ABSTRACT
Burkholderia thailandensis is a soil-dwelling bacterium that shares many metabolic pathways with the ecologically similar, but evolutionarily distant, Pseudomonas aeruginosa. Among the diverse nutrients it can utilize is choline, metabolizable to the osmoprotectant glycine betaine and subsequently catabolized as a source of carbon and nitrogen, similar to P. aeruginosa. Orthologs of genes in the choline catabolic pathway in these two bacteria showed distinct differences in gene arrangement as well as an additional orthologous transcriptional regulator in B. thailandensis. In this study, we showed that multiple glutamine amidotransferase 1 (GATase 1)-containing AraC family transcription regulators (GATRs) are involved in regulation of the B. thailandensis choline catabolic pathway (gbdR1, gbdR2, and souR). Using genetic analyses and sequencing the transcriptome in the presence and absence of choline, we identified the likely regulons of gbdR1 (BTH_II1869) and gbdR2 (BTH_II0968). We also identified a functional ortholog for P. aeruginosa souR, a GATR that regulates the metabolism of sarcosine to glycine. GbdR1 is absolutely required for expression of the choline catabolic locus, similar to P. aeruginosa GbdR, while GbdR2 is important to increase expression of the catabolic locus. Additionally, the B. thailandensis SouR ortholog (BTH_II0994) is required for catabolism of choline and its metabolites as carbon sources, whereas in P. aeruginosa, SouR function can by bypassed by GbdR. The strategy employed by B. thailandensis represents a distinct regulatory solution to control choline catabolism and thus provides both an evolutionary counterpoint and an experimental system to analyze the acquisition and regulation of this pathway during environmental growth and infection.
IMPORTANCE Many proteobacteria that occupy similar environmental niches have horizontally acquired orthologous genes for metabolism of compounds useful in their shared environment. The arrangement and differential regulation of these components can help us understand both the evolution of these systems and the potential roles these pathways have in the biology of each bacterium. Here, we describe the transcriptome response of Burkholderia thailandensis to the eukaryote-enriched molecule choline, identify the regulatory pathway governing choline catabolism, and compare the pathway to that previously described for Pseudomonas aeruginosa. These data support a multitiered regulatory network in B. thailandensis, with conserved orthologs in the select agents Burkholderia pseudomallei and Burkholderia mallei, as well as the opportunistic lung pathogens in the Burkholderia cepacia clade.
INTRODUCTION
Burkholderia thailandensis is a saprophytic, soil-dwelling bacterium common in tropical and subtropical regions, and it is an opportunistic pathogen of insects, plants, nematodes, and amoeba (1–3). B. thailandensis is used as a less virulent model for the select agents B. pseudomallei and B. mallei, the causative agents of melioidosis and glanders, respectively. The reduced virulence of B. thailandensis is due primarily to the absence of the major capsule locus, important for B. pseudomallei virulence in mammals (4), although it is used to study the effect of type III secretion systems on phagocytic escape and retains other genes associated with virulence (5, 6). Despite virulence differences, a great deal of the core genome is shared between B. thailandensis and its relatives, including many pathways for accessory metabolism.
Genes predicted to be involved in choline catabolism are found throughout the B. pseudomallei group (BPG) and Burkholderia cepacia complex (BCC) (7, 8). Many soil bacteria can use choline as a sole carbon and nitrogen source, and this catabolic pathway may be particularly important for bacteria associated with eukaryotes (9). Choline is part of the polar head group of both phosphatidylcholine and sphingomyelin, which together constitute the majority of lipids on the outer leaflet of eukaryotic cell membranes (10) and are also abundant in pulmonary surfactant (11). In addition to its role as a nutrient source, choline metabolism can generate glycine betaine (GB), an important osmoprotectant (12) and inducer of virulence factor production (13). The conversion of choline to GB has been shown to be important for Escherichia coli survival in urine (14) and Pseudomonas aeruginosa survival in the mammalian lung (15, 16). Many organisms, including Burkholderiales, maintain an intracellular GB pool, potentially as a hedge against future osmotic stress (17). Choline can also be found in the rhizosphere, exuded from plant roots, likely to influence populations of bacteria in this environment (18). In fact, many plants accumulate high levels of GB as an osmoprotectant, notably beets, from whence the appellation “betaine” was derived (19). In the environment, secreted or decaying organic matter provides a metabolic opportunity that holds much promise to microbial opportunists poised to exploit them.
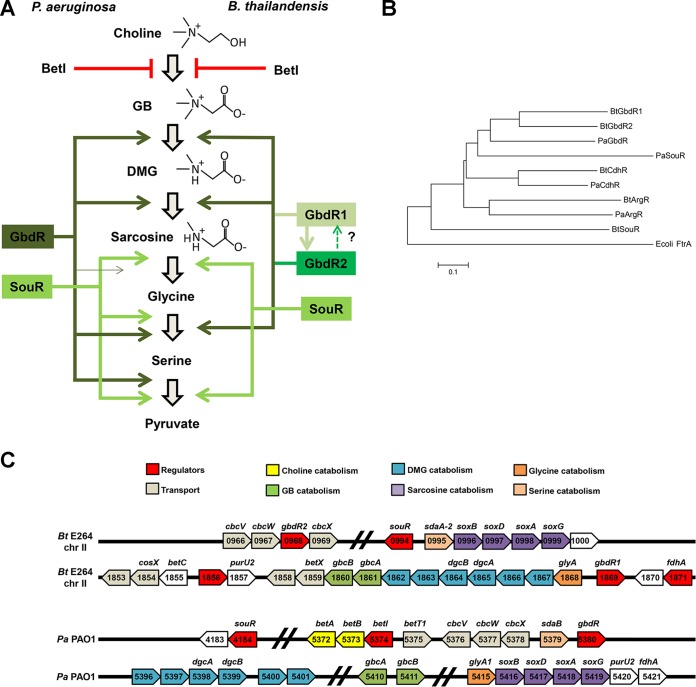
Our laboratory has previously studied the choline catabolic pathway and its regulation in Pseudomonas aeruginosa (20–22), and examination of orthologous genes in B. thailandensis suggested an alternative, perhaps more complex, regulatory strategy. In P. aeruginosa, the critical regulator of GB catabolism is GbdR, a glutamine amidotransferase 1 (GATase 1)-like AraC family transcription regulator (GATR) that regulates the genes required for sequential demethylation of GB to glycine in P. aeruginosa (Fig. 1A, left) (22). In contrast to P. aeruginosa, B. thailandensis and other Burkholderiales appeared to have two GbdR orthologs (Fig. 1B and C) whose cooccurrence and conserved synteny suggested that they play nonoverlapping roles in B. thailandensis choline catabolism. In this study, we have investigated the contributions of the two B. thailandensis GbdR orthologs to choline and GB catabolism and to global gene expression in response to choline. These data and follow-up analyses reveal that the choline catabolism pathway in B. thailandensis, while bearing many similarities to P. aeruginosa, contains key differences in organization and regulation.
FIG 1.
Comparison of B. thailandensis and P. aeruginosa choline metabolism and associated regulators. (A) Model of choline catabolic regulation in P. aeruginosa (left) and B. thailandensis (right). (B) Phylogenetic tree of relevant GATRs in B. thailandensis and P. aeruginosa rooted to FtrA, a GATR present in E. coli. (C) Diagram of the chromosomal context of gbdR, gbdR1, gbdR2, sour, and major catabolic genes for the catabolism of GB and subsequent metabolites for B. thailandensis and P. aeruginosa.
MATERIALS AND METHODS
Culture conditions.
Burkholderia thailandensis E264 (ATCC 700388) (23) cultures were inoculated from −80°C glycerol stocks onto LB (10 g/liter tryptone, 5 g/liter yeast extract, 5 g/liter NaCl) with 1.5% agar and incubated at 37°C overnight. For protein expression and molecular cloning, Escherichia coli DH5α or E. coli T7 Express cells (New England BioLabs [NEB]) were then grown in LB broth containing 100 μg/ml carbenicillin, kanamycin, or trimethoprim (Tp) as appropriate.
Strain construction.
Deletion strains (see Table S1 in the supplemental material) were generated as described in Thongdee et al., and the resistance markers were removed using the methods described in Choi et al. (24, 25). Briefly, a PCR product was amplified via splice overlap extension PCR that consisted of a trimethoprim resistance marker amplified from pUC18mini-TN7T-Tp, including the flanking Flp recombinase sites, and with regions of homology upstream and downstream of the gene of interest (see primers in Table S2). The resultant PCR product was then used to naturally transform B. thailandensis that had been grown in defined medium (DM) as previously described (24). To remove the resistance marker, pFLPe2 was electroporated into the Tp-resistant strain and plated onto low-salt LB (LSLB; 10 g/liter tryptone, 5 g/liter yeast extract, 3 g/liter NaCl, 1.5% agarose, pH 7.5, with 250 μg/ml zeocin and 0.2% l-rhamnose for Flp recombinase expression). Cells were cured of pFLPe2 by growth at 42°C. Resultant colonies were streaked onto plates with zeocin or Tp, or without antibiotics, to identify colonies where the trimethoprim marker and pFLPe2 had been lost. The ΔgbdR1, ΔgbdR2, and ΔsouR phenotypes were complemented by cloning each gene and ∼250 bp of the upstream regulatory region into pUC18mini-TN7T-Zeo and integration into one of the two attTn7 sites in B. thailandensis (26, 27) (see Table S1). Appropriate control strains were generated by attTn7 integration with the empty pUC18mini-TN7T-Zeo vector (see Table S1).
Growth assays.
Growth assays were performed by starting overnight cultures in morpholinepropanesulfonic acid (MOPS) minimal medium supplemented with 20 mM pyruvate and 5 mM glucose incubated at 37°C (28). Cultures were diluted in MOPS medium to an optical density at 600 nm (OD600) of 0.07, and 30 μl was used to inoculate 470 μl of MOPS medium supplemented with carbon sources as described in the figure legends in a flat-bottom 48-well plate (Costar). These plates were incubated at 37°C with agitation and OD600 measured using a Biotek Synergy 2 plate reader.
Alignments and phylogenetic tree construction.
Sequence alignments and phylogenetic analysis were preformed using MEGA version 6 (29). Amino acid sequences of the relevant GATRs were aligned using MUSCLE (30). The ftrA sequence, a GATR homolog from Escherichia coli, was included as an outgroup to root the phylogenetic tree.
RNA-Seq.
For RNA sequencing (RNA-Seq), wild-type (WT), ΔgbdR1, ΔgbdR2, and ΔgbdR1 ΔgbdR2 strains were streaked from frozen stocks onto LB agar plates and incubated overnight at 37°C. These plates were used to inoculate 3 ml MOPS with 20 mM pyruvate and 5 mM glucose cultures, which were incubated for 18 h at 37°C on a rotary wheel. To initiate the experiment, 1 ml of these cultures was added to 2 ml of prewarmed MOPS with 30 mM pyruvate or 2 ml MOPS with 30 mM pyruvate and 1.5 mM choline, resulting in final concentrations of 20 mM pyruvate and 1 mM choline. These cultures were incubated for 4 h at 37°C. Cells from 1.5 ml of culture were collected by centrifugation, and the resultant pellets were resuspended in RNA Protect bacterial reagent (Qiagen) and incubated at room temperature for 10 min. Cells were collected by centrifugation and resuspended in 50 μl 10 mM Tris, 1 mM EDTA (TE) buffer with 3 mg/ml lysozyme and incubated at room temperature for 5 min. To each of these resuspensions, 1 μl 20 mg/ml proteinase K, 0.5 μl of 10% SDS, 1 μl of DNase I (RNase free; Ambion), and 2 μl of 50 mM MgCl2 were added, and samples were incubated for 5 min at room temperature. RNA was then prepared using the Qiagen RNeasy kit according to the manufacturer's instructions. Eluate from this purification was further treated with 11 μl 10× DNase I buffer and 2 μl DNase I (RNase free; Ambion), incubated at 37°C for 20 min to remove contaminating DNA, and repurified using the Qiagen RNeasy protocol with the optional on-column DNase I step. The resulting purified RNA was assessed for purity by PCR and quantified by a Bioanalyzer chip, depleted of 16S and 23S rRNA by using MICROBExpress bacterial mRNA enrichment (Ambion), and subsequently reassessed by Bioanalyzer (Agilent Technologies). RNA-Seq DNA libraries were prepared by the Vermont Cancer Center–College of Medicine Massively Parallel Sequencing Facility using an Illumina TruSeq stranded total RNA library preparation kit and were run on an Illumina Hi-Seq 1500 to generate single end reads. Read quality was checked with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Samples were aligned to the NC_007650 and NC_007651 chromosomes using BWA (31), duplicate reads were marked using Picard (version 1.110; Broad Institute of MIT and Harvard [http://broadinstitute.github.io/picard/]), and gene counts were calculated using summarizeOverlaps from the GenomicRanges package (32) before differential expression was called using the DESeq package (33) in R (version 3.0.1; R Development Core Team [https://www.r-project.org/]), with assistance from the University of Vermont College of Medicine Bioinformatics Shared Resource group.
Quantitative reverse transcription-PCR (qRT-PCR) to confirm RNA-Seq findings.
Wild-type, ΔgbdR1, ΔgbdR2, and ΔgbdR1 ΔgbdR2 strains were grown and induced as described for the RNA-Seq experiment. RNA was extracted similarly to the procedure for RNA-Seq, except that a third DNase I treatment was performed using the Ambion DNA-free kit (AM1906) per the manufacturer's protocol. cDNA was generated using a Superscript IV first-strand synthesis system (Invitrogen) using 24 ng of RNA combined with the 5′-NSNSNSNSNS-3′ primer previously described (34). Quantitative PCR (qPCR) was performed as described in Willsey et al. (35). Briefly, 0.2× SYBR green I nucleic acid gel stain (Thermo Fisher Scientific [TFS]) was used with Q5 high-fidelity 2× master mix (NEB) and amplified using cycle conditions of 98°C for 2 min and 98°C for 20 s, annealing at 60°C for 30 s, and extension at 72°C for 20 s, repeated by going back to the second step 39 times. Reactions were performed in technical duplicate and biological triplicate with RT primers designed for BTH_I1140 (rplU), BTH_II1869 (gbdR1), BTH_II1861 (gbcA), BTH_II1853 (putative porin), BTH_II0968 (gbdR2), BTH_II0966 (cbcV), BTH_II0994 (souR), and BTH_II0996 (soxB). A dilution series was used to generate a standard curve for each primer set. Each reaction was normalized to its respective rplU value, and then induced values were divided by uninduced values to derive a fold effect value.
Purification of GbdR1, GbdR2, and SouR.
gbdR1, gbdR2, and souR were each cloned into the pMAL-C2X expression vector, generating an N-terminal maltose-binding protein (MBP) tag. The constructs were transformed into E. coli T7 Express, transformants were selected on LB agar plus 100 μg/ml carbenicillin, and positive colonies were used to inoculate 20-ml cultures of LB plus 100 μg/ml carbenicillin. After 18 h, these cultures were used to inoculate 500-ml flasks of LB plus 100 μg/ml LB carbenicillin, grown to an OD600 of 0.3, and induced for 4 h at 30°C with 500 μM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were collected by centrifugation. Cells were then washed and collected by centrifugation, and the resulting cell pellets were flash frozen in liquid nitrogen. Cell pellets were resuspended on ice in 10 ml of chilled chromatography and lysis buffer (20 mM Tris, pH 7.4, 500 mM NaCl, 20 mM imidazole, 1× HALT protease inhibitor [TFS]). Ten microliters of DNase I was added to the resuspended cell pellet, and the mixture was incubated for 10 min on ice. The resuspended cells were lysed with a French cell press using three 1,100-lb/in2 passages, and NP-40 was then added to a final concentration of 0.05%. The lysate was clarified by centrifugation for 15 min at 15,000 rpm at 4°C. The supernatant was passed over a column containing a 1-ml bed volume of amylose resin beads (NEB). The column was then washed with 10 ml of column buffer 300 (20 mM Tris, pH 7.4, 300 mM KCl, 1 mM EDTA, 0.05% NP-40, 0.7 μl/ml β-mercaptoethanol) and then 10 ml column buffer 150 (20 mM Tris, pH 7.4, 150 mM KCl, 1 mM EDTA, 0.05% NP-40, 0.7 μl/ml β-mercaptoethanol). Protein was eluted in column buffer 150 containing 10 mM maltose, and 12 300-μl fractions were collected. Fractions were analyzed by SDS-PAGE acrylamide gels stained with Coomassie. Since GATR proteins tend to precipitate at higher concentrations, fractions of lower concentration were pooled and dialyzed in Slide-A-Lyzer dialysis cassette G2 (TFS) against 2 liters of column buffer 150. Aliquots were flash frozen in liquid nitrogen and stored at −80°C.
EMSA.
Biotinylated or unlabeled primers were used to generate DNA probes from appropriate templates containing the putative promoter regions using Q5 high-fidelity DNA polymerase (NEB) and then subjected to a QIAquick PCR purification kit (Qiagen) and quantified using a NanoDrop 1000 (TFS). Twenty-microliter reactions were assembled using 2× binding buffer (10 mM Tris, pH 7.4, 200 mM KCl, 2 mM EDTA, 2 mM dithiothreitol [DTT], 10% glycerol, and 200 μg/μl bovine serum albumin [36]), 5 ng/μl poly(dI-dC), 25 to 75 nM purified MBP-tagged protein, 1 nM biotinylated DNA probe, and (optionally) 20 nM unlabeled competitor DNA probe. Reaction mixtures were incubated in a 30°C water bath for 20 min, 5 μl 5× electrophoretic mobility shift assay (EMSA) loading buffer (1× Tris-borate-EDTA [TBE], 20% glycerol, 0.01% xylene cyanol, 0.01% bromophenol blue) was added, and samples were loaded onto a 5% acrylamide 0.5× TBE gel. Gels were run for 1 h at 100 V and then transferred to a BioDyne-B nylon membrane (Pierce) for 1 h at 80 V in 0.5× TBE at 4°C. DNA was cross-linked to the membrane using a UV Stratalinker 2400 (Stratagene) on the auto-cross-link setting, and the biotin-labeled probe was visualized using the Pierce LightShift chemiluminescent EMSA kit (TFS) and imaged on a ChemiDoc XRS+ molecular imager (Bio-Rad). Density of resulting bands was quantified using Quantity One software from Bio-Rad.
Generation of reporter constructs and β-galactosidase assays.
Yeast homologous recombination was used to replace the kanamycin resistance gene in the shuttle vector pMQ131 (37) (apaHA3) with the Tp resistance gene (dhrFII), resulting in pAN1. Yeast recombination was also used to replace the lacZα fragment with the full lacZYA operon, resulting in pAN7. The intergenic region between gbdR1 and glyA, as well as truncations of this region, were ligated into the SphI site proximal to the lacZYA ribosomal binding site, and transformants were selected on LB agar plus 100 μg/ml Tp at 37°C. Tp-resistant colonies were used to inoculate 3 ml MOPS medium with 20 mM pyruvate, 5 mM glucose, and 100 μg/ml Tp, which were incubated for 48 h at 37°C. Cells from these cultures were collected by centrifugation at 13,000 rpm for 2 min at room temperature, washed with 1 ml MOPS medium, and collected again by centrifugation, and the pellets were resuspended in MOPS medium. Thirty microliters of this resuspension was used to inoculate each well in a 48-well plate that contained 470 μl MOPS medium with 20 mM pyruvate with or without 1 mM choline. These cultures were incubated for 48 h at 37°C on an orbital shaker at 170 rpm in a humidified chamber. The extended induction time was shown to be necessary for detectable β-galactosidase activity empirically in our hands and as indicated by Kang et al. (38). β-Galactosidase activity was quantified as previously described (21).
Accession number(s).
RNA-Seq data have been deposited in NCBI GEO under record GSE81652.
RESULTS
Organization of predicted B. thailandensis orthologs of P. aeruginosa choline catabolic genes.
The choline catabolic pathway and its regulation in P. aeruginosa have been described previously (20, 22). Briefly, in P. aeruginosa the TetR family transcription factor BetI represses expression of betIBA and is derepressed by choline, allowing production of the BetA and BetB enzymes that convert choline into GB (39–41). The GATase 1-like AraC family transcription regulator (GATR) GbdR responds to GB and dimethylglycine (DMG) by inducing expression of catabolic genes, including gbcA-B, the dgc operon (PA5376, PA5377, dgcA, dgcB, PA5400, and PA5401), and the sarcosine oxidase genes, which together contribute to the sequential demethylation of GB to DMG, sarcosine, and finally glycine (Fig. 1A and C). SouR, also a GATR, controls transcriptional induction of the sarcosine oxidase genes in response to sarcosine (35). Homologs of betI and betBA in B. thailandensis have been described previously (38).
Sequence analysis revealed two likely GbdR orthologs in B. thailandensis, BTH_II1869 (gbdR1) and BTH_II0968 (gbdR2), with strong amino acid sequence homology to P. aeruginosa GbdR (48.2% and 47.0% identity and 75.5% and 71.3% similarity, respectively, using BLOSUM50 along the entire protein length). Alignments of the characterized P. aeruginosa GATRs (ArgR, CdhR, and SouR), and their putative B. thailandensis orthologs, revealed that the B. thailandensis GbdR orthologs were more similar to each other than to P. aeruginosa GbdR or any other GATRs (Fig. 1B). This suggested that they also have similar functions or play separate roles within the same catabolic pathway. The presence of a more distantly related GATR divergently transcribed from the sarcosine oxidase operon (Fig. 1B and C) led us to predict that BTH_II0994 was a functional ortholog of P. aeruginosa SouR, although the phylogenetic tree suggests that they share no recent paralogy.
On the P. aeruginosa chromosome, gbdR is located one gene away from the cbcXWV transport genes (Fig. 1C), previously implicated in the transport of choline and its immediate metabolites into the cytosol from the periplasm (42) and adjacent to the sdaB gene known to participate in the pathway (43). In B. thailandensis, gbdR1 is located adjacent to a putative operon containing the bulk of predicted orthologous genes encoding enzymes needed for GB catabolism to sarcosine, while gbdR2 is present within a locus containing the putative cbcXWV orthologs (Fig. 1C). The homology of GbdR1 and GbdR2 with P. aeruginosa GbdR, combined with their chromosomal locations adjacent to genes involved in choline metabolism, strongly suggested that both transcription factors were involved in the regulation of this pathway. The synteny of the gbdR1 and gbdR2 loci in B. thailandensis E264 is conserved throughout the various clades of Burkholderiales, including species such as B. pseudomallei, B. mallei, B. cenocepacia, B. ambifaria, and B. multivorans. This conservation in synteny suggests that both GbdR1 and GbdR2 play roles in the regulation of choline metabolism in Burkholderia.
gbdR1 and gbdR2 contribute differentially to choline catabolism.
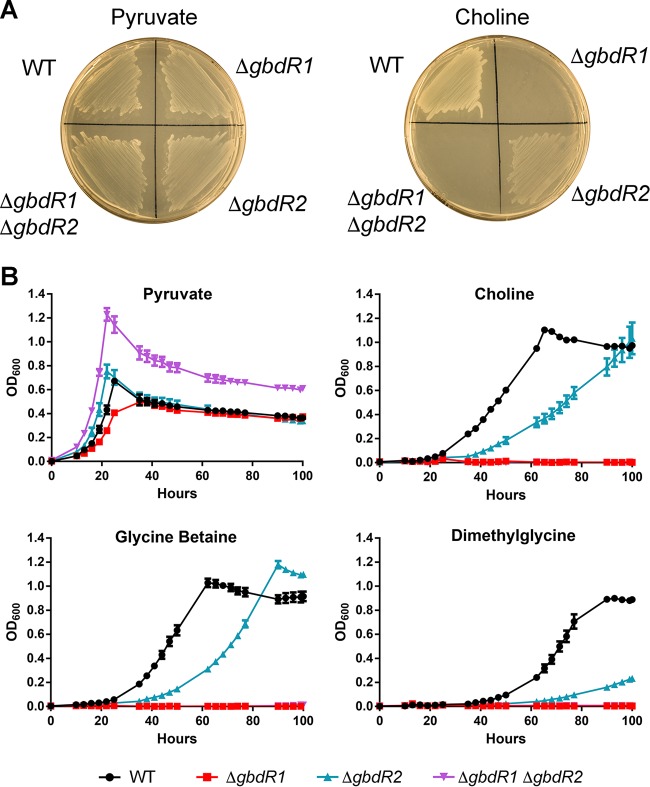
B. thailandensis, like P. aeruginosa (44), can use choline as a sole source of carbon and/or nitrogen (Fig. 2A). Based on our predictions of gbdR1 and gbdR2 involvement in choline metabolism, we tested the ability of each single-deletion strain and the double-deletion strain to grow using choline as a sole carbon source. Wild-type B. thailandensis grows on choline as a sole carbon source, while a ΔgbdR1 or ΔgbdR1 ΔgbdR2 strain cannot (Fig. 2A, right). Deletion of gbdR2 alone resulted in reduced growth on choline as a sole carbon source (Fig. 2A, right). On plates, none of the manipulations appeared to alter growth on pyruvate as a sole carbon source (Fig. 2A, left). The impaired but visible growth of the ΔgbdR2 strain led us to examine the growth kinetics of these strains on choline and its downstream metabolites, GB and DMG. Deletion of gbdR2 results in slower growth than that of the wild type on choline, GB, and DMG, comprised of both an extended lag phase and a lower growth rate during exponential phase (Fig. 2B). Deletion of these transcription factors did not substantially alter growth kinetics using pyruvate as a sole carbon source but did alter maximal yield in liquid culture in the double deletion strain (Fig. 2B), although this difference was not apparent on solid media (Fig. 2A, left). The ΔgbdR1 and ΔgbdR1 ΔgbdR2 strains showed no measurable growth on any of the choline-related metabolites tested within 100 h.
FIG 2.
Deletion of gbdR1 and gbdR2 results in altered growth on choline and its metabolites as sole carbon sources. (A) Growth of B. thailandensis deletion strains on MOPS minimal medium agar plates supplemented with either 20 mM pyruvate or 20 mM choline and incubated at 37°C for 72 h. ΔgbdR1 and ΔgbdR1 ΔgbdR2 strains fail to grow on choline, while the ΔgbdR2 strain exhibits less growth. (B) Growth curves reveal the inability of ΔgbdR1 and ΔgbdR1 ΔgbdR2 strains to grow on choline or its metabolites, while the ΔgbdR2 strain exhibits a slow-growth phenotype on choline, glycine betaine, and dimethylglycine. Growth curves are representative of at least three experiments per condition at each time point, each with biological triplicates, and the error bars represent standard deviations.
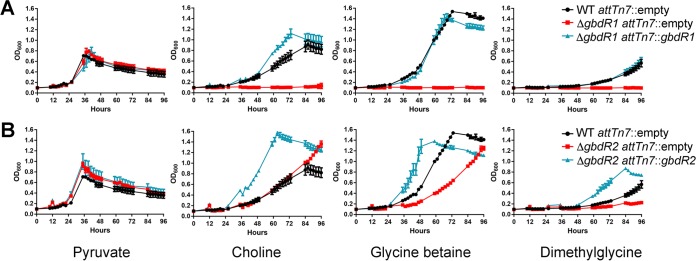
Integration of gbdR1 at the attTn7 site restored growth of a ΔgbdR1 strain on choline, GB, and DMG (Fig. 3A). Growth in GB and DMG was very similar to that of the wild type, while growth of the complemented strain on choline exceeded that of the wild type on choline (Fig. 3A). Similarly, integration of gbdR2 at the attTn7 site restored growth of a ΔgbdR2 strain on choline-related carbon sources and resulted in more rapid growth than that in the wild type for choline, GB, and DMG (Fig. 3B). We also tested complementation of the ΔgbdR1 ΔgbdR2 strain with either gbdR1 or gbdR2. The ΔgbdR1 ΔgbdR2 attTn7::gbdR1 strain was able to grow on choline as a sole carbon source in MOPS minimal medium, although not at wild-type levels, whereas the ΔgbdR1 ΔgbdR2 attTn7::gbdR2 strain was not capable of growth on choline (see Fig. S1 in the supplemental material). Taken together, these data suggest that gbdR1 and gbdR2 regulate the choline catabolic pathway, with gbdR1 absolutely required and gbdR2 playing an accessory role.
FIG 3.
Growth phenotypes of ΔgbdR1 and ΔgbdR2 strains can be complemented. (A) Complementation of ΔgbdR1 strain at attTn7 with gbdR1 and its putative promoter results in restoration of ability to grow on choline and its metabolites. (B) Complementation of ΔgbdR2 strain at attTn7 with gbdR2 results in an increased growth rate on choline and its metabolites, surpassing the growth rate of the wild type. Growth curves are representative of at least three experiments per condition at each time point, each with biological triplicates, and the error bars represent standard deviations.
GbdR1 and GbdR2 regulate transcription of genes involved in the choline catabolic pathway.
To identify the GbdR1 and GbdR2 regulons, we exposed our deletion strains to minimal medium with pyruvate as a primary carbon source and with or without 1 mM choline and analyzed the transcriptomes by RNA-Seq. Exposure of wild-type B. thailandensis to choline resulted in 57 transcripts being induced or repressed more than 2.3-fold, with many predicted to be involved in quaternary amine catabolism and transport (Table 1). A prominent cluster of induced genes from BTH_II1868 to BTH_II1853 includes orthologs to P. aeruginosa glyA (BTH_II1868), the dgc operon (BTH_II1867, BTH_II1866, BTH_II1865, BTH_II1864, BTH_II1863, and BTH_II1862), gbcA (BTH_II1861), gbcB (BTH_II1860), and betX (BTH_II1859). Based on these predictions, the putative operon consisting of glyA to BTH_II1858 (Fig. 1C) carry the genes likely to be responsible for the breakdown of GB to sarcosine. Also induced was a nearby predicted operon consisting of BTH_II1855-BTH_II1853, carrying likely orthologs of P. aeruginosa betC, cosX, and a predicted porin. Another GATR, BTH_II0994, and the divergently transcribed operon consisting of BTH_II0995 (sdaA-2), BTH_II0996 (soxB), BTH_II0997 (soxD), BTH_II0998 (soxA), BTH_II0999 (soxG), and BTH_II1000, were also induced, and we hypothesized that these are the B. thailandensis souR and sarcosine oxidase genes responsible for the breakdown of sarcosine to glycine (35).
TABLE 1.
Transcript changes in B. thailandensis in response to choline
| Locus ID | Geneb | Linear fold change in transcript over pyruvate alonea |
|||
|---|---|---|---|---|---|
| WT | ΔgbdR1 | ΔgbdR2 | ΔgbdR1 ΔgbdR2 | ||
| BTH_I0192 | −2.6 | NC | NC | NC | |
| BTH_I0393 | −4.8 | NC | NC | NC | |
| BTH_I0394 | −5.1 | NC | NC | NC | |
| BTH_I0395 | −8.3 | NC | NC | NC | |
| BTH_I0396 | −4.3 | NC | NC | NC | |
| BTH_I0398 | −3.9 | NC | NC | NC | |
| BTH_I0687 | 23.8 | NC | 10.5 | NC | |
| BTH_I0688 | 19.3 | NC | NC | NC | |
| BTH_I0698 | 11.3 | NC | NC | NC | |
| BTH_I0700 | 12.0 | NC | NC | NC | |
| BTH_I0866 | 5.8 | 5.3 | NC | NC | |
| BTH_I0959 | −2.6 | NC | −5.3 | −5.0 | |
| BTH_I1406 | −2.3 | NC | −4.4 | NC | |
| BTH_I1620 | 5.7 | NC | 4.4 | 4.4 | |
| BTH_I1621 | 61.2 | NC | 18.3 | NC | |
| BTH_I1622 | 52.7 | NC | 9.7 | NC | |
| BTH_I1623 | 40.0 | NC | 9.3 | NC | |
| BTH_I1624 | 43.7 | NC | 5.0 | NC | |
| BTH_I1625 | 5.5 | NC | NC | NC | |
| BTH_I3016 | −2.8 | NC | −3.6 | −3.4 | |
| BTH_I3017 | −3.1 | NC | −4.0 | −3.9 | |
| BTH_II0001 | 5.4 | NC | 9.0 | 6.5 | |
| BTH_II0643 | 2.4 | NC | NC | NC | |
| BTH_II0694 | −3.4 | NC | −2.4 | NC | |
| BTH_II0695 | −2.6 | NC | NC | NC | |
| BTH_II0964 | 2.9 | NC | NC | NC | |
| BTH_II0966 | cbcV | 45.4 | NC | NC | NC |
| BTH_II0967 | cbcW | 94.0 | NC | NC | NC |
| BTH_II0968 | gbdR2 | 81.7 | NC | NC | NC |
| BTH_II0969 | cbcX | 136.6 | NC | NC | NC |
| BTH_II0970 | 37.7 | NC | NC | NC | |
| BTH_II0971 | 8.8 | NC | NC | NC | |
| BTH_II0994 | souR | 3.9 | NC | 4.7 | NC |
| BTH_II0995 | sdaA-2 | 90.8 | NC | 32.3 | NC |
| BTH_II0996 | soxB | 130.6 | NC | 39.3 | NC |
| BTH_II0997 | soxD | 172.0 | NC | 102.3 | NC |
| BTH_II0998 | soxA | 122.7 | NC | 58.6 | NC |
| BTH_II0999 | soxG | 24.9 | NC | 11.7 | NC |
| BTH_II1000 | 7.6 | −3.2 | 8.7 | −3.5 | |
| BTH_II1546 | 4.3 | NC | 3.7 | NC | |
| BTH_II1853 | 5.4 | NC | NC | NC | |
| BTH_II1854 | cosX | 7.1 | NC | NC | NC |
| BTH_II1855 | betC | 3.8 | NC | NC | NC |
| BTH_II1856 | 11.5 | NC | NC | NC | |
| BTH_II1857 | purU2 | 8.4 | NC | NC | NC |
| BTH_II1858 | 6.5 | NC | NC | NC | |
| BTH_II1859 | betX | 175.6 | NC | 38.6 | NC |
| BTH_II1860 | gbcB | 115.8 | NC | 28.9 | NC |
| BTH_II1861 | gbcA | 110.8 | NC | 23.7 | NC |
| BTH_II1862 | 218.7 | NC | 44.4 | NC | |
| BTH_II1863 | 163.4 | NC | 97.2 | NC | |
| BTH_II1864 | dgcB | 154.1 | NC | 28.9 | NC |
| BTH_II1865 | dgcA | 199.7 | NC | 23.4 | NC |
| BTH_II1866 | 223.9 | NC | 29.8 | NC | |
| BTH_II1867 | 135.3 | NC | 47.4 | NC | |
| BTH_II1868 | glyA | 162.9 | NC | 20.7 | NC |
| BTH_II1869 | gbdR1 | 2.6 | NC | NC | NC |
Constraints on inclusion in the table were a ±2.3-fold change and P value of ≤0.005 based on the wild type. NC, no change due to significance, lower fold change, or experimental deletion of the gene.
Gene names are putative based on homology to P. aeruginosa.
Homologs of cbcXWV in P. aeruginosa, an operon important for the transport of choline, are present in B. thailandensis and are also induced by choline. The gbdR2 gene is situated in this operon between BTH_II0967 (cbcW) and BTH_II0969 (cbcX). The aforementioned genes are induced by choline in the wild-type strain but are not in either the ΔgbdR1 or ΔgbdR1 ΔgbdR2 strain, suggesting an absolute requirement of gbdR1 in the regulation of these genes. Cells lacking gbdR2 express all of the genes in the major catabolic operon, glyA-BTH_II1858, in response to choline but to a lower level than the wild type. Interestingly, the relative induction of the transcripts in the transport operon containing gbdR2 is unaffected by the absence of gbdR2, suggesting that there is no autoregulation of the gbdR2 operon.
To confirm our RNA-Seq results, we performed qRT-PCR on a subset of induced genes and the transcription factors of interest, normalized to rplU transcript levels (see Fig. S2 in the supplemental material). The wild-type B. thailandensis response to choline in our qRT-PCR experiment reflected our RNA-Seq results. gbcA, cbcV, and soxB are all induced in response to choline, suggesting overall induction of their putative operons. The qRT-PCR data also show that transcripts of the GATRs gbdR1 and souR were slightly induced in response to choline, while gbdR2 was induced at a higher level. This slight increase in gbdR1 expression observed may be negligible; therefore, it is unclear if induction of gbdR1 is a mechanism by which GbdR2 regulates choline catabolism. Overall, we interpret the matching trends in expression between the RNA-Seq and qRT-PCR experiments as a validation of our findings.
The glyA promoter is induced by choline under the control of GbdR1 and GbdR2.
pAN27 is a reporter plasmid containing the full 408-bp intergenic region between gbdR1 and glyA, such that the glyA promoter drives lacZYA expression. This reporter was used to assess the transcriptional control of the glyA promoter. This putative promoter likely controls the expression of BTH_II1868 to BTH_II1857 and thus governs the bulk of genes needed to convert GB to sarcosine. In a B. thailandensis wild-type background, pAN27 lacZ expression is induced by choline, GB, and DMG but not by ethylcholine (Fig. 4A). Ethylcholine is a nonmetabolizable inducer of GbdR-dependent transcription in P. aeruginosa (22). The inability of this choline analog to elicit similar effects in a GbdR/GbdR2-dependent promoter suggests specificity differences between GbdR and GbdR1 or GbdR2, a metabolite transport difference, or a difference in the specificity of choline oxidase. No β-galactosidase activity was detected when pAN27 in B. thailandensis was induced using 1 mM sarcosine, similar to findings in P. aeruginosa for gbdR-dependent induction of choE, a choline esterase (22), or for the phospholipase plcH (13, 45, and data not shown).
FIG 4.
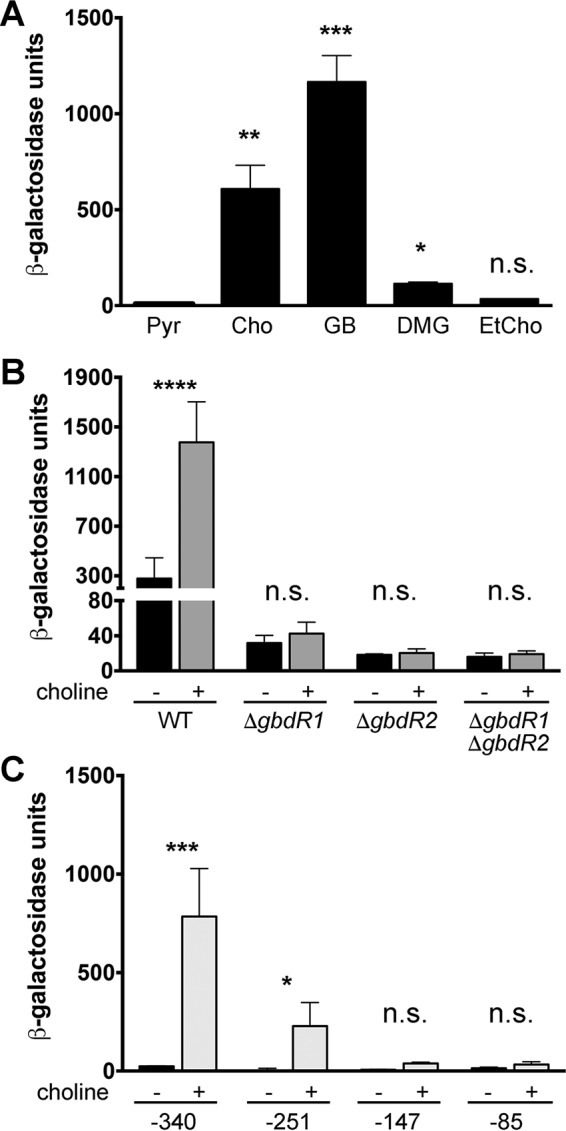
glyA promoter is induced by choline and its metabolites under the combined control of gbdR1 and gbdR2. (A) β-Galactosidase activity from pAN27 is induced in the presence of choline and its metabolites but not pyruvate. (B) gbdR1 and gbdR2 are both required for significant β-galactosidase induction in the presence of choline. (C) Promoter truncations of the glyA promoter reveal that the critical region is located between −147 and −340 with respect to the predicted translational start site of glyA. In each case, data presented represents the standard errors of the means determined from three independent experiments, each with three technical replicates. Statistical notations in panel A are based on one-way analysis of variance (ANOVA) with Dunnett's posttest using the pyruvate group as the comparator, while panels B and C were analyzed by two-way ANOVA with Sidak's posttest comparing conditions of with versus without choline. Abbreviations and symbols: Pyr, pyruvate; Cho, choline; GB, glycine betaine; DMG, dimethylglycine; EtCho, ethylcholine; n.s., not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
ΔgbdR1, ΔgbdR2, and ΔgbdR1 ΔgbdR2 deletion strains carrying pAN27 failed to show significant choline-induced β-galactosidase induction compared to the wild-type control (Fig. 4B), and basal expression was also low. This suggests that both gbdR1 and gbdR2 are required for robust induction of the glyA promoter. We would have predicted that the ΔgbdR2 strain would produce some β-galactosidase activity in response to choline, as the ΔgbdR2 strain can use choline as a sole carbon source (Fig. 2). The incongruity may be due to the unusually long time it takes to observe β-galactosidase activity in B. thailandensis combined with the long delay in growth of the ΔgbdR2 strain on choline (Fig. 2). As a follow-up experiment, we transformed pAN27 into a ΔsouR strain to determine if SouR is required for glyA promoter induction in B. thailandensis (see Fig. S3 in the supplemental material). β-Galactosidase activity in the ΔsouR strain in response to choline is robust, higher than that in the wild type, potentially due to blockage of the catabolic pathway and subsequent buildup of intermediate metabolites. This indicates that SouR is not required for induction of glyA but also could point to a positive feedback control between sarcosine metabolism and regulation of upstream steps.
Reporter plasmids derived from pAN27 with progressively smaller portions of the intergenic region between gbdR1 and glyA were produced (−340, −251, −147, and −85 with respect to the glyA putative translational start site) in order to map the choline-dependent portion of the glyA regulatory region. Exposure of cells containing these constructs to choline resulted in choline-dependent induction of β-galactosidase activity for −340 and −251 but not −147 or −85 (Fig. 4C). This suggests that the region from bp −251 to the translation start site of glyA is necessary for a response to choline.
GbdR1 and GbdR2 bind directly to the glyA promoter.
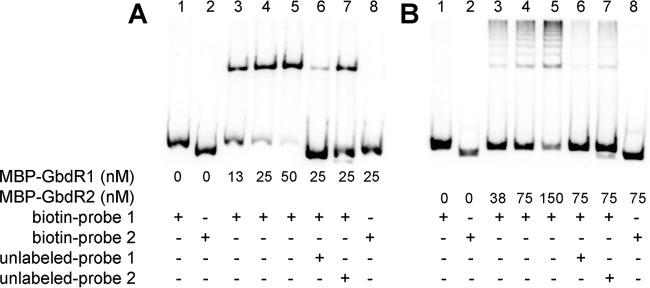
N-terminally tagged MBP-GbdR1 and MBP-GbdR2 were produced and assayed by EMSA using biotinylated DNA probes representing sections of the intergenic region between gbdR1 and glyA. Probe 1 is 216 bp, covering the bp −320 to −104 region with respect to the glyA translational start site, while probe 2 is 178 bp, covering the −160 to +18 region. MBP-GbdR1 could bind probe 1 in a concentration-dependent manner, and binding to biotinylated probe 1 could be significantly competed off with unlabeled probe 1 (Fig. 5A). MBP-GbdR1 did not interact with probe 2 (Fig. 5A, rightmost lane), and unlabeled probe 2 was markedly less able to compete with MBP-GbdR1 binding to labeled probe 1. These data support a GbdR1 binding site within probe 1. Similar results were obtained using MBP-GbdR2, which was also able to bind to probe 1, not probe 2, and could be similarly competed from labeled probe 1 using unlabeled probe 1 (Fig. 5B) and, again, much less so with the unlabeled probe 2. The shift produced by MBP-GbdR2 consists of multiple bands that are likely the result of oligomerization. Results using 6×His-N-terminally tagged versions of GbdR1 and GbdR2 produced the same results and banding patterns, although the solubility of the 6-His version was much lower than that of the MBP fusions (data not shown). These results provide evidence that both GbdR1 and GbdR2 can bind to the glyA promoter. When combined with results from our reporter assays (Fig. 4), we infer that the binding site for GbdR1 and GbdR2 is contained within the bp −251 to −160 region with respect to the glyA translational start site. As we have previously shown for P. aeruginosa GbdR and SouR, addition of their presumptive ligand did not impact binding to DNA (data not shown).
FIG 5.
MBP-GbdR1 and MBP-GbdR2 bind to the regulatory region of the major GB catabolic operon (BTH_II1868-BTH_II1858). (A) MBP-GbdR1 binds to biotin probe 1 in a dose-dependent manner (lanes 3 to 5). The interaction is specific, as it does not bind to biotin probe 2 (lane 8) and can be competed off biotin probe 1 with unlabeled probe 1 (compare lane 6 to lane 4, 17.2% shift versus 75.2% shift, respectively) but less so with unlabeled probe 2 (compare lane 7 to lane 4, 51.5% shift versus 75.2% shift, respectively). (B) MBP-GbdR2 also binds specifically to probe 1 in a dose-dependent manner (lanes 3 to 5) while failing to bind to biotin probe 2 (lane 8). The binding between MBP-GbdR2 and biotin probe 1 is also specific, as it can be competed off by unlabeled probe 1 (compare lane 6 to lane 4, 4.3% shift versus 16%, respectively) but less so by unlabeled probe 2 (compare lane 7 to lane 4, 7.7% shift versus 16%, respectively).
B. thailandensis SouR regulates sarcosine metabolism.
Sarcosine is a downstream metabolite of choline, and orthologs of P. aeruginosa sarcosine oxidase genes were identified in our RNA-Seq results (Table 1 and Fig. 1C). P. aeruginosa uses sarcosine as a sole carbon and nitrogen source, and this is regulated by the transcription factor SouR (35). While B. thailandensis can use choline as a sole carbon or sole carbon and nitrogen source, B. thailandensis is unable to grow on sarcosine as a sole carbon source or as a sole carbon and nitrogen source (see Fig. S4 in the supplemental material). We hypothesize that the ability of B. thailandensis to transport extracellular sarcosine is limited compared to that of P. aeruginosa, and the rate of import is insufficient to support growth as a sole carbon source. It is, however, able to utilize sarcosine as a sole nitrogen source when pyruvate is available as a primary carbon source, and it does so in an SouR-dependent manner (Fig. 6A). Deletion of gbdR1 or gbdR2 has no major effect on the ability of B. thailandensis to utilize sarcosine as a nitrogen source, but the ΔsouR strain does show a reduced level of growth.
FIG 6.
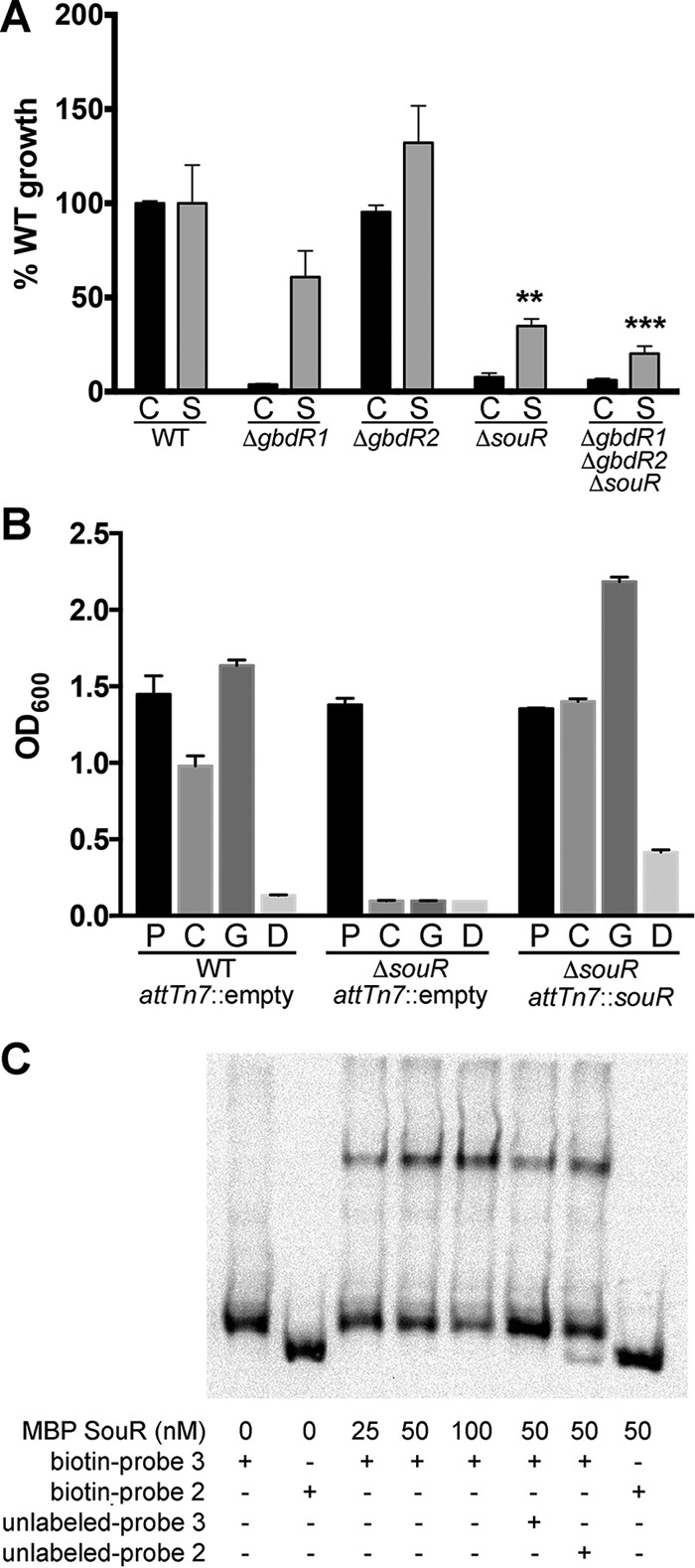
SouR is a critical regulator of the choline catabolic pathway. (A) B. thailandensis can utilize choline as a nitrogen source. Strains were grown on 20 mM pyruvate with either 5 mM choline (C) or sarcosine (S) for 72 h at 37°C. ΔgbdR1, ΔsouR, and ΔgbdR1 ΔgbdR2 ΔsouR strains were significantly diminished in their ability to utilize choline as a nitrogen source. The ΔgbdR2 strain utilized both choline and sarcosine as a nitrogen source at levels similar to that of the wild type. The ΔsouR strain used sarcosine as a nitrogen source at reduced rates compared to the wild type. OD600 measurements were normalized to basal growth without a nitrogen source and then normalized to the wild type and are represented as percent growth. (B) Deletion of souR prevents B. thailandensis from using choline as a carbon source. Complementation of souR at an attTn7 site with souR, under the control of its putative native promoter, restores the ability of B. thailandensis to utilize choline and its metabolites as carbon sources. Strains were grown with either 40 mM pyruvate (P), choline (C), GB (G), or DMG (D) as a sole carbon source in MOPS minimal medium for 72 h at 37°C. (C) MBP-SouR binds to the putative promoter of the sarcosine oxidase operon represented by biotin probe 3 in a dose-dependent manner (lanes 3 to 5). The interaction is specific, as MBP-SouR does not bind to biotin probe 2 (lane 8) and can be competed off by unlabeled probe 3 (compare lane 6 to lane 4, 22% shift versus 37.1%, respectively) but not by unlabeled probe 2 (compare lane 7 to lane 4, 38.2% shift versus 37.1%, respectively). Data in panels A and B are averaged from three experiments each with three biological replicates, and error bars represent SEM. These data were analyzed by two-way ANOVA with Tukey's posttest testing for strain effect for each carbon source separately, not comparing the carbon sources. For panel A, except for the ΔgbdR2 strain, all choline changes are different from the WT and are not noted. Only the sarcosine comparisons different from the WT are noted. For panel B, pyruvates were not significantly different but all other comparisons were, except for WT versus the mutant in dimethylglycine. **, P < 0.01; ***, P < 0.001.
BTH_II0994 is a GATR divergently transcribed from the sarcosine oxidase operon, and we hypothesized that it is functionally orthologous to souR in P. aeruginosa (Fig. 1C), despite BTH_II0994 and SouR not sharing a high degree of similarity compared to the other GATR orthologous pairs (Fig. 1B). To test if BTH_II0994 was a functional souR, we generated a B. thailandensis ΔsouR strain and used it to determine if souR is required for growth on choline. The ΔsouR strain fails to grow on choline, GB, or DMG as a sole carbon source (Fig. 6B). This is contrary to regulation in P. aeruginosa, where choline, GB, and DMG can still be used as sole carbon sources for growth in the absence of souR, although at a diminished growth rate (35). This result also suggests that GbdR1 and GbdR2 are insufficient to cause induction of the sarcosine oxidase genes by themselves or together, contrary to the regulation scheme in P. aeruginosa (Fig. 1A). This phenotype can be complemented by chromosomal integration of souR at an attTn7 site under the control of its native promoter (Fig. 6B).
MBP-SouR was produced and its DNA binding assessed using EMSA with a biotinylated 230-bp oligonucleotide designated probe 3, covering the region of bp −208 to +22 relative to the putative translation start site of sdaA-2, the first gene in the likely sarcosine oxidase operon of B. thailandensis. MBP-SouR was able to bind the biotinylated probe 3 but not the probe 2 negative control. The binding of MBP-SouR to biotinylated probe 3 could be competed off with unlabeled probe 3 but not unlabeled probe 2, suggesting that this interaction is specific (Fig. 6C). The binding of MBP-SouR to this putative promoter region coupled to the phenotype of the ΔsouR strain suggests that SouR is responsible for the regulation of the sarcosine oxidase genes in B. thailandensis.
DISCUSSION
The ability to utilize GB as a compatible solute, providing protection against osmotic and other stresses without creating significant disruption of normal cell processes, is nearly ubiquitous among bacteria; however, the ability to catabolize GB is not as widespread (46). B. thailandensis and other species in this genus possess the enzymes for utilizing choline and its metabolites as carbon and nitrogen sources, taking advantage of this widely available biomolecule as befits their description as metabolically adaptable bacteria. P. aeruginosa, another soil-dwelling and metabolically adaptable microorganism, shares this ability and was our original model for the study of the GB catabolic pathway and its regulation (20). B. thailandensis and P. aeruginosa inhabit similar environments, possess similarly sized genomes, and encode diverse metabolic pathways; thus, they are considered to have similar generalist strategies, despite B. thailandensis and P. aeruginosa belonging to the betaproteobacteria and gammaproteobacteria classes, respectively. The metabolic enzymes in the choline catabolic pathway are well conserved, and the orthologous genes in different species are readily identifiable. However, the identification of gbdR2 and the alternative gene organization in B. thailandensis prompted us to ask if the choline regulatory network was as well conserved. We show here that B. thailandensis uses an alternative regulatory solution to control this pathway compared to that of P. aeruginosa. Both B. thailandensis and P. aeruginosa can store pools of GB (17) and metabolize it as a carbon and nitrogen source; therefore, the different regulatory networks suggest evolution of alternative strategies to control the decision to store or catabolize.
Although the data presented here indicate that choline catabolism is regulated differently in B. thailandensis than in P. aeruginosa (Fig. 1A), the basic components of the pathway are conserved; thus, the overall scheme of the pathway is the same. In both organisms, choline catabolism begins with choline-dependent derepression of the catabolic genes responsible for the conversion of choline to GB, betBA, mediated by the choline-sensing TetR family transcription factor BetI. These genes have been described previously (39), and evidence suggests they function in B. thailandensis as they do in P. aeruginosa (38). In P. aeruginosa, GB is subsequently metabolized to DMG by GbcA and GbcB heterodimer (47, 48) and from DMG to sarcosine by enzymes in the dgc operon (49), all under the regulation of GbdR (20). In B. thailandensis the same enzymatic steps are under the control of GbdR1, modulated by GbdR2. Sarcosine is then demethylated to glycine by components of the sarcosine oxidase operon and glyA1, controlled in tandem by GbdR and SouR in P. aeruginosa and controlled separately by SouR as well as GbdR1 and GbdR2, respectively, in B. thailandensis as described below.
In this study, we determined that both GbdR1 and GbdR2 participate in the regulation of GB catabolism and characterized their regulons. Both regulators are GATRs with strong amino acid sequence similarity to GbdR in P. aeruginosa, and deletion strains confirmed that both play roles in the regulation of choline catabolism and catabolism of its downstream metabolites (Fig. 2). Combining the transcriptional data with the growth phenotypes of the deletion strains supports gbdR1 being required for expression of the major catabolic cluster of genes and the transport operon that includes gbdR2. gbdR2, on the other hand, is important for a robust response to choline, enhancing expression of many genes required to import and catabolize choline and its metabolites, particularly the major catabolic cluster (Fig. 1C and Table 1). The operon containing gbdR2 and the putative cbcWVX orthologs is likely regulated solely by GbdR1. Although the presence of gbdR1 is sufficient for growth (Fig. 2), reporter assays and growth studies showed that both gbdR1 and gbdR2 are required for a robust transcriptional response (Fig. 4A) and growth on choline (Fig. 2). In agreement with promoter-mapping results (Fig. 4C), EMSA data suggest that GbdR1 and GbdR2 share a similar binding site, raising the question of whether or not they compete for the same binding site or act synergistically. A possibility is that GbdR1 and GbdR2 possess differential DNA binding affinities that result in differential responses to choline and its metabolites under different physiological conditions.
In addition to gbdR1 and gbdR2, our transcriptome analysis revealed a third GATR induced in response to choline, BTH_II0994, which we confirmed as a functional souR ortholog in B. thailandensis. In P. aeruginosa, SouR regulates the expression of the sarcosine oxidase catabolic operon, the components of which are responsible for converting sarcosine to glycine (35). SouR in B. thailandensis is less similar to GbdR, GbdR1, and GbdR2 than is SouR in P. aeruginosa (Fig. 1B). B. thailandensis souR (BTH_II0994) is divergently transcribed from the sarcosine oxidase operon, whereas P. aeruginosa souR (PA4184) is distantly located and part of a two-gene operon. These factors suggest a divergence in how souR was acquired or evolved in both organisms. The inability of B. thailandensis to grow on sarcosine as a sole carbon source was surprising given that choline is readily catabolized and the enzymes for sarcosine catabolism are expressed (Fig. 6A). We found that souR in B. thailandensis is required for sarcosine catabolism, unlike in P. aeruginosa, where GbdR can induce the sarcosine oxidase operon in the absence of souR (35). The complementation analysis and EMSA for the sarcosine oxidase operon putative promoter provides further evidence that BTH_II0994 is indeed the functional souR ortholog in Burkholderia (Fig. 6B and C). It is not clear if SouR is capable of autoregulation of souR or if GbdR1 controls its expression directly.
GbdR1, GbdR2, and SouR are all GATRs implicated in the regulation of genes involved in the catabolism of quaternary amines or their metabolites. GATRs are members of the AraC transcription factor family, grouped by their canonical C-terminal DNA binding domain, but unlike classic AraC proteins they contain a GATase 1-like N-terminal domain. The GATase 1-like domain is characterized by its homology to class I glutamine amidotransferases, which bind glutamine or ammonia and participate in the amidation/deamidation reaction (50, 51). Many of the GATR N termini, notably excluding SouR in P. aeruginosa, also retain a bioinformatically identified cysteine residue that would be part of the functional catalytic triad of GATase 1 family enzymes. The N-terminal domain of AraC proteins typically is involved in ligand binding and dimerization, which is a characteristic important for their functionality, and dimerization is often affected by the binding of ligand (52). P. aeruginosa possesses other GATRs implicated in the catabolism of N-substituted amines, including argR, the regulator of arginine catabolism (53), and cdhR, the regulator of carnitine catabolism (20). Putative argR and cdhR orthologs are also present in B. thailandensis (unpublished data). We hypothesize that the GATase 1-like domain has been combined with the AraC-style DNA binding domain to enable detection of charged amine-containing compounds and that GATRs as a whole may represent a subfamily of transcription factors that regulate metabolism of accessory nitrogen sources, including choline and its metabolites.
The transport of choline and derivative molecules also appears to be handled differentially between B. thailandensis and P. aeruginosa based on genomic information. P. aeruginosa possesses an array of BCCT and ABC family transporters that have been implicated in choline and GB transport that are effective under different osmotic conditions or primarily utilized when choline is in such abundance that it can be used as a carbon or nitrogen source (42). B. thailandensis possesses fewer transporters, with only one putative homolog of the BCCT family transporter (BTH_II1109), compared to three in P. aeruginosa (betT1, betT2, and betT3). B. thailandensis possesses a putative amino acid permease, BTH_II1858, as part of the major GB catabolic operon with no obvious ortholog in P. aeruginosa. The placement of BTH_II1858 and conserved synteny among Burkholderia species suggests a role as a transporter for choline or its derivatives, but there is no functional evidence as of yet. Some pseudomonads have a putative ortholog of BTH_II1858 identified as an ethanolamine transporter, ethanolamine being similar to choline in its role as a head group for phospholipids, suggesting the possibility of a link to general fatty acid metabolism. Both B. thailandensis and P. aeruginosa also possess orthologs of the choline/GB transporter, opuC, that was described in P. syringae and found to function under hyperosmolar conditions (54). B. thailandensis orthologs to many of the P. aeruginosa periplasmic binding proteins mediate ABC transporter-dependent import of quaternary amines (cbcX [BTH_II0969], caiX [BTH_II1849], betX [BTH_II1859], and cosX [BTH_II1853]), but apparently these are not sufficient for efficient sarcosine transport (see Fig. S4 in the supplemental material). This result is perhaps not surprising, as limited current data suggest that sarcosine does not compete well with choline for these transporters in P. aeruginosa (55). To date, the sarcosine transporter has not been identified, and the functional differences between P. aeruginosa and B. thailandensis for sarcosine utilization in the background of a similar metabolic pathway may provide a platform for identification of this transporter.
In this study, we have identified an alternative model for the regulation of choline catabolism that incorporates multiple GATRs and examined their respective regulons. The conservation of gbdR1 and gbdR2 throughout the Burkholderia genus suggests that differences in regulation of choline catabolism outlined here represent a model that can be extrapolated to the more pathogenic strains, as well as those strains associated with the rhizosphere. Contrasting the models of choline catabolism in B. thailandensis and P. aeruginosa will serve as a useful tool to probe the remaining unanswered questions concerning the pathway.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jeff Bond, Julie Dragon, Robert Devins, and David J. Shirley for bioinformatics assistance. We also owe thanks to Graham Willsey and Lauren Hinkel for critical reading of the manuscript.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIGMS.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00372-16.
REFERENCES
- 1.Pilatova M, Dionne MS. 2012. Burkholderia thailandensis is virulent in Drosophila melanogaster. PLoS One 7:e49745. doi: 10.1371/journal.pone.0049745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inglis TJJ, Rigby P, Robertson TA, Dutton NS, Henderson M, Chang BJ. 2000. Interaction between Burkholderia pseudomallei and Acanthamoeba species results in coiling phagocytosis, endamebic bacterial survival, and escape. Infect Immun 68:1681–1686. doi: 10.1128/IAI.68.3.1681-1686.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee YH, Chen YH, Ouyang XZ, Gan YH. 2010. Identification of tomato plant as a novel host model for Burkholderia pseudomallei. BMC Microbiol 10:28. doi: 10.1186/1471-2180-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reckseidler SL, DeShazer D, Sokol PA, Woods DE. 2001. Detection of bacterial virulence genes by subtractive hybridization: identification of capsular polysaccharide of Burkholderia pseudomallei as a major virulence determinant. Infect Immun 69:34–44. doi: 10.1128/IAI.69.1.34-44.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angus AA, Agapakis CM, Fong S, Yerrapragada S, Estrada-de los Santos P, Yang P, Song N, Kano S, Caballero-Mellado J, de Faria SM, Dakora FD, Weinstock G, Hirsch AM. 2014. Plant-associated symbiotic Burkholderia species lack hallmark strategies required in mammalian pathogenesis. PLoS One 9:e83779. doi: 10.1371/journal.pone.0083779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haraga A, West TE, Brittnacher MJ, Skerrett SJ, Miller SI. 2008. Burkholderia thailandensis as a model system for the study of the virulence-associated type III secretion system of Burkholderia pseudomallei. Infect Immun 76:5402–5411. doi: 10.1128/IAI.00626-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu YT, Kim HS, Chua HH, Lin CH, Sim SH, Lin DX, Derr A, Engels R, DeShazer D, Birren B, Nierman WC, Tan P. 2006. Genomic patterns of pathogen evolution revealed by comparison of Burkholderia pseudomallei, the causative agent of melioidosis, to avirulent Burkholderia thailandensis. BMC Microbiol 6:46. doi: 10.1186/1471-2180-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim HS, Schell MA, Yu Y, Ulrich RL, Sarria SH, Nierman WC, DeShazer D. 2005. Bacterial genome adaptation to niches: divergence of the potential virulence genes in three Burkholderia species of different survival strategies. BMC Genomics 6:174. doi: 10.1186/1471-2164-6-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wargo MJ. 2013. Homeostasis and catabolism of choline and glycine betaine: lessons from Pseudomonas aeruginosa. Appl Environ Microbiol 79:2112–2120. doi: 10.1128/AEM.03565-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zachowski A. 1993. Phospholipids in animal eukaryotic membranes: transverse asymmetry and movement. Biochem J 294(Part 1):1–14. doi: 10.1042/bj2940001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernhard W, Hoffmann S, Dombrowsky H, Rau GA, Kamlage A, Kappler M, Haitsma JJ, Freihorst J, von der Hardt H, Poets CF. 2001. Phosphatidylcholine molecular species in lung surfactant: composition in relation to respiratory rate and lung development. Am J Respir Cell Mol Biol 25:725–731. doi: 10.1165/ajrcmb.25.6.4616. [DOI] [PubMed] [Google Scholar]
- 12.Chambers S, Kunin CM. 1985. The osmoprotective properties of urine for bacteria: the protective effect of betaine and human urine against low pH and high concentrations of electrolytes, sugars, and urea. J Infect Dis 152:1308–1316. doi: 10.1093/infdis/152.6.1308. [DOI] [PubMed] [Google Scholar]
- 13.Shortridge VD, Lazdunski A, Vasil ML. 1992. Osmoprotectants and phosphate regulate expression of phospholipase C in Pseudomonas aeruginosa. Mol Microbiol 6:863–871. doi: 10.1111/j.1365-2958.1992.tb01537.x. [DOI] [PubMed] [Google Scholar]
- 14.Kunin CM, Hua TH, White LV, Villarejo M. 1992. Growth of Escherichia coli in human urine–role of salt tolerance and accumulation of glycine betaine. J Infect Dis 166:1311–1315. doi: 10.1093/infdis/166.6.1311. [DOI] [PubMed] [Google Scholar]
- 15.Wargo MJ. 2013. Choline catabolism to glycine betaine contributes to Pseudomonas aeruginosa survival during murine lung infection. PLoS One 8:e56850. doi: 10.1371/journal.pone.0056850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun ZX, Kang Y, Norris MH, Troyer RM, Son MS, Schweizer HP, Dow SW, Hoang TT. 2014. Blocking phosphatidylcholine utilization in Pseudomonas aeruginosa, via mutagenesis of fatty acid, glycerol and choline degradation pathways, confirms the importance of this nutrient source in vivo. PLoS One 9:e103778. doi: 10.1371/journal.pone.0103778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fitzsimmons LF, Hampel KJ, Wargo MJ. 2012. Cellular choline and glycine betaine pools impact osmoprotection and phospholipase C production in Pseudomonas aeruginosa. J Bacteriol 194:4718–4726. doi: 10.1128/JB.00596-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Rudder KE, Sohlenkamp C, Geiger O. 1999. Plant-exuded choline is used for rhizobial membrane lipid biosynthesis by phosphatidylcholine synthase. J Biol Chem 274:20011–20016. doi: 10.1074/jbc.274.28.20011. [DOI] [PubMed] [Google Scholar]
- 19.Craig SA. 2004. Betaine in human nutrition. Am J Clin Nutr 80:539–549. [DOI] [PubMed] [Google Scholar]
- 20.Wargo MJ, Szwergold BS, Hogan DA. 2008. Identification of two gene clusters and a transcriptional regulator required for Pseudomonas aeruginosa glycine betaine catabolism. J Bacteriol 190:2690–2699. doi: 10.1128/JB.01393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fitzsimmons LF, Flemer S Jr, Wurthmann AS, Deker PB, Sarkar IN, Wargo MJ. 2011. Small-molecule inhibition of choline catabolism in Pseudomonas aeruginosa and other aerobic choline-catabolizing bacteria. Appl Environ Microbiol 77:4383–4389. doi: 10.1128/AEM.00504-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hampel KJ, Labauve AE, Meadows JA, Fitzsimmons LF, Nock AM, Wargo MJ. 2014. Characterization of the GbdR regulon in Pseudomonas aeruginosa. J Bacteriol 196:7–15. doi: 10.1128/JB.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brett PJ, DeShazer D, Woods DE. 1998. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int J Syst Bacteriol 48(Part 1):317–320. doi: 10.1099/00207713-48-1-317. [DOI] [PubMed] [Google Scholar]
- 24.Thongdee M, Gallagher LA, Schell M, Dharakul T, Songsivilai S, Manoil C. 2008. Targeted mutagenesis of Burkholderia thailandensis and Burkholderia pseudomallei through natural transformation of PCR fragments. Appl Environ Microbiol 74:2985–2989. doi: 10.1128/AEM.00030-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi KH, Mima T, Casart Y, Rholl D, Kumar A, Beacham IR, Schweizer HP. 2008. Genetic tools for select-agent-compliant manipulation of Burkholderia pseudomallei. Appl Environ Microbiol 74:1064–1075. doi: 10.1128/AEM.02430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi KH, Schweizer HP. 2006. Mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc 1:153–161. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- 27.Choi KH, DeShazer D, Schweizer HP. 2006. Mini-Tn7 insertion in bacteria with multiple glmS-linked attTn7 sites: example Burkholderia mallei ATCC 23344. Nat Protoc 1:162–169. doi: 10.1038/nprot.2006.25. [DOI] [PubMed] [Google Scholar]
- 28.LaBauve AE, Wargo MJ. 2012. Growth and laboratory maintenance of Pseudomonas aeruginosa. Curr Protoc Microbiol Chapter 6:Unit 6E.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol 9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wargo MJ, Gross MJ, Rajamani S, Allard JL, Lundblad LK, Allen GB, Vasil ML, Leclair LW, Hogan DA. 2011. Hemolytic phospholipase C inhibition protects lung function during Pseudomonas aeruginosa infection. Am J Respir Crit Care Med 184:345–354. doi: 10.1164/rccm.201103-0374OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willsey GG, Wargo MJ. 2016. Sarcosine catabolism in Pseudomonas aeruginosa is transcriptionally regulated by SouR. J Bacteriol 198:301–310. doi: 10.1128/JB.00739-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brutinel ED, Vakulskas CA, Brady KM, Yahr TL. 2008. Characterization of ExsA and of ExsA-dependent promoters required for expression of the Pseudomonas aeruginosa type III secretion system. Mol Microbiol 68:657–671. doi: 10.1111/j.1365-2958.2008.06179.x. [DOI] [PubMed] [Google Scholar]
- 37.Shanks RM, Kadouri DE, MacEachran DP, O'Toole GA. 2009. New yeast recombineering tools for bacteria. Plasmid 62:88–97. doi: 10.1016/j.plasmid.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang Y, Norris MH, Barrett AR, Wilcox BA, Hoang TT. 2009. Engineering of tellurite-resistant genetic tools for single-copy chromosomal analysis of Burkholderia spp. and characterization of the Burkholderia thailandensis betBA operon. Appl Environ Microbiol 75:4015–4027. doi: 10.1128/AEM.02733-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rkenes TP, Lamark T, Strom AR. 1996. DNA-binding properties of the BetI repressor protein of Escherichia coli: the inducer choline stimulates BetI-DNA complex formation. J Bacteriol 178:1663–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Velasco-Garcia R, Villalobos MA, Ramirez-Romero MA, Mujica-Jimenez C, Iturriaga G, Munoz-Clares RA. 2006. Betaine aldehyde dehydrogenase from Pseudomonas aeruginosa: cloning, over-expression in Escherichia coli, and regulation by choline and salt. Arch Microbiol 185:14–22. doi: 10.1007/s00203-005-0054-8. [DOI] [PubMed] [Google Scholar]
- 41.Andresen PA, Kaasen I, Styrvold OB, Boulnois G, Strom AR. 1988. Molecular cloning, physical mapping and expression of the bet genes governing the osmoregulatory choline-glycine betaine pathway of Escherichia coli. J Gen Microbiol 134:1737–1746. [DOI] [PubMed] [Google Scholar]
- 42.Malek AA, Chen C, Wargo MJ, Beattie GA, Hogan DA. 2011. Roles of three transporters, CbcXWV, BetT1, and BetT3, in Pseudomonas aeruginosa choline uptake for catabolism. J Bacteriol 193:3033–3041. doi: 10.1128/JB.00160-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diab F, Bernard T, Bazire A, Haras D, Blanco C, Jebbar M. 2006. Succinate-mediated catabolite repression control on the production of glycine betaine catabolic enzymes in Pseudomonas aeruginosa PAO1 under low and elevated salinities. Microbiology 152:1395–1406. doi: 10.1099/mic.0.28652-0. [DOI] [PubMed] [Google Scholar]
- 44.Kortstee GJ. 1970. The aerobic decomposition of choline by microorganisms. I. The ability of aerobic organisms, particularly coryneform bacteria, to utilize choline as the sole carbon and nitrogen source. Arch Mikrobiol 71:235–244. [PubMed] [Google Scholar]
- 45.Domenech CE, Garrido MN, Lisa TA. 1991. Pseudomonas aeruginosa cholinesterase and phosphorylcholine phosphatase: two enzymes contributing to corneal infection. FEMS Microbiol Lett 66:131–135. [DOI] [PubMed] [Google Scholar]
- 46.Collins RE, Deming JW. 2013. An inter-order horizontal gene transfer event enables the catabolism of compatible solutes by Colwellia psychrerythraea 34H. Extremophiles 17:601–610. doi: 10.1007/s00792-013-0543-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith LT, Pocard JA, Bernard T, Le Rudulier D. 1988. Osmotic control of glycine betaine biosynthesis and degradation in Rhizobium meliloti. J Bacteriol 170:3142–3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White RF, Kaplan L, Birnbaum J. 1973. Betaine-homocysteine transmethylase in Pseudomonas denitrificans, a vitamin B12 overproducer. J Bacteriol 113:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meskys R, Harris RJ, Casaite V, Basran J, Scrutton NS. 2001. Organization of the genes involved in dimethylglycine and sarcosine degradation in Arthrobacter spp.: implications for glycine betaine catabolism. Eur J Biochem 268:3390–3398. doi: 10.1046/j.1432-1327.2001.02239.x. [DOI] [PubMed] [Google Scholar]
- 50.Weng ML, Zalkin H. 1987. Structural role for a conserved region in the CTP synthetase glutamine amide transfer domain. J Bacteriol 169:3023–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nyunoya H, Lusty CJ. 1984. Sequence of the small subunit of yeast carbamyl phosphate synthetase and identification of its catalytic domain. J Biol Chem 259:9790–9798. [PubMed] [Google Scholar]
- 52.Schleif R. 2010. AraC protein, regulation of the l-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol Rev 34:779–796. doi: 10.1111/j.1574-6976.2010.00226.x. [DOI] [PubMed] [Google Scholar]
- 53.Hashim S, Kwon DH, Abdelal A, Lu CD. 2004. The arginine regulatory protein mediates repression by arginine of the operons encoding glutamate synthase and anabolic glutamate dehydrogenase in Pseudomonas aeruginosa. J Bacteriol 186:3848–3854. doi: 10.1128/JB.186.12.3848-3854.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen C, Beattie GA. 2007. Characterization of the osmoprotectant transporter OpuC from Pseudomonas syringae and demonstration that cystathionine-beta-synthase domains are required for its osmoregulatory function. J Bacteriol 189:6901–6912. doi: 10.1128/JB.00763-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen C, Malek AA, Wargo MJ, Hogan DA, Beattie GA. 2010. The ATP-binding cassette transporter Cbc (choline/betaine/carnitine) recruits multiple substrate-binding proteins with strong specificity for distinct quaternary ammonium compounds. Mol Microbiol 75:29–45. doi: 10.1111/j.1365-2958.2009.06962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.