ABSTRACT
Two-component systems (TCS) that employ histidine kinases (HK) and response regulators (RR) are critical mediators of cellular signaling in bacteria. In the model cyanobacterium Synechococcus elongatus PCC 7942, TCSs control global rhythms of transcription that reflect an integration of time information from the circadian clock with a variety of cellular and environmental inputs. The HK CikA and the SasA/RpaA TCS transduce time information from the circadian oscillator to modulate downstream cellular processes. Despite immense progress in understanding of the circadian clock itself, many of the connections between the clock and other cellular signaling systems have remained enigmatic. To narrow the search for additional TCS components that connect to the clock, we utilized direct-coupling analysis (DCA), a statistical analysis of covariant residues among related amino acid sequences, to infer coevolution of new and known clock TCS components. DCA revealed a high degree of interaction specificity between SasA and CikA with RpaA, as expected, but also with the phosphate-responsive response regulator SphR. Coevolutionary analysis also predicted strong specificity between RpaA and a previously undescribed kinase, HK0480 (herein CikB). A knockout of the gene for CikB (cikB) in a sasA cikA null background eliminated the RpaA phosphorylation and RpaA-controlled transcription that is otherwise present in that background and suppressed cell elongation, supporting the notion that CikB is an interactor with RpaA and the clock network. This study demonstrates the power of DCA to identify subnetworks and key interactions in signaling pathways and of combinatorial mutagenesis to explore the phenotypic consequences. Such a combined strategy is broadly applicable to other prokaryotic systems.
IMPORTANCE Signaling networks are complex and extensive, comprising multiple integrated pathways that respond to cellular and environmental cues. A TCS interaction model, based on DCA, independently confirmed known interactions and revealed a core set of subnetworks within the larger HK-RR set. We validated high-scoring candidate proteins via combinatorial genetics, demonstrating that DCA can be utilized to reduce the search space of complex protein networks and to infer undiscovered specific interactions for signaling proteins in vivo. Significantly, new interactions that link circadian response to cell division and fitness in a light/dark cycle were uncovered. The combined analysis also uncovered a more basic core clock, illustrating the synergy and applicability of a combined computational and genetic approach for investigating prokaryotic signaling networks.
INTRODUCTION
Bacterial two-component systems (TCS) are ubiquitous signaling modules consisting of at least one histidine kinase (HK) and one response regulator (RR) (1), which transduce signals via transfer of a phosphate group from the HK to a receiver domain on the RR. Over the years, studies have expanded the common paradigm of TCS signaling via phosphotransfer between single HK-RR pairs to encompass a wide network of interconnected pathways, integrating environmental inputs with several other cellular processes (2). The model cyanobacterium Synechococcus elongatus PCC 7942 harbors 37 TCS proteins, including hybrid kinases that combine functions of an HK and an RR (3). Although many interactions between TCS component proteins have been characterized, the functional connections between two-component systems remain largely unclear.
One of the most fascinating questions in biological signaling is the transduction of external time information to downstream cellular processes. Cyanobacteria are, thus far, unique among prokaryotes in possessing a robust circadian clock, an autonomous 24-hour oscillator (comprising the KaiA, KaiB, and KaiC proteins) that generates gene transcription rhythms synchronized with the rotation of the earth (4). The KaiC-interacting HKs CikA (circadian input kinase A) and SasA (Synechococcus adaptive sensor A) produce oscillations in activity of the RR RpaA (regulator of phycobilisome association A), which controls a wide regulon (5). CikA acts as a phosphatase on RpaA, an activity dependent on association with the KaiBC complex (6). CikA is an unusual multidomain kinase that functions in clock resetting and cell division in addition to its role in clock output (7). The circadian clock prevents cell division during a portion of the night period, a phenomenon termed gating (8). A cikA-null mutant has an extended duration of the “gate closure,” resulting in elongated cells (9); the penetrance of this phenotype is suppressed at moderate to high light intensities. Closing of the gate is correlated with high ATPase activity of KaiC (10) and peak output activity of the Kai complex (11), activities that are suppressed by deletion of sasA or rpaA, indicating that a negative signal from the clock regulates the cell division checkpoint.
Recent investigation has revised the view of the circadian clock from a linear model in which proteins play distinct input, oscillatory, and output functions to one wherein input and output functions themselves are products of integrated network dynamics between clock proteins and associated regulatory systems (12). Cross talk between the response regulator RpaB (regulator of phycobilisome association B) and RpaA demonstrated integration between the SasA-RpaA and environmentally sensitive NblS (nonbleaching sensor)-RpaB TCS (13). However, cross talk between other TCS and the SasA-RpaA circadian TCS remained unexplored, due in part to the complexity of the network and the likelihood that signal transfer pathways are redundant. To gain a more comprehensive understanding of the network, we utilized direct-coupling analysis (DCA), a sequence analysis method designed to uncover pairs of coevolving protein residues (14). DCA has been applied to coevolving interacting interfaces (15, 16), making it a useful tool to predict interactions between TCS proteins in S. elongatus with the goal of identifying important subnetworks on which to focus biological efforts. Major advantages of S. elongatus PCC 7942 include a low occurrence of gene duplication, several measurable phenotypes related to known clock components (light/dark sensitivity, cell length, protein expression via luciferase activity, and measurement of phosphorylation state of the RRs RpaA and RpaB), and an available library of gene knockouts (17). These features make the S. elongatus circadian clock an excellent paradigm for testing and validating sequence analysis tools to improve our systems-level understanding.
We used a score derived from DCA to predict a high probability of interaction specificity between the clock HKs SasA and CikA and the phosphate-sensing RR SphR (Synechococcus phosphate regulator). DCA between SphR/RpaA and HKs also revealed a mutual predicted interaction with the PAS/GAF domain HK Synpcc7942_0480 (here, circadian input kinase B, or CikB). Deletion of the gene for CikB (cikB) in a background lacking both SasA and CikA resulted in phenotypes similar to those of an rpaA-null mutant, suggesting that CikB kinase activity in vivo acts on RpaA. These results demonstrate the predictive power of DCA and reveal novel insights into the network architecture that connects the circadian clock to the wider signaling network in S. elongatus.
MATERIALS AND METHODS
Bacterial strains and culture.
The Synechococcus elongatus PCC 7942 strains used in this study are listed in Table 1 and were grown as previously described (13) with appropriate antibiotics. Samples for cell elongation assays and corresponding immunoblotting were grown under low light (10 μE/m2 s) in liquid BG-11 medium for 21 days. Strains for other assays were maintained under medium (100 μE/m2 s) or high (300 μE/m2 s) light conditions.
TABLE 1.
Synechococcus strains used in this study
| Strain name | Genotype | Antibiotic resistance | Reference |
|---|---|---|---|
| AMC541 | WT PkaiBC::luc | Cm | 41 |
| AMC1275 | PkaiBC::luc, sasA in-frame deletion | Cm | 42 |
| AMC2415 | PkaiBC::luc cikA::Gmr | Cm Gm | This study |
| AMC2416 | PkaiBC::luc sphR | Cm Nt | This study |
| AMC2417 | PkaiBC::luc sasA cikA | Cm Gm | This study |
| AMC2426 | PkaiBC::luc sphR sphS | Cm Nt Km | This study |
| AMC2385 | PkaiBC::luc sasA cikA sphR | Cm Gm Nt | This study |
| AMC2418 | PkaiBC::luc sasA cikA sphS | Cm Gm Km | This study |
| AMC2419 | PkaiBC::luc cikB | Cm Km | This study |
| AMC2428 | PkaiBC::luc sasA cikA sphR cikB | Cm Gm Nt Km | This study |
| AMC2429 | PkaiBC::luc sasA cikA sphR sphS | Cm Gm Nt Km | This study |
| AMC2425 | PkaiBC::luc sasA cikA cikB | Cm Gm Km | This study |
| AMC2420 | PkaiBC::luc cikA cikB | Cm Gm Km | This study |
| AMC2421 | PkaiBC::luc sasA cikA srrA | Cm Gm Km | This study |
| AMC2422 | PkaiBC::luc sasA cikA rr2466 | Cm Gm Km | This study |
| AMC2158 | WT PkaiBC::luc | Nt | This study |
| AMC2427 | PkaiBC::luc hk1872::Cmr | Nt Cm | This study |
| AMC2424 | PkaiBC::luc hk1872::Cmr cikA::Gmr | Nt Cm Gm | This study |
| AMC2423 | PkaiBC::luc hk1872::Cmr cikA::Gmr sasA::Kmr | Nt Cm Gm Km | This study |
| AMC2433 | PkaiBC::luc cikA cikB Ptrc::FLAG-CikB | Cm Gm Km Sp Sm | This study |
Plasmid and strain construction.
The plasmids used in this study are listed in Table 2. Transposon insertion strains were constructed by transformation of cyanobacterial strains with gene-specific cosmids from the S. elongatus PCC 7942 unigene set (UGS) library (17) by a protocol described previously (18). Gene replacement plasmids were designed using the CYANO_VECTOR server (http://golden.ucsd.edu/CyanoVECTOR/) and assembled via an isothermal assembly reaction using an Invitrogen GeneArt seamless cloning kit (Thermo Fisher, Carlsbad, CA). A 3× FLAG tag to generate FLAG-tagged CikB was amplified from p3XFLAG_CMV_14 (Sigma, St. Louis, MO) using primers FLAG frw (AAAAGAATTCATGGACTACAAAGACCATGACGGTG) and FLAG rev (AAAAGGATCCCTTGTCATCGTCATCCTTGTAGTC). Plasmid cloning was carried out in Escherichia coli strain DH5α using standard techniques.
TABLE 2.
Plasmids used in this study
| Plasmid name | Genotype | Antibiotic resistance | Reference or source |
|---|---|---|---|
| AM2241 | cikA::Gmr | Gm | This study |
| AM5172 | sphR::NAT1 | Nt | This study |
| AM5307 | Ptrc-FLAG-CikB | Sp Sm | This study |
| 3XFLAG_CMV_14 | 3×FLAG vector | Ap | Sigma |
| 8S10-E2 | Tn5 insertion in sasA | Km | 17 |
| 8S42-C8 | Tn5 insertion in sphS | Km | 17 |
| 8S17-H4 | Tn5 insertion in cikB | Km | 17 |
| 8S35-F4 | Tn5 insertion in srrA | Km | 17 |
| 8S35-P4 | Tn5 insertion in rr2466 | Km | 17 |
| 8G3-D2 | Tn5 insertion in hk1872 | Cm | 17 |
Bioluminescence assays.
Bioluminescence from firefly luciferase reporter strains was measured on a TopCount microplate luminescence counter (PerkinElmer, Waltham, MA) as described previously (19). Data were analyzed and graphed in Microsoft Excel using the Biological Rhythms Analysis Software System (BRASS) (http://millar.bio.ed.ac.uk/PEBrown/BRASS/BrassPage.htm).
DCA.
In order to calculate a score for interaction specificity between HKs and RRs, we performed direct-coupling analysis (DCA) (14, 20) on a collection of sequences of HK and RR partners. First, we focused on the multiple-sequence alignments (MSA) of the HK and RR, HiskA (PF00512) and Response_reg (PF00072) Pfam families, respectively. With the exception of CikA, which possesses a noncanonical pseudoreceiver domain, sequences of hybrid kinases, harboring both histidine kinase and receiver domains, were excluded from the analysis. We constructed our database of TCS partners from diverse species by using a genetic locus proximity criterion (i.e., chromosomal adjacency), obtaining more than 30,000 HK-RR partner sequences. Each sequence in our database of partners consisted of the concatenated MSA of an HK and an RR of the form “sequence” = (s1, s2, … sL−1, sL), where L is the total length of the concatenated HK-RR sequence. Next, we inferred a global statistical model that captures the coevolved interdomain residue interactions using DCA. We then applied a formulation similar to the one described previously (15), where a direct information score (DIS) is computed for each HK-RR pair of sequences by summing all the interdomain coupling scores of the amino acid pairs located at the interface using a representative HK-RR structure (21) as a template aligned to the sequences of interest. The equation quantifying the DIS score has the form
where si is the amino acid at position i belonging to the HisKA domain and sj is the amino acid at position j belonging to the Response_reg domain. The direct (disentangled) amino acid pair probability distribution of DCA obtained is noted as and ϴ is a step function used to include only residue pairs within a cutoff distance X = 12 Å in the representative structure. We computed DISs for all HK-RR pairs in S. elongatus that have a HisKA domain, except hybrid kinases. DISs were computed for a generic (“null”) model, where HK and RR sequences in the database of TCS partners had randomized HK-RR pairings, to capture the generic properties of HK-RR interaction. Next, a resulting specific DIS was obtained by subtracting the generic DIS from the DISs that were constructed from a model using the genetic locus adjacency criterion, obtaining a model that captured the determinants of interaction specificity in the TCS. The resulting specific DIS were used to rank HK-RR interactions and create a master matrix of specific DISs. Calculations were performed using custom-made data manipulation programs and Matlab scripts. Pfam sequences were obtained from version 27 (22).
Microscopy.
Cultures of S. elongatus were grown at a light intensity of 10 μE m−2 s−1 for 3 weeks in liquid BG-11 medium without added antibiotics. Samples of cultures (0.5 ml) were condensed by microcentrifugation (5,000 × g, 2.5 min), and supernatant was removed. Cells were resuspended in 30 to 50 μl medium, and 2 to 5 μl of condensed culture was spotted on 1% (wt/vol) agarose in filtered BG-11 medium on a deep-well slide and coverslipped. Cells were imaged using a DeltaVision inverted epifluorescence microscope (Applied Precision, Issaquah, WA). Images were captured using a CoolSnap HD charge-coupled device (CCD) camera (Photometrics, Tucson, AZ).
Immunoblotting.
Cultures of S. elongatus were grown with low or medium light (10 μE m−2 s−1 or 100 μE m−2 s−1, respectively) and sampled as described previously (23). Phos-tag SDS-PAGE was performed, and blots were probed with RpaA, RpaB, or KaiC antiserum or anti-FLAG antiserum according to the protocol described previously (13).
Image analysis.
Densitometry was performed, and cell length measurements were obtained from TIFF images using ImageJ software (National Institutes of Health) (24). Statistical analysis was carried out using Microsoft Excel.
Figure preparation.
Graphs were prepared using Excel and PowerPoint (Microsoft, Redmond, WA) and Matlab (MathWorks, Natick, MA), and final figures were prepared using Adobe Illustrator (Adobe Systems, San José, CA).
RESULTS AND DISCUSSION
DCA predicts coevolution between CikA/SasA and the response regulator SphR.
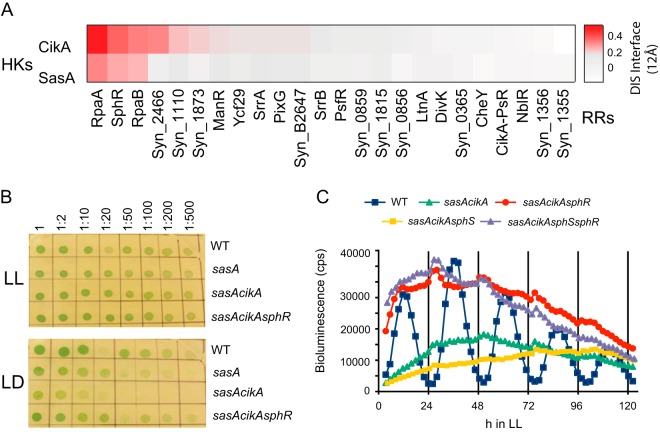
In order to validate the utility of DCA for network interactions, we computed coevolutionary coupling scores between the S. elongatus sequences of the CikA and SasA HK proteins with the family of cyanobacterial response regulators homologous to all S. elongatus RRs using sequences available in the Pfam database (http://pfam.xfam.org). As expected, RpaA, the cognate response regulator of both CikA and SasA, scored highly on the direct interaction scale to both HKs (Fig. 1A). The RR SphR, a known component of the phosphate signaling pathway (25) and homolog of Escherichia coli PhoB (26), also had a high interaction specificity score. Although the cognate HK of SphR has been identified as SphS, others have shown that SphR accepts phosphate from CikA and SasA in an in vitro phosphotransfer assay (6). Little background noise was apparent among the pairwise interaction coupling scores, suggestive that the DCA score is predictive of true cognate interactions between the protein pairs.
FIG 1.
SphR is a component of the clock interaction network. (A) Direct-coupling analysis showing different degrees of specificities between CikA/SasA and response regulators. (B) sphR inactivation suppresses the light-dark (LD) sensitivity phenotype of a sasA cikA null mutant. Serial dilutions of strains were spotted on solid BG-11 medium and grown at 100 μE m−2 s−1 for 5 days under conditions of constant light (LL) or 12 h of light and 12 h of dark (LD). (C) Bioluminescence traces of kaiBC-luc promoter activity over several days in constant light. The sasA cikA sphR triple mutant strain displays high promoter activity relative to that of the sasA cikA strain. Inactivation of sphS, encoding the canonical HK partner of SphR, does not affect kaiBC-luc activity in sasA cikA or sasA cikA sphR backgrounds.
To ascertain phenotypic effects of perturbing the interaction between SasA/CikA and SphR in vivo, the sphR coding sequence was replaced with a nourseothricin resistance cassette via homologous recombination in backgrounds in which sasA, cikA, or both sasA and cikA had been deleted. As expected from previous surveys of RR genes in S. elongatus, a sphR single null mutant strain had wild-type (WT) circadian rhythms, as assessed via transcriptional activity from a kaiBC-luciferase reporter (see Fig. S1 in the supplemental material). Both sasA and sasA cikA mutant strains exhibit a growth defect under a light-dark (LD) regime, a metabolic consequence of misregulation of genes that require phosphorylated RpaA for activation (27). Deletion of sphR largely rescued the LD defect of the sasA cikA null strain (Fig. 1B), and the sasA cikA sphR strain showed constitutively high baseline kaiBC-luc transcription, which consistently correlates with high phosphorylated RpaA (28). Expression of kaiBC correlates with that of the genes shown to be important for dark survival (27), consistent with the LD rescue. Insertional inactivation of the sphS gene, encoding the cognate HK of SphR, did not affect transcriptional activity of the kaiBC promoter in sasA cikA or sasA cikA sphR backgrounds (Fig. 1C), suggesting that the high transcriptional output state in the absence of SphR and SasA/CikA is not directly attributable to activity of the SphS-SphR system but rather is attributable to specific disruption of SphR interactions with the SasA/CikA/RpaA system.
Deletion of sphR greatly extends cell length in a sasA cikA double mutant background.
Another known phenotype of deleting CikA is cell elongation under low-light conditions (9). Thus, to test for potential interactions between this circadian-related HK and other RRs, we assessed cell lengths in a WT S. elongatus PCC 7942 strain and a series of deletion mutants. Deletion of sphR or sasA alone did not alter the cell elongation phenotype of cikA-null cells; however, deletion of sphR in a sasA cikA double mutant background greatly enhanced the severity of cell elongation (Fig. 2A and B). sasA cikA sphR triple mutants exhibited a much wider range of cell lengths than sasA cikA cells, having an average length four times that of sasA cikA cells and 15 times that of WT cells (Fig. 2B). Phosphate limitation has been shown to result in hyperelongated cells in aged (8-week-old) cultures (29). In principle, the hyperelongation of sasA cikA sphR cells could represent an exaggerated Pi deprivation response; however, cell elongation was not observed in the single sphR deletion strain (see Fig. S2 in the supplemental material). Thus, we conclude that the phenotype is more likely produced through a more global network perturbation.
FIG 2.
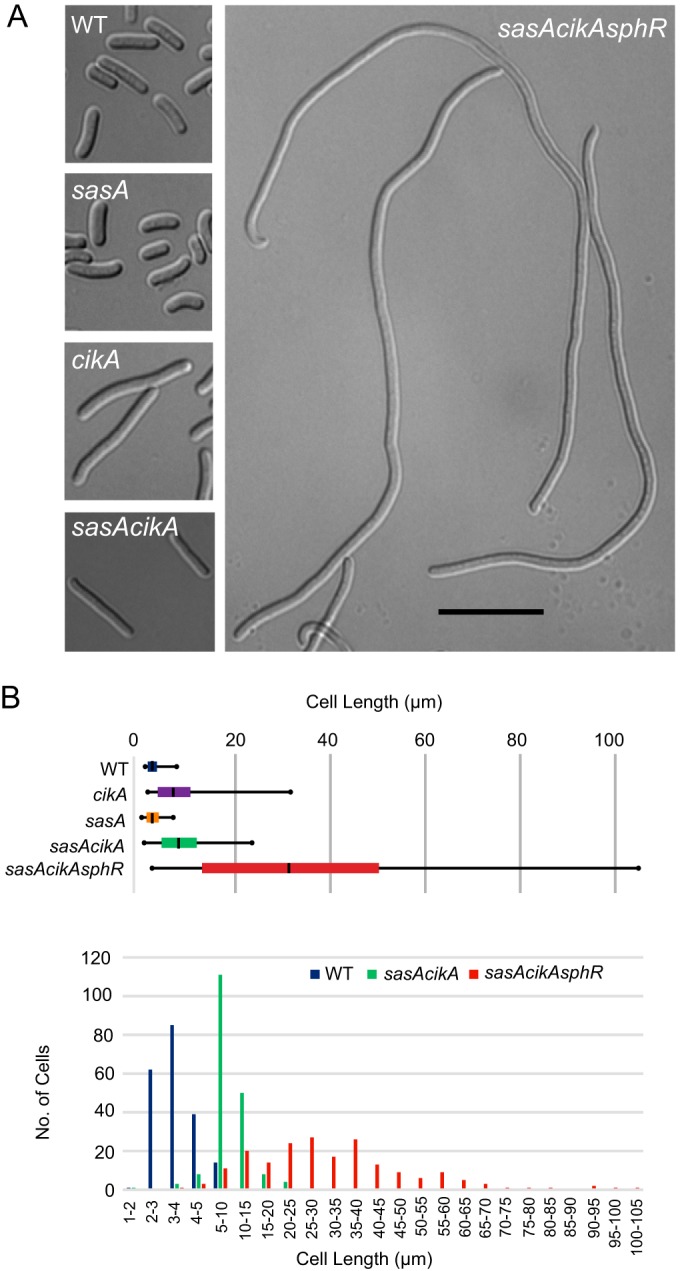
Deletion of sphR intensifies the cell elongation defect of a sasA cikA null mutant. (A) Representative bright-field micrographs of WT, sasA, cikA, sasA cikA, and sasA cikA sphR cells grown under low-light conditions for 21 days. (B) Quantitation of average cell lengths under low-light growth. The average length of sasA cikA sphR cells was 10 times the WT length under the conditions studied, and the cells exhibited a broad range of lengths, with the longest cells exceeding 100 μm.
RpaA and SphR interact genetically with CikB.
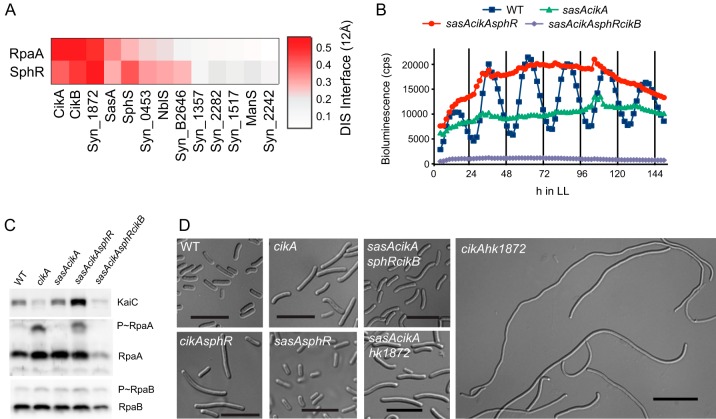
To determine additional possible interaction partners of RpaA and SphR, a coevolution score was calculated between these RRs and sequences of HKs in S. elongatus. Intriguingly, DCA predicted a high probability of coevolution of RpaA with CikB (Fig. 3A), an HK not previously mapped to a specific pathway. If CikB is responsible for hyperstimulation of RpaA and extension of the cell division gate when the RpaA cognate HKs (CikA and SasA) are genetically ablated, knockout of cikB might be expected to revert this phenotype. We constructed strains in which cikB was inactivated via Tn5 insertion and examined cell length and bioluminescence. Inactivation of cikB in a cikA-null background suppressed the cell elongation phenotype (Fig. 3D) in a similar manner as did inactivation of rpaA (see Fig. S3 in the supplemental material).
FIG 3.
CikB is responsible for RpaA activation in the absence of SasA and CikA. (A) Diagram showing direct coupling between RpaA/SphR and HKs in Synechococcus elongatus PCC 7942. The direct interaction score is displayed for the HK-RR interface. (B) Deletion of cikB abolishes kaiBC transcription in a sasA cikA background. (C) Immunoblots of low-light-grown strains probed with KaiC, RpaA, and RpaB antisera. RpaA phosphorylation and KaiC levels are elevated in the sasA cikA sphR strain, whereas RpaB phosphorylation levels are unchanged. (D) Representative bright-field micrographs of cells from strains grown under low-light conditions for 21 days. Deletion of cikB restores cell length in the sasA cikA sphR background. Knockout of hk1872 enhances the cell elongation phenotype of a cikA-null strain but does not affect cell length in a sasA cikA background. Scale bar, 10 μm.
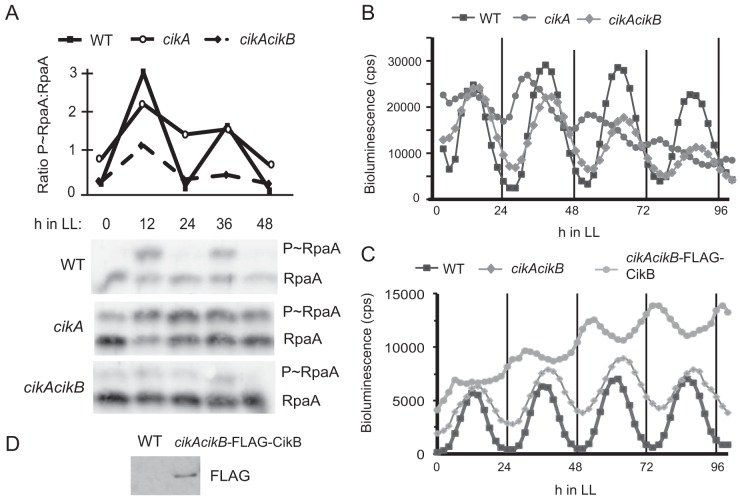
Astonishingly, interruption of the cikB gene in a cikA-null mutant restored near-WT amplitude and period of circadian transcriptional rhythms from a PkaiBC-luc reporter (Fig. 4B and Table 3). This finding is consistent with the demonstration that specific point mutations in SasA can enable the circadian gene expression network to operate normally in the absence of CikA (12). Ectopic expression of FLAG-tagged CikB from an uninduced trc promoter in a cikA cikB background was sufficient to revert expression rhythms to a cikA-null-like phenotype (Fig. 4C and D and Table 3), supporting the hypothesis that CikB activity modulates the clock output. Furthermore, the cikA cikB double mutant exhibited phase shifting in response to a dark pulse (Fig. 5), indicating restoration of environmental input functions. These results concur with those of a previous study that showed that the phase of the Kai oscillator can be reset directly by oxidized quinone (30), and they demonstrate that neither CikA nor a potential sensing function of CikB is necessary for resetting of the core oscillator in vivo. These results demonstrate that the KaiABC oscillator and SasA-RpaA output system together are sufficient to maintain WT rhythms and respond to external input cues in vivo when freed from other network interactions. The data also support a model in which the metabolic state of the cell, rather than the signaling system itself, is fundamental to determining the phase resetting response.
FIG 4.
Inactivation of CikB restores rhythms and resetting in a CikA knockout background. (A) Quantitation of RpaA phosphorylation and representative immunoblots of WT, cikA, and cikA cikB strains sampled every 12 h for 2 days and probed with RpaA antisera. RpaA phosphorylation is decreased in a cikA cikB strain compared to a cikA-null strain. (B and C) Bioluminescence traces showing transcriptional activity of a PkaiBC-luc reporter over several days after entrainment to three 12-h light-dark (LD) cycles. (B) Inactivation of CikB restores the amplitude and period of kaiBC transcription in a cikA-null background. (C) Expression of FLAG-tagged CikB in a cikA cikB background reverts rhythms to a cikA-null phenotype. (D) Immunoblot showing in vivo expression of FLAG-tagged CikB in a cikA cikB background.
TABLE 3.
Free-running periods
| Strain | Avg period (h) ± SD | Difference from WT period (h) |
|---|---|---|
| WT | 24.8 ± 0.34 | |
| cikA mutant | 22.7 ± 0.67 | −2.06 |
| cikB mutant | 24.8 ± 0.04 | 0 |
| cikA cikB mutant | 25.6 ± 0.22 | +0.82 |
| cikA cikB FLAG-CikB mutant | 23.3 ± 0.05 | −1.5 |
| sasA mutant | 23.6 ± 0.18 | −1.21 |
FIG 5.
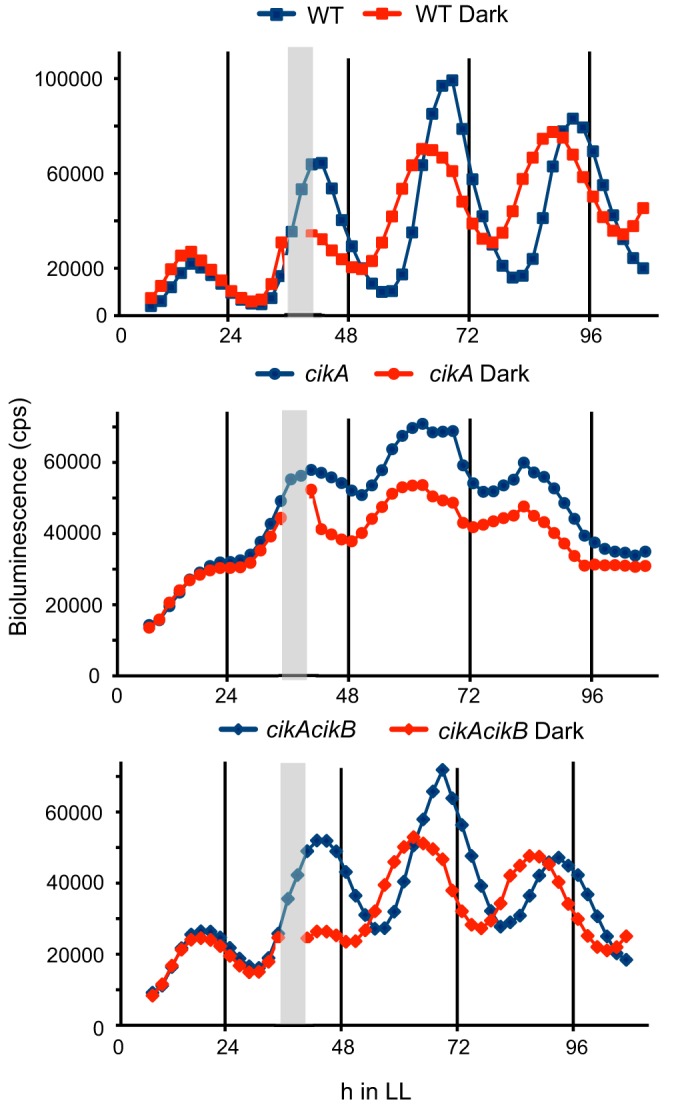
Inactivation of CikB restores resetting ability in a cikA knockout background. Bioluminescence traces of PkaiBC-luc reporter strains are shown. Inactivation of CikB restores the resetting ability of a cikA-null strain following a 5-h dark pulse, represented by gray bars, administered at 32 h after entry into constant light.
Inactivation of cikB in sasA cikA sphR and sasA cikA backgrounds also changed baseline kaiBC transcriptional activity from high to low (Fig. 3B; see Fig. S1 in the supplemental material), a level characteristic of an rpaA deletion strain. Phosphorylated RpaA was not detectable in extracts from sasA cikA sphR cikB cells (Fig. 4A), supporting the hypothesis that CikB is responsible for RpaA phosphorylation in the absence of SasA and CikA. Notably, levels of phosphorylated and nonphosphorylated RpaB remained similar among WT and elongated mutant strains, suggesting that RpaB regulation is not contributing to the cell division phenotype (Fig. 3C). The major cell division effect exhibited in the sasA cikA sphR strain likely does not result solely from an RpaA-mediated transcriptional mechanism, because rpaA mutants have normal cell lengths, and expression of a constitutively active RpaA results in only minor effects on cell length (5).
Although high transcription of kaiBC has been linked to cell elongation (10), the lack of the CikA/SasA output system in the elongated sasA cikA sphR cells makes it unlikely that hyperstimulated KaiC is directly influencing the cell division checkpoint under these conditions. The severity of the cell division phenotypes is comparable to those for ftn2 and ftn6 mutants, which are defective in cell division genes that encode components of the division machinery (9). In a yeast two-hybrid analysis, CikB interacted with a receiver protein annotated as a homolog of the cell division regulator DivK (3), but divK mutants did not exhibit cell division defects (see Fig. S2 in the supplemental material). Thus, it is likely that the link between the SasA-CikA-SphR pathway and cell division involves a connection not yet identified. SphR, and to a lesser extent RpaA, ranked highly in direct interaction scores with HK Synpcc7942_1872 (Fig. 3A), part of an operon with RR1873 and of unknown function. Insertional inactivation of the hk1872 gene alone resulted in moderate cell elongation under low-light culture conditions (see Fig. S2 in the supplemental material). Cell hyperelongation was still observed in a cikA hk1872 double-knockout strain, whereas knockout of sasA in the cikA hk1872 background suppressed the hyperelongation phenotype, resulting in cell lengths comparable to those of a cikA single mutant (Fig. 3D). Deletion of the hk1872 gene did not affect characteristic transcription rhythms of a cikA-null strain (see Fig. S4 in the supplemental material).
Insights into network architecture.
To obtain a more complete picture of the overall HK-RR network, the complete DCA matrix of S. elongatus HK and RR couplings was calculated. Strikingly, the circadian HK-RR system (CikA-SasA-RpaA) is part of a strongly coupled subnetwork involving three noncircadian HKs (CikB, HK1872, and SphS) and three RRs (SphR, RpaB, and RR2466) (Fig. 6). This subnetwork connects key circadian, nutrient, and environmental sensing regulators within the broader signaling network. The subnetwork organization can be perceived genetically by observing effects on transcriptional output: deletion of the RR2466 or SrrA, less highly coevolved with SasA and CikA, in a sasA cikA background does not affect transcription of the clock-controlled kaiBC promoter (see Fig. S1A in the supplemental material), in contrast to the dramatic transcriptional effects of deleting RpaA and SphR in a sasA cikA background. Although the possibility that the phenotypes observed are due to aberrant signaling cannot be ruled out, the high correlation of direct interaction scores with functional specificity in other model bacterial systems in which TCS have been well studied (31) favors a network model of functional connections. As our DCA model was constructed from a database of HK-RR partners which interact, the coevolution captured in our models reflects positive design; thus, if an HK-RR pair is found to highly coevolve based on our model, it is more likely to correspond to functional interaction probability. Further, if an interaction (e.g., phosphotransfer) exists under knockout conditions, the interaction should also exist in the presence of all proteins (no knockout). The most highly coevolved HK-RR pairs include CikB and HK1872, with the RRs RpaA and SphR, and RpaB, with the phosphate-sensing HK SphS. A high degree of coevolution between SphR and the TCS Synpcc7942_1872/Synpcc7942_1873 may imply that this HK-RR pair functions in micronutrient sensing and integrates with phosphate signaling in S. elongatus; likewise, the coevolution between HK1872 and SphS with RpaB is consistent with the role of RpaB as an essential transcription factor that responds to and integrates a variety of environmental cues (32). HK1872 possesses likely membrane-spanning domains, similar to ion-sensing HKs such as the manganese-sensing HK ManS (33, 34). It is possible that HK1872 senses a cation such as Zn2+ or Cu2+, but the striking light-dependent cell length defects of a cikA hk1872 mutant raise the possibility that the 1872/1873 TCS is directly integrated with cell division regulation. RR2466 is a homolog of NrrA, a nitrogen-regulated transcription factor that controls nitrogen and carbon balance in Synechocystis sp. strain PCC6803 (35). As an HK partner of NrrA has not yet been identified, our results support the hypothesis that NrrA interacts with multiple HK partners and that integration of input signals is a component of its function in metabolic regulation. Our DCA results also agree with in vivo studies that demonstrated the HK NblS interacting with both RpaB and SrrA in a branched pathway (36, 37).
FIG 6.
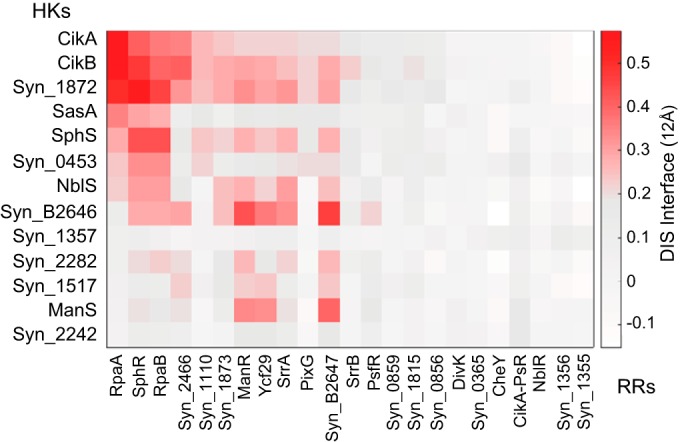
Direct-coupling matrix of two-component system proteins in S. elongatus. DIS specific scores showing direct correlations of amino acid sequences of histidine kinases (y axis) versus response regulators (x axis) are shown. Higher positive coevolutionary scores are represented by darker red shading, whereas gray represents low and white denotes negative coevolutionary coupling. Proteins without assigned names are designated by Syn_XXXX, abbreviated from numbered notation in the JGI genome database.
Identification of the core circadian oscillator.
DCA is a recent computational technique that takes advantage of the ever-increasing availability of sequence data, which is particularly abundant for the readily annotated TCS sequences. DCA has been used to characterize protein-protein interaction interfaces (15) and in this work as a convincing predictive tool for TCS specificity. As such, DCA provides a high-throughput computational method to identify potential subnetworks of interacting proteins within a vastly larger network of combinatorial possibilities. When integrated with experimental measurements, as in this study, it can reveal interactions important for key biological processes that would not be predicted a priori: DCA predicted the known HK-RR circadian interactions and several new interactions that we confirmed through biological experiments. Specifically, DCA revealed a more basic core circadian system, identified HKs and RRs that connect the circadian system to biological output, and changed our view of the role of CikA in phase resetting.
Surprisingly, we found that removal of CikB allowed robust circadian rhythms to persist in vivo via the SasA-RpaA system, in the absence of the phosphatase or redox-sensing functions of CikA. These results support a model wherein KaiABC-SasA-RpaA comprises the core circadian network, which is balanced by secondary interactors CikA and CikB. CikB possesses predicted PAS and GAF domains, implying a possible function as a direct light sensor that informs the circadian clock. The findings that WT oscillations and resetting occur in the absence of CikA led us to reevaluate the interpretation of the cikA knockout mutant. In the single deletion mutant, the alterations in the amplitude, period, and phase-resetting ability were attributed to the loss of necessary functions of the CikA protein. The results of this study suggest that CikA activity more likely offsets the activity of CikB, as the phenotypes of a cikA-null mutant are suppressed when CikB is removed together with CikA and restored when CikB is expressed in the cikA cikB double mutant; note that the single cikB deletion does not perturb the circadian clock (see Fig. S1 in the supplemental material). Thus, the alterations in the cikA-null mutant are due to the interference by CikB on circadian properties.
The power of DCA to predict signaling protein candidates worthy of analysis makes it a particularly useful methodology for the study of organisms that do not yet have tractable experimental genetics for extensive mutant hunts. Although this integrated computational and experimental approach proved to be a useful tool to disentangle molecular interactions in the circadian signaling system of S. elongatus, its applicability is much broader. The ability to infer function partners in a complex signaling network can be of use for understanding the stress response of Alphaproteobacteria, where the role of HRXXN kinases and response regulators has not been fully characterized (38, 39). Also, our framework might inform the study of organisms like myxobacteria, which have the largest number of TCS in any organism and a remarkably large number of unpaired TCS proteins (40). The ability to find coevolving residues across proteins and protein pairs is restricted not by system or organism but rather by the diversity of sequences that emerged from evolutionary pressure. The parameters of the DCA Hamiltonian can be used not only to find cognate or potential partners but also to estimate the effects of mutations at the interface (15) or to estimate structural complexes with high accuracy (16). It is notable that DCA has predicted several biologically important interactions that were not previously known. An approach that includes DCA-guided experiments should facilitate the discovery of new and important protein interactions and yield fresh insights into the complex interactions of the cell.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant R01GM062419 (to S.S.G.), Center for Theoretical Biological Physics National Science Foundation grants PHY-1427654 and MCB-1214457 (to F.M. and R.R.C.), and NSF INSPIRE grant MCB-1241332 (to R.R.C.).
J.S.B. and M.L.P. designed experiments, J.S.B., C.S., and F.M. conducted experiments and analyzed results, R.R.C. and F.M. contributed analytical tools, and J.S.B., F.M., M.L.P., and S.S.G. wrote the paper.
K. Gibfried, C. Lieu, and L. Welch provided technical support.
We declare that we have no financial interest from this work.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00235-16.
REFERENCES
- 1.Stock A, Robinson V, Godreau P. 2000. Two-component signaling systems. Annu Rev Biochemistry 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 2.Shultzaberger R, Boyd J, Diamond S, Greenspan RJ, Golden SS. 2015. Giving time purpose: the S. elongatus circadian clock in a broader network context. Annu Rev Genetics 49:21.21-21.21. doi: 10.1146/annurev-genet-112414-054823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kato H, Watanabe S, Nimura-Matsune K, Chibazakura T, Tozawa Y, Yoshikawa H. 2012. Exploration of a possible partnership among orphan two-component system proteins in cyanobacterium Synechococcus elongatus PCC 7942. Bio Biotechnol Biochem 76:1484–1491. doi: 10.1271/bbb.120172. [DOI] [PubMed] [Google Scholar]
- 4.Cohen S, Golden S. 2015. Circadian rhythms in cyanobacteria. Microbiol Mol Biol Rev 79:373–385. doi: 10.1128/MMBR.00036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Markson J, Piechura J, Puszynska A, O'Shea E. 2013. Circadian control of global gene expression by the cyanobacterial master regulator RpaA. Cell 155:1396–1408. doi: 10.1016/j.cell.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutu A, O'Shea E. 2013. Two antagonistic clock-regulated histidine kinases time the activation of circadian gene expression. Molecular Cell 50:288–294. doi: 10.1016/j.molcel.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmitz O, Katayama M, Williams S, Kondo T, Golden S. 2000. CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science 289:765–768. doi: 10.1126/science.289.5480.765. [DOI] [PubMed] [Google Scholar]
- 8.Yang Q, Pando B, Dong G, Golden S, van Oudenaarden A. 2010. Circadian gating of the cell cycle revealed in single cyanobacterial cells. Science 327:1522–1526. doi: 10.1126/science.1181759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyagishima S, Wolk C, Osteryoung K. 2005. Identification of cyanobacterial cell division genes by comparative and mutational analyses. Mol Microbiol 56:126–143. doi: 10.1111/j.1365-2958.2005.04548.x. [DOI] [PubMed] [Google Scholar]
- 10.Dong G, Yang Q, Wang Q, Kim Y-I, Wood T, Osteryoung K, van Oudenaarden A, Golden S. 2010. Elevated ATPase activity of KaiC applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell 140:529–539. doi: 10.1016/j.cell.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paddock M, Boyd J, Adin D, Golden S. 2013. Active output state of the Synechococcus Kai circadian oscillator. Proc Natl Acad Sci U S A 110:E3849–E3857. doi: 10.1073/pnas.1315170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shultzaberger R, Boyd J, Katsuki T, Golden S, Greenspan R. 2014. Single mutations in sasA enable a simpler ΔcikA network architecture with equivalent circadian properties. Proc Natl Acad Sci U S A 111:E5069–E7511. doi: 10.1073/pnas.1419902111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Espinosa J, Boyd J, Cantos R, Salinas P, Golden S, Contreras A. 2015. Cross-talk and regulatory interactions between the essential response regulator RpaB and cyanobacterial circadian clock output. Proc Natl Acad Sci U S A 112:2198–2203. doi: 10.1073/pnas.1424632112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morcos F, Pagnani A, Lunt B, Bertolino A, Marks DS, Sander C, Zecchina R, Onuchic JN, Hwa T, Weigt M. 2011. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc Natl Acad Sci U S A 108:E1293–E1301. doi: 10.1073/pnas.1111471108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng RR, Morcos F, Levine H, Onuchic JN. 2014. Toward rationally redesigning bacterial two-component signaling systems using coevolutionary information. Proc Natl Acad Sci U S A 111:E563–E571. doi: 10.1073/pnas.1323734111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.dos Santos RN, Morcos F, Jana B, Andricopulo AD, Onuchic JN. 2015. Dimeric interactions and complex formation using direct coevolutionary couplings. Sci Rep 5:13652. doi: 10.1038/srep13652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holtman C, Chen Y, Sandoval P, Gonzales A, Nalty M, Thomas T, Youderian P. 2005. High-throughput functional analysis of the Synechococcus elongatus PCC 7942 genome. DNA Res 12:103–115. doi: 10.1093/dnares/12.2.103. [DOI] [PubMed] [Google Scholar]
- 18.Clerico E, Ditty J, Golden S. 2007. Specialized techniques for site-directed mutagenesis in cyanobacteria. Methods Mol Biol 362:155–171. doi: 10.1007/978-1-59745-257-1_11. [DOI] [PubMed] [Google Scholar]
- 19.Mackey S, Ditty J, Clerico E, Golden S. 2007. Detection of rhythmic bioluminescence from luciferase reporters in cyanobacteria. Methods Mol Biol 362:115–129. doi: 10.1007/978-1-59745-257-1_8. [DOI] [PubMed] [Google Scholar]
- 20.Morcos F, Hwa T, Onuchic JN, Weigt M. 2014. Direct coupling analysis for protein contact prediction. Methods Mol Biol 1137:55–70. doi: 10.1007/978-1-4939-0366-5_5. [DOI] [PubMed] [Google Scholar]
- 21.Casino P, Rubio V, Marina A. 2009. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell 139:325–336. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 22.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. 2010. The Pfam protein families database. Nucleic Acids Res 38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim Y-I, Boyd J, Espinosa J, Golden S. 2015. Detecting KaiC phosphorylation rhythms of the cyanobacterial circadian oscillator in vitro and in vivo. Methods Enzymol 551:153–173. doi: 10.1016/bs.mie.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider C, Rasband W, Eliceiri K. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagaya M, Aiba H, Mizuno T. 1994. The sphR product, a two-component system response regulator protein, regulates phosphate assimilation in Synechococcus sp. strain PCC 7942 by binding to two sites upstream from the phoA promoter. J Bacteriol 176:2210–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santos-Beneit F. 2015. The pho regulon: a huge regulatory network in bacteria. Front Microbiol 6:402. doi: 10.3389/fmicb.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diamond S, Jun D, Rubin B, Golden S. 2015. The circadian oscillator in Synechococcus elongatus controls metabolite partitioning under diurnal growth. Proc Natl Acad Sci U S A 112:E1916–E1925. doi: 10.1073/pnas.1504576112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyd JS, Bordowitz JR, Bree AC, Golden SS. 2013. An allele of the crm gene blocks cyanobacterial circadian rhythms. Proc Natl Acad Sci U S A 110:13950–13955. doi: 10.1073/pnas.1312793110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goclaw-Binder H, Senderesky E, Shimoni E, Kiss V, Reich Z, Perelman A, Schwarz R. 2012. Nutrient-associated elongation and asymmetric division of the cyanobacterium Synechococcus PCC 7942. Environ Microbiol 14:680–690. doi: 10.1111/j.1462-2920.2011.02620.x. [DOI] [PubMed] [Google Scholar]
- 30.Kim Y-I, Vinyard D, Ananyev G, Dismukes G, Golden S. 2012. Oxidized quinones signal onset of darkness directly to the cyanobacterial circadian oscillator. Proc Natl Acad Sci U S A 109:17765–17769. doi: 10.1073/pnas.1216401109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Procaccini A, Lunt B, Szurmant H, Hwa T, Weigt M. 2011. Dissecting the specificity of protein-protein interaction in bacterial two-component signaling: orphans and crosstalk. PLoS One 6:e19729. doi: 10.1371/journal.pone.0019729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moronta-Barrios F, Espinsa J, Contreras A. 2012. In vivo features of signal transduction by the essential response regulator RpaB from Synechococcus elongatus PCC 7942. Microbiology 158:1229–1237. doi: 10.1099/mic.0.057679-0. [DOI] [PubMed] [Google Scholar]
- 33.Ogawa T, Bao D, Katoh H, Shibata M, Pakrasi H, Bhattacharyya-Pakrasi M. 2002. A two-component signal transduction pathway regulates manganese homeostasis in Synechocystis 6803, a photosynthetic organism. J Biol Chem 32:28981–28986. [DOI] [PubMed] [Google Scholar]
- 34.Yamaguchi K, Suzuki I, Yamamoto H, Lyukevich A, Bodrova I, Los D, Piven I, Zinchenko V, Kanehisa M, Murata M. 2002. A two-component Mn2+-sensing system negatively regulates expression of the mntCAB operon in Synechocystis. Plant Cell 14:2901–2913. doi: 10.1105/tpc.006262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu D, Yang C. 2014. The nitrogen-regulated response regulator NrrA controls cyanophycin synthesis and glycogen catabolism in the cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem 289:2055–2071. doi: 10.1074/jbc.M113.515270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez-Redondo M, Moronta F, Salinas P, Espinosa J, Cantos R, Dixon R, Marina A, Contreras A. 2010. Environmental control of phosphorylation pathways in a branched two-component system. Mol Microbiol 78:475–489. doi: 10.1111/j.1365-2958.2010.07348.x. [DOI] [PubMed] [Google Scholar]
- 37.Kato H, Kubo T, Hayashi M, Kobayashi I, Yagasaki T, Chibazakura T, Watanabe S, Yoshikawa H. 2011. Interactions between histidine kinase NblS and the response regulators RpaB and SrrA are involved in the bleaching process of the cyanobacterium Synechococcus elongatus PCC 7942. Plant Cell Physiol 52:2115–2122. doi: 10.1093/pcp/pcr140. [DOI] [PubMed] [Google Scholar]
- 38.Kaczmarczyk A, Hochstrasser R, Vorholt JA, Francez-Charlot A. 2014. Complex two-component signaling regulates the general stress response in Alphaproteobacteria. Proc Natl Acad Sci U S A 111:E5196–5204. doi: 10.1073/pnas.1410095111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Francez-Charlot A, Kaczmarczyk A, Fischer HM, Vorholt JA. 2015. The general stress response in Alphaproteobacteria. Trends Microbiol 23:164–171. doi: 10.1016/j.tim.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 40.Whitworth DE, Cock PJ. 2008. Two-component systems of the myxobacteria: structure, diversity and evolutionary relationships. Microbiology 154:360–372. doi: 10.1099/mic.0.2007/013672-0. [DOI] [PubMed] [Google Scholar]
- 41.Ditty J, Williams S, Golden S. 2003. A cyanobacterial circadian timing mechanism. Annu Rev Genet 37:513–543. doi: 10.1146/annurev.genet.37.110801.142716. [DOI] [PubMed] [Google Scholar]
- 42.Ditty J, Canales S, Anderson B, Williams S, Golden S. 2005. Stability of the Synechococcus elongatus PCC 7942 circadian clock under directed antiphase expression of the kai genes. Microbiology 151:2605–2613. doi: 10.1099/mic.0.28030-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.