ABSTRACT
Many pathogens use homologous recombination to vary surface antigens in order to avoid immune surveillance. Neisseria gonorrhoeae, the bacterium responsible for the sexually transmitted infection gonorrhea, achieves this in part by changing the sequence of the major subunit of the type IV pilus in a process termed pilin antigenic variation (Av). The N. gonorrhoeae chromosome contains one expression locus (pilE) and many promoterless, partial-coding silent copies (pilS) that act as reservoirs for variant pilin information. Pilin Av occurs by high-frequency gene conversion reactions, which transfer pilS sequences into the pilE locus. We have developed a 454 sequencing-based assay to analyze the frequency and characteristics of pilin Av that allows a more robust analysis of pilin Av than previous assays. We used this assay to analyze mutations and conditions previously shown to affect pilin Av, confirming many but not all of the previously reported phenotypes. We show that mutations or conditions that cause growth defects can result in Av phenotypes when analyzed by phase variation-based assays. Adapting the 454 sequencing to analyze pilin Av demonstrates the utility of this technology to analyze any diversity generation system that uses recombination to develop biological diversity.
IMPORTANCE Measuring and analyzing complex recombination-based systems constitute a major barrier to understanding the mechanisms used to generate diversity. We have analyzed the contributions of many gonococcal mutations or conditions to the process of pilin antigenic variation.
INTRODUCTION
Neisseria gonorrhoeae (the gonococcus [Gc]) is the sole causative agent of the sexually transmitted infection gonorrhea, with an estimated 800,000 new cases per year in the United States (1) and an estimated 106 million cases worldwide (2). Gc is a human-specific pathogen whose infection is characterized by a robust immune response and a purulent exudate composed of polymorphonuclear leukocytes (PMNs) and Gc cells (3, 4). While Gc primarily colonizes the urogenital epithelium, ocular gonorrhea can occur in the eyes of newborns as they pass through the birth canal, resulting in a leading cause of blindness in developing countries (5). Gc infections are often asymptomatic, which can lead to serious complications such as cervicitis, epididymitis, endometriosis, or pelvic inflammatory disease, and although rare, Gc can also disseminate to the bloodstream, causing infective arthritis or endocarditis (1, 2).
One of the major virulence factors of Gc is the type IV pilus (TFP), a long hair-like appendage involved in DNA transformation, twitching motility, adherence to epithelial cells, and protection from PMN killing (6–10). The TFP is composed primarily of repeating units of the pilin protein PilE, and the exposed regions of the pilus surface are recognized by human antibodies (11). PilE is encoded by the chromosomal locus pilE (Fig. 1A and B) (12). The process of pilin antigenic variation (Av) alters the sequence of the pilE gene as a mechanism of generating diversity that affects the pilus function and allows avoidance of immune surveillance (13).
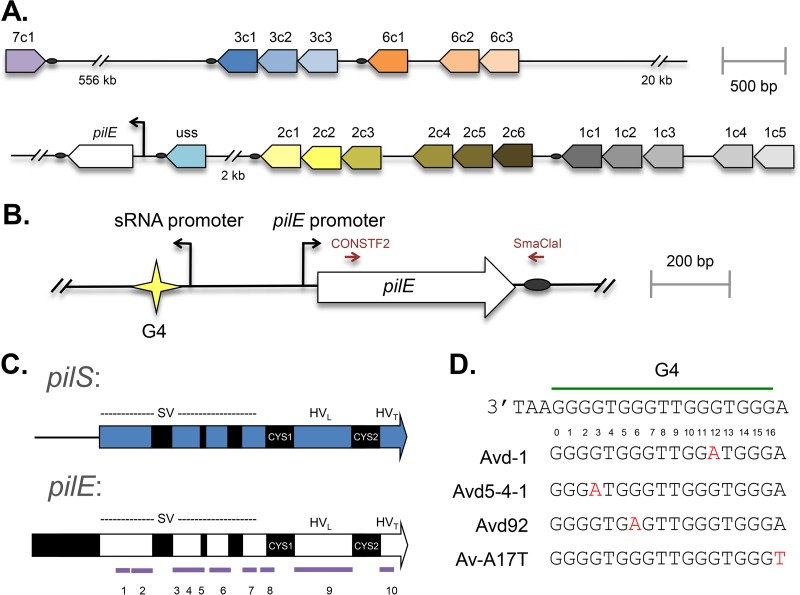
FIG 1.
Organization of pilin genes in N. gonorrhoeae. Representative maps of the pilE gene (white block arrows) and pilS loci (colored arrows), with the pointed ends showing the direction of the open reading frame (ORF), are shown. The black arrows represent functional promoters, the ovals represent the conserved Sma/Cla repeat (SCR), and the star indicates the G4 sequence. (A) Position, orientation, and relative distances between the pilin loci and the silent copies within each locus for strain FA1090. (B) The pilE gene and upstream region, with the positions of the primers used for 454 sequencing shown in red. (C) The constant (black) and variable (white or blue) regions of pilE and a representative pilS copy. The semivariable (SV), hypervariable loop (HVL), and hypervariable tail (HVT) regions are regions of heterology between the pilE gene and pilS copies, while cys1 and cys2 are conserved sequences among all pilin copies. The 10 purple lines represent the 10 regions used for silent-copy analysis. The figure is not drawn to scale. (D) The pilE G4 sequence with the G4 point mutations highlighted in red.
Pilin Av is mediated by a gene conversion process, a nonreciprocal transfer of DNA from a donor locus to a recipient locus without a change in the donor locus sequence. The pathogenic Neisseria species incorporate homologous DNA from numerous 5′-truncated silent pilS copies that act as a reservoir of variant pilin information. The pilE gene is approximately 500 bp long, with the first 150 bases encoding the conserved N-terminal domain of the protein that is not present in the pilS copies and therefore remains unchanged during pilin Av. Based on the level of homology with the silent copies, the remainder of the gene is defined as consisting of the semivariable (SV), cys1, hypervariable loop (HVL), cys2, and hypervariable tail (HVT) regions (Fig. 1C). In the FA1090 pilin variant expressed in human volunteer isolate 1-81-S2 (14), the SV region is from bp 150 to 360, with a highly variable region from bp 223 to 239. The cys1 and cys2 regions (bp 365 to 393 and 453 to 473) are identical in all pilin gene copies, each encoding a conserved cysteine residue that forms a disulfide bridge that is important to the structure of PilE (15). The sequences of the HVL and HVT show the greatest level of sequence divergence between the different silent copies. In N. gonorrhoeae strain FA1090, there are 19 pilS copies located at six separate loci (Fig. 1A) (16).
Pilin Av has one of the highest reported rates for a prokaryotic diversity generation system (17). A population of Gc cells grown for 19 generations will have a 10 to 13% frequency of pilE sequences that differ from the starting progenitor, approximately 6.8 × 10−3 recombination events per CFU per generation for FA1090 with the pilE variant 1-81-S2 (18). Anywhere from 1 bp to greater than 200 bp of heterology can be transferred during an Av event, and the transferred region is always bordered by regions of homology or microhomology that can range from 3 to 68 bp (18, 19). This frequency can be measured in different mutants and under different growth conditions to help determine the role of various proteins in pilin Av; the reduced Av frequency of a particular mutant generally means that its protein product is required for pilin Av.
There are many factors reported to be involved in pilin Av, particularly the proteins responsible for homologous recombination, RecA, RecX, RecO, RecR, RecJ, RecQ, RdgC, RecG, and RuvABC (20–26). Of these proteins, only RecA, RecO, and RecR are absolutely required for pilin Av, since loss-of-function mutations result in no measurable pilin Av, whereas loss-of-function mutations to the other genes produce an intermediate phenotype. RecA is the main bacterial protein for homologous recombination, responsible for strand invasion (27), and RecA's activity is modulated by the RecX and RdgC proteins in Neisseria (26, 28). RecO, RecR, RecJ, and RecQ are part of the RecF-like pathway in Neisseria (there is no RecF ortholog), which uses gapped DNA as a substrate for homologous recombination (29, 30). The RecFOR complexes assist RecA with loading onto DNA (31). RuvA and RuvB are helicases, which form a complex with RuvC to resolve Holliday junctions (32). RecG can also process Holliday junctions (33) and has roles in replication restart and DNA repair (reviewed in reference 29). Disruption of both RuvABC and RecG pathways results in a pilin Av-dependent synthetic lethality (25, 34, 35). Conditions that cause an increase in recombination, such as the absence of iron (36) or the loss of mismatch correction proteins (35, 37), lead to increased pilin Av. The RecBCD and RecN recombination proteins, which facilitate double-strand break repair, do not have a strong effect on pilin Av in FA1090 (24, 38).
There are two cis-acting sequences that have been implicated in pilin Av. At the 3′ end of each locus is a conserved 65-bp sequence termed the Sma/Cla repeat (SCR) (Fig. 1B). The SCR was found to be required for efficient pilin Av using a hybridization-based assay (39), and although proteins have been found to bind to the SCR (40), it is unclear whether the sequence provides a function other than an extended region of homology at the 3′ end of the pilE gene. Upstream of the pilE gene promoter is a 16-nucleotide (nt) G-rich sequence (Fig. 1D), which forms a guanine quartet (G4) structure in vitro (34). Mutation of the 12 GC base pairs that disrupt the G4 structure (numbered G1 to G3, G5 to G7, G10 to G12, and G14 to G16) results in loss of pilin Av, while mutation of the AT base pairs, which are not required for G4 structure formation, has no effect. Replacing the G4-forming sequence with alternative G4-forming sequences results in an Av-deficient (Avd) strain, while altering the orientation or strand of the G4 and associated transcript also leads to an Avd strain (34). Transcription of a noncoding RNA that initiates within the pilE G4-forming sequence (Fig. 1B) is also required for efficient pilin Av; however, expression of the noncoding RNA in trans does not complement a promoter mutant (41). Thus, this noncoding RNA is cis acting, its transcription is required to form the G4 DNA structure (17), and it is required for pilin Av. These findings have led to a model where the G4 structure is formed to disrupt replication and initiate this programmed recombination process (17).
Pilin Av has been measured by a variety of assays, including Southern blot hybridization and quantitative reverse transcription-PCR (RT-PCR) measurement of the transfer of specific HVL sequences into the pilE locus, counting the percentage of nonpiliated variants from piliated variants, quantifying the appearance of pilus-dependent colony morphology changes (PDCMC), or direct sequencing of pilE DNA amplified from random piliated progeny (18, 23, 39, 42, 43). The primary issue with many of these methods (Southern hybridization, RT-PCR, or colony phenotypes) is that only a subset of possible variants is measured. The most comprehensive means to measure pilin Av is by sequencing the pilE genes of potential variants, although the cost of individually sequencing hundreds of reads becomes impractical and the sample size is fairly small. We have therefore developed a next-generation sequencing assay to analyze pilin Av. Using 454 technology, we have assayed pilin Av in many mutants with mutations previously described to inhibit the process and under some growth conditions that have been reported to alter the process. A recently reported Illumina-based sequencing was used to assess pilin Av (44); however, there are many differences between that previous study and ours. Most importantly, we chose to use 454 sequencing because it can sequence a 400- to 600-bp amplicon, which allows the entire variable portion of each pilE gene to be determined. This allows an exact recording of each antigenic variation product, while the shorter reads in Illumina sequencing can record the changes that occur over the entire population but not each product. Although 454 sequencing technology is being phased out, other companies now offer amplicon sequencing for read lengths greater than 600 bp.
We have used 454 sequencing as an efficient and sensitive method for analyzing the frequency and character of N. gonorrhoeae pilin variants. The measured frequencies correlate well with previously reported values for many of the tested mutants. We confirm that (i) RecA, RecO, and the upstream G4 are critical factors in catalyzing pilin Av, (ii) RecQ, Rep, RecJ, RecX, RecG, RdgC, and iron sequestration are important factors in promoting pilin Av, but mutations still allow the process to occur at reduced frequencies, and (iii) RecB, RecN, and RuvB are dispensable for pilin Av. We also note that the donor silent-copy profile remains fairly consistent across different mutants, suggesting there is one central mechanism for pilin Av.
MATERIALS AND METHODS
Bacteria and growth conditions.
The strains used in this study (Table 1) were primarily derivatives of N. gonorrhoeae FA1090 recA6 (45) to prevent antigenic variation of the pilE gene in the absence of inducer. Cultures were revived from frozen stocks to GCB agar plates and routinely incubated for 22 h at 37°C with 5% CO2. GCB consists of 36.25 g GC medium base (Difco) containing 1.25 g of agar, Kellogg's supplements I (0.4% glucose [Sigma], 0.01% glutamine [Sigma], 0.0000002% cocarboxylase [Sigma]), and Kellogg's supplements II (0.0005 g ferric nitrate [Difco]) per liter. When required, isopropyl-β-d-thiogalactopyranoside (IPTG) (Diagnostic Chemicals) was added to 1 mM, and final concentrations of antibiotics were as follows: kanamycin, 50 μg/ml; chloramphenicol, 0.5 μg/ml; erythromycin, 2 μg/ml; and tetracycline, 0.2 μg/ml. Desferal (deferoxamine mesylate; Sigma) was added to plates at 7 μM.
TABLE 1.
Strains sequenced in this study
| Strain, mutant genotype, or conditiona | No. of reads | First reference |
|---|---|---|
| Parent 1 | 6,495 | 45 |
| Parent 2 | 5,977 | 45 |
| pilC1PL | 13,172 | 54 |
| recA9::ermC | 96,993 | 45 |
| recO::ermC | 42,107 | 21 |
| recQ::ermC | 9,907 | 21 |
| rep::tetM | 41,577 | 65 |
| recN::ermC | 17,769 | 24 |
| recB::ermC | 9,909 | 21 |
| recJ::kan | 6,458 | 24 |
| recX::ermC | 8,917 | 22 |
| rdgC1::ermC | 11,419 | 26 |
| recG::kan | 6,766 | 56 |
| ruvB::ermC | 10,261 | 25 |
| recAEC | 12,359 | 46 |
| recAXEC | 5,058 | 46 |
| Avd-1 | 126,968 | 34 |
| Avd92 | 119,079 | 34 |
| Avd-5-4-1 | 24,660 | 34 |
| Av-T17A | 7,575 | Laty Cahoon, unpublished data |
| −35 mut | 27,906 | 41 |
| Desferal | 10,010 | 36 |
| thrB::kan | 36,258 | 56 |
Except for the recA9, recAEC, and recAXEC mutants, strains are in the FA1090 recA6 background and all contain the pilE sequence 1-81-S2.
PDCMC assay.
The pilus-dependent colony morphology change (PDCMC) assay was performed as described previously (35). Each strain was spread onto a fresh GCB plate with IPTG, and colonies were examined under a stereomicroscope after 22 h. Ten colonies that were entirely piliated were selected, and the number of nonpiliated or underpiliated blebs was counted every 2 h, with each bleb receiving a score of 1, until 4 or more appeared, which was scored as a maximum of 4. The assay was repeated 6 to 11 times, and all colony scores were considered for the Student t test. The standard error of the mean (SEM) was provided for the average of each 10-colony PDCMC repeat.
Growth curve and colony titers.
The viability assays were performed as previously described (35). In brief, strains were spread onto plain GCB plates and GCB plates supplemented with IPTG. At different times points, e.g., 20 h or 40 h, 3 or 4 colonies were individually picked with a filter disk and dispersed into 500 μl GCBL (1.5% peptone protease no. 3 [Difco], 0.4% K2HPO4 [Fisher], 0.1% KH2PO4 [Fisher], 0.1% NaCl [Fisher]). Serial dilutions were plated onto GCB plates in duplicate, and the number of colonies was counted. Each assay was performed 4 to 6 times, and the SEM was provided for each of the averaged titers.
Gonococcal transformation.
Gonococcal genomic DNA was isolated from the donor strain by swabbing a half plate of a confluent lawn grown for 22 h into 1 ml GCBL and washed once in 1× phosphate-buffered saline (PBS). The pellet was resuspended in 180 μl ATL buffer from the Qiagen QIAamp DNA minikit, and total DNA was extracted following the manufacturer's instructions. The recipient strain FA1090 recA6 was transformed by plating a small patch on a GCB IPTG plate using 5 starting colonies. Ten microliters of supplemented GCBL containing 10 mM MgSO4 was mixed with 10 μl genomic DNA, which was placed on the lawn, dried, and grown for 22 h. The growth containing the spot was swabbed into GCBL and spread onto a GCB plate containing the antibiotic whose marker was linked to the mutation in the donor DNA. Colonies were streaked twice on plates with the antibiotic, and 5 or 6 candidates were saved for confirmation of the mutation (by the size of locus-specific PCR products) and for the identity of the pilE gene (by direct sequencing of the pilE PCR product) (Table 2).
TABLE 2.
Primers used
| Name | Sequence | Purpose |
|---|---|---|
| PILRBS | GGCTTTCCCCTTTCAATTAGGAG | pilE verification |
| SP3A | CCGGAACGGACGACCCCG | pilE verification |
| CONSTF2 | TACCAAGACTACACCGCCCG | 454 sequencing |
| SmaClaI | CAAACCCTTAAAAGACAAGC | 454 sequencing |
| LACPFOR | GAGCGGATAACAATTTCACA | recA6 verification |
| GcRecA-Rev2 | CAAAGCCGAAGAAACCGC | recA verification |
| GcRecA-For | ATGCGATAATGACCGACCG | recA verification |
| GCRECA-REV | AGATCTTTATTCTTCGGGTCGTTCGC | recA verification |
| ThrB-For | GACTTACCATGTGTTGTCGC | thrB verification |
| ThrB-Rev | AGCCGATATTCTGAACTGGG | thrB verification |
| RECO3 | GCATAATGTTTCCTTTTCTCTTCG | recO verification |
| RECO4 | TTCAGACGGCATTTCCACAAC | recO verification |
| RDGC3 | TTCAGCAAATAATTCCCGAC | rdgC verification |
| RDGC4 | AGGGCGAACATCATGGCG | rdgC verification |
| RUVBFOR2 | TGCCGTCTGAAACGCGCCG | ruvB verification |
| RUVBREV2 | CAAACGTCTGATAACAATGCCG | ruvB verification |
| ermCout1 | CAATTCTTATCTCTTTTCAATAGC | recB verification |
| ermCout2 | GAAGTAATAAAGTTTTGACTGTG | recB verification |
| RECB1 | ATTGCCGCCCTGTTTACGC | recB verification |
| RecJfor_N-His | AGCATACATATGTCAGTCAAAATCCAAACC | recJ verification |
| RecJrev_N-His | CGTCGCAAGCTTACGCCGCTTCCCAGTAG | recJ verification |
| RecN game primer S | ATGCTTCTAACACTTTCTTTGCG | recN verification |
| RecN3primeend | AAATAAACTATTTTTCGACGCAAG | recN verification |
| RECGREV | GTCCTGATTTTTGTTAATCCAC | recG verification |
| RECG1FOR | CCGCCGGGGCTTGATTCC | recG verification |
| GCRECXREV | TTAGAATTCTCAGCAGCTGTCTTCCC | recX verification |
| GCRECXFOR | GATCTTAATTAAATTTACGAAAACCTGCAA | recX verification |
| Repfor_N-His | GAATTCCATATGATGAAACTCAATGCCCAA | rep verification |
| Reprev_N-His | TTAATCCGCCGGGCCGCCCTAGGGCACGT | rep verification |
| RecQver_for | ACCGACCAACACCCTTAC | recQ verification |
| RecQver_rev2 | TTAAACAGAGGCAATGCCG | recQ verification |
| pilCdownstream | CCATCTTTGGCGGTACCCTCGCTG | pilC verification |
| PilCPLfor | GGCGGAGGTGGCGGGGCC | pilC verification |
Preparation of samples for deep sequencing.
Mutant strains were revived from frozen stock to fresh GCB plates and grown for 22 h. One colony was picked with a filter disk and dispersed into 500 μl GCBL. One microliter was diluted into 500 μl GCBL, and 30-, 40-, and 50-μl portions were spread onto fresh GCB IPTG plates and grown for 22 h. Between 265 and 370 colonies (average, 323.5; median, 321.5) were collected in 1.5 ml GCBL using a glass spreader. The cells were pelleted and resuspended in 180 μl ATL buffer (Qiagen), and total DNA was extracted by using the QIAamp DNA minikit (Qiagen) according to the manufacturer's instructions. The pilE region was amplified using 5 to 15 ng genomic DNA with high-pressure liquid chromatography (HPLC)-purified primers based on CONSTF2 and SmaClaI from IDT (Table 2).
Primer design was based on 454 sequencing application brief no. 001-2009: forward primer (primer A-Key), 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAG-(10-nt MID)-CONSTF2; reverse primer (primer B-Key), 5′-CCTATCCCCTGTGTGCCTTGGCAGTCTCAG-SmaClaI. Multiplex identifiers (MIDs) were designed using 454 sequencing technical bulletin no. 005-2009.
Primers were input into IDT's Oligo Analyzer website to ensure a ΔG greater than −10 kcal/mol (most were −6.75 kcal/mol). KOD Hot Start DNA polymerase (Toyobo) was chosen for its high fidelity. All polymerases tested produced a second faint band. Each reaction mixture was prepared in a 50-μl volume, split into 3 tubes of ∼17 μl during the reaction (95°C for 2 min; 29 cycles of 95°C for 20 s, 58°C for 10 s, and 70°C for 10 s; and 70°C for 5 min), and then combined back together for the 580-bp band to be excised from a 0.8% agarose gel using a gel extraction kit (Qiagen). Typical yields were 3 μg of PCR product. Sample concentrations were measured with a NanoDrop instrument (ND-1000), and the samples were visualized using an Experion Bioanalyzer (Bio-Rad).
Strains with growth defects.
FA1090 containing the Escherichia coli recAX genes (recAXEC) and FA1090 recA6 with ruvB and recG were grown for 26 h, FA1090 recA6 with 7 μM deferoxamine mesylate was grown for 28 h, and FA1090 recA6 recB1 was grown for 40 h. The titers (CFU/ml) of these strains at those time points was equivalent to that of the parent FA1090 recA6 grown on IPTG for 22 h. Iron sequestration of Desferal was confirmed by Western blotting of the TbpB protein as described previously (36).
Deep-sequencing assay.
One microgram of each sample was sent separately to 454 Life Sciences (Roche Diagnostics) at the 454 Sequencing Center in Branford, CT, where they were combined so that 27 samples with normal to high expected Av frequencies were run on one quarter of a plate, 16 reduced-frequency samples were run on a second quadrant, and 6 Av-deficient samples were run on the remaining half plate. Each sample was collected once, except for FA1090 recA6, which was sampled in triplicate (although only two provided sufficient reads for analysis). The samples were run using GS FLX with titanium chemistry, and a total of 1,173,399 reads were generated.
Analysis of variants.
GS Reference Mapper v2.7 (Roche) was used to map the percentage of variants at each nucleotide position. Default settings were chosen, with the exception of the “-srv” parameter, which allowed for all variants, even if they occurred only once. The data were exported to Microsoft Excel and plotted as the frequency of variants per nucleotide base of sequence. This work was assisted by the Northwestern University NGS Core Facility. The average number of nucleotide changes in the SV (positions 122 to 359) and HVL (positions 395 to 447) regions was calculated by taking the average percentage of detected variants divided by the length of the tract size according to the formula [(SV × 3.5) + (HVL)]/2.
In-house BioPerl program to identify silent copies.
The FA1090 1-81-S2 variant pilE gene was divided into 10 different regions (positions relative to the ATG starting nucleotide are as follows: R1, 159 to 181; R2, 187 to 210; R3, 223 to 239; R4, 253 to 273; R5, 274 to 288; R6, 289 to 305; R7, 315 to 332; R8, 347 to 365; R9, 394 to 447; and R10, 474 to 494), (Fig. 1C). The variant sequences for each region were defined for all 19 silent copies of FA1090. In many regions, a particular sequence was present in more than one silent copy. For example, CGTTACCGGGTATTGCCCGAATC in R1 is found in both 1c2 and 2c6. The sequences of each region were aligned based on the boundaries between the 10 regions and compared to the library of silent-copy sequences. If an exact match to one or more silent copies was found in a region, the potential donor(s) was identified. If an exact match to a donor silent copy was not found, the Wagner-Fischer algorithm (http://search.cpan.org/∼davidebe/Text-WagnerFischer-0.04/WagnerFischer.pm) was used to find the best match between the variant and one of the silent copies. If the Wagner-Fisher distance was 1 (e.g., one insertion or deletion relative to the donor), “mismatch” was returned for that region; otherwise, the sequence was returned for inspection. Mismatched and other unassigned reads were manually assigned to a donor until the total number of unassigned reads was approximately equal to 1% of the total reads. Single-nucleotide changes were found in the variants that could have originated from multiple silent copies, and these were identified as “var” to denote that the single-nucleotide change was part of a pilin antigenic variation event but could not be assigned to a particular donor or two. Once all 10 regions were analyzed, each region was mapped onto a 19-bit number. If a region could have come from several silent copies, a “1” appeared in each place in the 19-bit number. For example, if the sequence within a region could have come from silent copy 2c1 or 6c1, the binary number for that region was 0b0000100000000100000. The reference sequence was mapped to the 19-bit binary number 0b1111111111111111111. The binary “AND” was taken for the 10 regions to identify the silent copy. If there was a match to exactly one silent copy, that silent copy was identified as the donor for that variant. The verdict was the result of the binary AND, excluding “var” regions adjacent to named silent copies (indicating that the boundary of the silent copy fell in the middle of the region). If the verdict was all 0s, meaning the read contained two or more silent copies, then the verdict of a double crossover (meaning multiple recombination events) was returned; e.g., if region 2 came from silent copy 2c3 = 0b0000000000010000000, region 7 came from silent copy 6c1 = 0b0000100000000000000, and the rest of the regions were REF = 0b1111111111111111111, the binary AND of these numbers would be 0b0000000000000000000. One special sequence was identified as 2c1_6c1 (which matched the HVT region for pilS copies 2c1 and 6c1). Short reads (less than the full-length variable region) were excluded from analysis.
The program can be accessed at https://github.com/davidwebber/silent-dna.
RESULTS AND DISCUSSION
Selection of mutants and conditions.
In order to develop a pilin Av assay based on 454 sequencing, we chose a representative set of mutant strains and conditions that have all previously been shown to affect the frequency of pilin Av by the percentage of nonpiliated colonies or by using the PDCMC assay. A list of mutants and conditions analyzed can be found in Table 1. The recombination mutants used in this study had recA, recO, recJ, recX, rdgC, recG, recN, recB, ruvB, rep, or recQ inactivated. Strains expressing the Escherichia coli recA and recAX genes in place of the Gc recA were also tested, as were growth in iron-limited conditions. A series of G4-related mutants were also analyzed, including those with single-nucleotide changes to the pilE G4 sequence (Fig. 1D) and disruption of the −35 region in the promoter of the G4-associated small RNA (sRNA).
Strain and sample preparation.
Most mutants were backcrossed by transformation into N. gonorrhoeae strain FA1090 recA6 containing the pilE from human volunteer isolate 1-81-S2 (14) to ensure an isogenic background. The recAEC and recAXEC (46) and the recA9 null (45) mutations were transformed into FA1090 with the 1-81-S2 pilE sequence. The recA6 allele is under the control of a lac operator, allowing for the regulated expression of recA and pilin Av with IPTG (45). The more recently constructed mutants Avd1, Avd92, Avd5-4-1, and Av-T17A and the pilE G4-associated RNA −35 promoter mutant (34, 41) were analyzed without backcrossing.
Strains with a normal growth rate were grown for 22 h (19 to 20 generations) on medium with IPTG to allow for expression of recA and pilin Av, while strains with growth defects (recAXEC, ruvB, recG, and recB) were grown longer to achieve the same number of generations as the parent strain at 22 h. Total DNA was extracted from ∼300 combined colonies, and the variable portion of pilE was amplified with primers CONSTF2 and SmaClaI (Fig. 1B) (47). CONSTF2 anneals 90 bp into the pilE gene without homology to any pilS copies, while the SmaClaI primer anneals downstream of the gene in the SCR. Including the extra 40 nt on each primer, the PCR product is ∼580 bp, falling within the optimal range for 454 sequencing. The samples were gel purified to eliminate any contaminating PCR products and submitted for emulsion PCR and 454 sequencing. DNA concentrations in the sequencing reaction mixtures were adjusted so that mutants previously reported to have lower pilin Av were overrepresented in the sample to attempt to provide increased sequencing depth (Table 1).
Overview of the sequencing results.
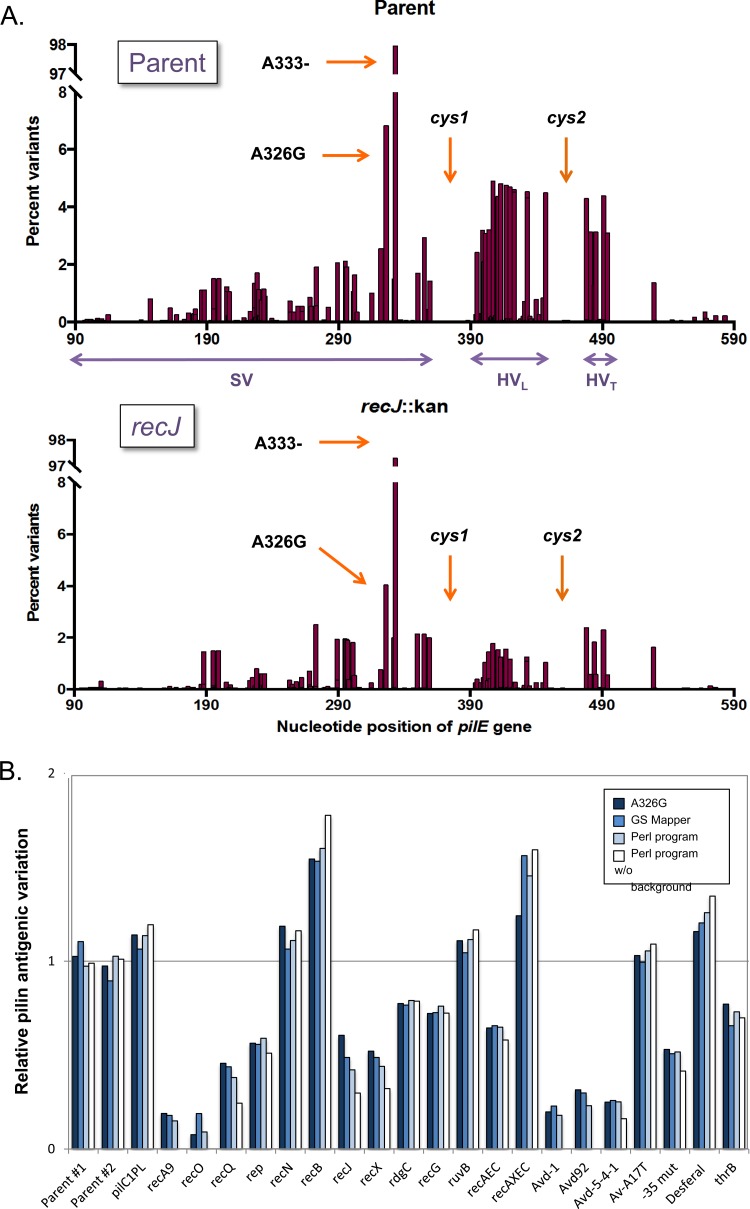
The number of reads per sample ranged from 5,000 to 125,000 (Table 1), with sufficient data for all mutants to perform a complete analysis. We measured the frequency of nucleotide changes for each variant at each base pair compared to the parental pilE sequence using GS Mapper (Roche), with examples of the parent and a mutant with reduced pilin Av shown in Fig. 2A. This analysis showed that sequence changes were found in all the variable regions but not in the conserved base pairs; as expected, the majority of variations were within the HVL and HVT regions, while the conserved cys1 and cys2 regions had relatively few changes present. One prominent change recorded in about 98% of all the reads was at a portion of the gene containing an A6 homopolymer (positions 328 to 333), which the sequencing reported as losing one nucleotide to create an A5 polynucleotide run (Fig. 2A). Insertion or deletion of nucleotides at a homopolymeric tract is a known issue of 454 sequencing (48), and this variation was not present using traditional sequencing; therefore, it was discounted as a sequencing artifact. The second most prominent change was A326G (Fig. 2A), a centrally located variation present in the SV region of 15/19 pilS copies but not in the starting pilE of 1-81-S2. This A326G change occurred both singly and as parts of longer recombination tracts (data not shown). In the two parental samples, the frequencies of reads containing the A326G transition were 6.84 and 6.5%, while a mutant with reduced pilin Av, such as a recJ mutant, gave 4.0% (Fig. 2A). It is interesting to note that this single change proved to be a good indicator of the overall Av frequency (Fig. 2B). The A326G value could be used as a simple method to quantify pilin Av, but it is not comprehensive and can be used only when the starting pilE sequence does not already contain a G at 326.
FIG 2.
Pilin antigenic variation of mutants. (A) Graphs of pilin Av plotted for FA1090 recA6 (parent) 1 and its recJ::kan derivative using the program GS Mapper. The distribution shows the percentage of variant nucleotide changes per base pair for each strain. The A333- change, appearing in 97 to 99% of reads, is indicated, as well as the second most common change A326G. The locations of the cys1 and cys2 regions are also indicated. (B) Comparison between the percentage of A326G changes, the average percent changes of the SV and HVL regions from the GS Mapper program, the calculated pilin Av frequencies from the Perl program (light blue), and the calculated pilin Av frequencies with the background subtracted, based on Table 3 (white). Values are represented relative to the average of the two parent strains set to 1.0.
We investigated whether further analysis of the GS Mapper data could determine the relative pilin Av frequency more accurately by looking at the average number of nucleotide changes in the SV and HVL regions (Fig. 2B). This analysis provided a relative measurement of the frequency of pilin Av on par with the level of A326G mutations and is compatible for either 454 or Illumina sequencing, but it is limited since it cannot determine the variant sequences of individual antigenic variants.
We created a BioPerl script to calculate the percentage of variant reads in regard to total reads and to identify the silent-copy donors for each variant. We divided the pilE sequence (from the SV through the HVT region) into 10 regions (Fig. 1C) and matched the DNA sequence of each region either to the parental sequence or to one or more donor silent-copy sequences. The output of the script reported which silent copy (or copies) was the donor for each region. If consecutive regions matched the same donor copy, this result was assumed to represent a recombination event from only that donor. Alternatively, if two adjacent regions had received two sequences originating from different donor silent copies, the variant was identified as having multiple recombination events. For every sample, including mutants previously reported as being totally deficient in pilin Av, there was a small percentage of variant sequences that could not be assigned to any silent-copy donor. In some cases, these unassigned reads contained simple insertion/deletions (indels) or substitutions that were not found in any silent donor copy and likely represented sequencing errors common to 454 sequencing (49, 50). Other variant sequences that could not be assigned to a silent-copy donor had a recombination crossover within one of the 10 arbitrarily defined regions. For example, there were several variants that had crossovers within the HVL region (region 9), which has been previously reported (18, 51). We manually identified common indels and crossover points to include these in the variant analysis. The overall number of the unassigned reads was reduced to between 0.5% and 1.5% in the different samples. The results of the BioPerl analysis are presented in Table 3, graphically depicted in Fig. 2B, and further discussed below.
TABLE 3.
Comparison of pilin antigenic variation frequencies
| Strain, mutant genotype, or conditiona | Relative pilin Avb determined by: |
% Av determined from 454 sequencing | |
|---|---|---|---|
| Previous methodsc | 454 sequencing | ||
| Parent 1 | +++ | +++ | 10.58 |
| Parent 2 | +++ | +++ | 10.81 |
| pilC1PL | +++ | +++ | 12.77 |
| recA9d | − | − | − |
| recO | − | − | − |
| recQ | + | + | 2.61 |
| rep | ++ | ++ | 5.46 |
| recN | +++ | +++ | 12.43 |
| recB | +++ | ++++ | 19.00 |
| recJ | + | + | 3.19 |
| recX | + | + | 3.44 |
| rdgC | ++ | ++ | 8.42 |
| recG | + | ++ | 7.74 |
| ruvB | + | +++ | 12.48 |
| recAECd | ++ | ++ | 6.21 |
| recAXECd | ++++ | ++++ | 17.04 |
| Avd-1 | − | − | − |
| Avd92 | − | − | − |
| Avd-5-4-1 | + | + | 1.73 |
| Av-A17T | ND | +++ | 11.67 |
| −35 mut | ++ | + | 4.44 |
| Desferal | ++++ | ++++ | 14.40 |
| thrB | ++ | ++ | 7.47 |
All strains are in the FA1090 recA6 background, except as noted.
−, Av deficient; +, severely deficient; ++, moderately deficient; +++, parental levels of variation; ++++, increased levels of Av; ND, not determined.
Previously analyzed by PDCMC assay, PCR, RT-PCR, percent nonpiliated, and direct sequencing (18, 21, 22, 24–26, 34–36, 38, 41, 43, 46, 56, 61, 62, 65).
FA1090 recA+ background.
Analysis of pilin Av of the parental strain and Avd mutants.
We were puzzled that strains that had previously been reported to be pilin Av deficient (Avd) showed low levels of variation (e.g., the recA9, recO, Avd-1, and Avd92 strains). One silent copy, pilS7 copy 1 (7c1), was the most common donor sequence recorded in the Avd strains (the recO construct was missing pilS 7c1 and did not have this issue), followed by the donor sequences 1c1, 3c1, and 2c1/6c1 (Table 4). Since we did not detect pilS 7 copy 1 variants at this level using traditional sequencing-based assays, we reasoned that these “variants” might have been artifacts produced during the PCR amplification of the pilE gene. Recombination can occur between extension products produced from the reverse primer in the copy 1 silent copies and extension intermediates produced from the CONSTF2 primer at pilE to produce a spurious signal (47). To test this hypothesis, we used an oligonucleotide probe specific for the pilS 7 copy 1 HVL region to probe for this sequence in PCR products produced from recA9 DNA (data not shown). The PCR conditions were varied by (i) lowering the PCR template DNA to <1 ng per reaction mixture, (ii) lowering the PCR cycling to <25 rounds, and (ii) using Phusion or FastStart polymerase. The combination of these conditions reduced the presence of the 7c1 HVL sequence in the PCR products to undetectable levels (data not shown). Future uses of this technique will use the conditions that suppress PCR recombination.
TABLE 4.
Examples of the silent-copy breakdown by number of reads
| Donor copy | No. of reads for: |
||||
|---|---|---|---|---|---|
| Parent 1 | recA9 mutant | recO mutant | Avd-1 | Avd92 | |
| Unknown nonparental | 108 | 571 | 286 | 737 | 916 |
| pilS1c1 | 100 | 95 | 75 | 127 | 195 |
| pilS 1c2 | 7 | 2 | 3 | 10 | 14 |
| pilS 1c3 | 3 | 2 | 2 | 3 | 8 |
| pilS 1c4 | 0 | 1 | 0 | 3 | 1 |
| pilS 1c5 | 7 | 2 | 0 | 254 | 2 |
| pilS 2c1 | 21 | 0 | 3 | 4 | 0 |
| pilS 2c1/6c1 | 157 | 11 | 9 | 81 | 34 |
| pilS 2c2 | 3 | 0 | 0 | 0 | 1 |
| pilS 2c3 | 5 | 0 | 0 | 1 | 2 |
| pilS 2c4 | 3 | 0 | 0 | 1 | 0 |
| pilS 2c5 | 0 | 0 | 0 | 0 | 0 |
| pilS 2c6 | 10 | 0 | 0 | 0 | 0 |
| pilS 3c1 | 198 | 68 | 59 | 171 | 155 |
| pilS 3c2 | 21 | 2 | 4 | 5 | 16 |
| pilS 3c3 | 22 | 23 | 13 | 30 | 41 |
| pilS 6c1 | 26 | 13 | 9 | 37 | 43 |
| pilS 6c2 | 3 | 16 | 19 | 40 | 25 |
| pilS 6c3 | 0 | 0 | 0 | 0 | 0 |
| pilS 7c1 | 87 | 1,041 | 1 | 1,396 | 2,089 |
| Multiple events | 26 | 13 | 3 | 22 | 33 |
| Parental | 5,645 | 95,041 | 41,594 | 123,903 | 115,392 |
| uss | 2 | 2 | 0 | 3 | 3 |
| Unassigned variant | 41 | 90 | 27 | 140 | 109 |
| Totala | 6,495 | 96,993 | 42,107 | 126,968 | 119,079 |
Number of reads counted, excluding short or truncated reads.
The recA9::ermC, recO::ermC, Avd-1, and Avd92 mutants have been previously reported to be Avd (21, 34, 45). To adjust the results to account for the background of in vitro recombination artifacts, the average percent variation (variant reads per total reads) of these four Avd strains was subtracted from all samples. This factor estimates the contribution of the in vitro recombination to the calculated pilin Av frequencies but weakens the conclusions we can make for the true Avd mutants. However, since the in vitro recombination background is a small proportion of the error in the other data, we are confident that those pilin Av frequencies are close to the actual frequencies.
The two FA1090 recA6 parental strain samples showed 10.58% and 10.81% variant pilE sequences after background correction (Table 3). These values are consistent with results reported in previous assays using traditional sequencing of only piliated progeny, which reported frequencies of between 10 and 13% variants using the identical strain with the identical starting pilE grown under the same conditions (18, 38).
Effect of pilC phase variation.
Previous sequencing assays showed different spectra of donor pilS copies when piliated and nonpiliated progeny were analyzed separately (18). Loss of piliation can occur by several mechanisms: (i) insertion of a stop codon encoded in a subset of pilS copies, (ii) PilE variants that are poorly assembled into pili, (iii) deletion of the pilE gene (52), or (iv) slipped-strand phase variation at one of the two accessory pilC genes (53). The first two mechanisms are the result of pilin antigenic variation and will be recorded in the overall frequency. Strains with pilE deletions will not amplify with the primers used and will not be present in our analysis. To determine the contribution of slipped-strand phase variation, we included a phase-locked pilC mutant (54).
The phase-locked pilC mutant showed a pilin Av frequency of 12.77% variants/pilE, which was slightly higher than that of the isogenic pilC phase-variable parent (Table 3). This slight increase in the frequency can most likely be attributed to the fact that nonpiliated Gc (of which a pilC mutant is a subtype) grow faster than piliated Gc (Fig. 3). A majority of the cells that are nonpiliated due to the loss of PilC would still retain the parental pilE sequence and result in an effective increase in the number of parental reads relative to the phase-locked pilC mutant.
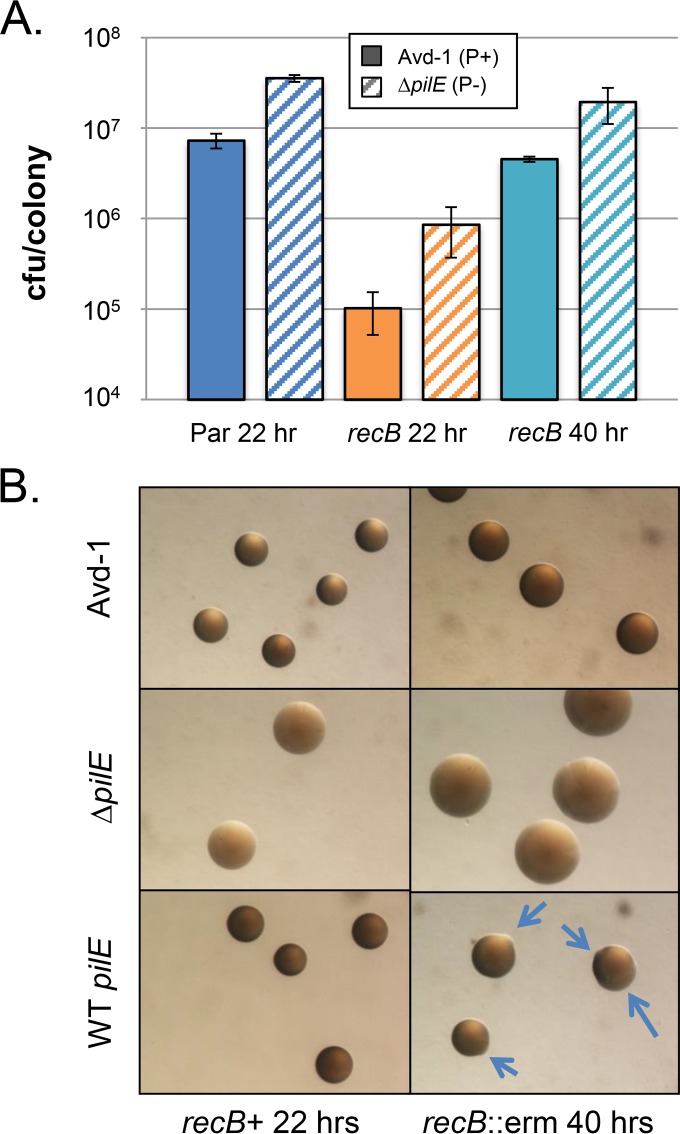
FIG 3.
Effect of piliation on the growth of a recB mutant. (A) Growth of piliated versus nonpiliated parental and recB mutants. The titer of the parent FA1090 recA6 strain (Par) after 22 h of growth is compared to that of its recB::ermC derivative grown for 22 and 40 h. The solid bars indicate a completely piliated background (Avd-1), while the hatched bars indicate a completely nonpiliated background (ΔpilE). P+, piliated; P−, nonpiliated. (B) Colony appearance of the parental background strain (left) and the recB::ermC mutant (right) grown in the presence of IPTG for the indicated times. Top row, piliated Avd-1 background; middle row, nonpiliated ΔpilE background; bottom row, pilin Av-proficient background. The arrows point to emerging nonpiliated blebs.
Pilin Av of recombination mutants.
RecJ is a 5′-to-3′ single-strand exonuclease involved in recombination and repair (55) and contributes to the RecF-like pathway of single-strand gapped repair that is required for pilin Av (24). A recJ::kan mutant was reported to have a low level of pilin Av as measured by the ratio of nonpiliated to piliated colonies (24), and an independent transposon insertion in recJ was isolated in a screen for pilin Av mutants using the PDCMC assay (56). The small amount of variants in the recJ::kan mutant was confirmed by this sequencing assay, showing a frequency of 3.19% variants/pilE (Table 3).
RecQ is an evolutionarily conserved 3′-to-5′ DNA helicase that is responsible for proper repair of DNA damage by unwinding duplex DNA (57). RecQ family members are also important helicases for dissolving G4 structures (58–60). The gonococcal RecQ has a unique triplication of the C-terminal HRDC domain that is required for pilin Av (61, 62). An insertion in recQ was originally identified using a PCR-based assay (21, 63) and a PDCMC screen for pilin Avd mutants (56). Using the deep-sequencing assay, we confirmed that the recQ::ermC mutant was necessary for pilin Av, with 2.61% variants/pilE (Table 3).
The Rep helicase in E. coli is required for replication progression and replication restart and prevents illegitimate recombination at short regions of homology (64). In Gc, the Rep protein has been shown to participate in DNA transformation and pilin Av, as measured by the percentage of nonpiliated progeny colonies and the PDCMC assay (65). The deep-sequencing assay confirmed the intermediate phenotype of the rep::tetM mutant, with a pilin Av frequency of 5.46% variants/pilE (Table 3). This result is consistent with the previous assays, and it is possible that Rep is partially redundant for pilin Av with another helicase.
The E. coli RecX and RdgC proteins are inhibitors of RecA activity (66–68). In contrast to that of E. coli, the gonococcal RecX acts as a recombination enhancer by facilitating RecA disassembly (28), and a recX::ermC mutant was shown to have reduced pilin Av levels by a reduced percentage of colonies with the nonpiliated phenotype (22). In the deep-sequencing assay, the recX::ermC allele yielded a variant/pilE level of 3.44%. The RdgC protein of E. coli inhibits the strand exchange activity of RecA (67), but in Gc, an rdgC mutant was shown to have reduced frequencies of pilin Av by the PDCMC assay (26). It is possible that while RdgC in E. coli physically blocks RecA polymerization by competitively binding to double-stranded DNA (dsDNA) (67), it could act similarly to RecX to limit RecA activity to enhance pilin Av. The rdgC1::ermC mutant produced 8.42% of pilE variants by the deep-sequencing assay (Table 3), confirming an enhancing role for RdgC in pilin Av. These data confirm that these negative regulators of RecA activity both act as positive modulators of pilin Av in Gc.
The Gc RecA and RecX proteins show many differences compared to the E. coli orthologs. Like in many bacteria, the E. coli recA gene is upstream of the recX gene in an operon that is regulated by the SOS regulon, while in Gc, the two genes are carried separately, and this organism does not have a traditional SOS regulon (69, 70). Replacing the Gc recA with the E. coli ortholog partially restored pilin Av, as measured by the production of nonpiliated variants from a piliated progenitor (46). Curiously, when the Gc recA locus was replaced with the E. coli recAX operon, pilin Av was increased over that of the same strain expressing the Gc recA gene (46). Consistent with earlier results, the recAEC strain showed 6.21% pilin Av by the deep-sequencing assay (Table 3), confirming that the E. coli RecA cannot completely substitute for the Gc RecA for pilin Av. The strain with the E. coli recAX operon replacing the Gc recA showed an increased pilin Av frequency (17.04% variants/pilE) by the deep-sequencing assay (Table 3), confirming that coexpression of RecXEC with RecAEC elevates the frequency of pilin Av by enhancing the ability of the E. coli RecA protein to mediate pilin Av. It is likely that the stabilization of E. coli RecA by RecX in Gc (46) results in the increased frequencies, which suggests that the amount of RecA is limiting during pilin Av. However, the enhancement could also be due to the limitation of E. coli RecA activity by E. coli RecX, similar to the effect of Gc RecX on Gc RecA (22, 28).
RecN is part of the RecF-like pathway of DNA repair in Gc (24). Although RecN is required for efficient transformation and repair of UV damage in Gc, the recN mutant showed the same level of pilin Av as its parent strain by PDCMC (24). Consistent with this earlier result, the recN::ermC mutant showed 12.43% variants/pilE in the deep-sequencing assay (Table 3), confirming that the RecN pathway is not required for pilin Av.
The frequency of pilin Av measured for the recB mutant was increased over that for the parent strain with a frequency of 19.00% variants/pilE (Table 3). While there have been reports of recD mutants having increased pilin Av (71) and recB mutants having decreased pilin Av (72), the sequencing data here suggest that pilin Av does not depend on RecBCD activity. There are two possible explanations for the increase in variants. First, if the RecBCD complex is destroying nonproductive intermediates with double-strand breaks, this would increase the frequency. However, we do not favor this explanation but rather believe that the enhanced pilin Av is explained by the severe growth defect of the recB mutant and the higher growth rate of nonpiliated or underpiliated cells compared to piliated variants (Fig. 3). The recB mutant takes 40 h to grow to the same CFU/colony as its recB+ parent in 22 h (Fig. 3A), and the size and amount of cell matter per colony are greater in the recB mutants (Fig. 3B). The faster-growing nonpiliated cells outcompete their piliated siblings, resulting in higher cell numbers during the same time period, independent of recB status (Fig. 3B). This can be seen by the increase of nonpiliated (and underpiliated) blebs that emerged and outcompeted the slower-growing piliated variants (Fig. 3B, bottom) and the increase in known nonpiliated pilS donors (2c1, 2c4, and 3c2) (Table 5). This view is substantiated by the previous report using standard sequencing technology on only piliated recB mutant progeny, which did not show an increased frequency of pilin Av (38).
TABLE 5.
Silent-copy usage in strains with elevated nonpiliated variantsa
| Donor copy | Usage (%) in: |
|||
|---|---|---|---|---|
| Combined parent strains | recB mutant | ruvB mutant | recAXEC mutant | |
| Unknown nonparental | 8.46 | 10.89 | 7.87 | 10.08 |
| pilS 1c1 | 13.37 | 7.31 | 7.90 | 5.70 |
| pilS 1c2 | 0.51 | 0.81 | 0.88 | 1.93 |
| pilS 1c3 | 0.20 | 0.09 | 0.00 | 0.09 |
| pilS 1c4 | 0.07 | 0.05 | 0.23 | 0.23 |
| pilS 1c5 | 0.89 | 1.70 | 2.48 | 0.51 |
| pilS 2c1 | 2.74 | 5.72 | 3.34 | 15.30 |
| pilS 2c1/6c1 | 26.27 | 22.19 | 29.11 | 16.17 |
| pilS 2c2 | 0.45 | 0.58 | 0.70 | 0.23 |
| pilS 2c3 | 0.51 | 0.26 | 0.15 | 0.11 |
| pilS 2c4 | 0.22 | 1.59 | 1.17 | 1.74 |
| pilS 2c5 | 0.00 | 0.21 | 0.08 | 0.12 |
| pilS 2c6 | 0.80 | 1.70 | 1.72 | 1.97 |
| pilS 3c1 | 27.31 | 21.78 | 20.91 | 8.93 |
| pilS 3c2 | 1.99 | 5.64 | 3.77 | 11.10 |
| pilS 3c3 | 2.42 | 3.41 | 4.54 | 3.43 |
| pilS 6c1 | 2.34 | 2.21 | 1.83 | 2.99 |
| pilS 6c2 | 0.08 | 0.12 | 0.24 | 1.81 |
| pilS 6c3 | 0.00 | 0.11 | 0.08 | 0.00 |
| pilS 7c1 | 1.85 | 4.37 | 1.38 | 7.75 |
| Multiple events | 3.21 | 4.48 | 2.52 | 4.89 |
| uss | 0.21 | 0.10 | 0.06 | 0.22 |
| Unassigned variant | 6.10 | 4.68 | 9.04 | 4.69 |
| Total | 100.00 | 100.00 | 100.00 | 100.00 |
Bold indicates silent-copy incorporation known to cause a nonpiliated or underpiliated colony morphology.
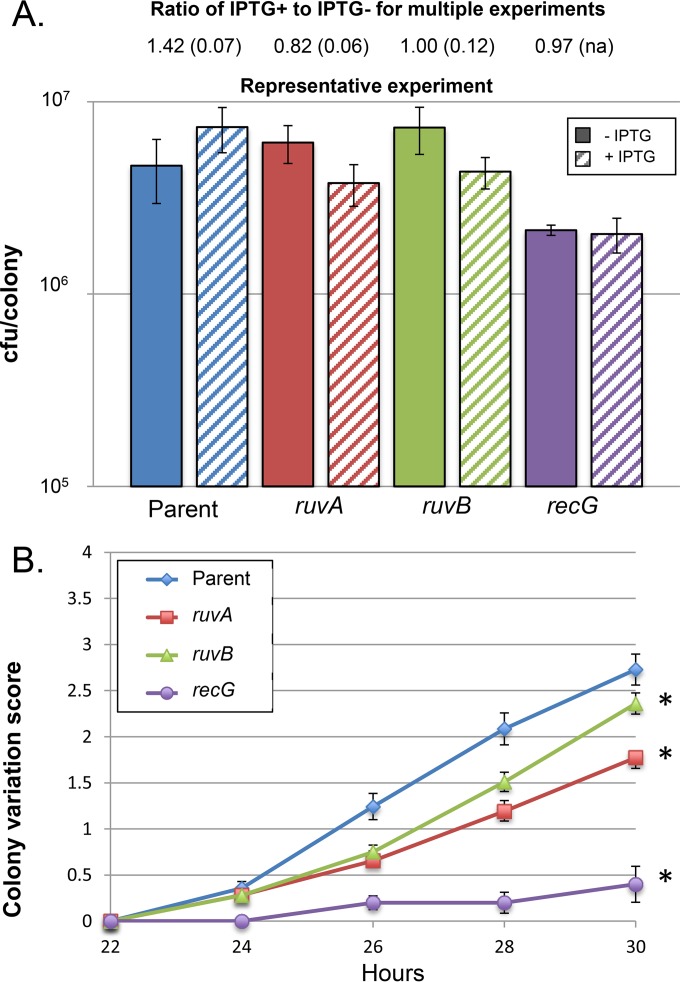
RuvABC and RecG resolve Holliday junctions that form during recombination (29, 32, 73). An insertion in ruvA and two insertions in ruvB were initially found by the transposon screen to mildly reduce pilin Av by PDCMC (56). Subsequent analysis of complete deletions of ruvA, ruvB, ruvC, and recG showed significant decreases in pilin Av for all four mutants by PDCMC (25). However, the ruvA, ruvB, and recG mutants all show a growth defect in the presence of IPTG-induced homologous recombination (Fig. 4A). Both the ruvB::ermC and recG::kan strains were grown for 26 h to account for the lower growth rate. The deep-sequencing data reveal that the recG::kan mutant was moderately decreased for pilin Av, showing a frequency of 7.74% variants; however, the ruvB::kan mutant did not show decreased levels of pilin Av, with a frequency of 12.48% variants/pilE (Table 3). Repeating the analysis of ruvA::kan, ruvB::ermC, and recG::kan mutants by PDCMC assay showed the recG mutant to be greatly reduced for pilin Av, but the ruvA and ruvB mutants had only a slight defect in Av that was proportional to the decrease in growth (Fig. 4B). The ruvB mutant also showed an increased use of nonpiliated or underpiliated donor silent copies (Table 5). Although the ruv mutants are involved in recombination (and other aspects thereof), they appear not to be directly required for pilin Av, and their identification as Avd was due to the reduced growth rate. This finding appears to be in contrast to the reported synthetic lethality of the Ruv system and RecA when pilin Av is allowed (25, 34). These results strongly suggest that the RuvABC complex is necessary to resolve intermediates that are formed during pilin Av when RecG is inactivated but that the RuvABC system is not by itself required for the recombination process.
FIG 4.
Growth rate and colony variation scores of ruvA, ruvB, and recG mutants. (A) CFU/colony for the parent FA1090 recA6, ruvA::kan derivative, ruvB::ermC derivative, and recG::kan derivative grown without IPTG and in the presence of IPTG. Shown are results from a representative experiment, where the error bars represent the SEM for 3 to 5 colonies. Above the graph is the ratio of the CFU/colony of cells grown with IPTG (recA+) to that of cells grown without IPTG (recA) for each pair of samples. Shown are the averages from five experiments for the parent, ruvA::kan, and ruvB::ermC strains and from two experiments for the recG::kan strain, with parentheses representing the SEM. (B) Pilin-dependent colony morphology changes (PDCMC) of the parent FA1090 recA6 strain, ruvA::kan derivative, ruvB::ermC derivative, and recG::kan derivative grown in the presence of IPTG. Shown are the averages of results from 6 to 11 assays (of 10 colonies each), with error bars representing the SEM. Statistical significance was determined using Student's two-tailed t test. Asterisks indicate strains with P values of <0.05 relative to the parent strain for the time points 26, 28, and 30 h.
In summary, the proteins involved in general recombination processes (RecA, RdgC, RecX, and RecG) and the proteins of the single-strand repair pathway (RecJ, RecO, RecQ, and Rep) were confirmed to be involved in pilin Av by decreasing the frequency of variant pilE sequences in their respective mutants. The Holliday junction resolution helicase subunit RuvB was not required for pilin Av, and the double-strand break repair recombinases RecB and RecN were dispensable for pilin Av. These data also show that the different growth rates of piliated and nonpiliated Gc and the increased differential in some mutant backgrounds can be an important parameter in measuring pilin Av.
Pilin Av of G4 mutants.
Formation of the G4 structure upstream of pilE is absolutely required for pilin Av, as analyzed by PDCMC assay and a sequencing assay (34). The G4-related mutants used in this study were Avd-1 (G12 → A), Avd5-4-1 (G3 → A), Avd92 (G6 → A), Av-A17T (A17 → T) (Fig. 1C), and −35 mut (with a change of AGAGTT → CTCACC in the sRNA promoter). The mutations in Avd-1 and Avd92 were reported to abolish pilin Av due to the disruption of the critical G residues in the G4 sequence. While Avd5-4-1 also has a disrupted canonical G, the flanking G0 outside the core sequence can compensate for disruption to the G3 nucleotide, leading to residual pilin Av (34). Indeed, Avd5-4-1, at 1.73% variants/pilE, had a low level of pilin Av that was above the background of the Avd-1 and Avd92 G4-null mutants (Table 3). The A17 → T G4 substitution caused the same level of pilin Av as the wild-type G4 sequence, with 11.67% variants/pilE (Table 3), confirming the requirement of the Gs alone for proper pilin Av.
Another requirement for pilin Av is the transcription of a cis-acting RNA that initiates within the G4 region (41). Mutation of the required −10 and −35 promoter elements disrupts the pilE sRNA promoter, reducing its expression and the level of pilin Av (41). Consistent with earlier results, the −35 mutant had an intermediate level of pilin Av in the deep-sequencing assay with 4.44% variants/pilE (Table 3). Thus, this sequencing confirms the requirement of the pilE G4-forming sequence and its associated sRNA to promote pilin Av.
Other genes and conditions.
Pilin Av was also investigated in the thrB mutant and in cultures grown with the iron chelator Desferal. The threonine biosynthesis genes thrB and thrC were found in a transposon screen looking for insertions that reduce pilin Av (56). ThrB and ThrC convert homoserine to threonine. Both genes caused a modest decrease in the appearance of nonpiliated colonies over time. This deep-sequencing assay confirms the reduction in pilin Av in the thrB::kan mutant with 7.47% variant pilE sequences, although the mechanism remains unknown.
Desferal sequesters iron to create an iron-starved environment similar to that found within the host. Previously, it was reported that iron starvation led to an increase in pilin Av as measured by a quantitative RT-PCR assay (36). FA1090 grown under iron-limited conditions produced a pilin Av frequency of 14.40% variants/pilE by the next-generation sequencing assay. This result confirms that pilin Av is increased when the bacteria are grown under iron-limiting conditions and suggests that this may be a way to regulate pilin Av in vivo. Since high Desferal concentrations can affect the growth rate of Gc, it would be interesting to measure pilin Av at different levels of iron sequestration inhibition.
Donor silent-copy repertoire of variant pilE sequences.
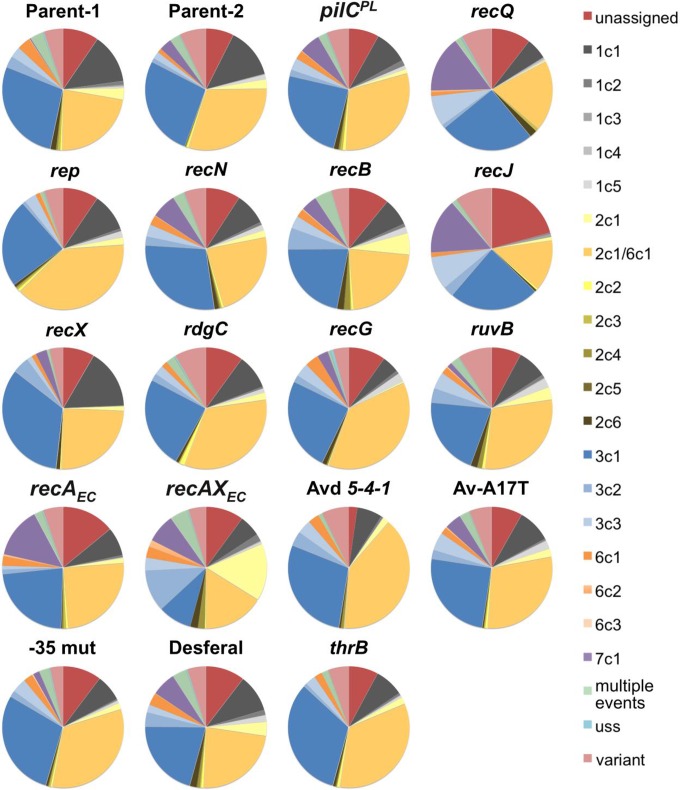
Previous sequencing assays using the FA1090 1-81-S2 variant have shown that there is a nonrandom incorporation of silent-copy donor DNA used for pilin Av, with the top donor silent copies being 2c1/6c1 and 3c1 and other prominent donor copies being 1c1, 3c2, 1c5, 1c3, and 7c1 (Table 6) (18, 34, 38). Some donor silent copies are rarely detected, despite sharing significant homology to the 1-81-S2 pilE (e.g., uss, 6c3, or 2c5). It has also been suggested that the sequence of the initial pilE can affect the pilin Av frequency (43). We analyzed the donor pilS copies used in the mutants from this study. Consistently, the top two silent donor copies used, regardless of the mutant background, were 2c1/6c1 and 3c1 (Fig. 5; Table 6). The spectrum of donor silent copies recorded in the deep-sequencing data was largely unchanged for most of the mutant strains (Fig. 5). This result implies that the recombination proteins that participate in pilin Av but are not essential for the process are not involved in the selection of the pilS donor DNA. There were three exceptions to the pattern of silent-copy incorporation among the mutants. The recJ mutant barely showed any 1c1 donors but did have an increase in 3c3, as did the recQ mutant. The recAXEC mutant exhibited a broader spectrum of silent-copy donors, with enhanced identification of 2c1 and 3c2. Since the profile of the recAEC mutant did not deviate far from the canonical pattern (except for an increase in possible artifactual 7c1), this argues that the modulation of RecAEC by RecXEC is affecting the process of pilin Av.
TABLE 6.
Comparison between traditional and 454 sequencing of silent-copy profiles for starting sequence pilE 1-81-S2
| pilS copy | % determined by: |
|||
|---|---|---|---|---|
| Criss et al. (18) | Cahoon and Seifert (34) | Helm and Seifert (38) | This study | |
| pilS 2c1 or pilS 6c1 | 34.8 | 41.7 | 40.7 | 26.27 |
| pilS 3c1 | 21.7 | 20.8 | 18.5 | 27.31 |
| pilS 3c2 | 17.4 | 8.3 | 7.4 | 1.99 |
| pilS 1c1 | 8.3 | 8.3 | 11.1 | 13.37 |
| pilS 2c4 or pilS 6c1 | 4.3 | 8.3 | ||
| pilS 3c3 | 3.3 | 2.42 | ||
| pilS 1c5 | 3.3 | 7.4 | 0.89 | |
| pilS 2c4 | 1.1 | 0.22 | ||
| pilS 1c4 | 1.1 | 0.07 | ||
| pilS 7c1 | 1.1 | 8.3 | 3.7 | 1.85 |
| pilS 6c3 | 1.1 | |||
| pilS 2c5 | 1.1 | |||
| pilS 6c1 | 1.1 | 2.34 | ||
| pilS 2c1 | 2.7 | |||
| pilS 6c2 | 0.08 | |||
| pilS 1c3 | 4.1 | 7.4 | 0.2 | |
| pilS 1c1 or pilS 3c1 | 3.7 | |||
| pilS 1c2 | 0.51 | |||
| pilS 2c2 | 0.45 | |||
| pilS 2c3 | 0.51 | |||
| pilS 2c6 | 0.8 | |||
| uss | 0.21 | |||
| Multiple events | 3.21 | |||
| Unassigned | 6.1 | |||
| Unknown | 8.46 | |||
FIG 5.
Silent-copy repertoire of different strains. Pie charts represent the different pilS copies identified in the nonparental pilin variant sequences of different mutants. Each pilS copy is represented by a different color that corresponds to the diagram in Fig. 1A. On the right the silent copies are indicated by their position (e.g., 2c1 represents pilS 2 copy 1). variant, recombinant DNA donated from more than one unique silent copy; multiple events, copies with two or more identified pilS DNAs; 2c1/6c1, the common 2c1/6c1 variant that could have arisen from either pilS 2c1 or pilS 6c1.
The recAXEC complement had a high rate of pilin Av (17.04% variants/pilE), and the increased number of donor silent copies may reflect this higher frequency. However, since mismatch correction mutants did not display the same increased donor spectrum even though they have an increased frequency (35), the heightened frequency cannot in itself explain the greater use of donors. Of note, the recAXEC strain displayed abnormal colony morphologies and had a mild growth defect (data not shown), and the variants produced had a higher percentage of donor silent copies known to produce nonpiliated variants (Table 5).
Summary.
Pilin Av is an important process used by the Neisseria that varies the sequence of the pilE gene as a method of generating diversity both to effect functional changes and to avoid the immune surveillance. Pilin Av is dependent on homologous recombination proteins and the RecF-like pathway of single-strand gap repair, which were confirmed in the deep-sequencing analysis performed in this study. Equally important to pilin Av is the intact G4 sequence upstream of pilE and expression of its accompanying small RNA. With 19 different silent copies to use as donor DNA, the silent-copy profiles of all the different mutants remain remarkably consistent, implying that there is one central mechanism that governs which copies are chosen and that it is independent of the factors required for pilin Av to occur.
ACKNOWLEDGMENTS
We are grateful to Jing Xu for critical reading of the manuscript, to Matt Schipma for initial analysis, to Brian Budke for assistance with figures, and to members of the Seifert lab for helpful discussions.
REFERENCES
- 1.CDC. 2013. Sexually transmitted disease surveillance 2012. Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 2.WHO. 2012. Global incidence and prevalence of selected curable sexually transmitted infections—2008. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Shafer WM, Rest RF. 1989. Interactions of gonococci with phagocytic cells. Annu Rev Microbiol 43:121–145. doi: 10.1146/annurev.mi.43.100189.001005. [DOI] [PubMed] [Google Scholar]
- 4.Criss AK, Seifert HS. 2012. A bacterial siren song: intimate interactions between Neisseria and neutrophils. Nat Rev Microbiol 10:178–190. doi: 10.1038/nrmicro2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Workowski KA, Berman S, Centers for Disease Control and Prevention. 2010. Sexually transmitted diseases treatment guidelines, 2010. MMWR Recomm Rep 59:1–110. [PubMed] [Google Scholar]
- 6.Sparling PF. 1966. Genetic transformation of Neisseria gonorrhoeae to streptomycin resistance. J Bacteriol 92:1364–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swanson J. 1973. Studies on gonococcus infection. IV. Pili: their role in attachment of gonococci to tissue culture cells. J Exp Med 137:571–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolfgang M, Lauer P, Park HS, Brossay L, Hebert J, Koomey M. 1998. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol Microbiol 29:321–330. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]
- 9.Stohl EA, Chan YA, Hackett KT, Kohler PL, Dillard JP, Seifert HS. 2012. Neisseria gonorrhoeae virulence factor NG1686 is a bifunctional M23B family metallopeptidase that influences resistance to hydrogen peroxide and colony morphology. J Biol Chem 287:11222–11233. doi: 10.1074/jbc.M111.338830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stohl EA, Dale EM, Criss AK, Seifert HS. 2013. Neisseria gonorrhoeae metalloprotease NGO1686 is required for full piliation, and piliation is required for resistance to H2O2- and neutrophil-mediated killing. mBio 4:e00399-13. doi: 10.1128/mBio.00399-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forest KT, Bernstein SL, Getzoff ED, So M, Tribbick G, Geysen HM, Deal CD, Tainer JA. 1996. Assembly and antigenicity of the Neisseria gonorrhoeae pilus mapped with antibodies. Infect Immun 64:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perry AC, Nicolson IJ, Saunders JR. 1987. Structural analysis of the pilE region of Neisseria gonorrhoeae P9. Gene 60:85–92. doi: 10.1016/0378-1119(87)90216-2. [DOI] [PubMed] [Google Scholar]
- 13.Vink C, Rudenko G, Seifert HS. 2011. Microbial antigenic variation mediated by homologous DNA recombination. FEMS Microbiol Rev doi: 10.1111/j.1574-6976.2011.00321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seifert HS, Wright CJ, Jerse AE, Cohen MS, Cannon JG. 1994. Multiple gonococcal pilin antigenic variants are produced during experimental human infections. J Clin Invest 93:2744–2749. doi: 10.1172/JCI117290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giltner CL, Nguyen Y, Burrows LL. 2012. Type IV pilin proteins: versatile molecular modules. Microbiol Mol Biol Rev 76:740–772. doi: 10.1128/MMBR.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamrick TS, Dempsey JA, Cohen MS, Cannon JG. 2001. Antigenic variation of gonococcal pilin expression in vivo: analysis of the strain FA1090 pilin repertoire and identification of the pilS gene copies recombining with pilE during experimental human infection. Microbiology 147:839–849. doi: 10.1099/00221287-147-4-839. [DOI] [PubMed] [Google Scholar]
- 17.Cahoon LA, Seifert HS. 2011. Focusing homologous recombination: pilin antigenic variation in the pathogenic Neisseria. Mol Microbiol 81:1136–1143. doi: 10.1111/j.1365-2958.2011.07773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Criss AK, Kline KA, Seifert HS. 2005. The frequency and rate of pilin antigenic variation in Neisseria gonorrhoeae. Mol Microbiol 58:510–519. doi: 10.1111/j.1365-2958.2005.04838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howell-Adams B, Seifert HS. 2000. Molecular models accounting for the gene conversion reactions mediating gonococcal pilin antigenic variation. Mol Microbiol 37:1146–1158. doi: 10.1046/j.1365-2958.2000.02067.x. [DOI] [PubMed] [Google Scholar]
- 20.Koomey M, Gotschlich EC, Robbins K, Bergstrom S, Swanson J. 1987. Effects of recA mutations on pilus antigenic variation and phase transitions in Neisseria gonorrhoeae. Genetics 117:391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mehr IJ, Seifert HS. 1998. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol Microbiol 30:697–710. doi: 10.1046/j.1365-2958.1998.01089.x. [DOI] [PubMed] [Google Scholar]
- 22.Stohl EA, Seifert HS. 2001. The recX gene potentiates homologous recombination in Neisseria gonorrhoeae. Mol Microbiol 40:1301–1310. doi: 10.1046/j.1365-2958.2001.02463.x. [DOI] [PubMed] [Google Scholar]
- 23.Hill SA, Grant CC. 2002. Recombinational error and deletion formation in Neisseria gonorrhoeae: a role for RecJ in the production of pilE (L) deletions. Mol Genet Genomics 266:962–972. doi: 10.1007/s00438-001-0618-5. [DOI] [PubMed] [Google Scholar]
- 24.Skaar EP, Lazio MP, Seifert HS. 2002. Roles of the recJ and recN genes in homologous recombination and DNA repair pathways of Neisseria gonorrhoeae. J Bacteriol 184:919–927. doi: 10.1128/jb.184.4.919-927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sechman EV, Kline KA, Seifert HS. 2006. Loss of both Holliday junction processing pathways is synthetically lethal in the presence of gonococcal pilin antigenic variation. Mol Microbiol 61:185–193. doi: 10.1111/j.1365-2958.2006.05213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehr IJ, Long CD, Serkin CD, Seifert HS. 2000. A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics 154:523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.West SC, Cassuto E, Howard-Flanders P. 1981. recA protein promotes homologous-pairing and strand-exchange reactions between duplex DNA molecules. Proc Natl Acad Sci U S A 78:2100–2104. doi: 10.1073/pnas.78.4.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gruenig MC, Stohl EA, Chitteni-Pattu S, Seifert HS, Cox MM. 2010. Less is more: Neisseria gonorrhoeae RecX protein stimulates recombination by inhibiting RecA. J Biol Chem 285:37188–37197. doi: 10.1074/jbc.M110.171967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michel B, Leach D. 11 September 2012, posting date Homologous recombination—enzymes and pathways. sEcoSal Plus 2012 doi: 10.1128/ecosalplus.7.2.7. [DOI] [PubMed] [Google Scholar]
- 30.Kuzminov A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol Mol Biol Rev 63:751–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Umezu K, Kolodner RD. 1994. Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J Biol Chem 269:30005–30013. [PubMed] [Google Scholar]
- 32.Tsaneva IR, Muller B, West SC. 1992. ATP-dependent branch migration of Holliday junctions promoted by the RuvA and RuvB proteins of E. coli. Cell 69:1171–1180. doi: 10.1016/0092-8674(92)90638-S. [DOI] [PubMed] [Google Scholar]
- 33.Whitby MC, Vincent SD, Lloyd RG. 1994. Branch migration of Holliday junctions: identification of RecG protein as a junction specific DNA helicase. EMBO J 13:5220–5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cahoon LA, Seifert HS. 2009. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science 325:764–767. doi: 10.1126/science.1175653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rotman E, Seifert HS. 2015. Neisseria gonorrhoeae MutS affects pilin antigenic variation through mismatch correction and not by pilE guanine quartet binding. J Bacteriol 197:1828–1838. doi: 10.1128/JB.02594-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serkin CD, Seifert HS. 2000. Iron availability regulates DNA recombination in Neisseria gonorrhoeae. Mol Microbiol 37:1075–1086. doi: 10.1046/j.1365-2958.2000.02058.x. [DOI] [PubMed] [Google Scholar]
- 37.Criss AK, Bonney KM, Chang RA, Duffin PM, LeCuyer BE, Seifert HS. 2010. Mismatch correction modulates mutation frequency and pilus phase and antigenic variation in Neisseria gonorrhoeae. J Bacteriol 192:316–325. doi: 10.1128/JB.01228-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Helm RA, Seifert HS. 2009. Pilin antigenic variation occurs independently of the RecBCD pathway in Neisseria gonorrhoeae. J Bacteriol 191:5613–5621. doi: 10.1128/JB.00535-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wainwright LA, Pritchard KH, Seifert HS. 1994. A conserved DNA sequence is required for efficient gonococcal pilin antigenic variation. Mol Microbiol 13:75–87. doi: 10.1111/j.1365-2958.1994.tb00403.x. [DOI] [PubMed] [Google Scholar]
- 40.Wainwright LA, Frangipane JV, Seifert HS. 1997. Analysis of protein binding to the Sma/Cla DNA repeat in pathogenic Neisseriae. Nucleic Acids Res 25:1362–1368. doi: 10.1093/nar/25.7.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cahoon LA, Seifert HS. 2013. Transcription of a cis-acting, noncoding, small RNA is required for pilin antigenic variation in Neisseria gonorrhoeae. PLoS Pathog 9:e1003074. doi: 10.1371/journal.ppat.1003074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Serkin CD, Seifert HS. 1998. Frequency of pilin antigenic variation in Neisseria gonorrhoeae. J Bacteriol 180:1955–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rohrer MS, Lazio MP, Seifert HS. 2005. A real-time semi-quantitative RT-PCR assay demonstrates that the pilE sequence dictates the frequency and characteristics of pilin antigenic variation in Neisseria gonorrhoeae. Nucleic Acids Res 33:3363–3371. doi: 10.1093/nar/gki650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davies JK, Harrison PF, Lin YH, Bartley S, Khoo CA, Seemann T, Ryan CS, Kahler CM, Hill SA. 2014. The use of high-throughput DNA sequencing in the investigation of antigenic variation: application to Neisseria species. PLoS One 9:e86704. doi: 10.1371/journal.pone.0086704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seifert HS. 1997. Insertionally inactivated and inducible recA alleles for use in Neisseria. Gene 188:215–220. doi: 10.1016/S0378-1119(96)00810-4. [DOI] [PubMed] [Google Scholar]
- 46.Stohl EA, Blount L, Seifert HS. 2002. Differential cross-complementation patterns of Escherichia coli and Neisseria gonorrhoeae RecA proteins. Microbiology 148:1821–1831. doi: 10.1099/00221287-148-6-1821. [DOI] [PubMed] [Google Scholar]
- 47.Wright CJ, Jerse AE, Cohen MS, Cannon JG, Seifert HS. 1994. Nonrepresentative PCR amplification of variable gene sequences in clinical specimens containing dilute, complex mixtures of microorganisms. J Clin Microbiol 32:464–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilles A, Meglecz E, Pech N, Ferreira S, Malausa T, Martin JF. 2011. Accuracy and quality assessment of 454 GS-FLX titanium pyrosequencing. BMC Genomics 12:245. doi: 10.1186/1471-2164-12-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. 2007. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol 8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Howell-Adams B, Seifert HS. 1999. Insertion mutations in pilE differentially alter gonococcal pilin antigenic variation. J Bacteriol 181:6133–6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swanson J, Bergstrom S, Barrera O, Robbins K, Corwin D. 1985. Pilus-gonococcal variants. Evidence for multiple forms of piliation control. J Exp Med 162:729–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jonsson AB, Nyberg G, Normark S. 1991. Phase variation of gonococcal pili by frameshift mutation in pilC, a novel gene for pilus assembly. EMBO J 10:477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng Y, Johnson MD, Burillo-Kirch C, Mocny JC, Anderson JE, Garrett CK, Redinbo MR, Thomas CE. 2013. Mutation of the conserved calcium-binding motif in Neisseria gonorrhoeae PilC1 impacts adhesion but not piliation. Infect Immun 81:4280–4289. doi: 10.1128/IAI.00493-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morimatsu K, Kowalczykowski SC. 2014. RecQ helicase and RecJ nuclease provide complementary functions to resect DNA for homologous recombination. Proc Natl Acad Sci U S A 111:E5133–E5142. doi: 10.1073/pnas.1420009111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sechman EV, Rohrer MS, Seifert HS. 2005. A genetic screen identifies genes and sites involved in pilin antigenic variation in Neisseria gonorrhoeae. Mol Microbiol 57:468–483. doi: 10.1111/j.1365-2958.2005.04657.x. [DOI] [PubMed] [Google Scholar]
- 57.Bernstein KA, Gangloff S, Rothstein R. 2010. The RecQ DNA helicases in DNA repair. Annu Rev Genet 44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun H, Bennett RJ, Maizels N. 1999. The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucleic Acids Res 27:1978–1984. doi: 10.1093/nar/27.9.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun H, Karow JK, Hickson ID, Maizels N. 1998. The Bloom's syndrome helicase unwinds G4 DNA. J Biol Chem 273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 60.Han H, Bennett RJ, Hurley LH. 2000. Inhibition of unwinding of G-quadruplex structures by Sgs1 helicase in the presence of N,N′-bis[2-(1-piperidino)ethyl]-3,4,9,10-perylenetetracarboxylic diimide, a G-quadruplex-interactive ligand. Biochemistry 39:9311–9316. doi: 10.1021/bi000482r. [DOI] [PubMed] [Google Scholar]
- 61.Cahoon LA, Manthei KA, Rotman E, Keck JL, Seifert HS. 2013. Neisseria gonorrhoeae RecQ helicase HRDC domains are essential for efficient binding and unwinding of the pilE guanine quartet structure required for pilin antigenic variation. J Bacteriol 195:2255–2261. doi: 10.1128/JB.02217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Killoran MP, Kohler PL, Dillard JP, Keck JL. 2009. RecQ DNA helicase HRDC domains are critical determinants in Neisseria gonorrhoeae pilin antigenic variation and DNA repair. Mol Microbiol 71:158–171. doi: 10.1111/j.1365-2958.2008.06513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mehr IJ, Seifert HS. 1997. Random shuttle mutagenesis: gonococcal mutants deficient in pilin antigenic variation. Mol Microbiol 23:1121–1131. doi: 10.1046/j.1365-2958.1997.2971660.x. [DOI] [PubMed] [Google Scholar]
- 64.Shiraishi K, Imai Y, Yoshizaki S, Ikeda H. 2005. Rep helicase suppresses short-homology-dependent illegitimate recombination in Escherichia coli. Genes Cells 10:1015–1023. doi: 10.1111/j.1365-2443.2005.00901.x. [DOI] [PubMed] [Google Scholar]
- 65.Kline KA, Seifert HS. 2005. Role of the Rep helicase gene in homologous recombination in Neisseria gonorrhoeae. J Bacteriol 187:2903–2907. doi: 10.1128/JB.187.8.2903-2907.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stohl EA, Brockman JP, Burkle KL, Morimatsu K, Kowalczykowski SC, Seifert HS. 2003. Escherichia coli RecX inhibits RecA recombinase and coprotease activities in vitro and in vivo. J Biol Chem 278:2278–2285. doi: 10.1074/jbc.M210496200. [DOI] [PubMed] [Google Scholar]
- 67.Drees JC, Chitteni-Pattu S, McCaslin DR, Inman RB, Cox MM. 2006. Inhibition of RecA protein function by the RdgC protein from Escherichia coli. J Biol Chem 281:4708–4717. doi: 10.1074/jbc.M513592200. [DOI] [PubMed] [Google Scholar]
- 68.Drees JC, Lusetti SL, Chitteni-Pattu S, Inman RB, Cox MM. 2004. A RecA filament capping mechanism for RecX protein. Mol Cell 15:789–798. doi: 10.1016/j.molcel.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 69.Schook PO, Stohl EA, Criss AK, Seifert HS. 2011. The DNA-binding activity of the Neisseria gonorrhoeae LexA orthologue NG1427 is modulated by oxidation. Mol Microbiol 79:846–860. doi: 10.1111/j.1365-2958.2010.07491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Black CG, Fyfe JA, Davies JK. 1998. Absence of an SOS-like system in Neisseria gonorrhoeae. Gene 208:61–66. doi: 10.1016/S0378-1119(97)00653-7. [DOI] [PubMed] [Google Scholar]
- 71.Chaussee MS, Wilson J, Hill SA. 1999. Characterization of the recD gene of Neisseria gonorrhoeae MS11 and the effect of recD inactivation on pilin variation and DNA transformation. Microbiology 145:389–400. doi: 10.1099/13500872-145-2-389. [DOI] [PubMed] [Google Scholar]
- 72.Hill SA, Woodward T, Reger A, Baker R, Dinse T. 2007. Role for the RecBCD recombination pathway for pilE gene variation in repair-proficient Neisseria gonorrhoeae. J Bacteriol 189:7983–7990. doi: 10.1128/JB.00980-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Whitby MC, Ryder L, Lloyd RG. 1993. Reverse branch migration of Holliday junctions by RecG protein: a new mechanism for resolution of intermediates in recombination and DNA repair. Cell 75:341–350. doi: 10.1016/0092-8674(93)80075-P. [DOI] [PubMed] [Google Scholar]