ABSTRACT
Thiopeptides represent one of several families of highly modified peptide antibiotics that hold great promise for natural product engineering. These macrocyclic peptides are produced by a combination of ribosomal synthesis and extensive posttranslational modification by dedicated processing enzymes. We previously identified a compact, plasmid-borne gene cluster for the biosynthesis of micrococcin P1 (MP1), an archetypal thiopeptide antibiotic. In an effort to genetically dissect this pathway, we have reconstituted it in Bacillus subtilis. Successful MP1 production required promoter engineering and the reassembly of essential biosynthetic genes in a modular plasmid. The resulting system allows for rapid pathway manipulation, including protein tagging and gene deletion. We find that 8 processing proteins are sufficient for the production of MP1 and that the tailoring enzyme TclS catalyzes a C-terminal reduction step that distinguishes MP1 from its sister compound micrococcin P2.
IMPORTANCE The emergence of antibiotic resistance is one of the most urgent human health concerns of our day. A crucial component in an integrated strategy for countering antibiotic resistance is the ability to engineer pathways for the biosynthesis of natural and derivatized antimicrobial compounds. In this study, the model organism B. subtilis was employed to reconstitute and genetically modularize a 9-gene system for the biosynthesis of micrococcin, the founding member of a growing family of thiopeptide antibiotics.
INTRODUCTION
Maintaining an arsenal of effective antibiotics is a tremendous biomedical challenge, as antibiotic discovery is outpaced by the evolution of resistance (1–3). Ribosomally synthesized and posttranslationally modified peptides (RiPPs) are natural compounds with the appealing attributes of being derived directly from a genetic template and possessing numerous exotic chemical features that contribute to stability and antimicrobial activity (4). Thiopeptides, characterized by posttranslationally formed sulfur- and nitrogen-containing heterocycles, constitute a rapidly expanding class of RiPP antibiotics (5). Over 100 different thiopeptides have been chemically identified from numerous culturable bacterial producers, and the mining of genomic and metagenomic data promises to uncover many more chemical species that have eluded discovery by conventional means (6–8).
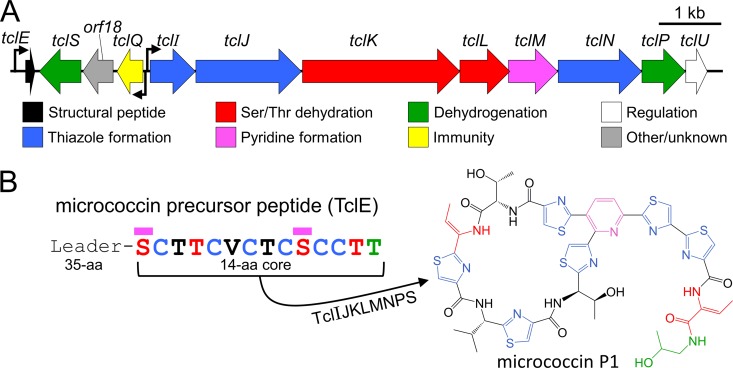
Thiopeptides are generally encoded by chromosomal or plasmid-localized gene clusters (example in Fig. 1A) that consist of approximately 6 to 25 protein-coding genes (7). These include a gene encoding the precursor peptide that is modified to become the thiopeptide product as well as an assemblage of posttranslational modification genes that are required for thiopeptide maturation and genes for host immunity and other functions. The precursor peptide for thiopeptides and other RiPPs consists of an N-terminal leader that acts as a required docking site for the biosynthetic machinery as well as a C-terminal core peptide that is the direct substrate for posttranslational modifications (9, 10). The leader peptide is ultimately cleaved from the modified core during posttranslational processing, releasing the active compound (example in Fig. 1B). In 2009, multiple independent researchers reported this general scheme for thiopeptide synthesis (11–15). In the years since, several additional biosynthetic principles have been elucidated (16–18). The process appears to involve three core steps: (i) cyclodehydration/oxidation of Cys residues to form thiazole heterocycles (Ser and Thr residues may also be modified to oxazoline or oxazole), (ii) dehydration of Ser and Thr residues yielding dehydroalanines and dehydrobutyrines, respectively, and (iii) cycloaddition of two of the dehydrated residues to yield the final macrocyclic thiopeptide scaffold (Fig. 1B).
FIG 1.
The M. caseolyticus strain 115 tcl gene cluster is responsible for the production of the antibiotic MP1. (A) The tcl gene cluster from M. caseolyticus (GenBank accession number KM613043) with predicted functions. Bent arrows represent putative promoters. (B) Sequence of the micrococcin precursor peptide TclE. The 14-amino-acid core peptide gives rise to the bioactive thiopeptide MP1. Magenta bars indicate residues joined to form the macrocycle.
Specific physical interactions between thiopeptide biosynthetic enzymes and cognate leader peptides are generally thought to license each processing enzyme to act relatively promiscuously on multiple substrate moieties within the core (19–21). This means that a single biosynthetic active site can work processively at multiple locations on the core peptide. For a typical thiopeptide gene cluster in nature, one often encounters numerous additional genes that obscure the simplicity of central thiopeptide chemistry. The recent in vitro reconstitution of the thiomuracin biosynthetic pathway illustrates this simplicity (17). In that study, five different protein enzymes, along with an additional RiPP precursor peptide recognition element (RRE)-containing protein, were found to be sufficient for the synthesis of a bioactive thiopeptide scaffold. This and other studies pursuing minimal thiopeptide biosynthetic pathways (7) inspire efforts to create easily manipulated in vivo platforms for the investigation of thiopeptide biology and for the derivation of novel antimicrobial compounds.
In this paper, we address the biosynthesis of micrococcin, the founding member of the thiopeptide antibiotic family (22–24). The genetic basis of its synthesis was first discovered in the producer strain Bacillus cereus ATCC 14579 (12). The cluster of 24 tcl genes in this strain controls the production of a mixture of structurally similar compounds, including several thiocillins (hence the tcl gene designation), micrococcin P1 (MP1), and micrococcin P2 (MP2). MP1 and MP2 share a structure and intracellular target but differ by a pair of hydrogen atoms as will be described later. A number of incisive studies have delineated key players from the 24-gene tcl cluster and have provided insights into how the Tcl-processing machinery tolerates alterations in the core peptide sequence (25–28). More recently, we identified a second micrococcin gene cluster in a strain of Staphylococcus epidermidis (now classified as Macrococcus caseolyticus) strain 115 (29). This gene cluster is considerably smaller (12 genes) (Fig. 1) and was found to produce only a single thiopeptide product, MP1. We report here our effort to minimize and modularize the M. caseolyticus micrococcin gene cluster for expression in the laboratory model organism Bacillus subtilis, enabling rapid genetic analysis and pathway engineering.
MATERIALS AND METHODS
Basic bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are summarized in Table 1. The micrococcin-producing strain 115, previously identified as S. epidermidis (30, 31), was investigated by multilocus sequence typing (MLST) and reassigned as M. caseolyticus. MLST primers and results are summarized in Tables S1 and S2 in the supplemental material. Plasmid DNA from M. caseolyticus 115 was used as the template for tcl gene amplification. The thiopeptide expression chassis B. subtilis 168 was used for promoter analysis, and a micrococcin-resistant derivative (BS 168R) with a single-codon deletion in the rplK gene (ΔP22) was used for micrococcin expression experiments. The indicator strain used in all spot-on-lawn bioassays was Staphylococcus aureus ATCC 6538. All plasmids in this study were selected and maintained in Escherichia coli DH5α. All strains listed above were maintained in liquid or solidified lysogeny broth (LB) unless described otherwise. The antibiotics used were ampicillin (Ap, 100 μg/ml), chloramphenicol (Cm, 5 μg/ml), and spectinomycin (Sp, 80 μg/ml). All bacterial cultures were grown at 37°C.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmida | Descriptionb |
|---|---|
| Strains | |
| M. caseolyticus 115 | MP1 producer, source of all tcl genes used in this study |
| B. subtilis 168 | Heterologous host for promoter activity assays |
| B. subtilis BS 168R | Micrococcin-resistant (rplKΔP22) derivative of B. subtilis 168 used for heterologous expression |
| S. aureus ATCC 6538 | Indicator strain for spot-on-lawn bioassays |
| Plasmids | |
| pDG1661 | spoVG-lacZ reporter for amyE integration; Cmr in B. subtilis and Apr or Spr in E. coli |
| pLacZ_E | pDG1661 with PtclE-spoVG-lacZ |
| pLacZ_Q | pDG1661 with PtclQ-spoVG-lacZ |
| pLacZ_I | pDG1661 with PtclI-spoVG-lacZ |
| pBS4S | B. subtilis thrC integration vector; Spr in B. subtilis and Apr in E. coli |
| pPxyl_U | pBS4S derivative with PxylA-tclU |
| pTclE | pBS4S derivative with Pstrong-tclE; Spr |
| pTclE_His | pTclE with His6-tclE (N-terminal tag) |
| pTclE_GST | pTclE with GST-tclE (N-terminal tag) |
| pDG1662 | amyE integration vector; Cmr in B. subtilis and Apr or Spr in E. coli |
| pLEGO_proto | pDG1662 derivative with p15A ori, PxylA promoter, and new MCS; Cmr |
| pLEGO | pLEGO_proto with PxylA-tclIJKLMNPS |
| pLEGO_strepI | pLEGO with Strep-tclI (N-terminal tag) |
| pLEGO_strepJ | pLEGO with Strep-tclJ (N-terminal tag) |
| pLEGO_strepK | pLEGO with Strep-tclK (N-terminal tag) |
| pLEGO_Kstrep | pLEGO with tclK-Strep (C-terminal tag) |
| pLEGO_strepL | pLEGO with Strep-tclL (N-terminal tag) |
| pLEGO_Lstrep | pLEGO with tclL-Strep (C-terminal tag) |
| pLEGO_strepM | pLEGO with Strep-tclM (N-terminal tag) |
| pLEGO_Nstrep | pLEGO with tclN-Strep (C-terminal tag) |
| pLEGO_strepP | pLEGO with Strep-tclP (N-terminal tag) |
| pLEGO_Pstrep | pLEGO with tclP-Strep (C-terminal tag) |
| pLEGO_strepS | pLEGO with Strep-tclS (N-terminal tag) |
| pLEGO_Sstrep | pLEGO with tclS-Strep (C-terminal tag) |
| pLEGO_ΔS | pLEGO with tclS removed |
| pLEGO_ΔP | pLEGO with tclP removed |
Strains and plasmids not from this study are cited in the Materials and Methods.
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Spr, spectinomycin resistance.
B. subtilis transformation.
Transformation of B. subtilis was accomplished by using the previously described starvation-induced competence protocol (29). Following selection, colonies were verified for correct integration of transgenes using a modified colony PCR protocol as follows: 1 ml of a saturated overnight culture was pelleted and resuspended in 200 μl of B. subtilis lysis buffer (50 mM Tris with 0.5 mg/ml lysozyme). Cells were then incubated at 30°C for 20 min and vortexed briefly before boiling for 3 min to complete cell lysis. Lysates were cleared by microcentrifugation (16,100 × g, 5 min), and 1 μl of supernatant was used as the template for PCR with primers flanking the integration junctions.
Strain construction for tcl promoter analysis.
To generate the reporter strains that were used in the promoter analysis experiments, a two-step integration strategy was used. First, integration of the thrC::PxylA-tclU insert was accomplished using a derivative of the thrC integrative plasmid pBS4S (32), in which the plasmid was modified to include the xylose-responsive PxylA promoter and its cognate repressor (xylR) upstream of the putative regulatory gene tclU (full sequences provided in Fig. S1 in the supplemental material). The correctly assembled construct was transformed into B. subtilis 168, and the resulting strain was used as the recipient strain for subsequent transformations. To generate the promoter-lacZ fusion plasmids pLacZ_E, pLacZ_Q, and pLacZ_I, the amyE integrative vector pDG1661 (33) was modified to include promoters PtclE, PtclQ, and PtclI. These promoters were amplified from M. caseolyticus plasmid DNA and were ligated into pDG1661 within the polylinker upstream of the spoVG-lacZ reporter gene (full sequences provided in Fig. S2 in the supplemental material). The promoter inserts consisted of approximately 500 bp of sequence immediately upstream of the start codons of the corresponding genes. Once constructed, each plasmid was transformed into the B. subtilis strain carrying thrC::PxylA-tclU described above to yield the strains used in β-galactosidase assays.
β-Galactosidase assays.
B. subtilis strains were grown overnight in LB-Sp and were then diluted 1:100 into fresh antibiotic-free broth and cultured until late-exponential phase (optical density at 600 nm [OD600] ≈ 1.0). Cultures were then placed on ice for 10 min to arrest growth before quantifying OD600 values. A total of 150 μl of culture was added to 750 μl of buffer Z (60 mM Na2HPO4, 40 mM NaH2PO4, 20 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol, pH 7.0) before adding 10 μl of toluene to permeabilize cells. Suspensions were vortexed for 15 s, warmed to 30°C for 5 min, combined with 150 μl of o-nitrophenyl-β-d-galactopyranoside (ONPG) solution (4 mg/ml in buffer Z), and incubated at 30°C for 30 to 40 min. Reactions were stopped by adding 400 μl of stop buffer (1 M Na2CO3). Insoluble debris was sedimented by microcentrifugation (16,100 × g, 5 min) before measuring OD420 values. A sample processed in an identical manner but omitting cells was used as a blank. Miller units were calculated according to reference 34.
Construction of pLEGO and derivatives.
To construct pLEGO, numerous alterations were made to the amyE integrative vector pDG1662 (33). In brief, the high-copy-number ColE1 replication origin was replaced with the lower-copy-number p15A origin from pACYC184 (35) followed by removal of the β-lactamase (bla) gene and destruction of many backbone restriction sites. The small polylinker within the integrating region of the plasmid was expanded to include the following restriction sites: BamHI, AvrII, SphI, KpnI, PstI, SacI, SalI, AatII, XhoI, BglII, and EcoRI. Finally, the xylR-PxylA promoter cassette from pHCMC04 (36) was introduced into the polylinker between BamHI and AvrII sites. We disrupted XhoI, EcoRI, and SalI sites within xylR in the process. This construct, termed pLEGO_proto, served as the base upon which pLEGO was constructed. To complete the assembly of pLEGO, the eight selected tcl processing genes (tclIJKLMNPS) were amplified from M. caseolyticus strain 115 and introduced between appropriate restriction sites in the pLEGO_proto polylinker. In several cases, mutagenic PCR was employed to disrupt restriction sites within tcl genes without altering the amino acid sequence of gene products. The complete sequence of pLEGO is provided in Fig. S3A in the supplemental material. Epitope-tagged derivatives of pLEGO-encoded enzymes were made by reamplifying tcl genes from pLEGO with primers designed to append the 8-amino-acid (Trp-Ser-His-Pro-Gln-Phe-Glu-Lys) Strep tag (sequences given in Fig. S3B). For pLEGO derivatives lacking tclP or tclS, the genes were removed by digesting pLEGO at the appropriate flanking restriction sites followed by treatment with Klenow polymerase and religation.
Construction of pTclE and derivatives.
The plasmid pTclE was engineered as a derivative of the thrC integrative vector pBS4S (32). The strong constitutive B. subtilis promoter Pstrong (iGEM biological parts registry BBa_K780003) was introduced between the NotI and XbaI sites of pBS4S using hybridizing oligonucleotides, and the tclE gene was cloned downstream between the XbaI and PstI sites to generate the final construct, pTclE. The complete sequence of pTclE is provided in Fig. S4A in the supplemental material. Plasmid derivatives for appending hexahistidine (His6) or glutathione S-transferase (GST) tags to the N terminus of TclE were generated in a similar manner by incorporating the tag in the primer (His6) or using overlap extension PCR (GST). The GST tag was amplified from pGEX-4T-1 (GE Life Sciences, Marlborough, MA). Detailed sequences are provided in Fig. S4B.
Micrococcin susceptibility bioassays.
Methanolic extracts from B. subtilis strains expressing tcl genes were obtained as follows. After overnight growth in antibiotic-supplemented LB, cells were diluted 1:60 into 6 ml of LB supplemented with xylose (1%, wt/vol). After an additional 8 h of growth, 4.8 ml of each culture was pelleted using iterative microcentrifugation (16,100 × g, 2 min for each run) and pellets were stored at −20°C prior to extraction. Extractions were accomplished by suspending thawed cell pellets in 1 ml of high-pressure liquid chromatography (HPLC)-grade methanol and vortexing vigorously for 3 min. Suspensions were left to incubate at room temperature for 5 min, followed by vortexing for an additional 3 min. Insoluble material was sedimented by microcentrifugation (16,100 × g, 5 min), and supernatants were transferred to fresh microcentrifuge tubes. These extracts were concentrated at room temperature to 80-μl volumes using a DNA120 SpeedVac (Savant Instruments Inc., Holbrook, NY) and stored at 4°C until assayed. The bioactivity of each extract was evaluated using a spot-on-lawn bioassay as described previously (29). The spot-on-lawn indicator strain S. aureus ATCC 6538 was grown overnight and diluted 1:20 into fresh LB before 200 μl was spread onto LB agar and allowed to dry. Once dry, 5-μl spots of concentrated extracts were placed upon bacterial lawns, allowed to dry, and then incubated at 37°C. After 8 h, plates were assessed for zones of inhibition and imaged.
Extraction and electrospray ionization mass spectrometric analysis of MP1 and MP2.
For each MP1- or MP2-producing strain, a single colony was grown overnight in LB-Sp and used to inoculate LB-Sp supplemented with 1% xylose (1 liter, 37°C, 200 rpm, 16 h). The cells were harvested by centrifugation (7,878 × g, 15 min), resuspended in 25 ml of methanol, vortexed for 30 s, and gently agitated on a benchtop shaker for 20 min at room temperature. Sodium sulfate (10 g) was added to the suspended cells before centrifugation (16,420 × g, 5 min). The methanol fraction was evaporated to <1 ml, and 30 ml of ethyl acetate was added. The mixture was transferred to a separatory funnel, washed with 30 ml of water, and subsequently washed with 30 ml of saturated sodium chloride. The combined aqueous washes were back extracted with ethyl acetate. The ethyl acetate layers were combined, dried with sodium sulfate, and filtered, and the volatiles were removed under reduced pressure. The resulting oily residue was redissolved in 50 μl of methanol, and 5 μl was transferred to a 96-well sample plate and evaporated under a stream of air. The residue was taken up in 20 μl of dimethyl sulfoxide (DMSO), injected (10 μl) into a Waters XBridge C18 column (3.5 μm, 4.6 by 50 mm), and separated using a Waters Alliance 2795 HPLC system (Waters, Milford, MA). The following linear gradient was used: 5% to 100% B over 8 min (A = water/0.1% formic acid, B = acetonitrile/0.1% formic acid, 1 ml/min). The eluent was analyzed by UV absorbance (Waters 2996 diode array UV detector) and electrospray ionization (ESI) positive-mode mass analysis (Waters ZQ-4000 quadrupole mass spectrometer). MP1 and MP2 products were eluted between 7.3 and 7.6 min.
RESULTS
Identification of the essential elements of an engineered micrococcin expression system.
To aid in the initial design of a minimized B. subtilis micrococcin expression system, we combined bioinformatic analyses (29), structural modeling, and a detailed literature survey to identify essential elements within the M. caseolyticus tcl gene cluster (shown in Fig. 1A). Homology arguments (12, 17, 29) suggest that TclI, TclJ, and TclN are involved in converting each of the core peptide Cys residues to thiazoles (Fig. 1B, blue), with TclI as the Ocin-ThiF-like RRE-containing protein, TclJ performing ATP-dependent heterocyclization (to thiazoline), and TclN oxidizing the thiazoline groups to thiazoles. TclK and TclL are likely responsible for the four Ser/Thr dehydration reactions (Fig. 1B, red), with TclM catalyzing peptide macrocyclization via the two dehydrated serines (magenta). TclP and TclS have homology with short-chain dehydrogenases, and we hypothesized that these enzymes are involved in decarboxylation of the C-terminal Thr residue (Fig. 1B, green) as has also been proposed for the homologous proteins in the B. cereus tcl cluster (12). TclQ is a ribosomal L11 homologue that we have shown to be involved in immunity (29), TclU is a putative transcriptional regulator (discussed below), and the function of the protein potentially encoded by orf18 is unclear.
Analysis of tcl promoter activity in B. subtilis.
The tcl gene cluster is organized into three apparent transcriptional units that are controlled by promoters upstream of tclE, tclQ, and tclI (Fig. 1A). The tclE unit is monocistronic, encoding only the TclE precursor peptide; the tclQ unit is a three-gene operon encoding TclQ, Orf18, and TclS; and the tclI transcriptional unit encodes eight proteins, most of which are predicted to participate in posttranslational processing of the TclE core peptide. The final gene in the tclI operon (tclU) encodes a putative MerR-type transcriptional regulator. Sequence alignments and structural modeling of TclU (not shown) indicate the presence of a conserved helix-turn-helix DNA-binding domain; however, the effector domain typically found in MerR regulators appears to be truncated or absent in TclU, making it difficult to predict its regulatory role with respect to tcl gene expression.
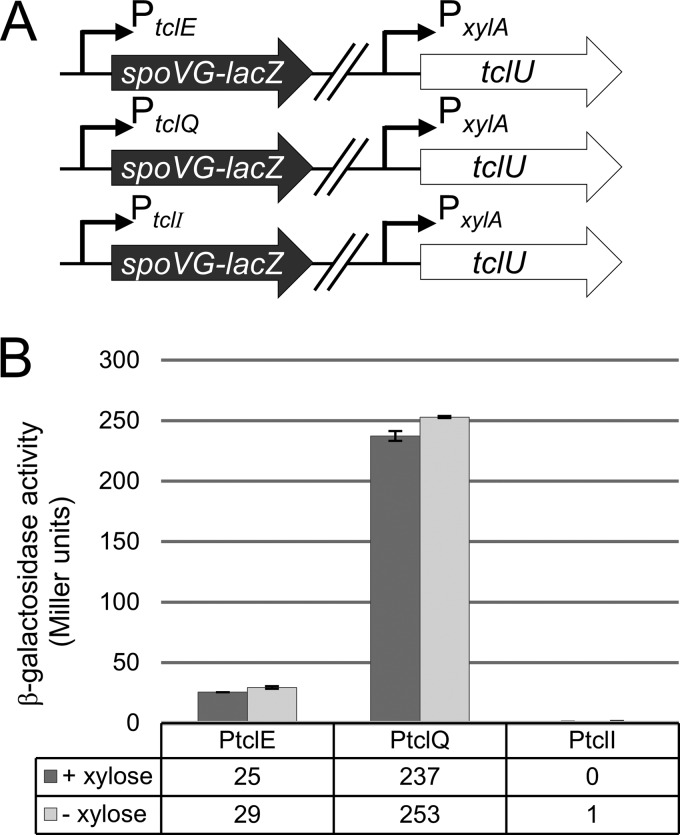
In transferring a functional tcl gene cluster into B. subtilis, transcriptional compatibility was a critical consideration. We sought to test whether the three tcl promoters described above are active in B. subtilis and whether TclU influences their activity. As an initial step, we employed a lacZ reporter assay to monitor the activity of the three predicted tcl promoters in B. subtilis. The 500-bp regions immediately upstream of the tclE, tclQ, and tclI start codons were fused to the spoVG-lacZ reporter, and these fusions were integrated into the B. subtilis chromosome. In each of these reporter strains, tclU (controlled by the xylose-inducible promoter PxylA) was integrated at a second chromosomal locus (Fig. 2A). Promoter strength was quantified on the basis of β-galactosidase activity in the presence or absence of TclU induction (Fig. 2B). From these results, we conclude that PtclE and PtclQ exhibit modest to strong activity, whereas PtclI is inactive. TclU appears to play only a minor role, if any, in the regulation of these promoters in B. subtilis, justifying its exclusion from the heterologous expression system.
FIG 2.
Analysis of M. caseolyticus tcl promoter activity in B. subtilis strain 168. (A) The promoter regions upstream of tclE, tclQ, and tclI were fused to a lacZ reporter and integrated into the B. subtilis chromosome. PxylA-tclU was similarly integrated at a separate locus for inducible expression of tclU. (B) β-Galactosidase activity was measured to quantify tcl promoter strength in the presence (+ xylose) or absence (− xylose) of tclU expression. Error bars represent the standard deviation from the mean using three biological replicates per condition.
Genetic refactoring of the tcl gene cluster supports production of MP1 in B. subtilis.
We next turned to the construction of a functional tcl gene cluster in the laboratory model organism B. subtilis strain 168. We sought to accomplish three primary objectives: (i) minimization of the ensemble of tcl genes required for micrococcin production, (ii) modularization of these genes to permit rapid manipulation of the pathway, and (iii) transcriptional optimization of the precursor peptide-encoding gene tclE and the tcl biosynthetic operon.
We decided on a two-plasmid system, allowing independent manipulation of different components of the pathway. In a preliminary expression system, the naturally cotranscriptionally arranged genes tclIJKLMNP were introduced downstream of PxylA in a vector designed for transgene integration at the B. subtilis amyE locus. The tclU gene was omitted in this construct. The remaining four genes (tclE, tclS, orf18, and tclQ) were introduced into a second vector that was designed for integration at the thrC locus. These genes were maintained in their original configuration (Fig. 1A) under the control of their native promoters (PtclE and PtclQ). Integration of these two preliminary plasmids into B. subtilis, followed by growth in the presence of xylose, led to the production of a methanol-extractable product with bioactivity against wild-type B. subtilis, based on a qualitative spot-on-lawn assay (not shown).
With the success of the proof-of-concept experiment described above, we further reduced and modularized our two-plasmid heterologous expression system. This system is summarized in Fig. 3. For the amyE integration plasmid, the putative biosynthetic genes were all arranged as a single operon under the control of the xylose-inducible PxylA promoter. To accomplish this, tclS was moved from the thrC integration plasmid described above, completing the eight-gene tclIJKLMNPS biosynthetic operon. This operon was punctuated by unique intergenic restriction sites, multiple overlapping regions between tcl open reading frames were resolved, and artificial ribosome binding sites were installed. As such, each biosynthetic gene constitutes a translationally independent unit that can be altered or removed while minimally affecting the expression of neighboring genes. This final plasmid (pLEGO) is represented in Fig. 3A, and its full sequence is provided in Fig. S3 in the supplemental material. The thrC integration plasmid was dedicated to tclE expression. This required the removal of tclS (moved to pLEGO), orf18 (a gene of unknown function), and the immunity gene tclQ. We alleviated the requirement for tclQ by selecting a spontaneous micrococcin-resistant B. subtilis mutant, which was used for all subsequent micrococcin expression experiments. This strain (BS 168R) was found to possess a single-codon deletion in rplK, which encodes the known ribosomal protein target of micrococcin (37). Given the relatively weak expression level of the native tclE promoter in B. subtilis (Fig. 2B), we replaced it with a strong constitutive promoter (Pstrong). The resulting Pstrong-tclE expression plasmid (termed pTclE) is represented in Fig. 3B. Its complete sequence is given in Fig. S4 in the supplemental material.
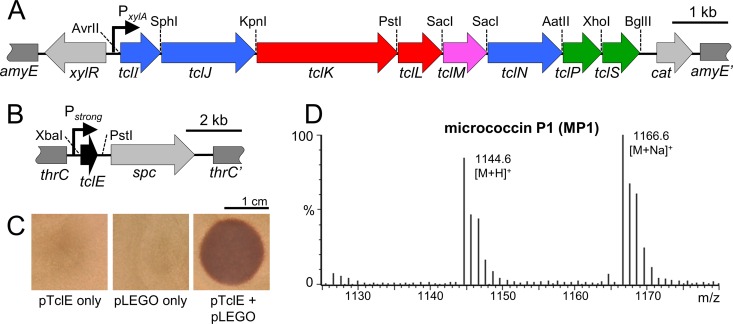
FIG 3.
Minimization and modularization of the tcl biosynthetic pathway in B. subtilis. (A) Gene map of the tcl region of pLEGO showing the xylose-inducible promoter (bent arrow), relevant processing genes (colored arrows), unique restriction sites, and homology regions (dark gray) for recombination into the B. subtilis chromosome. Light gray arrows indicate the xylose-responsive regulatory gene (xylR) and the chloramphenicol resistance determinant (cat). (B) Gene map of the TclE-encoding portion of pTclE showing the constitutive promoter (bent arrow), tclE, homology regions for integration into the B. subtilis chromosome (dark gray), and the spectinomycin resistance determinant (spc). (C) Spot-on-lawn bioassays with methanolic extracts from strains transformed with pTclE only, pLEGO only, or both. (D) ESI-MS analysis of the bioactive extract from the pTclE + pLEGO strain. MP1: observed 1,144.6 [M+H]+, calculated 1,144.2 [M+H]+; observed 1,166.6 [M+Na]+, calculated 1,166.2 [M+Na]+.
The pLEGO and pTclE plasmids were transformed into BS 168R, and transformants were evaluated for micrococcin production (Fig. 3C and D). Methanolic extracts from strains transformed with pTclE only, pLEGO only, or pTclE + pLEGO were tested for bioactivity against the S. aureus reference strain ATCC 6538. Only the strain with both transgenic components supported the production of a bioactive compound (Fig. 3C). Mass analysis of this methanol-extractable product was consistent with its identity as MP1 (expected m/z 1,144.2 [M+H]+; observed m/z 1,144.6 [M+H]+), which is in agreement with earlier work characterizing the product of this pathway (29). In addition, methanolic extracts from strains that were missing either TclI or TclP lacked bioactivity and were deficient in compounds detectable by mass spectrometry (MS) within the expected mass range for MP1 and MP2 (not shown). Taken together, these results support the identity of the antibacterial compound as MP1 and demonstrate the success of this highly engineered system in reconstituting the M. caseolyticus micrococcin pathway in B. subtilis.
Testing the utility of the heterologous, modularized genetic system.
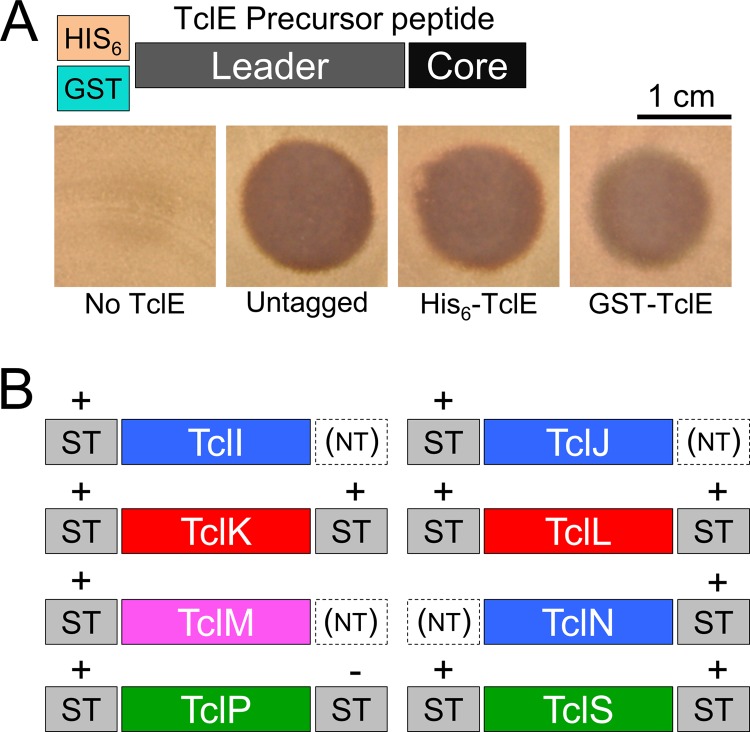
The successful reconstitution of this pathway in B. subtilis opens the door for detailed mechanistic studies of this pathway and its constituents. Such studies will inevitably require purification of substrate, enzymes, or both and would be benefitted by a better understanding of how Tcl proteins in this pathway tolerate N- or C-terminal affinity tags. To address this, we tagged each pathway component at one or both termini. As illustrated in Fig. 1B, the TclE precursor peptide consists of a 35-amino-acid N-terminal leader (which presumably recruits the biosynthetic machinery) and the C-terminal 14-amino-acid core peptide. Because this core peptide undergoes C-terminal decarboxylation as part of its maturation process, we did not attempt to fuse tags to this end of TclE. We were gratified to observe that fusions of glutathione S-transferase (GST) and hexahistidine (His6) to the N terminus of TclE result in the production of a bioactive compound (Fig. 4A), which suggests that such tags do not interfere with posttranslational processing and would be suitable for purifying TclE intermediates at various stages of maturation. We also tested each of the biosynthetic Tcl proteins for their ability to tolerate the 8-amino-acid Strep tag at their N or C termini. Based on the ability to produce biologically active micrococcin, all eight proteins are compatible with this tag in at least one orientation (Fig. 4B). Of the 12 Strep-tagged variants tested, only one (TclP-Strep) failed to support micrococcin biosynthesis. Taken together, these results demonstrate that every protein component in this pathway can be modified in a manner that would allow for affinity purification and detection with commercial reagents. These results also demonstrate the relative ease with which alterations can be made to individual components in this expression system without perturbing the rest of the pathway.
FIG 4.
Each component of the micrococcin biosynthetic pathway is amenable to affinity tagging. (A) Graphical representation of the affinity tags fused to the N terminus of the precursor peptide TclE. Spot-on-lawn assays were used to test the bioactivity resulting from tagged TclE. (B) Representations of the various locations in which Strep tags (ST) were fused to Tcl-processing proteins. The effect of these tags on bioactivity is also indicated; +, the fusion protein was functional; −, the fusion abolished bioactivity; NT, the tag was not tested.
Accumulation of MP1 versus MP2 is determined by the activity of the putative C-terminal dehydrogenase TclS.
The modularity of this micrococcin production system also allows one to rapidly perform specific tcl gene removal experiments. Of the eight processing proteins in this system, six have predictable roles in thiazole installation (TclI/TclJ/TclN), Ser/Thr dehydration (TclK), and macrocyclization (TclM). We preserved the two putative short-chain dehydrogenases (TclP and TclS) in our expression system with the assumption that they are somehow involved in the decarboxylation of the core peptide C-terminal Thr residue; however, defined roles for these enzymes have remained unclear. To address this, either tclP or tclS was removed from pLEGO by simple restriction digestion and religation. The resulting plasmids were integrated into a B. subtilis strain background harboring the GST-tclE cassette. Methanol extracts from these strains were then tested for bioactivity by spot-on-lawn assay (Fig. 5A) and the presence of thiopeptide compounds by electrospray ionization mass spectrometry (ESI-MS). Interestingly, the extract from the tclP deletion strain showed no bioactivity and no detectable thiopeptide product; however, the tclS deletion mutant produced a bioactive compound with a mass consistent with MP2 rather than MP1 (Fig. 5B). MP2 has a methyl ketone group at the C terminus, whereas MP1 is reduced to the alcohol at the same position. Therefore, we conclude that TclS is responsible for the conversion of MP2 to MP1 (Fig. 5C). TclP appears to play an essential role in micrococcin production, acting at some point prior to TclS in the biosynthetic pathway. Its presumed function as a short-chain dehydrogenase points to a possible role for TclP in the initial oxidative decarboxylation of the C-terminal Thr as illustrated in Fig. 5C.
FIG 5.
Analysis of the role of TclS in micrococcin C-terminal processing. (A) Spot-on-lawn assays with methanolic extracts from B. subtilis strains transformed with pLEGO and pLEGO derivatives lacking tclS and tclP. In these experiments, the pTclE plasmid had been modified to express the GST-TclE fusion. (B) ESI-MS analysis of purified extracts from pLEGO and pLEGOΔtclS strains. MP1: observed 1,144.2 [M+H]+, calculated 1,144.2 [M+H]+. MP2: observed 1,142.2 [M+H]+, calculated 1,142.2 [M+H]+. (C) Proposed roles of TclP and TclS in C-terminal processing of MP1 and MP2.
DISCUSSION
Heterologous thiopeptide expression serves several purposes. (i) It allows confirmation of the sufficiency of a given gene cluster to facilitate the production of a specific thiopeptide product. (ii) With the gene cluster cloned on a plasmid, it allows the researcher to probe the biosynthetic logic of the pathway with greater facility. (iii) It opens the door to the rational design and large-scale production of new thiopeptide analogs with improved pharmacological or bioactivity properties. Attempts to heterologously express diverse thiopeptide gene clusters have been met with mixed success (7, 15, 38–40). In most cases, Streptomyces species, such as Streptomyces lividans and Streptomyces coelicolor, have been employed. These expression hosts can be limiting due to low growth rate and potential incompatibility with genes derived from distantly related organisms, such as the low-GC firmicutes. It is remarkable that B. subtilis has rarely (if ever) been utilized for thiopeptide production. The B. subtilis strain used in this study is fast-growing and straightforward to genetically manipulate, with its natural competence and high recombinogenicity as well as the wealth of genetic tools developed by a large community of researchers. B. subtilis is particularly attractive for the production of thiopeptides, whose native producers (such as Bacillus, Staphylococcus, and Macrococcus spp.) share close evolutionary relatedness.
Several levels of potential incompatibility should be considered with respect to heterologous thiopeptide production. These include codon bias, transcriptional efficiency, self-intoxication by the bioactive product, and cofactor compatibility. Worthy of special mention is the tRNAGlu glutamate donor that is employed by the RiPP Ser/Thr dehydratase systems (TclK and TclL in the work presented here). Initially demonstrated for the two-domain NisB lantibiotic dehydratase (41), tRNAGlu is presumably the source of the glutamyl group used to activate Ser/Thr residues for subsequent dehydration in many RiPP pathways. In the in vitro reconstituted thiomuracin biosynthetic system (17), all thiopeptide biosynthetic components can be produced and purified from E. coli; however, the E. coli tRNAGlu was not a suitable substrate for Ser/Thr dehydration, so a hybrid system that utilized the E. coli aminoacyl tRNAGlu synthetase and Thermobispora bispora tRNAGlu was required. tRNAGlu is therefore a crucial consideration in transferring thiopeptide biosynthetic systems across bacterial taxa. In moving the micrococcin pathway from M. caseolyticus to B. subtilis, we were satisfied with the similar codon preferences of the two organisms, and we were able to override transcriptional incompatibilities by using promoters with predictable behavior in the producer strain. We were encouraged by the close similarity in the tRNAGlu sequence (a 1-bp difference located in the acceptor stem) (see Fig. S5 in the supplemental material), and successful micrococcin production in B. subtilis indicates that there was not a problem with tRNAGlu incompatibility.
Our functional genetic reconstruction of the M. caseolyticus micrococcin pathway in B. subtilis defines a minimally sufficient gene set for micrococcin biosynthesis. By preselecting a micrococcin-resistant variant of the producer strain, we could omit immunity functions from the engineered gene cluster. We also excluded genes for micrococcin export; thus, it is not surprising that micrococcin can be successfully extracted from B. subtilis cell pellets. It is not known whether our strains secrete residual micrococcin into the medium either passively or via endogenous efflux systems. Our data show that the minimal genetic requirement for conversion of the TclE precursor peptide to micrococcin (MP2) is the seven-gene set tclIJKLMNP, with tclS responsible for the reduction of MP2 to MP1. TclI, which we predict to be an Ocin-ThiF-like RRE-containing protein, is required for micrococcin production (not shown), TclJ and TclN constitute a putative thiazole installation module, TclK and TclL constitute a Ser/Thr dehydration system, TclM is predicted to catalyze the [4 + 2] cycloaddition required for macrocycle formation, and TclP is likely involved in oxidative decarboxylation of the core peptide C terminus.
The genetic refactoring described here enables unprecedented control over the properties of this system as demonstrated by our affinity tagging of every protein component and our ability to easily remove the processing enzymes TclP and TclS. These experiments constitute the beginning of an ongoing series of tests that are allowing us to dissect the entire micrococcin biosynthetic pathway in great detail. These experiments capitalize on the ability to genetically block the pathway and rapidly purify the affinity-tagged precursor peptide TclE at various stages of maturation. This approach may be employed to assess the biosynthetic consequences of numerous tcl gene deletions and point mutations, all of which are easily incorporated using standard molecular cloning techniques. We hope to be able to leverage this system to study physical interactions between pathway components, to explore how well the biosynthetic proteins tolerate core peptide sequence variation, and to create a system for combinatorial biosynthesis of never-seen-by-nature bioactive compounds.
Supplementary Material
ACKNOWLEDGMENTS
We thank Paul Price and J. C. Price for useful discussions and insights.
We acknowledge the Bacillus Genetic Stock Center for access to critical research tools.
This work was funded by a grant from the BYU College of Life Sciences Vaccine Royalties Fund (to R.A.R.), a BYU Graduate Studies Fellowship Award (to P.R.B.), and private donations through the UCSF Foundation (to S.M.M.).
Funding Statement
The research conducted at UCSF received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00396-16.
REFERENCES
- 1.Tegos GP, Hamblin MR. 2013. Disruptive innovations: new anti-infectives in the age of resistance. Curr Opin Pharmacol 13:673–677. doi: 10.1016/j.coph.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lewis K. 2013. Platforms for antibiotic discovery. Nat Rev Drug Discov 12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 3.Wright GD. 2015. Solving the antibiotic crisis. ACS Infect Dis 1:80–84. doi: 10.1021/id500052s. [DOI] [PubMed] [Google Scholar]
- 4.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach MA, Garavelli JS, Goransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Muller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, et al. 2013. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30:108–160. doi: 10.1039/C2NP20085F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Just-Baringo X, Albericio F, Alvarez M. 2014. Thiopeptide antibiotics: retrospective and recent advances. Mar Drugs 12:317–351. doi: 10.3390/md12010317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, Clardy J, Linington RG, Fischbach MA. 2014. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158:1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashi S, Ozaki T, Asamizu S, Ikeda H, Omura S, Oku N, Igarashi Y, Tomoda H, Onaka H. 2014. Genome mining reveals a minimum gene set for the biosynthesis of 32-membered macrocyclic thiopeptides lactazoles. Chem Biol 21:679–688. doi: 10.1016/j.chembiol.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Qu X, He X, Duan L, Wu G, Bi D, Deng Z, Liu W, Ou HY. 2012. ThioFinder: a web-based tool for the identification of thiopeptide gene clusters in DNA sequences. PLoS One 7(9):e45878. doi: 10.1371/journal.pone.0045878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burkhart BJ, Hudson GA, Dunbar KL, Mitchell DA. 2015. A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat Chem Biol 11:564–570. doi: 10.1038/nchembio.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunbar KL, Tietz JI, Cox CL, Burkhart BJ, Mitchell DA. 2015. Identification of an auxiliary leader peptide-binding protein required for azoline formation in ribosomal natural products. J Am Chem Soc 137:7672–7677. doi: 10.1021/jacs.5b04682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morris RP, Leeds JA, Naegeli HU, Oberer L, Memmert K, Weber E, LaMarche MJ, Parker CN, Burrer N, Esterow S, Hein AE, Schmitt EK, Krastel P. 2009. Ribosomally synthesized thiopeptide antibiotics targeting elongation factor Tu. J Am Chem Soc 131:5946–5955. doi: 10.1021/ja900488a. [DOI] [PubMed] [Google Scholar]
- 12.Wieland Brown LC, Acker MG, Clardy J, Walsh CT, Fischbach MA. 2009. Thirteen posttranslational modifications convert a 14-residue peptide into the antibiotic thiocillin. Proc Natl Acad Sci U S A 106:2549–2553. doi: 10.1073/pnas.0900008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelly WL, Pan L, Li C. 2009. Thiostrepton biosynthesis: prototype for a new family of bacteriocins. J Am Chem Soc 131:4327–4334. doi: 10.1021/ja807890a. [DOI] [PubMed] [Google Scholar]
- 14.Liao R, Duan L, Lei C, Pan H, Ding Y, Zhang Q, Chen D, Shen B, Yu Y, Liu W. 2009. Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem Biol 16:141–147. doi: 10.1016/j.chembiol.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelhardt K, Degnes KF, Kemmler M, Bredholt H, Fjaervik E, Klinkenberg G, Sletta H, Ellingsen TE, Zotchev SB. 2010. Production of a new thiopeptide antibiotic, TP-1161, by a marine Nocardiopsis species. Appl Environ Microbiol 76:4969–4976. doi: 10.1128/AEM.00741-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunbar KL, Mitchell DA. 2013. Revealing nature's synthetic potential through the study of ribosomal natural product biosynthesis. ACS Chem Biol 8:473–487. doi: 10.1021/cb3005325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hudson GA, Zhang Z, Tietz JI, Mitchell DA, van der Donk WA. 2015. In vitro biosynthesis of the core scaffold of the thiopeptide thiomuracin. J Am Chem Soc 137:16012–16015. doi: 10.1021/jacs.5b10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wever WJ, Bogart JW, Baccile JA, Chan AN, Schroeder FC, Bowers AA. 2015. Chemoenzymatic synthesis of thiazolyl peptide natural products featuring an enzyme-catalyzed formal [4 + 2] cycloaddition. J Am Chem Soc 137:3494–3497. doi: 10.1021/jacs.5b00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thibodeaux CJ, Ha T, van der Donk WA. 2014. A price to pay for relaxed substrate specificity: a comparative kinetic analysis of the class II lanthipeptide synthetases ProcM and HalM2. J Am Chem Soc 136:17513–17529. doi: 10.1021/ja5089452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melby JO, Dunbar KL, Trinh NQ, Mitchell DA. 2012. Selectivity, directionality, and promiscuity in peptide processing from a Bacillus sp. Al Hakam cyclodehydratase. J Am Chem Soc 134:5309–5316. doi: 10.1021/ja211675n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koehnke J, Bent AF, Zollman D, Smith K, Houssen WE, Zhu X, Mann G, Lebl T, Scharff R, Shirran S, Botting CH, Jaspars M, Schwarz-Linek U, Naismith JH. 2013. The cyanobactin heterocyclase enzyme: a processive adenylase that operates with a defined order of reaction. Angew Chem Int Ed Engl 52:13991–13996. doi: 10.1002/anie.201306302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su TL. 1948. Micrococcin, an antibacterial substance formed by a strain of Micrococcus. Br J Exp Pathol 29:473–481. [PMC free article] [PubMed] [Google Scholar]
- 23.Walker J. 1977. Total structure of the polythiazole-containing antibiotic micrococcin P. A 13C nuclear magnetic resonance study. J Chem Soc Chem Commun 20:706–708. [Google Scholar]
- 24.Ciufolini MA, Lefranc D. 2010. Micrococcin P1: structure, biology and synthesis. Nat Prod Rep 27:330–342. doi: 10.1039/b919071f. [DOI] [PubMed] [Google Scholar]
- 25.Acker MG, Bowers AA, Walsh CT. 2009. Generation of thiocillin variants by prepeptide gene replacement and in vivo processing by Bacillus cereus. J Am Chem Soc 131:17563–17565. doi: 10.1021/ja908777t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowers AA, Acker MG, Koglin A, Walsh CT. 2010. Manipulation of thiocillin variants by prepeptide gene replacement: structure, conformation, and activity of heterocycle substitution mutants. J Am Chem Soc 132:7519–7527. doi: 10.1021/ja102339q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowers AA, Acker MG, Young TS, Walsh CT. 2012. Generation of thiocillin ring size variants by prepeptide gene replacement and in vivo processing by Bacillus cereus. J Am Chem Soc 134:10313–10316. doi: 10.1021/ja302820x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowers AA, Walsh CT, Acker MG. 2010. Genetic interception and structural characterization of thiopeptide cyclization precursors from Bacillus cereus. J Am Chem Soc 132:12182–12184. doi: 10.1021/ja104524q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennallack PR, Burt SR, Heder MJ, Robison RA, Griffitts JS. 2014. Characterization of a novel plasmid-borne thiopeptide gene cluster in Staphylococcus epidermidis strain 115. J Bacteriol 196:4344–4350. doi: 10.1128/JB.02243-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilkinson DM, Jensen MM. 1987. Staphylococcosis of turkeys. 4. Characterization of a bacteriocin produced by an interfering Staphylococcus. Avian Dis 31:80–84. [PubMed] [Google Scholar]
- 31.Meyers CM, Jensen MM. 1987. Staphylococcosis of turkeys. 3. Bacterial interference as a possible means of control. Avian Dis 31:74–79. [PubMed] [Google Scholar]
- 32.Radeck J, Kraft K, Bartels J, Cikovic T, Dürr F, Emenegger J, Kelterborn S, Sauer C, Fritz G, Gebhard S, Mascher T. 2013. The Bacillus BioBrick box: generation and evaluation of essential genetic building blocks for standardized work with Bacillus subtilis. J Biol Eng 7:29. doi: 10.1186/1754-1611-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guerout-Fleury AM, Frandson N, Stragier P. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57–61. doi: 10.1016/S0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- 34.Miller JH. 1972. Assay for β-galactosidase. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 35.Chang AC, Cohen SN. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol 134:1141–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen HD, Nguyen QA, Ferreira RC, Ferreira LC, Tran LT, Schumann W. 2005. Construction of plasmid-based expression vectors for Bacillus subtilis exhibiting full structural stability. Plasmid 54:241–248. doi: 10.1016/j.plasmid.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 37.Porse BT, Cundliffe E, Garrett RA. 1999. The antibiotic micrococcin acts on protein L11 at the ribosomal GTPase centre. J Mol Biol 287:33–45. doi: 10.1006/jmbi.1999.2600. [DOI] [PubMed] [Google Scholar]
- 38.Young TS, Walsh CT. 2011. Identification of the thiazolyl peptide GE37468 gene cluster from Streptomyces ATCC 55365 and heterologous expression in Streptomyces lividans. Proc Natl Acad Sci U S A 108:13053–13058. doi: 10.1073/pnas.1110435108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malcolmson SJ, Young TS, Ruby JG, Skewes-Cox P, Walsh CT. 2013. The posttranslational modification cascade to the thiopeptide berninamycin generates linear forms and altered macrocyclic scaffolds. Proc Natl Acad Sci U S A 110:8483–8488. doi: 10.1073/pnas.1307111110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flinspach K, Kapitzke C, Tocchetti A, Sosio M, Apel AK. 2014. Heterologous expression of the thiopeptide antibiotic GE2270 from Planobispora rosea ATCC 53733 in Streptomyces coelicolor requires deletion of ribosomal genes from the expression construct. PLoS One 9(3):e90499. doi: 10.1371/journal.pone.0090499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ortega MA, Hao Y, Zhang Q, Walker MC, van der Donk WA, Nair SK. 2015. Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature 517:509–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.