naturally transformable Vibrio cholerae cells. PLoS Genet 10:e1004066. doi: 10.1371/journal.pgen.1004066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen I, Gotschlich EC. 2001. ComE, a competence protein from Neisseria gonorrhoeae with DNA-binding activity. J Bacteriol 183:3160–3168. doi: 10.1128/JB.183.10.3160-3168.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaussee MS, Hill SA. 1998. Formation of single-stranded DNA during DNA transformation of Neisseria gonorrhoeae. J Bacteriol 180:5117–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allemand JF, Maier B, Smith DE. 2012. Molecular motors for DNA translocation in prokaryotes. Curr Opin Biotechnol 23:503–509. doi: 10.1016/j.copbio.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maier B, Chen I, Dubnau D, Sheetz MP. 2004. DNA transport into Bacillus subtilis requires proton motive force to generate large molecular forces. Nat Struct Mol Biol 11:643–649. doi: 10.1038/nsmb783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seifert HS. 1997. Insertionally inactivated and inducible recA alleles for use in Neisseria. Gene 188:215–220. doi: 10.1016/S0378-1119(96)00810-4. [DOI] [PubMed] [Google Scholar]
- 32.Tonjum T, Freitag NE, Namork E, Koomey M. 1995. Identification and characterization of pilG, a highly conserved pilus-assembly gene in pathogenic Neisseria. Mol Microbiol 16:451–464. doi: 10.1111/j.1365-2958.1995.tb02410.x. [DOI] [PubMed] [Google Scholar]
- 33.Winther-Larsen HC, Hegge FT, Wolfgang M, Hayes SF, van Putten JP, Koomey M. 2001. Neisseria gonorrhoeae PilV, a type IV pilus-associated protein essential to human epithelial cell adherence. Proc Natl Acad Sci U S A 98:15276–15281. doi: 10.1073/pnas.261574998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aas FE, Lovold C, Koomey M. 2002. An inhibitor of DNA binding and uptake events dictates the proficiency of genetic transformation in Neisseria gonorrhoeae: mechanism of action and links to type IV pilus expression. Mol Microbiol 46:1441–1450. doi: 10.1046/j.1365-2958.2002.03265.x. [DOI] [PubMed] [Google Scholar]
- 35.Vink C, Rudenko G, Seifert HS. 2012. Microbial antigenic variation mediated by homologous DNA recombination. FEMS Microbiol Rev 36:917–948. doi: 10.1111/j.1574-6976.2011.00321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolfgang M, Lauer P, Park HS, Brossay L, Hebert J, Koomey M. 1998. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol Microbiol 29:321–330. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]
- 37.Overballe-Petersen S, Harms K, Orlando LA, Mayar JV, Rasmussen S, Dahl TW, Rosing MT, Poole AM, Sicheritz-Ponten T, Brunak S, Inselmann S, de Vries J, Wackernagel W, Pybus OG, Nielsen R, Johnsen PJ, Nielsen KM, Willerslev E. 2013. Bacterial natural transformation by highly fragmented and damaged DNA. Proc Natl Acad Sci U S A 110:19860–19865. doi: 10.1073/pnas.1315278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aathavan K, Politzer AT, Kaplan A, Moffitt JR, Chemla YR, Grimes S, Jardine PJ, Anderson DL, Bustamante C. 2009. Substrate interactions and promiscuity in a viral DNA packaging motor. Nature 461:669–673. doi: 10.1038/nature08443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kouzel N, Oldewurtel ER, Maier B. 2015. Gene transfer efficiency in gonococcal biofilms: role of biofilm age, architecture, and pilin antigenic variation. J Bacteriol 197:2422–2431. doi: 10.1128/JB.00171-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zweig M, Schork S, Koerdt A, Siewering K, Sternberg C, Thormann K, Albers SV, Molin S, van der Does C. 2014. Secreted single-stranded DNA is involved in the initial phase of biofilm formation by Neisseria gonorrhoeae. Environ Microbiol 16:1040–1052. doi: 10.1111/1462-2920.12291. [DOI] [PubMed] [Google Scholar]
- 41.Stein DC. 1991. Transformation of Neisseria gonorrhoeae: physical requirements of the transforming DNA. Can J Microbiol 37:345–349. doi: 10.1139/m91-056. [DOI] [PubMed] [Google Scholar]
- 42.Ambur OH, Frye SA, Tonjum T. 2007. New functional identity for the DNA uptake sequence in transformation and its presence in transcriptional terminators. J Bacteriol 189:2077–2085. doi: 10.1128/JB.01408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.