ABSTRACT
Bacteroides is a major component of the human gut microbiota which has a broad impact on the development and physiology of its host and a potential role in a wide range of disease syndromes. The predominance of this genus is due in large part to expansion of paralogous gene clusters, termed polysaccharide utilization loci (PULs), dedicated to the uptake and catabolism of host-derived and dietary polysaccharides. The nutritive value and availability of polysaccharides in the gut vary greatly; thus, their utilization is hierarchical and strictly controlled. A typical PUL includes regulatory genes that induce PUL expression in response to the presence of specific glycan substrates. However, the existence of additional regulatory mechanisms has been predicted to explain phenomena such as hierarchical control and catabolite repression. In this report, a previously unknown layer of regulatory control was discovered in Bacteroides fragilis. Exploratory transcriptome sequencing (RNA-seq) analysis revealed the presence of cis-encoded antisense small RNAs (sRNAs) associated with 15 (30%) of the B. fragilis PULs. A model system using the Don (degradation of N-glycans) PUL showed that the donS sRNA negatively regulated Don expression at the transcriptional level, resulting in a decrease in N-glycan utilization. Additional studies performed with other Bacteroides species indicated that this regulatory mechanism is highly conserved and, interestingly, that the regulated PULs appear to be closely linked to the utilization of host-derived glycans rather than dietary plant polysaccharides. The findings described here demonstrate a global control mechanism underlying known PUL regulatory circuits and provide insight into regulation of Bacteroides physiology.
IMPORTANCE The human gut is colonized by a dense microbiota which is essential to the health and normal development of the host. A key to gut homeostasis is the preservation of a stable, diverse microbiota. Bacteroides is a dominant genus in the gut, and the ability of Bacteroides species to efficiently compete for a wide range of glycan energy sources is a crucial advantage for colonization. Glycan utilization is mediated by a large number of polysaccharide utilization loci (PULs) which are regulated by substrate induction. In this report, a novel family of antisense sRNAs is described whose members repress gene expression in a distinct subset of PULs. This repression downregulates PUL expression in the presence of energy sources that are more readily utilized such as glucose, thereby allowing efficient glycan utilization.
INTRODUCTION
The human gut microbiota plays diverse roles in the normal functioning of host physiology and under pathological conditions. These activities include but are not limited to the extraction of energy in undigested dietary components, normal development of the intestinal tract and the immune system, and the onset of obesity and diabetes (1–5). Species of Bacteroidetes constitute a numerically predominate phylum in the human gut, and the genus Bacteroides has been linked to many of the microbiota's normal activities, but, in addition, they are significant opportunistic pathogens (6–8). Bacteroides species, and especially Bacteroides fragilis, are the anaerobes most frequently isolated from a wide range of extraintestinal infections. The most common infections generally arise when the intestinal wall is compromised and the peritoneum is contaminated with colonic contents. This can lead to peritonitis, intra-abdominal abscess, and bacteremia (9, 10). The dominance of B. fragilis in these infections can be attributed to an array of virulence factors that include polysaccharide capsule phase variation, a striking resistance to oxidative stress, and the ability to exploit novel host nutrient sources (11–14).
The ability to effectively compete for limited nutrients is essential for Bacteroides colonization in the gut as well as for success in extraintestinal infections. The key element of nutrient acquisition is the presence of a family of outer membrane protein complexes dedicated to the uptake and catabolism of polysaccharides (15–17). These multiprotein complexes are coded by a large family of paralogous genes and are organized in substrate-specific polysaccharide utilization loci (PULs). The central role of PULs in Bacteroides biology is indicated by their frequency in the genome. Bacteroides thetaiotaomicron, an abundant species in the gut, is considered a polysaccharide generalist, and nearly 18% of its genome is dedicated to the PULs (18).
The Bacteroidetes PULs have a unique and well-characterized operon structure that is shared across the phylum. The model PUL is a starch utilization system (Sus) found in B. thetaiotaomicron. The Sus PUL codes for two highly conserved outer membrane proteins which are homologs of SusC, the TonB-dependent transporter, and of SusD, a substrate binding/sensing protein crucial for substrate recognition (19, 20). Other members of the PUL are not conserved but are required for binding and hydrolysis of specific glycan substrates. Regulatory genes usually appear in positions adjacent to and upstream of the SusC homolog location and frequently occur in the form of a sigma factor/anti-sigma factor pair or a hybrid two-component regulatory system (HTCS). These regulatory systems are responsible for the induction of PUL gene expression when a substrate is present, but recent studies suggested that a more robust regulatory model is needed to explain catabolite repression and how glycan use is prioritized (21, 22). The results from these studies indicate the existence of mechanisms that repress specific PUL gene transcription in media when a preferred carbon source is present and that the extent of this repression is dependent on where the particular substrate fits into the overall hierarchy of utilization. There is evidence that the HTCSs can mediate both the induction of PUL expression and repression of lower-priority PULs, but there is no similar mechanism associated with the sigma/anti-sigma systems (23).
In this report, we describe the discovery of a family of antisense small RNA (sRNA) molecules that can repress the expression of 15 PULs in B. fragilis, 14 of which have the sigma/anti-sigma regulatory organization. These sRNAs act primarily on their cognate PULs, providing a significant level of specificity to the system. Finally, we present evidence suggesting that this mechanism is not restricted to B. fragilis but that at least three other Bacteroides species have a similar genetic organization associated with a subset of their PULs.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth.
Bacterial strains and plasmids are listed in Table 1 (24–26). Throughout the manuscript, sRNAs are named according to the cognate susC homolog in the PUL and the names are preceded by the letters “sr” (e.g., sr3597). Bacteroides strains were routinely cultured in an anaerobic chamber in brain heart infusion-supplemented (BHIS) broth or in defined medium (DM) with specific carbon sources, and Escherichia coli strains were grown in Luria-Bertani broth (27, 28). Ampicillin (100 μg/ml), spectinomycin (50 μg/ml), rifampin (20 μg/ml), gentamicin (100 μg/ml), erythromycin (10 μg/ml), tetracycline (5 μg/ml), trimethoprim (100 μg/ml), and thymine (50 μg/ml) were added to media as indicated. Mucin glycans were prepared by proteolysis of porcine gastric mucin followed by alkaline β-elimination to release free glycans as described previously (16). Media containing human transferrin (25 mg/ml) as a carbon source contained 50 μM FeSO4 to ensure a readily available iron source for growth (11).
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Description and/or genotypea | Reference or source |
|---|---|---|
| Bacterial strains | ||
| 638R | B. fragilis clinical isolate, Rifr | 26 |
| ADB77 | 638R ΔthyA Rifr Tpr | 24 |
| DH10B | E. coli F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara leu)7697 galU galK rpsL nupG λ− | Invitrogen |
| IB558 | B. fragilis strain 638R with pYC1:donA, donA overexpression strain, Rifr Ermr | This study |
| IB559 (ΔdonA) | B. fragilis strain 638R with donA deletion, Rifr | This study |
| IB560 (ΔdonB) | B. fragilis strain 638R with donB deletion, Rifr | This study |
| IB561 (donS null mutant) | B. fragilis strain 638R with donS null mutation created by mutation of the donS promoter sequence, Rifr | This study |
| IB563 | B. fragilis strain 638R with pYC2:donS, donS overexpression strain, Rifr Ermr | This study |
| IBpYC4:sr3597 | B. fragilis strain 638R with sr3597 overexpressed on pYC4:sr3597, Rifr Ermr | This study |
| IBpYC5:srCcfC | B. fragilis strain 638R with srCcfC overexpressed on pYC5:srCcf, Rifr Ermr | This study |
| Plasmids | ||
| pYT102 | P15A ori, RP4 oriT; B. fragilis suicide vector, thyA+ (Cmr) Tetr | 24 |
| pFD340 | Bacteroides-E. coli expression shuttle vector, (Ampr) Ermr | 25 |
| pYC1:donA | B. fragilis expression vector pFD340 containing the donA gene with the ahpC ribosomal binding sequence cloned into BamHI and SacI sites downstream of the IS4351 promoter | This study |
| pYC2:donS | B. fragilis expression vector generated by replacing the IS4351 promoter on pFD340 with an 82-bp 16S rRNA promoter sequence followed by the donS gene | This study |
| pYC3:rdm | B. fragilis expression vector generated by replacing the IS4351 promoter on pFD340 with an 82-bp 16S rRNA promoter sequence followed by the random RNA sequence rdm | This study |
| pYC4:sr3597 | B. fragilis expression vector generated by replacing the IS4351 promoter on pFD340 with an 82-bp 16S rRNA promoter sequence followed by the sRNA sr3597 | This study |
| pYC5:srCcf | B. fragilis expression vector generated by replacing the IS4351 promoter on pFD340 with an 82-bp 16S rRNA promoter sequence followed by the sRNA srCcfC | This study |
Rifr, rifampin resistant; Tetr, tetracycline resistant; Ermr, erythromycin resistant; Tpr, trimethoprim resistant; Cmr, chloramphenicol resistant; Ampr, ampicillin resistant. For Bacteroides-E. coli shuttle vectors, parentheses indicate antibiotic resistance expression in E. coli.
Construction of mutant strains.
All oligonucleotide primers used in this report are listed in Table S1 in the supplemental material. The ΔdonA and ΔdonB deletion mutants IB559 and IB560 were the products of unmarked, in-frame deletions. Chromosomal fragments flanking donA were PCR amplified using primer pairs sigOK+2kL/sigOK+2kR and sigOK-2kL/sigOK-2kR. The fragments were cloned into the pYT102 allelic exchange vector (24) and then mobilized into B. fragilis mutant ADB77 (lacking thyA). Transconjugants were selected on BHIS plates containing rifampin, gentamicin, and tetracycline. Transconjugants were resolved in media supplemented with thymine and then plated on BHIS plates with trimethoprim to select for the double-crossover allelic exchange. Mutants were reverted to thyA+ (24). Construction of the donB mutant was similar except that the primer pairs anti-sigOK+2kL/anti-sigOK+2kR and anti-sigOK-2kL/anti-sigOK-2kR were used. donA overexpression strain IB558 was constructed by PCR amplification of donA using primers sigOK-340L/sigOK-340R. The amplified fragment was inserted in the pFD340 expression shuttle vector (25) downstream of the constitutive IS4351 promoter sequence. The donS null mutation was constructed by mutating the consensus promoter sequence TTTG to AAAC. First, a 1,301-bp chromosomal fragment covering the entire donS region was amplified and the TTTG sequence was replaced with AAAC by using overlapping PCR with the mutagenic primer pairs sRNA117+L/sRNA117+R(AAACmut) and sRNA117-L(AAACmut)/sRNA117-R. The mutated fragment was cloned into pYT102 and mobilized into strain ADB77. Following resolution, the mutant allele was identified by colony PCR followed by nucleotide sequencing. The DonS overexpression strain IB563 contained the donS gene downstream of a 16S rRNA promoter sequence. Briefly, the primer pair sRNA117-L/sRNA117-R1 was used to amplify donS such that 36 bp of the 16S rRNA promoter sequence was engineered upstream of the donS sequence. This PCR product was then used as a template in a PCR with primer pair 16SPR-36-82/sRNA117-R1, which engineered the remaining base pairs of the 16S rRNA promoter upstream of the 36-bp sequence. This PCR product was cloned into pFD340 between the SacI site and the PstI site, replacing the existing IS4351 promoter. Other sRNA overexpression strains (strains overexpressing sRNAs sr3597 and srCfcC) were constructed using the same strategy as was used for donS overexpression. An overexpression strain containing a random sRNA sequence was made by fusion of a random DNA sequence (described in Table S2 in the supplemental material) to the 82-bp 16S rRNA promoter sequence. The 5S rRNA termination sequence was positioned at the 3′ end of the random sequence, resulting in a sequence consisting of 131 nucleotides. This fragment was cloned into pFD340, replacing the IS4351 promoter.
RNA extraction and qRT-PCR.
Total RNA was extracted from cell pellets using the hot phenol method and stored in 50% formamide at −80°C (27). DNA contamination was determined by PCR using primers for the 16S rRNA gene. The quantitative reverse transcription-PCR (qRT-PCR) procedure was performed as previously described (11). Primer pair omp117rtL and omp117rtR was used to amplify a 140-bp fragment for the donC gene, and all reactions were performed in triplicate. Expression values were normalized to 16S rRNA, and results represent at least two independent experiments.
Differential RNA-seq.
Differential transcriptome sequencing (differential RNA-seq) analysis was performed as previously described (29, 30). Total RNA was treated with DNase I to remove contamination, and processed RNAs were then removed by treatment with terminator 5′-phosphate-dependent exonuclease (TEX) (Epicentre catalog no. TER51020) for 60 min at 30°C. Identical samples were treated in buffer alone. cDNA libraries were prepared from TEX-treated and untreated RNA samples. The cDNA libraries were sequenced using an Illumina GA IIx genome analyzer with 100 cycles. The resulting reads in FASTQ format were trimmed with a cutoff phred score of 20 by the program fastq_quality_trimmer from FASTX toolkit version 0.0.13 (http://hannonlab.cshl.edu/fastx_toolkit/). The downstream analyses were conducted using the subcommands “create,” “align,” and “coverage” of the READemption tool (31) (version 0.3.5) with default parameters. Sequences shorter than 12 nucleotides were eliminated, and the collections of remaining reads were mapped to the reference genome sequences (NC_016776.1) using segemehl (32). Coverage plots in wiggle format representing the number of aligned reads per nucleotide were generated based on the aligned reads and inspected in the Integrated Genome Browser (33). Each graph was normalized to the total number of reads that could be aligned from the respective library. To restore the original data range and prevent rounding of small errors to zero by genome browsers, each graph value was then multiplied by the minimum number of mapped reads calculated over all libraries. RNA-seq data are available at the NCBI Gene Expression Omnibus database.
Northern hybridization.
Northern hybridization analyses of sRNAs have been previously described (34). Briefly, 10 μg of RNA was separated on a 10% denaturing polyacrylamide gel containing 7 M urea. A low-molecular-weight DNA ladder labeled with [γ-32P]ATP was used as the size standard. Nucleic acids were transferred to an Amersham Hybond-N+ membrane using a Bio-Rad Mini Trans-Blot Cell. Membranes were prehybridized for 3 h at 43°C. Oligonucleotide probes were end labeled with [γ-32P]ATP and polynucleotide kinase and incubated with the membranes at 43°C overnight. The membranes were then washed, exposed to X-ray film, and visualized by autoradiography.
Northern hybridization analysis of the don operon utilized two probes, donC and donG, that were labeled by PCR using a Prime-a-Gene labeling system (Promega catalog no. U1100) and primers described in Table S1 in the supplemental material. RNA samples (30 μg of total RNA) were separated on RNA denaturing gels and then transferred, blotted, and hybridized as previously described (27). 16S rRNA was used as the loading control.
Whole-cell deglycosylation assays.
Cell suspensions were prepared from mid-logarithmic-phase cultures grown in DM-mucin glycan media as previously described (11). The cell suspensions (100 μl) were mixed with an equal volume of purified human transferrin (1 mg/ml)–phosphate-buffered saline [PBS]) and then incubated anaerobically at 37°C for 3 h. Cells were removed by centrifugation, and the supernatants were mixed with loading buffer and subjected to electrophoresis on 12% SDS-PAGE gels. The gels were stained with Coomassie blue, or the proteins were transferred to polyvinylidene difluoride (PVDF) membranes for glycan determinations using Sambucus nigra agglutinin (SNA) according to the manufacturer's instructions (Roche; digoxigenin [DIG] glycan differentiation kit).
Accession number.
RNA-seq data are available at the NCBI Gene Expression Omnibus database under accession number GSE73107.
RESULTS
Identification of sRNA-associated PULs.
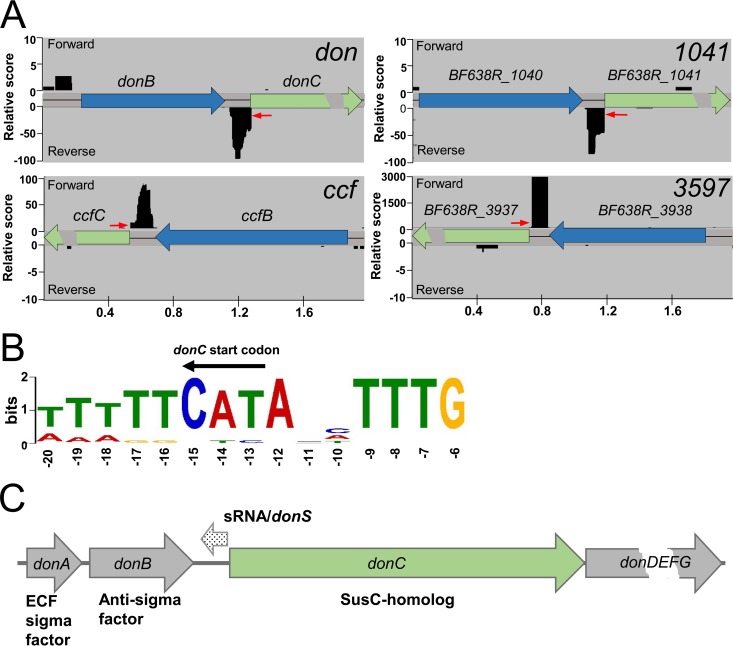
Genome analyses show that strains of B. fragilis have about 69 susC-susD pairs arranged in 47 PULs and 22 pairs not obviously associated with a classic PUL. In a recent study, one of these PULs, Don, was found to be highly expressed in a model of extraintestinal infection and to be responsible for the rapid deglycosylation of N-glycans from transferrin and other glycoproteins in serous fluid (11). During the course of a study performed to examine the regulation of Don gene expression, a differential RNA-seq analysis (29, 30) was performed to identify primary transcripts in mid-logarithmic-phase cells grown in complex media with glucose. Results from this exploratory study revealed the presence of an sRNA signal in the intergenic region upstream of the susC homolog donC. Upon further examination, antisense sRNA signals were observed between the anti-sigma factor genes and the SusC-like genes in 14 additional PULs. Results shown in Fig. 1A are representative of cDNA reads from the primary transcript library (terminator exonuclease [TEX] treated) for 4 PULs, and in each case there was a strong antisense signal that initiated just prior to the start codon of the SusC homolog. The sRNA transcripts were estimated to be 78 to 128 nucleotides in length and were initiated from a highly conserved promoter sequence that precisely overlapped the A residue of the ATG translational start site of the cognate SusC homolog. A conserved promoter sequence extended 7 nucleotides upstream of the consensus Bacteroidetes promoter sequence, TANNTTTG (35, 36) (Fig. 1B). Shown schematically in Fig. 1C, the position and orientation of the sRNA within the operon indicate that it is a cis-encoded antisense sRNA that likely regulates expression of its cognate PUL (37, 38).
FIG 1.
Identification of sRNAs associated with the susC homologs of B. fragilis PULs by RNA-seq. (A) Representative Illumina reads of cDNA from the TEX (terminator exonuclease)-treated libraries enriched for primary transcripts. Each locus is designated by the corresponding susC homolog (shown in green), and the donC and ccfC genes are BF638R_3439 and BF638R_3604, respectively. Data from the forward strand (+) are shown above the sequence line and, the reverse strand (−) reads are below. (B) The conserved promoter sequence for the sRNA-associated PULs is shown in the orientation of sRNA transcription (opposite donC). The conservation extends beyond the consensus housekeeping promoter recognition site of TANNTTTG. The 15 sRNA transcription start sites (TSS) were analyzed by MEME and compared to 1,200 TSSs in the transcriptome with the consensus promoter TANNTTTG. Conservation arises due to overlapping the susC homolog translation start codon which is indicated above the sequence logo. (C) Genetic organization of the Don PUL showing the location of the sRNA relative to the susC homolog and the sigma factor/anti-sigma factor genes. DonD is homologous to the highly conserved SusD protein, and the donEFG genes code for the PUL-specific hydrolases.
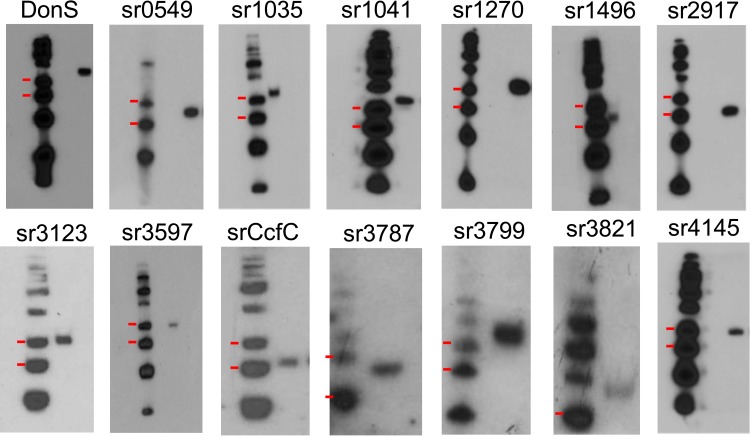
Nearly a third of B. fragilis 638R PULs demonstrated the presence of an sRNA signal in the transcriptome data. The existence of these sRNAs was confirmed by Northern hybridization using strand-specific oligonucleotide probes (Fig. 2). In the case of the Don PUL, an ∼125 nucleotide RNA molecule was observed and designated DonS. Further Northern analyses demonstrated an sRNA that was the approximate size predicted for every sRNA-associated PUL identified in the RNA-seq data (Fig. 2). In addition, these sRNAs and promoters are highly conserved in the genomes of the three most studied B. fragilis strains, NCTC-9343, YC46, and 638R, with only 0.3% mismatches (see Table S3 in the supplemental material for details of each sRNA and the gene designations for the cognate susC homologs in the different B. fragilis strains).
FIG 2.
Confirmation of sRNAs in B. fragilis PULs. Autoradiographs of Northern hybridization analyses of total RNA from mid-logarithmic-phase cultures grown in BHIS medium are shown. Left lanes, γ-32P-labeled low-molecular-mass DNA ladder with the 100-bp and 75-bp markers indicated by the red marks. Right lanes, sRNAs detected by hybridization to specific γ-32P-labeled probes. Except for the Don sRNA (DonS), the label above each panel indicates the BF638R gene designation of the SusC-like gene associated with the sRNA (sr).
sRNA control of don gene expression.
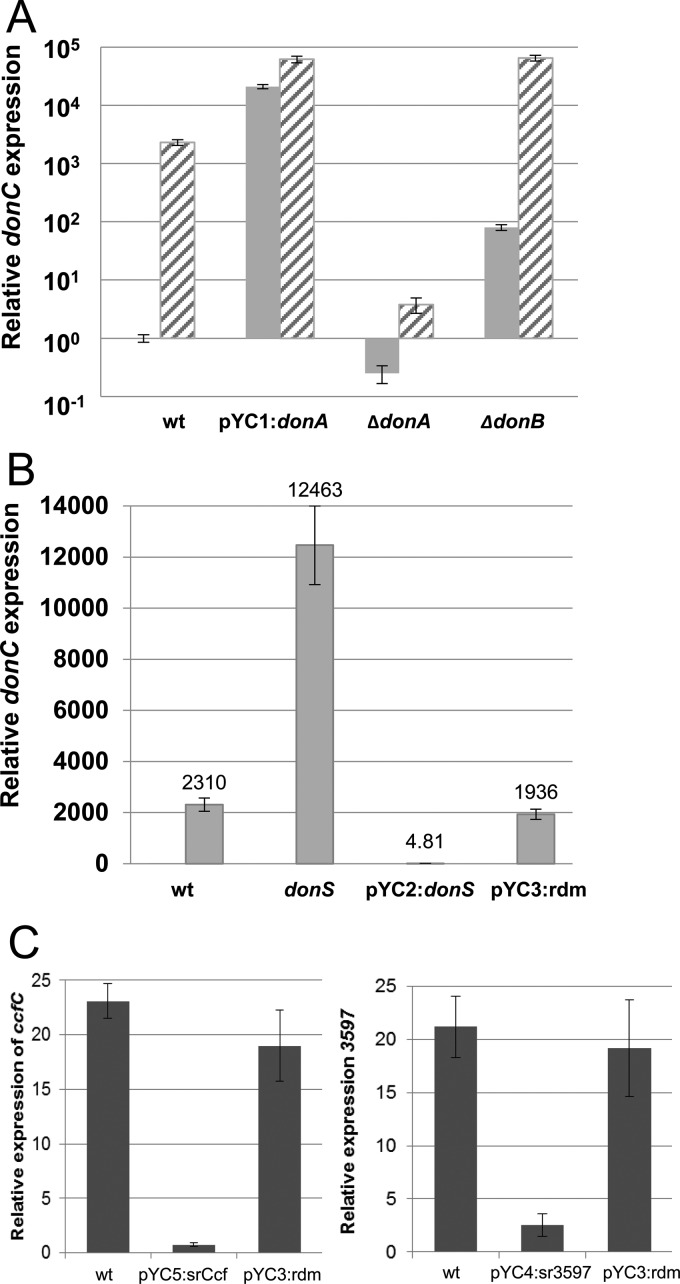
The Don PUL was used as a model to test the regulatory role of the sRNAs in PUL gene expression. Don has a conventional PUL organization, with sigma factor and anti-sigma homologs, donA and donB, followed by the susC and susD homologs and then three glycan processing genes, donEFG (Fig. 1C). The Don system can be highly induced in defined media (DM) with either mucin glycans or transferrin as the sole carbon/energy source (11). The regulatory activity of DonA/DonB was examined to establish their roles in induction of the donBCDEFG operon. First, qRT-PCR was used to measure the expression of donC during growth in noninducing media (DM-glucose) and in inducing media with mucin glycans as the sole carbon source (DM-mucin glycan). Results presented in Fig. 3A demonstrate approximately 1,000-fold induction in the presence of the substrate for the wild-type strain. Overexpression of sigma factor gene donA resulted in constitutive high-level expression of donC in media with or without the inducing substrates. The positive regulatory activity of the sigma factor was confirmed with a donA deletion mutant in which donC expression was severely repressed in both DM-glucose and DM-mucin glycan media. The anti-sigma factor is a negative regulator of the system; consequently, donC was induced nearly 100-fold in a donB deletion strain in the repressing DM-glucose medium (Fig. 3A). These results establish that the sigma/anti-sigma factors control substrate induction.
FIG 3.
Regulation of PUL gene expression. (A) Sigma factor pair DonA/DonB control substrate induction of the PUL. Expression of donC was measured by qRT-PCR using RNA samples from mid-logarithmic-phase cultures of the wild-type strain (wt), the strain overexpressing donA (pYC1:donA), donA deletion mutant IB559 (ΔdonA), and donB deletion mutant IB560 (ΔdonB). The striped bars represent cultures with expression induced by growth in DM-mucin glycan media, and the gray bars represent cells grown in the noninducing DM-glucose medium. The wt donC expression level under noninducing conditions was the baseline control. Results are from three independent experiments. (B) DonS has a significant effect on the expression of donC. Expression of donC was measured in cells grown in the inducing DM-mucin glycan medium using qRT-PCR. wt, wild type; donS, donS null mutant (IB561); pYC2:donS, donS overexpression strain; pYC3:rdm, the random RNA overexpression strain. The expression level of donC in the wild-type strain grown in DM-glucose medium was used as a baseline. (C) Expression analysis of the 3597 PUL and the Ccf PUL in the wild type (wt), sr3597 overexpression strain (pYC4:sr3597), and the srCcfC overexpression strain (pYC5:srCcf). RNA was extracted from mid-logarithmic-phase cultures grown in DM-mucin glycan medium to induce the PUL. The expression level of the susC homologs (ccfC, or BF638R_3597) in the wild-type strain grown in DM-glucose medium was used as a baseline. All qRT-PCRs were performed in triplicate, and the results are averages from two independent experiments. Error bars represent the standard errors of the means (SEM).
To test the regulatory role of the DonS sRNA, three strains were constructed: a donS null mutant lacking a functional donS promoter; a donS overexpression strain; and a strain overexpressing a random sRNA sequence (see Fig. S1 in the supplemental material). The abundances of donC mRNA were then compared for the three strains grown in the inducing DM-mucin glycan medium. The results clearly showed that DonS had a significant repressing effect on transcription (Fig. 3B). Relative to the wild-type results, the donS null mutant “overinduced” donC by more than 5-fold. This compares to >400-fold repression of the Don locus when donS was constitutively overexpressed (pYC2:donS). In a control experiment, constitutive expression of a random sRNA sequence (pYC3:rdm) had no effect on the PUL. The DonS-mediated repression of the operon was confirmed by Northern hybridization using donC or donG probes. The results showed that the ∼10-kb donBCDEFG mRNA was absent in the donS overexpression strain compared to a strong signal in the wild type (see Fig. S2).
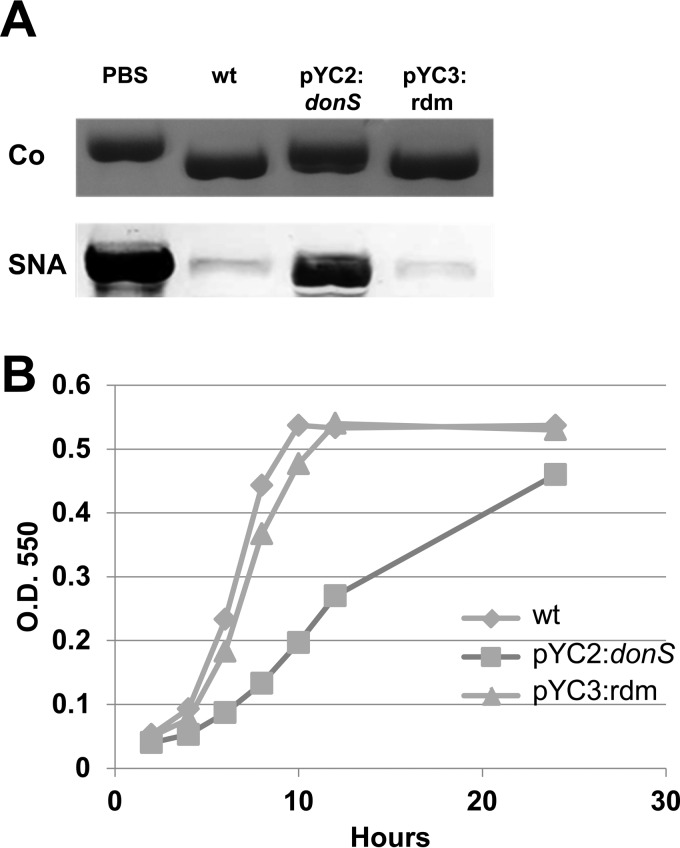
DonS overexpression resulted in a functional defect in the ability to harvest N-glycans from the Don PUL substrate human transferrin. When cells overexpressing donS were incubated with transferrin, there was little measurable deglycosylation activity (Fig. 4A). By comparison, the wild-type strain and the random sRNA control strain were able to efficiently remove the glycans as indicated by the increased migration of transferrin in the Coomassie-stained SDS-PAGE gel (Fig. 4A, upper panel) and by the loss of the glycan staining in the SNA Western blot (lower panel). The loss of deglycosylation activity also resulted in a growth defect for cells grown in DM medium with transferrin but not glucose as the sole carbon source (Fig. 4B; see also Fig. S3 in the supplemental material). Overall, these results indicate that DonS is a cis-encoded antisense sRNA that modulates expression of the Don PUL.
FIG 4.
The effect of donS overexpression on deglycosylation and on growth with human transferrin as the sole carbon source. (A) Deglycosylation analysis of human transferrin incubated overnight with wild-type B. fragilis (wt), the donS overexpression strain (pYC2:donS), a random RNA overexpression strain (pYC3:rdm), or no cells (PBS). Cells were induced prior to assay by growth in DM-mucin glycan medium. Samples were analyzed by SDS-PAGE followed by Coomassie blue staining (Co) or SNA glycan staining (SNA) to detect N-linked glycans. (B) Growth analysis of the wild-type strain (wt), the donS overexpression strain (pYC2:donS), and a random RNA overexpression strain (pYC3:rdm) in DM-transferrin medium. Growth was monitored by optical density at 550 nm (O.D. 550), and results represent triplicate measurements of two biological repeats. Error bars are standard deviations (SD).
Other sRNA-associated PULs.
Including DonC, there are 15 sRNA-associated SusC homologs in B. fragilis. Only one of these PULs had been previously studied; thus, to assign some prospective substrate targets, bioinformatics analysis of the cognate glycan hydrolases was performed using Phyre2 and InterPro protein modeling software (39, 40). The results from these analyses suggest that the PULs have a substrate range limited to host-derived polymers with a preponderance of enzymes active on gylcosaminoglycans, N- or O-linked glycans, and nucleic acids (see Table S4 in the supplemental material). Further, 14 of the 15 are regulated by sigma/anti-sigma factor pairs, with the FecR family being the predominate anti-sigma factor type. In order to determine if PUL repression was a conserved function for these sRNAs, two PULs that could be induced in mucin glycan media were tested in overexpression experiments. Cells overexpressing the srCcfC and sr3597 sRNAs were grown in DM-mucin glycan medium, and qRT-PCR was used to measure expression of the ccfC and BF638R_3597 susC homologs. The results in Fig. 3C show that there was a substantial repressing effect on expression of the cognate susC homolog in cells that overexpressed the sRNAs but not the random sRNA sequence. The repression was not as great as that seen with donS overexpression, which may have been due in part to their higher baseline expression in DM-glucose, but, notably, the Don PUL also was induced to higher levels of expression in the DM-mucin glycan medium.
DonS partially mediates a catabolite repression-like mechanism of PUL regulation.
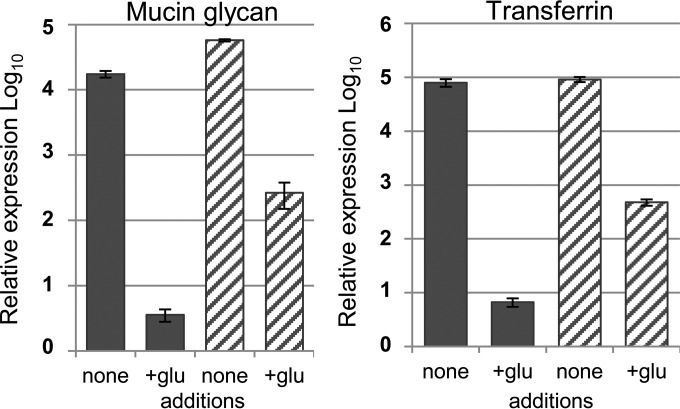
In B. thetaiotaomicron, there is a catabolite repression-like process in which preferred carbohydrate substrates cause repression of PULs for other substrates (21, 22). Transcriptional repression of a PUL by its associated sRNA would allow a second level of highly specific PUL control that complements substrate induction. Evidence for this was obtained by comparing levels of DonC expression in cultures grown with mucin glycans or human transferrin as the sole carbon source to those in cultures with the same carbon sources plus the addition of glucose (Fig. 5). In these experiments, the addition of glucose clearly repressed Don PUL expression in the wild-type strain. In contrast, the donS null mutant strain was 20-fold to 60-fold less capable of repressing donC expression when glucose was present, suggesting that the sRNA plays a role in the response to the carbon source.
FIG 5.
Catabolite repression of donC expression in the wild type and the donS null mutant. RNA was extracted from cultures of wild-type strain 638R (solid blue bars) and donS null mutant IB561 (striped bars) grown to mid-logarithmic phase in DM-mucin glycan medium (left panel) or DM-transferrin medium (right panel) with or without glucose added. Expression of donC was measured by qRT-PCR in triplicate, and the wild-type strain grown in a noninducing medium (DM-glucose) was used as a baseline. Carbon sources: mucin glycans (15 mg/ml), human transferrin (25 mg/ml), and glucose (glu; 4 mg/ml). Results represent averages of triplicate measurements of two biological repeats. Error bars represent the SEM.
Conservation of sRNA-associated PULs in Bacteroides species.
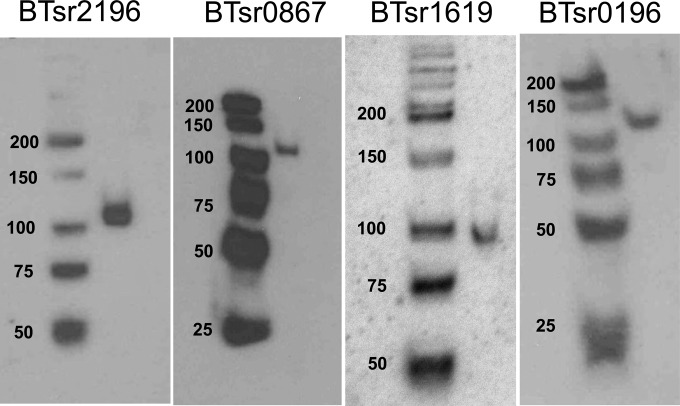
To determine if sRNA repression was conserved in the genus, the genomes of three additional Bacteroides species were screened for the sRNA signature, which is characterized by a susC homolog that contains a consensus promoter sequence overlapping the translational start site in the antisense orientation. Using these criteria, candidate sRNAs were observed for multiple PULs in each species tested (see Table S5 in the supplemental material). In Bacteroides vulgatus, Bacteroides ovatus, and B. thetaiotaomicron, 8 of 78, 8 of 121, and 7 of 111 susC homologs displayed the predicted organization of the promoter, respectively. Notably, the identified PULs all coded glycoside hydrolase enzymes, consistent with the utilization of host-derived glycans and not plant polysaccharides. Four of the sRNA predictions for B. thetaiotaomicron were tested by Northern analyses (Fig. 6), and the results showed a single antisense sRNA molecule in the size range of 90 to 110 nucleotides for each sample tested. This suggests that modulation of PUL gene expression by sRNAs is conserved in the gut Bacteroides.
FIG 6.
Confirmation of PUL-associated sRNAs in B. thetaiotaomicron. Autoradiographs of Northern hybridization analyses of total RNA from mid-logarithmic-phase cultures grown in BHIS medium are shown. Left lanes, γ-32P-labeled low-molecular-mass DNA ladder with the sizes marked in base pairs. Right lanes, sRNAs detected by hybridization to specific γ-32P-labeled oligonucleotide probes. Labeling above each panel indicates the B. thetaiotaomicron 5482 gene designation of the susC-like gene of each sRNA-associated PUL.
DISCUSSION
In the gut, there is an extensive diversity of glycan substrates that are potential energy sources for the microbiota. Likewise, the Bacteroides PULs demonstrate an equally broad range of substrate specificities that allow efficient utilization of these polysaccharides. Given this diversity, it is necessary to differentiate between substrates by giving preference to a greater nutritional (energy) value or greater abundance in the environment (22). It is not surprising that two or more types of regulatory mechanism are required to effectively manage the utilization of such a broad spectrum of substrates; thus, we see hybrid two-component systems, sigma/anti-sigma factor pairs, and other regulatory mechanism associated with PUL control. In this regard, the Don locus was first discovered in the context of growth in an artificial abscess using a rat tissue cage model where there was an abundance of N-linked glycans in the serous fluid (11). Arguably, N-glycans provide a high-quality energy source, and Don expression was highly induced in this environment, where there are few other available substrates. However, Don most likely did not evolve to benefit growth in extraintestinal habitats, and the availability of these glycans in the gut may be more transient; thus, there would be a need to rapidly control synthesis of the Don protein complex as these substrates become scarce. In the traditional model of sigma/anti-sigma factor PUL regulation, there is no known mechanism for rapid downregulation of expression, but the data described here provide a new, testable regulatory model that implicates a unique family of cis-encoded antisense sRNAs in the repression of PUL gene expression.
The model for Don regulation is based on the results shown in Fig. 3A that establish that this PUL responds to substrate induction via the activity of sigma factor DonA and that the FecR superfamily anti-sigma factor DonB acts as a negative regulator, likely by sequestration of DonA (41). We hypothesize that DonS has a significant role in preventing overexpression of the outer membrane protein complex and can respond rapidly as the substrate levels decline (see more detail in Fig. S4 in the supplemental material). This hypothesis is based on the observation that levels of induced Don mRNA can either be greatly reduced by overexpression of DonS or be greatly increased when DonS is absent (Fig. 3B). Further, in the absence of DonS, there is a tendency toward overexpression of Don mRNA when cells are in the presence of a more favorable substrate such as glucose (Fig. 5). In this model, as inducing substrate concentrations decline, the constitutive transcription of donS exceeds the amount of new DonBCDEFG mRNA being produced, effectively preventing accumulation of the message. There are two possible mechanisms by which the antisense sRNA could repress Don mRNA levels. The first is a simple one by which the duplex RNA formed with DonS is rapidly targeted for degradation. The second potential mechanism is transcriptional interference by RNA polymerase collision (38). The antisense sRNA also may repress Don expression at the level of translation. Given that DonS is located at the start codon of DonC and that it completely overlaps the DonC ribosome binding site, it is likely to prevent the initiation of DonC translation (42). This negative effect on Don translation may turn out to be more important than the transcriptional effect, but this will need to be tested. The addition of sRNA control to the Don sigma/anti-sigma factor regulatory pathway provides a mechanism to rapidly repress expression when the concentration of the substrate becomes too low, but additional work is needed to determine how DonS mediates repression when more-beneficial substrates are present.
The discovery of these PUL-associated sRNAs will impact our overall understanding of PUL regulation and substrate prioritization. These PULs specialize in the utilization of host-derived glycans or other nonplant glycan polymers (see Tables S4 and S5 in the supplemental material), many of which are prioritized in B. fragilis but not in “gylcan generalists” such as B. thetaiotaomicron (22). Currently, only two of these PULs, Ccf and Don, have been studied, and in both cases they are involved with the utilization of or binding glycans on host-secreted glycoproteins (11, 43). Arylsulfatases are found in 3 other sRNA-associated PULs. Although B. fragilis is not known to grow on any glycosaminoglycan (44), they may provide some nutritional benefit in complex mixtures of glycans or in a syntrophic relationship. Equally puzzling is the identity of substrates for the four sRNA-associated PULs containing enzymes with nuclease/phosphodiesterase domains. Although B. fragilis has been reported to have nuclease activity, there are no reports indicating that nucleic acids can function as the sole source of carbon and energy. Overall, the prevalence of PULs targeting host polymers suggests that these substrates are more abundant in the B. fragilis niche but does not explain the need for the tight control of PUL expression modulated by the sRNAs. Perhaps these substrates are transient in the normal habitat or do not provide a high return for energy extraction and it is necessary to rapidly shut down utilization when better energy sources become available. There is clearly a need to better understand substrate utilization in the gut, and elucidation of the sRNA regulatory model will be important for understanding Bacteroides host interactions.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. C. Caswell and R. M. Roop for discussions and A. C. Parker for technical assistance.
Funding Statement
The NIAID had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00381-16.
For a commentary on this article, see doi:10.1128/JB.00514-16.
REFERENCES
- 1.McNeil NI. 1984. The contribution of the large intestine to energy supplies in man. Am J Clin Nutr 39:338–342. [DOI] [PubMed] [Google Scholar]
- 2.Nobel YR, Cox LM, Kirigin FF, Bokulich NA, Yamanishi S, Teitler I, Chung J, Sohn J, Barber CM, Goldfarb DS, Raju K, Abubucker S, Zhou Y, Ruiz VE, Li H, Mitreva M, Alekseyenko AV, Weinstock GM, Sodergren E, Blaser MJ. 2015. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat Commun 6:7486. doi: 10.1038/ncomms8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Round JL, Mazmanian SK. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sommer F, Backhed F. 2013. The gut microbiota—masters of host development and physiology. Nat Rev Microbiol 11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 5.Tremaroli V, Backhed F. 2012. Functional interactions between the gut microbiota and host metabolism. Nature 489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 6.Brook I. 1988. Recovery of anaerobic bacteria from clinical specimens in 12 years at two military hospitals. J Clin Microbiol 26:1181–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. 2013. The long-term stability of the human gut microbiota. Science 341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lassmann B, Gustafson DR, Wood CM, Rosenblatt JE. 2007. Reemergence of anaerobic bacteremia. Clin Infect Dis 44:895–900. doi: 10.1086/512197. [DOI] [PubMed] [Google Scholar]
- 10.Vena A, Munoz P, Alcala L, Fernandez-Cruz A, Sanchez C, Valerio M, Bouza E. 2015. Are incidence and epidemiology of anaerobic bacteremia really changing? Eur J Clin Microbiol Infect Dis 34:1621–1629. doi: 10.1007/s10096-015-2397-7. [DOI] [PubMed] [Google Scholar]
- 11.Cao Y, Rocha ER, Smith CJ. 2014. Efficient utilization of complex N-linked glycans is a selective advantage for Bacteroides fragilis in extraintestinal infections. Proc Natl Acad Sci U S A 111:12901–12906. doi: 10.1073/pnas.1407344111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krinos CM, Coyne MJ, Weinacht KG, Tzianabos AO, Kasper DL, Comstock LE. 2001. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 414:555–558. doi: 10.1038/35107092. [DOI] [PubMed] [Google Scholar]
- 13.Polk BF, Kasper DL. 1977. Bacteroides fragilis subspecies in clinical isolates. Ann Intern Med 86:569–571. doi: 10.7326/0003-4819-86-5-569. [DOI] [PubMed] [Google Scholar]
- 14.Sund CJ, Rocha ER, Tzianabos AO, Wells WG, Gee JM, Reott MA, O'Rourke DP, Smith CJ. 2008. The Bacteroides fragilis transcriptome response to oxygen and H2O2: the role of OxyR and its effect on survival and virulence. Mol Microbiol 67:129–142. [DOI] [PubMed] [Google Scholar]
- 15.Koropatkin NM, Cameron EA, Martens EC. 2012. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol 10:323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martens EC, Chiang HC, Gordon JI. 2008. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. 2010. Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell 141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 19.Tancula E, Feldhaus MJ, Bedzyk LA, Salyers AA. 1992. Location and characterization of genes involved in binding of starch to the surface of Bacteroides thetaiotaomicron. J Bacteriol 174:5609–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. 2009. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J Biol Chem 284:24673–24677. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogers TE, Pudlo NA, Koropatkin NM, Bell JS, Moya Balasch M, Jasker K, Martens EC. 2013. Dynamic responses of Bacteroides thetaiotaomicron during growth on glycan mixtures. Mol Microbiol 88:876–890. doi: 10.1111/mmi.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pudlo NA, Urs K, Kumar SS, German JB, Mills DA, Martens EC. 2015. Symbiotic human gut bacteria with variable metabolic priorities for host mucosal glycans. mBio 6(6):e01282-15. doi: 10.1128/mBio.01282-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lynch JB, Sonnenburg JL. 2012. Prioritization of a plant polysaccharide over a mucus carbohydrate is enforced by a Bacteroides hybrid two-component system. Mol Microbiol 85:478–491. doi: 10.1111/j.1365-2958.2012.08123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baughn AD, Malamy MH. 2002. A mitochondrial-like aconitase in the bacterium Bacteroides fragilis: implications for the evolution of the mitochondrial Krebs cycle. Proc Natl Acad Sci U S A 99:4662–4667. doi: 10.1073/pnas.052710199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith CJ, Rogers MB, McKee ML. 1992. Heterologous gene expression in Bacteroides fragilis. Plasmid 27:141–154. doi: 10.1016/0147-619X(92)90014-2. [DOI] [PubMed] [Google Scholar]
- 26.Privitera G, Dublanchet A, Sebald M. 1979. Transfer of multiple antibiotic resistance between subspecies of Bacteroides fragilis. J Infect Dis 139:97–101. doi: 10.1093/infdis/139.1.97. [DOI] [PubMed] [Google Scholar]
- 27.Rocha ER, Smith CJ. 1997. Regulation of Bacteroides fragilis katB mRNA by oxidative stress and carbon limitation. J Bacteriol 179:7033–7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varel VH, Bryant MP. 1974. Nutritional features of Bacteroides fragilis sp. fragilis. Appl Microbiol 28:251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, Stadler PF, Vogel J. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 30.Sharma CM, Vogel J. 2014. Differential RNA-seq: the approach behind and the biological insight gained. Curr Opin Microbiol 19:97–105. doi: 10.1016/j.mib.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 31.Förstner KU, Vogel J, Sharma CM. 2014. READemption—a tool for the computational analysis of deep-sequencing-based transcriptome data. Bioinformatics 30:3421–3423. doi: 10.1093/bioinformatics/btu533. [DOI] [PubMed] [Google Scholar]
- 32.Hoffmann S, Otto C, Kurtz S, Sharma CM, Khaitovich P, Vogel J, Stadler PF, Hackermuller J. 2009. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput Biol 5:e1000502. doi: 10.1371/journal.pcbi.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicol JW, Helt GA, Blanchard SG Jr, Raja A, Loraine AE. 2009. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caswell CC, Gaines JM, Ciborowski P, Smith D, Borchers CH, Roux CM, Sayood K, Dunman PM, Roop RM II. 2012. Identification of two small regulatory RNAs linked to virulence in Brucella abortus 2308. Mol Microbiol 85:345–360. doi: 10.1111/j.1365-2958.2012.08117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bayley DP, Rocha ER, Smith CJ. 2000. Analysis of cepA and other Bacteroides fragilis genes reveals a unique promoter structure. FEMS Microbiol Lett 193:149–154. doi: 10.1111/j.1574-6968.2000.tb09417.x. [DOI] [PubMed] [Google Scholar]
- 36.Chen S, Bagdasarian M, Kaufman MG, Bates AK, Walker ED. 2007. Mutational analysis of the ompA promoter from Flavobacterium johnsoniae. J Bacteriol 189:5108–5118. doi: 10.1128/JB.00401-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brantl S. 2007. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr Opin Microbiol 10:102–109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 38.Georg J, Hess WR. 2011. cis-Antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev 75:286–300. doi: 10.1128/MMBR.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, Sangrador-Vegas A, Scheremetjew M, Rato C, Yong SY, Bateman A, Punta M, Attwood TK, Sigrist CJ, Redaschi N, Rivoire C, Xenarios I, Kahn D, Guyot D, Bork P, Letunic I, Gough J, Oates M, Haft D, Huang H, Natale DA, Wu CH, Orengo C, Sillitoe I, Mi H, Thomas PD, Finn RD. 2015. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res 43:D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martens EC, Roth R, Heuser JE, Gordon JI. 2009. Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont. J Biol Chem 284:18445–18457. doi: 10.1074/jbc.M109.008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wegmann U, Horn N, Carding SR. 2013. Defining the bacteroides ribosomal binding site. Appl Environ Microbiol 79:1980–1989. doi: 10.1128/AEM.03086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. 2013. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salyers AA, West SE, Vercellotti JR, Wilkins TD. 1977. Fermentation of mucins and plant polysaccharides by anaerobic bacteria from the human colon. Appl Environ Microbiol 34:529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.