ABSTRACT
DgcZ is the main cyclic dimeric GMP (c-di-GMP)-producing diguanylate cyclase (DGC) controlling biosynthesis of the exopolysaccharide poly-β-1,6-N-acetylglucosamine (poly-GlcNAc or PGA), which is essential for surface attachment of Escherichia coli. Although the complex regulation of DgcZ has previously been investigated, its primary role and the physiological conditions under which the protein is active are not fully understood. Transcription of dgcZ is regulated by the two-component system CpxAR activated by the lipoprotein NlpE in response to surface sensing. Here, we show that the negative effect of a cpxR mutation and the positive effect of nlpE overexpression on biofilm formation both depend on DgcZ. Coimmunoprecipitation data suggest several potential interaction partners of DgcZ. Interaction with FrdB, a subunit of the fumarate reductase complex (FRD) involved in anaerobic respiration and in control of flagellum assembly, was further supported by a bacterial-two-hybrid assay. Furthermore, the FRD complex was required for the increase in DgcZ-mediated biofilm formation upon induction of oxidative stress by addition of paraquat. A DgcZ-mVENUS fusion protein was found to localize at one bacterial cell pole in response to alkaline pH and carbon starvation. Based on our data and previous knowledge, an integrative role of DgcZ in regulation of surface attachment is proposed. We speculate that both DgcZ-stimulated PGA biosynthesis and interaction of DgcZ with the FRD complex contribute to impeding bacterial escape from the surface.
IMPORTANCE Bacterial cells can grow by clonal expansion to surface-associated biofilms that are ubiquitous in the environment but also constitute a pervasive problem related to bacterial infections. Cyclic dimeric GMP (c-di-GMP) is a widespread bacterial second messenger involved in regulation of motility and biofilm formation, and plays a primary role in bacterial surface attachment. E. coli possesses a plethora of c-di-GMP-producing diguanylate cyclases, including DgcZ. Our study expands the knowledge on the role of DgcZ in regulation of surface attachment and suggests that it interconnects surface sensing and adhesion via multiple routes.
INTRODUCTION
The Gram-negative bacterium Escherichia coli is a common inhabitant of the human intestine, where it usually prevents colonization by pathogenic bacteria. Nevertheless, certain pathogenic E. coli strains cause severe diarrheal and extraintestinal diseases (1). In the industrialized world, most chronic bacterial infections are caused by pathogens growing in biofilms (2). These are communities of bacterial cells embedded in an extracellular matrix, mainly composed of exopolysaccharides and flagella, and in some cases of extracellular DNA (3–5).
Biofilm formation is a gradual process starting with surface sensing and attachment (6). In E. coli K-12, recognition of the surface is mediated by the outer membrane lipoprotein NlpE that activates the two-component system CpxAR (7–9). Activation of this complex results in phosphorylation of the transcriptional regulator CpxR, which controls the expression of several target genes, ultimately leading to surface attachment (10–13). However, knowledge of the regulatory events downstream of CpxR phosphorylation resulting in surface attachment is still very incomplete.
To efficiently attach to different kind of surfaces, E. coli exploits its flagella as structural components for adhesion and regulates the flagellar activity during surface attachment (14, 15). In E. coli flagellar speed is controlled by cyclic dimeric GMP (c-di-GMP) through the so-called flagellar brake YcgR (16, 17). Upon c-di-GMP binding, YcgR interacts with the flagellar basal body, leading to a slowdown of rotational speed (16, 17).
The exopolysaccharide poly-β-1,6-N-acetylglucosamine (poly-GlcNAc or PGA) (18, 19) produced by different bacterial species, such as E. coli, Staphylococcus epidermidis, and Yersinia pestis (18), is another important factor in bacterial attachment to abiotic surfaces. E. coli strains deficient in PGA production are impaired in permanent attachment (18). PGA production is mediated by the PgaABCD complex and controlled by c-di-GMP (19, 20).
c-di-GMP is a well-characterized secondary messenger and a key regulator of biofilm formation, motility, and virulence in many bacterial species (21, 22). Intracellular c-di-GMP concentrations are controlled by two different classes of enzymes: diguanylate cyclases (DGCs) that synthesize the second messenger from two GTP molecules, and phosphodiesterases (PDEs) that degrade it to pGpG. DGCs harbor a distinctive GGDEF domain, while PDEs contain EAL or HD-GYP domains (23–26). In E. coli K-12, 29 proteins harbor a GGDEF and/or an EAL domain, but for most of them the physiological role is unknown (21). Here, we use the systematic nomenclature for E. coli DGCs and PDEs that was recently proposed by Hengge et al. (27). The corresponding traditional designations are highlighted in brackets.
DgcZ (previously named YdeH) is the main DGC involved in PGA production in E. coli (28, 29). Abundance and activity of DgcZ are regulated at several levels. dgcZ transcription is activated by the transcriptional regulator CpxR (12, 13, 30, 31). In carbon-rich media, CsrA, a global regulator acting at posttranscriptional level on a large variety of targets, prevents translation of dgcZ and pgaABCD mRNAs (32–35). Beyond transcriptional and translational regulation of protein concentration, DgcZ activity is regulated by Zn2+ (29). In addition to the characteristic GGDEF domain, DgcZ possesses a zinc-binding domain (CZB), common in many bacterial chemoreceptors (36). Upon Zn2+ binding DgcZ activity strongly decreases, while in the zinc-free conformation the protein is active (29).
The putative integral role of DgcZ in a regulatory network linking surface sensing to processes mediating adhesion led us to further investigate the role of DgcZ in this network. Here, we report (i) a DgcZ-dependent effect of cpxR deletion and NlpE overproduction on biofilm formation, (ii) a hitherto-unknown interaction between DgcZ and the fumarate reductase complex (FRD) involved in anaerobic respiration and in control of flagellum assembly and activity (37), (iii) an FRD-dependent increase in DgcZ-mediated biofilm formation upon induction of oxidative stress, and (iv) polar localization of DgcZ in response to alkaline pH and carbon starvation. Our data reveal new aspects of DgcZ-related regulation and suggest a more integrative role of DgcZ throughout the process of surface attachment.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The bacterial strains and plasmids examined in this study are listed in Tables S1 and S2 in the supplemental material, respectively. All of the oligonucleotide primers used here are listed in Table S3 in the supplemental material. Bacteria were grown in lysogeny broth (LB) at 37°C, unless specifically indicated. Unless otherwise indicated, bacterial cultures were inoculated with 1:100 dilutions of stationary overnight cultures grown at 37°C in LB medium. Minimal medium agar plates contained 15 g/liter agar, MgSO4 (1 mM), 1× MMA (38), and 0.2% l-rhamnose. When needed, antibiotics were used at the following concentrations: ampicillin (Amp; 100 μg/ml), kanamycin (Kan; 50 μg/ml), and streptomycin (Str; 75 μg/ml). A stock solution of 400 mM paraquat (N,N′-dimethyl-4,4′-bipyridinium dichloride) was prepared in distilled water. We briefly describe the construction of strains in this study. Additional details are given in the supplemental material. All genetic constructs and chromosomal integrations were validated by DNA sequencing.
The csrA::Tn5 background mutation was introduced by P1 transduction using AB400 as a donor strain (28) prior to each experiment to avoid the occurrence of suppressor mutations. P1 transduction was also applied to replace the native pgaD allele with pgaD-3×Flag-kan and to introduce the deletion of the cpxR gene derived from strain JW3883 of the Keio collection (39). The Δ7 and the Δ9 strains were generated by several cycles of P1 transductions using as a donor the strains of the Keio collection; each cycle was followed by removal of the kanamycin resistance cassette using the pCP20 plasmid (40).
Chromosomal replacement of the corresponding wild-type gene by mVENUS fusions to dgcZ wild-type or mutant variants, as well as by mCHERRY fusions to frdA and frdB, were achieved by standard λRED-mediated recombineering (41). In each case, the PRham-ccdB-kan element replaced the wild-type locus in the precursor strain. The dgcZ(+) and frdA(+) control strains were obtained by replacing the PRham-ccdB-kan element with the dgcZ and frdA genes amplified from E. coli MG1655. The same approach was used to generate the ΔfrdAFrt strain by replacing the PRham-ccdB-kan element in the precursor strain with frdA::Frt amplified from the ΔfrdA strain of the Keio collection after removal of the kanamycin resistance cassette.
The fluorescent protein mVENUS differs from VENUS by mutation A206K that abolishes protein dimerization (42, 43). A DNA sequence of 93 nucleotides coding for a flexible linker of 31 amino acids (see Table S3 in the supplemental material) was inserted between the dgcZ and mVENUS coding regions in the dgcZ-mVENUS construct. The plasmid carrying yfbR-mCHERRY was obtained by cloning yfbR and mCHERRY amplified from pWBP20911 (44) into pCJ30 (45). For bacterial two-hybrid (BACTH) assays, bait and prey sequences were inserted into vectors pKT25, pKNT25, pUT18, and pUT18C (46).
Quantification of intracellular c-di-GMP concentrations.
Bacterial cultures were grown under shaking conditions at 37°C in LB medium for 9 h (optical densities at 600 nm [OD600] of 3.1 to 3.2) and 48 h (OD600 of 4.5). Bacteria were harvested by centrifugation from 2-ml cultures, and extracts were essentially prepared and analyzed by liquid chromatography-tandem mass spectrometry as described previously (47).
Western blots.
Bacteria were cultured as indicated in the figure legends. At the indicated time points, the optical densities of bacterial cultures were measured, and 200- to 500-μl portions of the cultures were centrifuged at 13,000 rpm for 1 min. Supernatants were discarded, and the pellets were resuspended in adequate volumes of 1× SDS sample buffer. Sample concentrations were normalized to an OD600 of 5.0. Samples were vortexed for 10 s and boiled for 5 min. Gel electrophoresis and Western blotting were carried out according to standard protocols. PgaD-3×Flag was detected using a mouse monoclonal anti-M2 antibody and horseradish peroxidase (HRP)-conjugated rabbit anti-mouse immunoglobulins (DakoCytomation), both diluted 1:10,000 in 5% nonfat milk. For the detection of DgcZ and GroEL in Western blots, a primary anti-DgcZ antibody (dilution 1:2,000; T. Schirmer and U. Jenal, unpublished data) and anti-GroEL (dilution 1:10,000; Sigma) were used, respectively. As a secondary antibody for DgcZ and GroEL Western blots, an HRP-conjugated anti-rabbit antibody (1:10,000; Sigma) was used. In every case, the primary antibody was incubated at 4°C overnight, and the secondary antibody was incubated at room temperature for 1 h. After incubation with the primary and secondary antibodies, the membranes were washed three times for 5 min each time with a solution of 1× PBS containing 0.1% Tween. Blots were developed with an ECL kit (Pierce) and documented using an ImageQuant LAS 4000 (GE Healthcare) gel documentation system.
Attachment assay.
Bacterial biofilm formation in liquid cultures was measured as described before (28, 29). Briefly, bacteria were grown on LB agar plates at 30°C overnight, single colonies were inoculated into LB medium in a 96-well plate (BD Falcon, catalog no. 353072) and incubated statically for 24 h at 30°C. The cell density was recorded using an Infinite M200 Pro plate reader (Tecan), the supernatant was discarded, and the wells were rinsed with distilled water. After drying, the surface-attached biomass was stained with 0.1% crystal violet in water, 1-propanol, and methanol (96.7:1.66:1.66), the wells were rinsed two times with water, and the remaining crystal violet was dissolved in a solution containing 20% acetic acid. The OD600 was measured and normalized to the total cell density, and the results are indicated as arbitrary units.
Microscopy.
Bacterial cultures were grown as indicated in the figure legends. A 4-μl portion of each culture was placed on 1% agarose pads (in 1× PBS) for observation. Differential interference contrast and total internal reflection fluorescence (TIRF) microscopy using a Nikon Eclipse Ti-E inverse microscope was used to examine individual bacterial cells. The microscope was equipped with ×100 CFI Apo TIRF Oil objective lens (numerical aperture of 1.49) and AHF HC filter sets F36-528 (excitation band pass, 500/24 nm; beam splitter, 520 nm; emission band pass, 542/27 nm) and F36-504 (excitation band pass, 562/40 nm; beam splitter, 593 nm; emission band pass, 624/40 nm). A Multiline argon laser (Melles Griot; 65 mW, MHX00171) and a Sapphire laser (561 nm, 50 mW; MHX00146) were used as light sources for mVENUS and mCHERRY microscopy, respectively. Exposure times ranged from 50 to 300 ms. Images were acquired with an Andor iXon3 885 EMCCD camera. Image acquisition and adjustment were done with the Nikon NIS elements 4.0 software and with ImageJ. For time-lapse fluorescence microscopy, bacterial stationary-phase cultures (4 μl) were spotted onto agarose pads (LB medium, 1% agarose) and grown at 30°C under the microscope. Images were acquired every 15 min.
Microfluidics.
Details about the preparation and the set up of the microfluidic system used for this experiments have been described elsewhere (48, 49). A 50-ml culture of the E. coli dgcZ-mVENUS ibpA-mCHERRY csrA::Tn5 strain AB3324 (see Table S1 in the supplemental material) was grown overnight to an OD600 of 5.0. Portions (5 μl) of this culture were diluted ×1,000 into fresh LB medium, while the remainder was centrifuged for 10 min at 4,000 rpm. The supernatant was then sterile filtered using a 0.22-μm-pore-size PES syringe filter (Sarstedt) to obtain sterile spent medium.
Bacteria were cultured in LB medium to an OD600 of 0.2, and salmon sperm DNA and bovine serum albumin were added to the preculture to final concentrations of 75 and 25 μg/ml, respectively, to prevent excessive biofilm formation within the flow chamber device. Then, 1 ml of this cell suspension was spun down for 1 min at 8,000 rpm, and the resulting cell pellet was recovered in 20 μl of the residual LB medium. Next, 1 μl of this suspension was transferred into the main channel, and the applied medium flow flushed the cells into the side channels. With the help of two programmable syringe pumps (NE-1000; New Era Pump Systems, Inc.), a medium gradient from fresh (F) to spent (S) LB medium was driven. The following program was applied: (i) 2-h constant flow of F 0.7 ml/h and S 0.1 ml/h, (ii) 3 h of a gradient from F 0.7 m/h to F 0.0 ml/h and from S 0.1 ml/h to S 0.7 ml/h, (iii) 16-h constant flow of S 0.3 ml/h, (iv) 1 h of a gradient from S 0.3 m/h to S 0.0 ml/h and from F 0.0 ml/h to F 0.7 ml/h, and (v) 20-h constant flow of F 0.7 ml/h.
The microfluidic device was mounted on an automated Olympus BX81 microscope equipped with an UPLFN100O2PH/1.3 phase-contrast oil objective and an incubator chamber set to 37°C (Cube and Box incubation system; Life Imaging Services, Reinach, Switzerland). mVENUS (U-N41028 filter [Chroma]; excitation band pass, 500/20 nm; beam splitter, 515 nm; emission band pass, 535/30 nm) and mCHERRY (U-N49008 [Chroma]; excitation band pass, 560/40 nm; beam splitter, 585 nm; emission band pass, 630/75 nm) fluorescence and phase-contrast images were taken every 12 min. Image analysis was finally performed using CellM software (Olympus) and Fiji/ImageJ.
Coimmunoprecipitation (CoIP).
The dgcZ-3×Flag and dgcZ(wt) strains, both in the csrA::Tn5 background, were cultured in 200 ml of LB broth at 30°C under shaking conditions to an OD600 of 0.65. For the stationary-phase condition, bacteria were cultured in 50 ml of LB medium for 48 h (OD600 of 4.5). Then, 37% formaldehyde was added to a final concentration of 0.2%, and the cultures were shaken for 15 min. Glycine (1.875 M) was added to a final concentration of 0.375 M, and the cultures were shaken for 5 min. Bacteria were collected in 50-ml tubes and centrifuged at 4,000 × g for 10 min at 4°C. The bacterial pellets were then resuspended in 24 ml (6 ml for the stationary-phase condition) Tris-EDTA buffer supplemented with 0.1% Sarkosyl and centrifuged as described above. The pellets were washed twice with 50 ml (12.5 ml for the stationary-phase condition) cold 1× PBS, frozen in liquid nitrogen, and stored at −80°C. Before use, the pellets were resuspended in 4 ml of lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.2 mM phenylmethylsulfonyl fluoride) and disrupted using a French press. The cell lysates were then centrifuged at 100,000 × g for 1 h at 4°C. Next, 40 μl of anti-Flag M2 resin (Sigma, catalog no. A2220) was added to the cleared lysates, followed by incubation overnight with shaking at 4°C.
The suspensions were centrifuged, and the beads were resuspended in 500 μl of wash buffer (Flag immunoprecipitation kit; Sigma) and transferred to spin columns, followed by six washes in 500 μl of the same buffer. The samples were then incubated with a 150-ng/μl solution of 3×Flag peptide (Sigma; catalog no. F4799) in 100 μl of wash buffer at 4°C for 30 min with occasional gentle shaking. Proteins were collected by centrifugation at 8,200 × g for 1 min at 4°C and analyzed by mass spectrometry (as described in the supplemental material). Lysates from the dgcZ(wt) strain grown in the same conditions as the dgcZ-3×Flag strain were used as negative controls. The initial list of coimmunoprecipitated proteins was filtered by removing the proteins present in the negative controls. In two additional filtering steps, (i) all hits identified by fewer than three peptides were discarded, and (ii) only hits identified by at least two unique peptides were retained. The complete data set, including the low-scoring hits not present in the negative controls, is provided in the supplemental material (see Table S4 in the supplemental material).
Bacterial two-hybrid and β-galactosidase assay.
For all of the BACTH experiments, the E. coli BTH101 strain (46) was grown at 30°C on LB plates supplemented with Amp (100 μg/ml), Kan (50 μg/ml), Str (75 μg/ml), X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 40 μg/ml), and 500 μM IPTG (isopropyl-β-d-thiogalactopyranoside). The BTH101 strain was cotransformed with plasmids encoding the T25 and T18 protein fusions and grown overnight on LB agar plates (Amp, Kan, Str, X-Gal, and IPTG) at 30°C. Single colonies of each combination were streaked on new medium plates, followed by incubation overnight at 30°C. Subsequently, bacteria from these plates were resuspended in 1× PBS to an OD600 of 0.1, and then 10-μl portions of each solution were spotted onto new plates for detection. Images were acquired after growing the bacteria for 40 h at 30°C. Bacteria from the same medium plates were resuspended in 1× PBS to an OD600 of 1 and used for the β-galactosidase assays (38). The Miller units were determined as 1,000 × (OD405 × [T × V × OD600]−1), where T represents the time of the reaction in minutes and V is the volume in milliliters (in this case, 0.2 ml).
Motility assay.
The swimming behavior of bacteria was assayed on motility agar plates containing 1% tryptone, 0.5% NaCl, and 0.3% agar supplemented with 100 μg/ml Amp and 200 μM IPTG. Bacteria were grown overnight on LB plates (Amp) at 37°C. The following day, single colonies were inoculated on motility agar plates, followed by incubation at 37°C for 5 to 6 h before images were acquired. The assay was repeated at least three times.
RESULTS
DgcZ and CpxR contribute to biofilm formation in the csrA(wt) strain MG1655.
In E. coli MG1655, dgcZ mRNA levels have been shown to be low and constant during growth in LB medium at 37°C (32). Boehm et al. (28) reported that under conditions permissive for biofilm formation (static incubation in LB at 28°C) DgcZ protein levels are very low, as detected by Western blotting. We confirmed this observation and examined by attachment assays of dgcZ(wt) and ΔdgcZ strains whether DgcZ contributes to biofilm formation in the MG1655 strain in this assay (Fig. 1). This method is commonly used to test the efficiency with which E. coli strains adhere to abiotic surfaces, and it has been shown that strains producing more c-di-GMP attach more efficiently (28, 29, 50). In addition, biofilm formation of the ΔcpxR and ΔcpxR ΔdgcZ strains was tested. Our results show that all mutant strains formed about 20% less biofilm than the dgcZ(wt) cpxR(wt) strain (Fig. 1B), indicating that DgcZ and CpxR contribute to biofilm formation in the MG1655 wild-type strain.
FIG 1.
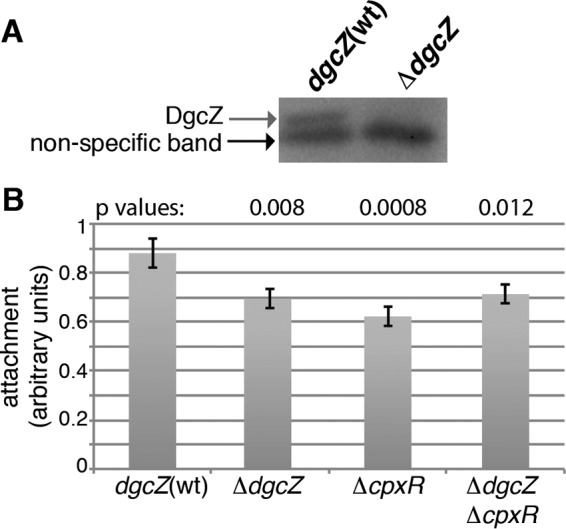
Effects of dgcZ and cpxR deletions on attachment of E. coli MG1655. (A) Western blot detection of DgcZ in E. coli MG1655 dgcZ(wt) and ΔdgcZ strains grown for 24 h at 30°C under static conditions. (B) Attachment assay of MG1655, ΔdgcZ, ΔcpxR, and ΔdgcZ ΔcpxR strains grown as described for panel A. The results are averages for 12 wells inoculated with single colonies in two different experiments. Error bars indicate the standard errors of the mean (SEM). Statistic analysis was performed using a Student t test for two independent means, where each P value was obtained by comparing the mutant to the MG1655 strain.
Effects of NlpE on biofilm formation depend on DgcZ.
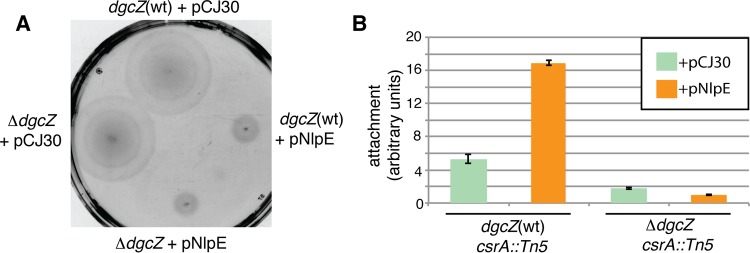
Since NlpE activates CpxR in response to surface sensing (7–9) and CpxR inhibits motility and stimulates dgcZ transcription (30, 51), the effects of NlpE overproduction on DgcZ-dependent regulation of motility and biofilm formation were analyzed (Fig. 2). Either the pNlpE overexpression plasmid or the empty vector pCJ30 were introduced to E. coli MG1655 and a ΔdgcZ mutant. NlpE overproduction repressed motility of the dgcZ(wt) and the ΔdgcZ strain (Fig. 2A). This effect on motility is probably due to the Cpx-mediated inhibition of motAB-cheAW transcription (51).
FIG 2.
Effects of NlpE overproduction on motility and biofilm formation. (A) Motility of the dgcZ(wt) E. coli strain MG1655 and the dgcZ mutant transformed either with the pNlpE overexpression plasmid or with the control plasmid pCJ30. (B) Attachment assay of the dgcZ(wt) and ΔdgcZ strains in the csrA::Tn5 background transformed either with pNlpE or with pCJ30. The medium was supplemented with 100 μg/ml ampicillin and 20 μM IPTG. Error bars indicate the SEM.
Since the partial loss of function csrA::Tn5 allele mitigates the negative regulatory effect of CsrA on translation of the dgcZ mRNA (18, 20, 28) it was used as mutant background in the following experiments. To examine whether nlpE overexpression affects biofilm formation, attachment assays of dgcZ(wt) and ΔdgcZ strains were performed in the csrA::Tn5 mutant background (Fig. 2B). The dgcZ(wt) strain overexpressing nlpE adhered three times more efficiently compared to the strain transformed with the empty plasmid. In contrast, the ΔdgcZ strain did not display enhanced adherence upon NlpE overproduction (Fig. 2B), indicating that NlpE affects this process in a DgcZ-dependent manner.
DgcZ localization and c-di-GMP concentrations change between transition and stationary phase.
PGA production, the main target process of DgcZ-derived c-di-GMP, occurs either at the bacterial cell poles or all around the bacterial cell under shaking or static cultivation conditions, respectively (19). To test whether DgcZ localization undergoes similar alterations upon changes in growth conditions a chromosomal dgcZ deletion was replaced by a dgcZ-mVENUS fusion in the E. coli csrA::Tn5 mutant (34). A dgcZ(+) control strain was constructed by restoring the dgcZ gene in the parent of the dgcZ-mVENUS fusion strain (see Materials and Methods).
In LB medium under shaking conditions (250 rpm) at 37°C, both the csrA::Tn5 and the csrA::Tn5 dgcZ-mVENUS strains grew exponentially until reaching an OD600 of 1.7 to 1.8 and then growth slowed down and stopped at an OD600 of 4.5 to 5.2 (Fig. 3A). Throughout this study, cultures with an OD600 of 1.8 to 5.1 are therefore indicated as being in the transition phase and afterward, when the OD600 remained constant (4.5 to 5.2), as being in stationary phase.
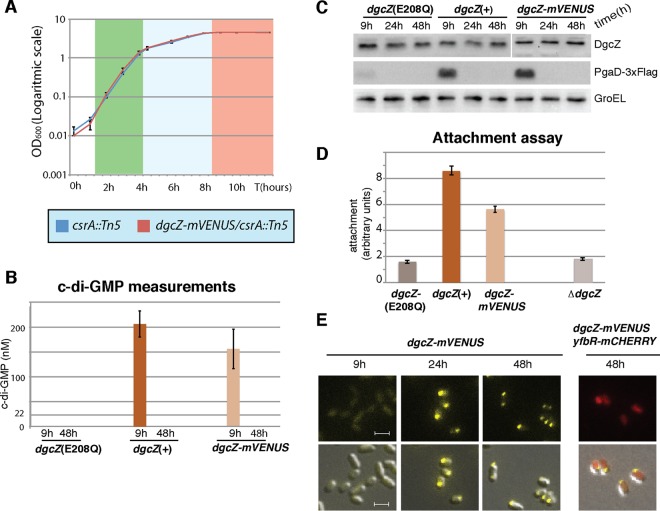
FIG 3.
Activity and localization of the DgcZ-mVENUS protein in the csrA::Tn5 mutant background. (A) Growth curves of the dgcZ-mVENUS csrA::Tn5 and csrA::Tn5 strains in LB medium at 37°C. The OD600 values were determined at the indicated time points. Error bars represent the SEM from three biological replicates. The green, blue, and red colors on the chart indicate the exponential, transition, and stationary growth phases, respectively. (B) E. coli csrA::Tn5 cells carrying either dgcZ-mVENUS, the dgcZ(E208Q) mutant gene (coding for an active-site mutant), or the complemented dgcZ(+) gene were grown for 9 h (transition phase) and 48 h (stationary phase) in LB medium at 37°C under shaking conditions. c-di-GMP was extracted, and the concentration was determined by mass spectrometry. Three biological replicates were used for each strain. The 22 nM line indicates the limit of detection of c-di-GMP in this experiment. Error bars indicate the SEM. (C) Western blots of the strains from panel A, with pgaD(wt) replaced by pgaD-3×Flag. Protein samples were extracted at 9 h, 24 h (stationary phase), and 48 h of growth under the same conditions as in panel A. (D) Strains from panel A and the ΔdgcZ strain were grown at 30°C for 24 h in 96-well plates, and surface attachment was assayed. For each strain, the average derived from measurements of six wells is shown. Error bars indicate the SEM. (E) Fluorescence microscopy images of the dgcZ-mVENUS csrA::Tn5 strain with or without plasmid pYfbR-mCHERRY. Growth conditions were as described for panel A. Cultures of the strain carrying pYfbR-mCHERRY were supplemented with ampicillin (100 μg/ml) and IPTG (100 μM). Size bars indicate 2 μm and are identical in each picture.
To validate activity of the DgcZ-mVENUS protein, cellular c-di-GMP levels of the dgcZ-mVENUS fusion strain and the dgcZ(+) control strain, both carrying the csrA::Tn5 mutation, were compared during the transition (9 h, OD600 of 3.5) and stationary (48 h, OD600 of 4.5) phases. c-di-GMP concentrations in transition phase (9 h) were about 25% lower in the strain carrying the dgcZ-mVENUS fusion compared to the strain with wild-type dgcZ (Fig. 3A). c-di-GMP concentrations were below the limit of detection of 22 nM in a strain carrying the active-site mutation E208Q that destroys the catalytic activity of DgcZ (29), suggesting that in the csrA::Tn5 background DgcZ is the main active DGC (Fig. 3A).
Since protein levels of PgaD-3×Flag have been shown to correlate with c-di-GMP concentrations (20, 29), and no differences were found in biofilm formation of strains carrying either the pgaD-3×Flag or the pgaD(wt) allele (28), levels of PgaD-3×Flag were used as indicator for intracellular c-di-GMP concentrations. Western blot analysis showed comparable levels of PgaD-3×Flag in the dgcZ(+) strain and in the dgcZ-mVENUS strain in the transition phase (9 h) (Fig. 3B). Under the same conditions, just a weak signal of PgaD-3×Flag was detected in the dgcZ active-site mutant (Fig. 3B). PgaD was not detected in any of these strains in the stationary phase (24 and 48 h).
The ability of the dgcZ-mVENUS strain to form biofilms was measured by attachment assays. The dgcZ-mVENUS strain adhered to the surface with approximately 70% efficiency compared to the dgcZ(+) strain and about three times better than both the dgcZ deletion mutant and the strain carrying the active-site mutation dgcZ(E208Q) (Fig. 3C). Altogether, this indicates that the chimeric DgcZ-mVENUS protein is functional, although slightly less active than the wild-type protein.
Aliquots from the bacterial cultures used for c-di-GMP quantification (Fig. 3) were used to analyze DgcZ-mVENUS localization. The protein was evenly distributed in the cytoplasm in transition phase (9 h), while in stationary phase (24 and 48 h) it localized almost entirely at one cell pole in the whole population (Fig. 3D). Occasionally, DgcZ localization was observed at both cell poles. The localization of the cytoplasmic protein YfbR fused to mCHERRY and overproduced from the pCJ30 plasmid was used as a control. The YfbR-mCHERRY signal displayed dispersed cytoplasmic localization in stationary phase (48 h) in the same bacteria where DgcZ-mVENUS was localized at the cell pole (Fig. 3D).
Polar localization of DgcZ is independent of protein activity and concentration.
We further investigated whether polar localization of DgcZ is affected by protein activity or concentration. DgcZ mutant proteins displaying different levels of enzymatic activity were fused to mVENUS, and protein localization was analyzed. Both the inactive DgcZ(H79L,H83L,E208Q) and the hyperactive DgcZ(H79L,H83L) mutant proteins showed the same localization phenotype as the wild-type protein (see Fig. S1A in the supplemental material). A detailed description of these mutant proteins is given in Zähringer et al. (29).
To test whether polar localization of DgcZ-mVENUS correlates with an increase in protein concentration, DgcZ levels were determined in a time-lapse study. The dgcZ-mVENUS strain was grown in LB medium at 37°C until it reached the stationary phase (OD600 of 4.5). Starting from 1 h later, microscopic images and protein samples were taken every 30 min. Although the DgcZ protein concentration remained constant, the proportion of cells showing polar localization increased over time (see Fig. S1B to C in the supplemental material), implying that an increase in DgcZ concentration does not correlate with polar localization.
Polar localization of DgcZ is observed in nondividing bacteria and is not detectable after cells resumed growth.
To further analyze the effect of growth state and nutritional conditions on the spatiotemporal pattern of DgcZ, cell growth and DgcZ-mVENUS localization were monitored in a microfluidic device that allows tracking of single bacterial cells over time and triggering changes in medium composition (48, 49). Here, we monitored DgcZ-mVENUS derived signals applying the following regime of medium supply: (i) cultivation in nutrient-sufficient medium (LB medium for 2 h), (ii) cultivation in a gradient from fresh to nutrient-depleted (spent) medium (for 3 h), (iii) cultivation in spent medium (for 16 h), (iv) cultivation in a gradient from spent to fresh medium (for 1 h), and (v) cultivation in nutrient-sufficient medium (LB medium for 20 h). DgcZ-mVENUS derived fluorescence appeared toward the end of the gradient phase and was dispersed (see Fig. S2A and Video S1 in the supplemental material). DgcZ localization at the bacterial cell pole started after 5 h of the spent medium phase, when bacteria ceased growth and division (see Fig. S2A and Video S1 in the supplemental material). When fresh medium was applied to the microfluidic chamber, the polar DgcZ-mVENUS foci disappeared, and cells resumed growth (see Video S1 in the supplemental material). The same phenomenon was observed in stationary-phase bacteria spotted onto LB agarose-coated microscopy slides. DgcZ foci vanished in the individuals that restarted growth, whereas cells that did not resume growth maintained polar localization of DgcZ (see Fig. S2B in the supplemental material). Hence, in both experimental systems the disappearance of polar DgcZ foci correlated with the restart of growth.
Polar localization of DgcZ is induced by carbon starvation and alkaline pH.
Nutrient starvation or the accumulation of by-products in the medium may result in polar localization of DgcZ in stationary phase. To discriminate between these two possibilities, the csrA::Tn5 dgcZ-mVENUS strain was grown to an OD600 of 2.5 in LB medium and subsequently transferred to spent LB medium or 1× MMA without carbon sources. Only bacteria transferred to spent LB medium showed polar localization of DgcZ starting 3 h after the transfer (Fig. 4A). This implies that one or more factors present in spent LB medium, but not in 1× MMA without carbon sources, cause this localization.
FIG 4.
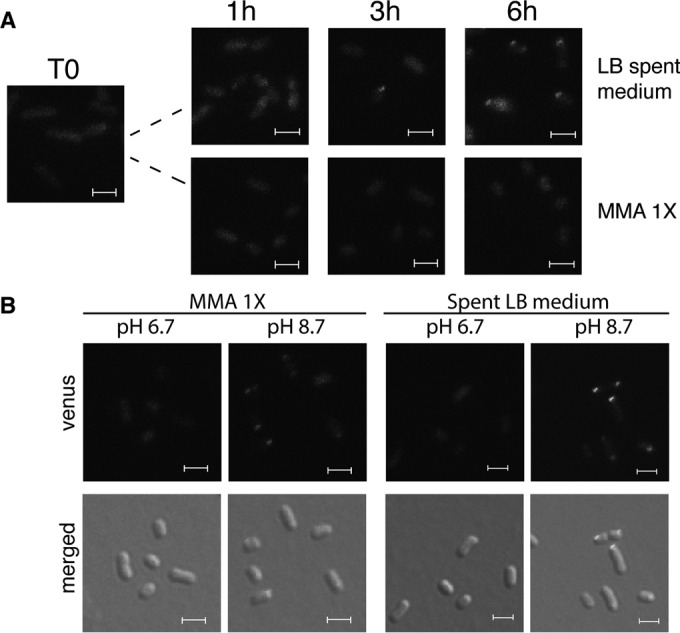
Effect of pH on DgcZ-mVENUS localization. (A) E. coli MG1655 dgcZ-mVENUS csrA::Tn5 was grown in LB to an OD600 of 2.5 and then resuspended in spent LB medium or in 1× MMA without carbon sources. Fluorescence microscopy images were taken after 1, 3, and 6 h. (B) The strain from panel A was grown as described above and transferred to buffered (50 mM K2HPO4, 15 mM KH2PO4) spent LB medium or 1× MMA without carbon sources. Pictures were taken after 6 h. Size bars indicate 2 μm and are identical in each picture.
Since spent LB medium and 1× MMA without carbon sources have pH values of 8.7 and 6.9, respectively, the effect of pH on localization of DgcZ was tested. Bacteria grown in LB medium to an OD600 of 2.5 were resuspended in 1× MMA without carbon sources or in buffered spent LB medium, both at pH 6.7 and at 8.7. After 5 h, DgcZ-mVENUS showed dispersed localization at pH 6.7 and polar localization at pH 8.7 in both media (Fig. 4B). Alkaline pH failed to induce polar localization of DgcZ-mVENUS in fresh LB medium (see Fig. S3A to C in the supplemental material), suggesting that alkaline pH in combination with carbon starvation induces this localization. At alkaline pH, the DgcZ-mVENUS signal was much stronger than at pH 6.7 (Fig. 4B; see Fig. S3A in the supplemental material), thus confirming previous observations of higher dgcZ expression at pH 8.7 than at pH 5.0 or at pH 7.0 (52) and activation of the Cpx two-component system by alkaline pH (52, 53). Since at alkaline pH the Cpx-mediated activation of dgcZ transcription is stronger, we tested whether CpxR is also needed for polar localization of DgcZ-mVENUS (see Fig. S3D in the supplemental material). However, DgcZ-mVENUS localization was independent of CpxR (see Fig. S3D in the supplemental material).
Restoring nutrient-sufficient conditions results in decreased levels and dispersed localization of DgcZ.
We investigated changes in the localization and the amount of DgcZ after the transfer of stationary-phase cells exposed to different pH values to nutrient-sufficient conditions. As previously observed, the DgcZ-mVENUS derived signal was dispersed in stationary-phase cells at pH 6.7 and polarly localized at pH 8.7. After restoring nutrient-sufficient conditions, in both cases the DgcZ-mVENUS signal was dispersed (see Fig. S4A and S5 in the supplemental material), and the levels of the fusion protein decreased, as detected by Western blots (see Fig. S4B in the supplemental material).
Multiple PDEs contribute to the decrease in c-di-GMP concentration in the csrA::Tn5 mutant during transition phase.
The drop in c-di-GMP concentrations observed in csrA::Tn5 cells (Fig. 3) might have been caused by PDE activity or by inactivation of DgcZ. To differ between these possibilities, PgaD levels, indicative of c-di-GMP concentration, and DgcZ levels were monitored over time to determine the onset of decrease in c-di-GMP (Fig. 5A and B). Furthermore, we tested whether PDEs are responsible for the declining levels of c-di-GMP.
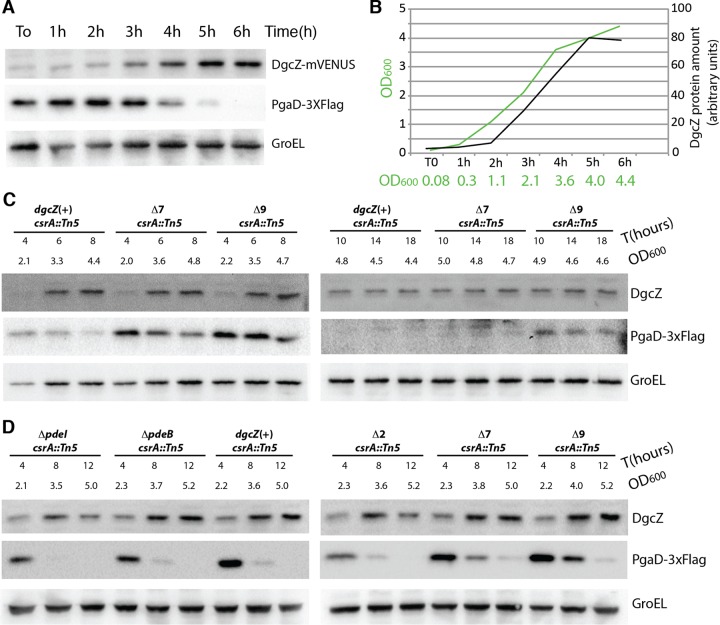
FIG 5.
Decrease in PgaD protein levels in transition phase is caused by PDEs. (A) Western blot detection of DgcZ-mVENUS, PgaD-3×Flag, and GroEL protein extracted from the dgcZ-mVENUS pgaD-3×Flag csrA::Tn5 strain grown in LB medium at 37°C. Proteins were extracted every hour until bacteria reached the stationary phase. (B) Graph indicating OD600 of the bacterial culture and DgcZ-mVENUS protein concentrations deduced from the experiment shown in panel A. For protein quantification, the intensities of the DgcZ bands were quantified using ImageJ. (C) Western blot detection of DgcZ, PgaD-3×Flag, and GroEL proteins in the dgcZ(+), Δ7, and Δ9 mutants in PDE-encoding genes in a pgaD-3×Flag csrA::Tn5 background. Strains were grown as described for panel A. (D) Western blot detection of DgcZ, PgaD-3×Flag, and GroEL proteins in the ΔpdeI, ΔpdeB, Δ2, Δ7, and Δ9 strains with the PDE-encoding genes deleted and in the dgcZ(+) strain in a pgaD-3×Flag csrA::Tn5 background. Strains were grown as described for panel A.
The csrA::Tn5 dgcZ-mVENUS pgaD-3×Flag strain was grown at 37°C under shaking conditions. In this strain, PgaD levels decreased from transition phase (OD600 of 2.1) on, whereas DgcZ levels increased until bacteria reached the stationary phase (Fig. 5A), implying that degradation of DgcZ is unlikely to be responsible for this drop in PgaD levels. To determine whether this decrease was caused by PDEs, PgaD levels were determined in the Δ7 strain (ΔpdeH [yhjH], ΔpdeL [yahA], ΔpdeA [yfeA], ΔpdeR [yciR], ΔpdeN [rtn], ΔpdeC [yjcC], and ΔpdeF [yfgF]) and the Δ9 strain (Δ7, ΔpdeI [yliE], and ΔpdeB [ylaB]), characterized by 7 and 9 deletions of PDE-encoding genes, respectively. The Δ7 and the Δ9 strains, both in the pgaD-3×Flag csrA::Tn5 background, were grown in LB medium at 37°C, and protein samples were extracted at different times. PgaD levels slightly decreased in the Δ9 strain, whereas a strong decrease was observed in the wild-type and in the Δ7 strain (Fig. 5C). Further, we tested whether PgaD detected in the transition phase in the Δ9 strain resulted from DgcZ-derived c-di-GMP. Although PgaD levels in the Δ9 ΔdgcZ strain are high during early transition phase (4 h, OD600 of 2.9), possibly because of the reduction in PDE activity, there was a marked decrease at later stages, and after 12 h (OD600 of 5.2) PgaD was not detectable (see Fig. S6 in the supplemental material). This implies that in the Δ9 strain at the late transition phase PgaD levels result from DgcZ-derived c-di-GMP.
We also tested whether PdeI (YliE) and/or PdeB (YlaB) reduced the c-di-GMP concentration at the entry of stationary phase in the csrA::Tn5 background (Fig. 5D). Neither the deletion of pdeI and pdeB individually nor the deletion of both genes together affected PgaD production (Fig. 5D). This suggests a role of multiple PDEs in controlling c-di-GMP levels in the csrA::Tn5 strain background.
It was further tested whether PgaD levels also decrease in the csrA::Tn5 dgcZ(H79L,H83L) pgaD-3×Flag strain carrying a hyperactive variant of DgcZ that cannot be inactivated by binding of Zn2+ (see Fig. S6B and C in the supplemental material) (29). The observed decrease in PgaD levels suggests that the drop in c-di-GMP levels is not caused by Zn2+-dependent inactivation of DgcZ.
DgcZ interacts with the FrdB subunit of the fumarate reductase (FRD) complex.
To gain further information about function and regulation of DgcZ, we screened for protein interaction partners by CoIP applying DgcZ-3×Flag as bait. Samples were taken from exponential-phase (OD600 of 0.6) and stationary-phase (OD600 of 4.5) cultures because we had previously shown that cells in these growth phases differ in c-di-GMP content and DgcZ localization. Although cells at an OD600 of ∼0.6 have a high c-di-GMP content and DgcZ is dispersed, cells at an OD600 of ∼4.5 have a low c-di-GMP content and show polarly localized DgcZ (Fig. 3 and 5A and B).
In these CoIP experiments, FrdB, AlaS, FrdA, GabD, YgfK, PspD, YgeY, TdcB, PflB, TdcE, and GrcA were identified with a significant score as putative interaction partners of DgcZ (Table 1; see also Table S4 in the supplemental material). Two subunits (FrdA and FrdB) of the fumarate reductase complex (FRD) were among the proteins identified in the CoIP from stationary-phase cells. This complex is involved in anaerobic respiration and also needed for the production of flagella and the control of flagellar activity (37). Interestingly, overproduction of DgcZ has been reported to reduce the number of flagella in surface-attached E. coli MG1655 through an unknown mechanism (32). Identification of two subunits of one protein complex and the high score of FrdB identification (see Table S4 in the supplemental material) motivated further experiments to support interactions between DgcZ and FrdA-B in the present study.
TABLE 1.
Proteins identified by coimmunoprecipitation with DgcZ-3×Flag
| Protein | Function | Phase |
|---|---|---|
| FrdB | Fumarate reductase, anaerobic, iron-sulfur protein subunit | Stationary |
| AlaS | Alanyl-tRNA synthetase | Stationary |
| FrdA | Fumarate reductase | Stationary |
| GabD | Succinate-semialdehyde dehydrogenase I, NADP dependent | Stationary |
| YgfK | Predicted oxidoreductase, Fe-S subunit | Exponential |
| PspD | Phage shock protein, inner membrane protein | Exponential |
| YgeY | Hypothetical protease | Exponential |
| TdcB | Threonine dehydratase | Exponential |
| PflB | Pyruvate formate lyase I | Exponential |
| TdcE | Pyruvate formate-lyase 4/2-ketobutyrate formate-lyase | Exponential |
| GrcA | Pyruvate formate lyase subunit | Exponential |
A bacterial two-hybrid assay indicated a direct interaction of DgcZ with FrdB, but not with FrdA (see Fig. S7 in the supplemental material). This interaction was more clearly detected when the frdB fusion genes were expressed from high-copy-number plasmids (see Fig. S7 in the supplemental material). Since the DgcZ protein is composed of the GGDEF domain and of the zinc binding domain (CZB), we tested which of these domains interacts with FrdB. In the bacterial two-hybrid assay, the CZB domain interacted with FrdB as efficiently as the full-length protein, whereas no interaction was detected using the GGDEF domain (Fig. 6).
FIG 6.
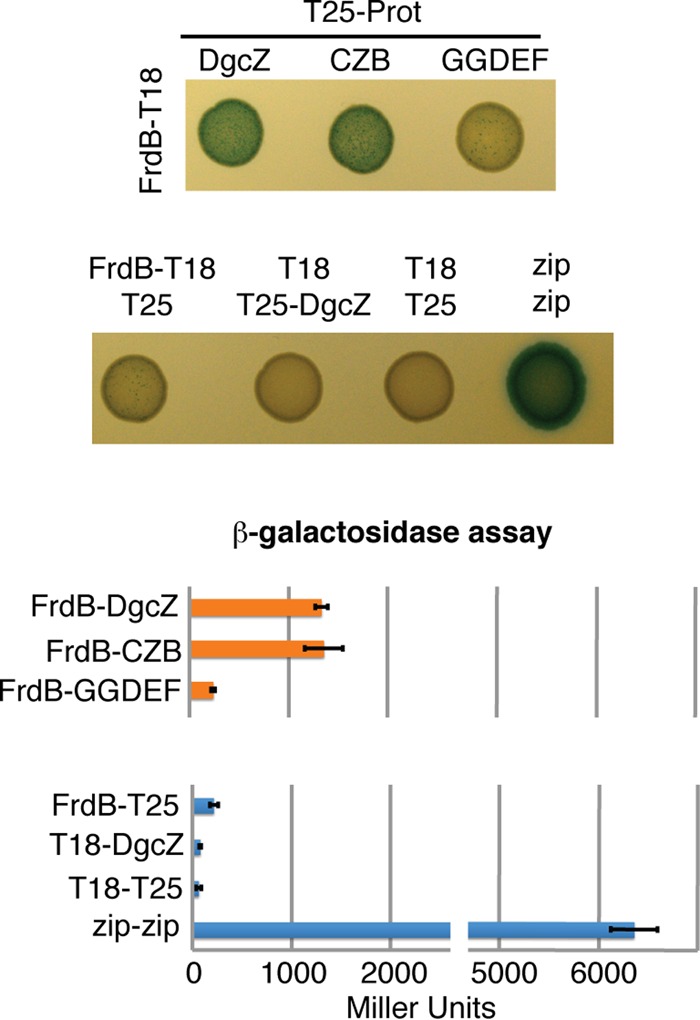
Validation of interactions between FrdB and DgcZ full-length protein, as well as between CZB and GGDEF domains, using bacterial two-hybrid assays. High-copy-number plasmid pUT18 encoding FrdB C-terminally fused to T18 and low-copy-number plasmid pKT25 encoding full-length DgcZ or the CZB or GGDEF domain N-terminally fused to T25 were combined. As negative controls, pUT18-FrdB and pKT25-DgcZ were combined with the empty plasmids pKT25 and pUT18, respectively. Images of cultures spotted onto LB agar plates supplemented with X-Gal and IPTG are shown on the left, with the relative β-galactosidase activities indicated on the right. The experiment was repeated three times, and one representative plate is shown. Error bars indicate the standard deviations of three biological replicates. The growth conditions are described in Materials and Methods.
Localization of FrdA, FrdB, and DgcZ was analyzed by fluorescence microscopy applying DgcZ-mVENUS and FrdA- or FrdB-mCHERRY fusion proteins (see Fig. S8 in the supplemental material). Both FrdA- and FrdB-mCHERRY localized all around the membrane, thus showing no correlation with the polar localization of DgcZ (see Fig. S8 in the supplemental material).
Superoxide stimulates biofilm formation in a DgcZ- and FRD-dependent manner.
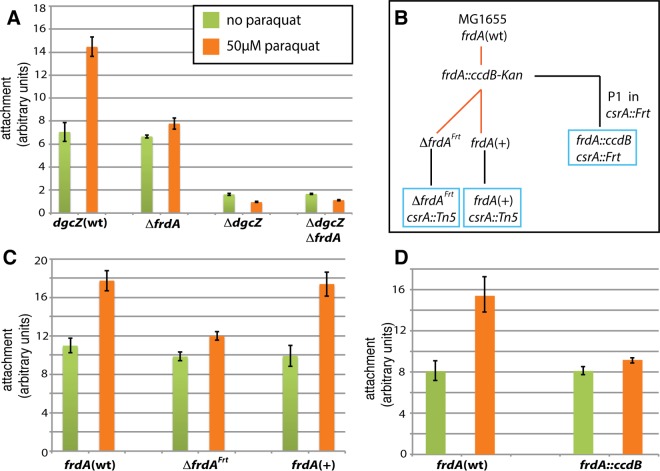
The FRD complex has been shown to produce superoxide in E. coli (54). Paraquat (N,N′-dimethyl-4,4′-bipyridinium dichloride) is an organic compound known to generate superoxide in bacteria, including E. coli (55). To test the effect of FRD on DgcZ-mediated biofilm formation in absence or presence of paraquat, attachment assays were conducted in a csrA::Tn5 background (Fig. 7A). In the absence of paraquat, the ΔfrdA strain showed similar levels of biofilm as the frdA(wt) strain. Upon the addition of 50 μM paraquat, the levels of biofilm formation of the frdA(wt) strain increased ∼2-fold, whereas a very small increase (ca. 10%) was detected in case of the ΔfrdA strain (Fig. 7A). The same pattern was observed for the ΔfrdB strain (see Fig. S9 in the supplemental material). However, this strain carries a longer frdA gene, as deduced from the primers used for generation of this strain in the Keio collection (39). Reconstituting frdA in the ΔfrdA strain restored paraquat-stimulated biofilm formation. This confirms that the decrease in paraquat-stimulated biofilm formation was caused by the lack of frdA (Fig. 7B to D).
FIG 7.
Paraquat enhances DgcZ-dependent biofilm formation in an FRD-dependent fashion. (A) Attachment assay of dgcZ(wt), ΔfrdA, ΔdgcZ, and ΔdgcZ ΔfrdA strains carrying the csrA::Tn5 mutation. LB medium was supplemented with 0 or 50 μM paraquat. (B) Schematic representation of strain construction. The frdA gene was first deleted in E. coli MG1655. The resulting frdA::ccdB-kan strain was then used as the ancestor for the ΔfrdAFrt and frdA(+) strains. Both strains were transduced using the P1 lysate from the crsA::Tn5 strain. The frdA::ccdB csrA::Frt strain was obtained by P1 transduction of the csrA::Frt strain using the lysate from the frdA::ccdB-kan strain. Black lines indicate P1 transductions, and red lines indicate λRed-mediated recombineering. Boxes indicate strains used for attachment assays. (C) Attachment assay of frdA(wt), ΔfrdAFrt, and frdA(+) strains in a csrA::Tn5 background. (D) Attachment assay of frdA(wt) and frdA::ccdB strains carrying the csrA::Tn5 mutation. The results in panels A, C, and D are averages for five wells inoculated with single colonies. Experiments were repeated three times with the same results. Error bars indicate the SEM.
DISCUSSION
Many bacteria are able to respond to alterations in environmental conditions by switching between different lifestyles. A frequently observed pattern is the change from a motile, planktonic state to a surface attached state, and subsequent biofilm formation. In E. coli, an intricate regulatory network controls flagellum biosynthesis and activity as well as production of matrix components to promote efficient attachment to surfaces and biofilm formation (16, 19, 32, 56, 57). Our study expands the knowledge on the role of DgcZ in this regulation.
Role of upstream processes controlling DgcZ.
Regulation of dgcZ expression is rather complex involving the transcriptional activator CpxR (12, 13, 30) and the translational inhibitor CsrA (32). These regulators control transcription and translation of various targets and are themselves regulated at many levels. It is therefore difficult to decipher under which physiological conditions their influence on DgcZ is manifested. In the present study, we showed that DgcZ and its regulator CpxR contribute to surface attachment in a csrA(wt) strain at 30°C, whereas Agladze et al. (18) reported that deletion of cpxR did not affect this process at 26°C.
In E. coli, the lipoprotein NlpE, together with the Cpx complex, mediates surface recognition and stimulate attachment (7, 8). Surface sensing activates CpxR mostly in stationary-phase bacteria (8). We observed that enhanced biofilm formation upon overproduction of NlpE requires dgcZ, suggesting that upon transcriptional activation of dgcZ, attachment occurs more efficiently.
Role of PDEs in reducing c-di-GMP levels in late growth phases.
Previously, a decrease in c-di-GMP levels, commencing with transition phase, was observed in the E. coli K-12 MG1655 wild-type strain (58). We found that, in the csrA::Tn5 background, the decrease in c-di-GMP content observed in the transition phase is mostly caused by multiple PDEs.
Alkaline pH and carbon starvation induce polar accumulation of DgcZ.
PGA plays an important role in initial adhesion to abiotic surfaces in Escherichia coli, Pseudomonas aeruginosa, and Pseudomonas fluorescens (18, 33). In several bacterial species, initial adhesion mediated by the bacterial cell pole is followed by the so-called irreversible attachment that can occur all around the bacterial cell (18, 59, 60). In E. coli, the bacterial cell pole has been shown to be the production site of cellulose (61) and PGA under shaking conditions (19). Whether polar localization of DgcZ observed in the late stationary phase at alkaline pH is responsible for activation of PGA biosynthesis at the cell pole remains unclear. c-di-GMP production is not required for DgcZ polar localization, since an inactive protein variant accumulated at the pole as well. After restoring nutrient-sufficient conditions, restart of growth of previously starved cells generally correlated with a decrease in DgcZ levels, and polar accumulation of DgcZ was no longer observable, probably because of protein degradation. The observed increase in DgcZ levels at alkaline pH is in line with the previously reported activation of the Cpx two-component system under alkaline conditions (52, 53).
Functional implication of DgcZ-FRD interaction.
A functional relationship between DgcZ and the fumarate reductase (FRD) complex is suggested by the interaction of DgcZ with the FrdB subunit of the FRD complex, mediated by the CZB domain of DgcZ. The FRD complex has been shown to produce superoxide (54), to be associated with FliG (37), and to be involved in regulation of flagellum assembly and activity in E. coli (37). An E. coli frd deletion mutant displays fewer flagella than does the wild type and very rarely switches the direction of flagellum rotation (37). Bacteria rotating flagella in just one direction attach to surfaces less efficiently than does the wild type (14). In E. coli, overproduction of DgcZ results in a drastic reduction of flagella and pili in surface-attached bacteria, implying involvement of DgcZ in flagellum regulation (32). Overproduction of the DGC DgcT (formerly YcdT) leads to higher c-di-GMP levels than those detected upon dgcZ overexpression but does not affect flagella, implying a DgcZ-specific effect (32).
Considering the role of the FRD complex in the assembly and control of flagella, it is tempting to speculate that the interaction of DgcZ with the FRD complex may affect this regulation, although direct evidence for such a regulatory link is missing. The requirement of FRD for the increase in DgcZ-mediated biofilm formation upon induced production of superoxide further hints at a functional link between DgcZ and the FRD complex. Furthermore, a decrease in respiration within the first 15 min after cell-surface and cell-cell contacts were observed (62). Since FRD is involved in anaerobic respiration, we speculate that FRD, together with DgcZ, may promote biofilm formation upon surface contact.
Recently, polar localization of TlpD, a CZB domain protein in Helicobacter pylori, has been reported in media of high energy, whereas in low-energy conditions, such as oxidative stress, localization changed toward the cell body (63). TlpD mediates repellent responses to oxidative stress (64). Polar localization was also lost in mutants in genes encoding chemotaxis histidine kinase CheAY2, the central metabolic enzyme aconitase AcnB and the detoxifying enzyme catalase KatA, all identified as putative interaction partners of TlpD in CoIP experiments (63). Like FrdB, AcnB contains an iron-sulfur cluster, and both proteins contribute to the tricarboxylic acid cycle. This suggests that CZB domains may mediate polar protein localization related to oxidative or metabolic stress in conjunction with proteins containing iron-sulfur clusters, such as AcnB and FrdB.
DgcZ may play an interlinking regulatory role during the entire process of surface attachment.
Previously, an involvement of DgcZ in biofilm formation in the csrA::Tn5 strain has been described (28, 29). Based on the following evidence, we suggest a physiological role for DgcZ in linking surface sensing and attachment: (i) CpxR transcriptionally activates DgcZ (12, 13, 30), (ii) DgcZ plays an important role in permanent attachment to abiotic surfaces in csrA(wt) (this study) and csrA::Tn5 (28, 29) strains, (iii) attachment of the csrA::Tn5 mutant increases upon NlpE overproduction in a DgcZ-dependent way (this study), and (iv) DgcZ interacts with the FRD complex, and both are needed for the increase of biofilm formation observed upon induction of oxidative stress (this study). However, we note that most data contributing to this model have not been obtained from surface-attached cells.
We speculate that upon surface contact under permissive conditions, e.g., nutrient starvation, DgcZ is produced and interferes with flagellar activity via YcgR (16, 17) in a c-di-GMP-dependent fashion. This hypothesis is supported by the evidence that overexpression of dgcZ affects bacterial motility (32). Interference with flagellar activity may impede escape of the bacterial cells from the surface and PGA production may then stabilize adhesion. Interestingly, motility is inhibited upon CpxR activation that represses transcription of the motAB-cheAW operon (7). CsrA possibly plays an important role in determining whether, after surface sensing, bacteria will attach or not. Sequestration of CsrA by the CsrB-C sRNAs (65, 66) and by the McaS sRNA (67, 68) at the entry of stationary phase and in nutrient-poor medium supports this hypothesis. Surface sensing and CsrA have been reported to also influence adhesion in other bacteria. In Pseudomonas aeruginosa, a CsrA homologue (RsmA) inhibits translation of the mRNA of the DGC SadC (69), activated in response to surface sensing (70). This parallel suggests a common role for c-di-GMP in signal transduction from surface sensing to adhesion.
Supplementary Material
ACKNOWLEDGMENTS
We thank Daniella Cavalcanti de Lucena for providing the mVENUS template (pDL-mVENUS) and, along with Benjamin Frage, for initial support in microscopy. We also thank Matthew McIntosh for pWBP20911, Kevin Dale Young for the ibpA-mCHERRY strain, Tilman Schirmer and Urs Jenal for the anti-DgcZ antibody, Uwe Linne for the service of the mass spectrometry facility (University of Marburg) in protein identification, Annette Garbe for assistance with the liquid chromatography-tandem mass spectrometry, Urs Jenal for helpful discussions, and Vera Bettenworth and Martin Ackermann for critical reading of the manuscript.
Alex Boehm passed away in November 2012. We express our most sincere thanks to him for initiating the studies on DgcZ.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00320-16.
REFERENCES
- 1.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. 2013. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev 26:822–880. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fux CA, Costerton JW, Stewart PS, Stoodley P. 2005. Survival strategies of infectious biofilms. Trends Microbiol 13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 3.Serra DO, Richter AM, Klauck G, Mika F, Hengge R. 2013. Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. mBio 4:e00103-13. doi: 10.1128/mBio.00103-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 5.Tetz GV, Artemenko NK, Tetz VV. 2009. Effect of DNase and antibiotics on biofilm characteristics. Antimicrob Agents Chemother 53:1204–1209. doi: 10.1128/AAC.00471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beloin C, Roux A, Ghigo JM. 2008. Escherichia coli biofilms. Curr Top Microbiol Immunol 322:249–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snyder WB, Davis LJ, Danese PN, Cosma CL, Silhavy TJ. 1995. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J Bacteriol 177:4216–4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otto K, Silhavy TJ. 2002. Surface sensing and adhesion of Escherichia coli controlled by the Cpx signaling pathway. Proc Natl Acad Sci U S A 99:2287–2292. doi: 10.1073/pnas.042521699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiGiuseppe PA, Silhavy TJ. 2003. Signal detection and target gene induction by the CpxRA two-component system. J Bacteriol 185:2432–2440. doi: 10.1128/JB.185.8.2432-2440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raivio TL, Silhavy TJ. 1997. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J Bacteriol 179:7724–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorel C, Vidal O, Prigent-Combaret C, Vallet I, Lejeune P. 1999. Involvement of the Cpx signal transduction pathway of E. coli in biofilm formation. FEMS Microbiol Lett 178:169–175. doi: 10.1111/j.1574-6968.1999.tb13774.x. [DOI] [PubMed] [Google Scholar]
- 12.Raivio TL, Leblanc SK, Price NL. 2013. The Escherichia coli Cpx envelope stress response regulates genes of diverse function that impact antibiotic resistance and membrane integrity. J Bacteriol 195:2755–2767. doi: 10.1128/JB.00105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raivio TL. 2014. Everything old is new again: an update on current research on the Cpx envelope stress response. Biochim Biophys Acta 1843:1529–1541. doi: 10.1016/j.bbamcr.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 14.McClaine JW, Ford RM. 2002. Reversal of flagellar rotation is important in initial attachment of Escherichia coli to glass in a dynamic system with high- and low-ionic-strength buffers. Appl Environ Microbiol 68:1280–1289. doi: 10.1128/AEM.68.3.1280-1289.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedlander RS, Vlamakis H, Kim P, Khan M, Kolter R, Aizenberg J. 2013. Bacterial flagella explore microscale hummocks and hollows to increase adhesion. Proc Natl Acad Sci U S A 110:5624–5629. doi: 10.1073/pnas.1219662110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. 2010. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141:107–116. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 17.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell 38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agladze K, Wang X, Romeo T. 2005. Spatial periodicity of Escherichia coli K-12 biofilm microstructure initiates during a reversible, polar attachment phase of development and requires the polysaccharide adhesin PGA. J Bacteriol 187:8237–8246. doi: 10.1128/JB.187.24.8237-8246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itoh Y, Rice JD, Goller C, Pannuri A, Taylor J, Meisner J, Beveridge TJ, Preston JF, Romeo T. 2008. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-β-1,6-N-acetyl-d-glucosamine. J Bacteriol 190:3670–3680. doi: 10.1128/JB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steiner S, Lori C, Boehm A, Jenal U. 2013. Allosteric activation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein-protein interaction. EMBO J 32:354–368. doi: 10.1038/emboj.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyd CD, O'Toole GA. 2012. Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu Rev Cell Dev Biol 28:439–462. doi: 10.1146/annurev-cellbio-101011-155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Römling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tal R, Wong HC, Calhoon R, Gelfand D, Fear AL, Volman G, Mayer R, Ross P, Amikam D, Weinhouse H, Cohen A, Sapir S, Ohana P, Benziman M. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J Bacteriol 180:4416–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simm R, Morr M, Kader A, Nimtz M, Römling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol 53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- 25.Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, He YW, Zhang LH, Heeb S, Cámara M, Williams P, Dow JM. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci U S A 103:6712–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Schirmer T, Jenal U. 2009. Structural and mechanistic determinants of c-di-GMP signaling. Nat Rev Microbiol 7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 27.Hengge R, Galperin MY, Ghigo JM, Gomelsky M, Green J, Hughes KT, Jenal U, Landini P. 2015. Systematic nomenclature for GGDEF and EAL domain-containing cyclic Di-GMP turnover proteins of Escherichia coli. J Bacteriol 198:7–11. doi: 10.1128/JB.00424-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boehm A, Steiner S, Zaehringer F, Casanova A, Hamburger F, Ritz D, Keck W, Ackermann M, Schirmer T, Jenal U. 2009. Second messenger signaling governs Escherichia coli biofilm induction upon ribosomal stress. Mol Microbiol 72:1500–1516. doi: 10.1111/j.1365-2958.2009.06739.x. [DOI] [PubMed] [Google Scholar]
- 29.Zähringer F, Lacanna E, Jenal U, Schirmer T, Boehm A. 2013. Structure and signaling mechanism of a zinc-sensory diguanylate cyclase. Structure 21:1149–1157. doi: 10.1016/j.str.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto K, Ishihama A. 2006. Characterization of copper-inducible promoters regulated by CpxA/CpxR in Escherichia coli. Biosci Biotechnol Biochem 70:1688–1695. doi: 10.1271/bbb.60024. [DOI] [PubMed] [Google Scholar]
- 31.Price NL, Raivio TL. 2009. Characterization of the Cpx regulon in Escherichia coli strain MC4100. J Bacteriol 191:1798–1815. doi: 10.1128/JB.00798-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jonas K, Edwards AN, Simm R, Romeo T, Römling U, Melefors Ö. 2008. The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol Microbiol 70:236–257. doi: 10.1111/j.1365-2958.2008.06411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T. 2005. CsrA posttranscriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol 56:1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- 34.Romeo T, Gong M, Liu MY, Brun-Zinkernagel AM. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J Bacteriol 175:4744–4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Timmermans J, Van Melderen L. 2010. Posttranscriptional global regulation by CsrA in bacteria. Cell Mol Life Sci 67:2897–2908. doi: 10.1007/s00018-010-0381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Draper J, Karplus K, Ottemann KM. 2011. Identification of a chemoreceptor zinc-binding domain common to cytoplasmic bacterial chemoreceptors. J Bacteriol 193:4338–4345. doi: 10.1128/JB.05140-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen-Ben-Lulu GN, Francis NR, Shimoni E, Noy D, Davidov Y, Prasad K, Sagi Y, Cecchini G, Johnstone RM, Eisenbach M. 2008. The bacterial flagellar switch complex is getting more complex. EMBO J 27:1134–1144. doi: 10.1038/emboj.2008.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 39.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-A. [DOI] [PubMed] [Google Scholar]
- 41.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zacharias DA, Violin JD, Newton AC, Tsien RY. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 43.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 44.Charoenpanich P, Soto MJ, Becker A, McIntosh M. 2015. Quorum sensing restrains growth and is rapidly inactivated during domestication of Sinorhizobium meliloti. Environ Microbiol Rep 7:373–382. doi: 10.1111/1758-2229.12262. [DOI] [PubMed] [Google Scholar]
- 45.Bibikov SI, Biran R, Rudd KE, Parkinson JS. 1997. A signal transducer for aerotaxis in Escherichia coli. J Bacteriol 179:4075–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Battesti A, Bouveret E. 2012. The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods 58:325–334. doi: 10.1016/j.ymeth.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 47.Burhenne H, Kaever V. 2013. Quantification of cyclic dinucleotides by reversed-phase LC-MS/MS. Methods Mol Biol 1016:27–37. doi: 10.1007/978-1-62703-441-8_3. [DOI] [PubMed] [Google Scholar]
- 48.Arnoldini M, Vizcarra IA, Peña-Miller R, Stocker N, Diard M, Vogel V, Beardmore RE, Hardt WD, Ackermann M. 2014. Bistable expression of virulence genes in Salmonella leads to the formation of an antibiotic-tolerant subpopulation. PLoS Biol 12:e1001928. doi: 10.1371/journal.pbio.1001928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang P, Robert L, Pelletier J, Dang WL, Taddei F, Wright A, Jun S. 2010. Robust growth of Escherichia coli. Curr Biol 20:1099–1103. doi: 10.1016/j.cub.2010.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Toole GA, Pratt LA, Watnick PI, Newman DK, Weaver VB, Kolter R. 1999. Genetic approaches to study of biofilms. Methods Enzymol 310:91–109. doi: 10.1016/S0076-6879(99)10008-9. [DOI] [PubMed] [Google Scholar]
- 51.De Wulf P, Kwon O, Lin EC. 1999. The CpxRA signal transduction system of Escherichia coli: growth-related autoactivation and control of unanticipated target operons. J Bacteriol 181:6772–6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maurer LM, Yohannes E, Bondurant SS, Radmacher M, Slonczewski JL. 2005. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J Bacteriol 187:304–319. doi: 10.1128/JB.187.1.304-319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Danese PN, Silhavy TJ. 1998. CpxP, a stress-combative member of the Cpx regulon. J Bacteriol 180:831–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Imlay JA. 1995. A metabolic enzyme that rapidly produces superoxide, fumarate reductase of Escherichia coli. J Biol Chem 270:19767–19777. [PubMed] [Google Scholar]
- 55.Hassan HM, Fridovich I. 1978. Superoxide radical and the oxygen enhancement of the toxicity of paraquat in Escherichia coli. J Biol Chem 253:8143–8148. [PubMed] [Google Scholar]
- 56.Lindenberg S, Klauck G, Pesavento C, Klauck E, Hengge R. 2013. The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signaling cascade in E. coli biofilm control. EMBO J 32:2001–2014. doi: 10.1038/emboj.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mika F, Hengge R. 2014. Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli. RNA Biol 11:494–507. doi: 10.4161/rna.28867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spangler C, Böhm A, Jenal U, Seifert R, Kaever V. 2010. A liquid chromatography-coupled tandem mass spectrometry method for quantitation of cyclic di-guanosine monophosphate. J Microbiol Methods 81:226–231. doi: 10.1016/j.mimet.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 59.Caiazza NC, O'Toole GA. 2004. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J Bacteriol 186:4476–4485. doi: 10.1128/JB.186.14.4476-4485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hinsa SM, Espinosa-Urgel M, Ramos JL, O'Toole GA. 2003. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol 49:905–918. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- 61.Le Quéré B, Ghigo JM. 2009. BcsQ is an essential component of the Escherichia coli cellulose biosynthesis apparatus that localizes at the bacterial cell pole. Mol Microbiol 72:724–740. doi: 10.1111/j.1365-2958.2009.06678.x. [DOI] [PubMed] [Google Scholar]
- 62.Geng J, Beloin C, Ghigo JM, Henry N. 2014. Bacteria hold their breath upon surface contact as shown in a strain of Escherichia coli, using dispersed surfaces and flow cytometry analysis. PLoS One 9:e102049. doi: 10.1371/journal.pone.0102049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Behrens W, Schweinitzer T, McMurry JL, Loewen PC, Buettner FF, Menz S, Josenhans C. 2016. Localisation and protein-protein interactions of the Helicobacter pylori taxis sensor TlpD and their connection to metabolic functions. Sci Rep 6:23582. doi: 10.1038/srep23582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Collins KD, Andermann TM, Draper J, Sanders L, Williams SM, Araghi C, Ottemann KM. 2016. The Helicobacter pylori CZB cytoplasmic chemoreceptor TlpD forms an autonomous polar chemotaxis signaling complex that mediates a tactic response to oxidative stress. J Bacteriol 198:1563–1575. doi: 10.1128/JB.00071-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jonas K, Melefors Ö. 2009. The Escherichia coli CsrB and CsrC small RNAs are strongly induced during growth in nutrient-poor medium. FEMS Microbiol Lett 297:80–86. doi: 10.1111/j.1574-6968.2009.01661.x. [DOI] [PubMed] [Google Scholar]
- 66.Liu MY, Gui G, Wei B, Preston JF, Oakford L, Yüksel U, Giedroc DP, Romeo T. 1997. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem 272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- 67.Thomason MK, Fontaine F, De Lay N, Storz G. 2012. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol Microbiol 84:17–35. doi: 10.1111/j.1365-2958.2012.07965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jørgensen MG, Thomason MK, Havelund J, Valentin-Hansen P, Storz G. 2013. Dual function of the McaS small RNA in controlling biofilm formation. Genes Dev 27:1132–1145. doi: 10.1101/gad.214734.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moscoso JA, Jaeger T, Valentini M, Hui K, Jenal U, Filloux A. 2014. The diguanylate cyclase SadC is a central player in Gac/Rsm-mediated biofilm formation in Pseudomonas aeruginosa. J Bacteriol 196:4081–4088. doi: 10.1128/JB.01850-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luo Y, Zhao K, Baker AE, Kuchma SL, Coggan KA, Wolfgang MC, Wong GC, O'Toole GA. 2015. A hierarchical cascade of second messengers regulates Pseudomonas aeruginosa surface behaviors. mBio 6:e02456-14. doi: 10.1128/mBio.02456-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.