Abstract
We show how enhancers of macrophage-specific genes are rendered accessible in differentiating macrophages to allow their induction in mature cells in response to an appropriate stimulus. Using a lentiviral knockdown approach in primary differentiating macrophages from mouse bone marrow, we demonstrate that enhancers of Il12b and Il1a are kept relatively lowly occupied by nucleosomes and accessible through recruitment of the nucleosome remodeler BAF/PBAF. Our results using an inducible cell line that expresses an estrogen receptor fusion of the macrophage-specific transcription factor PU.1 (PUER) show that BAF/PBAF recruitment to these enhancers is a consequence of translocation of PUER to the nucleus in the presence of tamoxifen, and we speculate that remodeler recruitment may be directly mediated by PU.1. In the absence of BAF/PBAF recruitment, nucleosome occupancy at the enhancer of Il12b (and to a lesser extent at Il1a) reaches high levels in bone marrow-derived macrophages (BMDMs), and the enhancers are not fully cleared of nucleosomes upon LPS induction, resulting in impaired gene expression. Analysis of Il12b expression in single cells suggests that recruitment of the remodeler is necessary for high levels of transcription from the same promoter, and we propose that remodelers function by increasing nucleosome turnover to facilitate transcription factor over nucleosome binding in a process we have termed “remodeler-assisted competition.”
Keywords: chromatin, chromatin remodeling, gene expression, gene regulation, macrophage, nucleosome occupancy, chromatin remodeler
Introduction
Lineage-specific transcription factors (TFs)3 play a crucial role in cellular differentiation. These TFs are often pioneer TFs that have been suggested to control access to cis-regulatory elements, in particular gene enhancers, by other ubiquitously expressed TFs (1). The idea that access to regulatory elements is controlled in a cell type-specific manner is supported by the finding that sensitivity of enhancers to nucleases such as DNase I or micrococcal nuclease (MNase) is cell type-specific (for recent studies, see Refs. 2 and 3), but how lineage-specific TFs render enhancers accessible during differentiation is unknown. Moreover, what constitutes accessible or “open” chromatin has remained unclear. Although regulatory regions of constitutively expressed genes are often completely nucleosome-free, we recently showed that the enhancers of inducible genes are occupied by intermediate levels of nucleosomes in resting macrophages, and these nucleosomes are evicted when the genes are induced (4). Furthermore, before induction, these enhancers are already bound by the macrophage-specific pioneer TF PU.1 and primed for activation as indicated by the presence of certain histone marks (i.e. H3K4me1) (5). Binding of PU.1 to enhancers was found to lead to a decrease in nucleosome binding (6, 7), and we showed that in the absence of PU.1 binding, macrophage-specific enhancers become associated with the polycomb repressive complex (PRC2) and with highly occupied, H3K27me3-marked nucleosomes as cells differentiate (8). These results indicated that the pioneer TF PU.1 keeps enhancers accessible and prevents heterochromatin formation at cell type-specific genes, but the underlying mechanism has remained unclear.
We sought to investigate whether nucleosome remodelers are involved in priming of enhancers. Remodelers of the SWI/SNF family have been shown to facilitate gene expression in many organisms, and SWI/SNF function is best understood in the yeast Saccharomyces cerevisiae, where studies showed that SWI/SNF remodelers remove nucleosomes from promoters or partially unwrap nucleosomes to expose TF binding sites (9–13). Mammals have two related SWI/SNF complexes, BAF and PBAF, which share certain subunits but also contain unique subunits that are thought to play a role in recruitment of either complex to specific sites. Both BAF and PBAF use the catalytic subunit BRG1, but BAF can also use the alternate catalytic subunit BRM. BRG1 deletion results in early embryonic lethality, but BRM−/− mice develop normally, and it has been suggested that up-regulation of BRG1 may, in part, compensate for the loss of BRM (14, 15). BRG1 is required for differentiation, including that of lymphoid and myeloid cells, and BRG1 is recruited to cell type-specific genes during differentiation of erythrocytes, suggesting that a BRG1-containing BAF/PBAF complex may prime gene regulatory regions during hematopoiesis (16–18). That BAF/PBAF may play a general role in cellular differentiation is further supported by the finding that BRG1 and other BAF/PBAF subunits are frequently mutated in diverse human cancers (19). The core subunit SNF5, for example, is mutated in malignant rhabdoid tumors, a rare aggressive cancer affecting young children, and SNF5 mutation is sufficient to induce such tumors in mice (20, 21). Rhabdoid tumor cells are unable to proliferate when BRG1 is inactivated, and it has been suggested that these cells may become dependent on an altered BAF/PBAF complex that still relies on the presence of BRG1 (22). Previous studies showed that BAF/PBAF is required for induction of pro-inflammatory genes in mouse macrophages, because simultaneous knockdown of both BRG1 and BRM impaired induction of a subset of pro-inflammatory genes in a macrophage cell line by bacterial lipopolysaccharides (LPS) (23). These investigators suggested a role for BAF/PBAF in remodeling non-CpG island promoters but did not investigate whether the remodeler creates accessible chromatin at the enhancers of these genes to prime them for later gene induction. These investigators also determined whether primary and secondary response genes show differential dependence on the BAF/PBAF remodelers, and concluded that secondary genes and a subset of primary response genes require the remodeler, whereas other primary response genes are largely independent.
Here, we show how regulatory regions of two representative macrophage-specific genes (i.e. Il1a, a primary response gen, and Il12b, a secondary response gene) are rendered accessible during differentiation through recruitment of BAF/PBAF, presumably as a consequence of PU.1 binding. This allows induction of these genes in mature macrophages in response to an appropriate signal. We find that both genes depend on BAF/PBAF for induction and nucleosome eviction at their enhancers, but the effects on Il1a are less pronounced. Our analysis of gene expression in single cells suggests that remodelers function by “remodeler-assisted competition” to facilitate TF binding over nucleosome formation at cell type-specific gene enhancers.
Results
BAF/PBAF Is Recruited to the Il12b and Il1a Enhancers in BMDMs
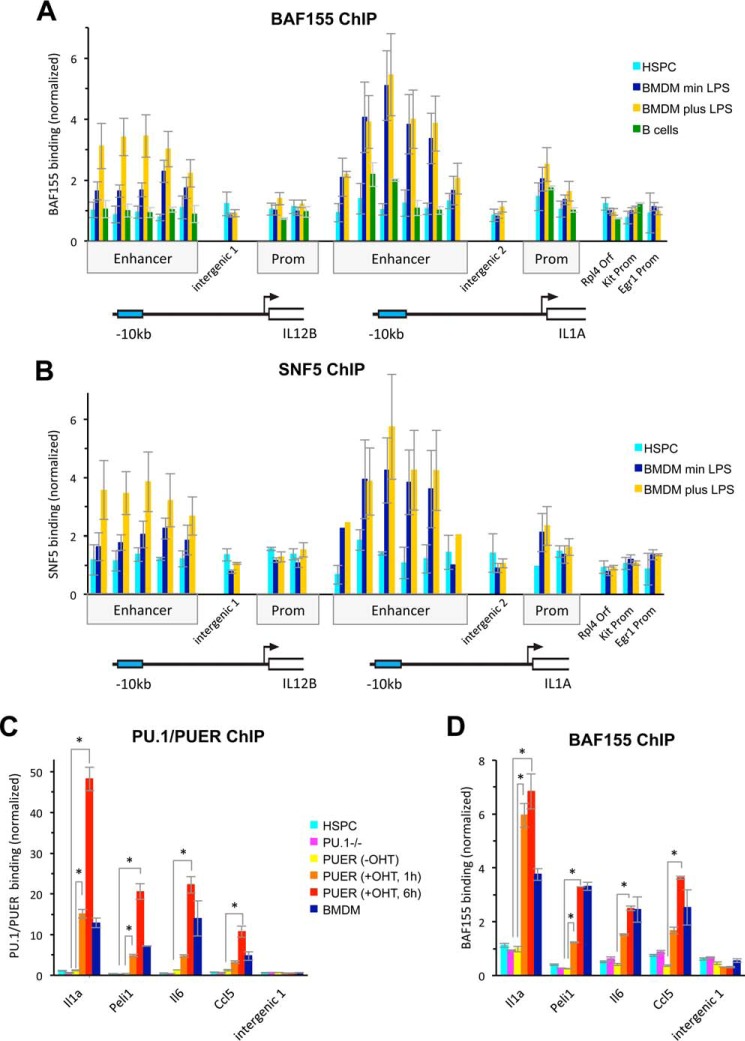
To investigate how the enhancers of Il12b and Il1a are kept accessible and occupied only by intermediate levels of nucleosomes in BMDMs, we investigated whether the BAF/PBAF complex is involved in the process. We determined binding of BAF/PBAF to Il12b and Il1a by ChIP and detected the core subunits BAF155 and SNF5 at both enhancers in resting macrophages (Fig. 1, A and B, dark blue bars). Recruitment of the remodeler to the Il12b enhancer further increased upon LPS induction (yellow bars), but the levels of remodeler at Il1a were already high in resting BMDMs and did not increase significantly upon induction. We found little binding of BAF/PBAF to the enhancers in hematopoietic stem and progenitor cells (HSPCs; isolated by Lin− selection from bone marrow) or B-cells (cyan and green bars, respectively), demonstrating that recruitment of the remodeler to these genes is macrophage-specific. Together our results indicate that BAF/PBAF is recruited to the enhancers of Il12b and Il1a at some time during macrophage differentiation, and that gene induction leads to further remodeler recruitment to Il12b. Binding of SNF5 and BAF155 to the promoters of both genes was low, suggesting that the nucleosome remodeler functions predominantly at the enhancers of these genes.
FIGURE 1.
Recruitment of BAF/PBAF to macrophage-specific enhancers. A, a ChIP experiment probing for BAF155 was performed in HSPCs (cyan) and in BMDMs grown without (dark blue) and with (yellow) LPS for 1.5 h, and in splenic B-cells (green). BAF155 binding to the enhancers, promoters (Prom), and intervening sequences of Il12b and Il1a, and at control regions, is shown. ChIP experiments were performed three times, and error bars indicate the S.E. One-way ANOVA shows that differences at the enhancers are statistically significant (at the p < 0.05 level) between different cell types, whereas differences at control locations, the promoters, and the intervening regions are not statistically significant. A post hoc Tukey HSD test confirmed that differences between uninduced BMDMs and HSPCs or B-cells at the enhancers were statistically significant. At the Il12b enhancer, differences between uninduced and induced BMDMs were also statistically significant, whereas those at the Il1a enhancer were not. B, a SNF5 ChIP was performed in HSPCs and in BMDMs grown with and without LPS, and a statistical analysis confirmed the significance of differences as for the BAF155 ChIP shown in A. C, a ChIP experiment using an antibody that recognizes both PU.1 and PUER was performed in HSPCs (cyan), in the PU.1−/− cell line (magenta), and in PUER cells grown in the absence of tamoxifen (-OHT, yellow), and for 1 h (orange) and 6 h (red) in the presence of tamoxifen (+OHT). All cells were grown in the absence of LPS, and resting BMDMs are shown as controls (blue). PU.1/PUER binding at LPS-inducible enhancers of Il1a, Peli1, Il6, and Ccl5 is shown (for genomic coordinates of the enhancers, see “Experimental Procedures”). ChIP experiments were performed twice, and error bars indicate the S.E. A one-way ANOVA shows statistically significant differences (p < 0.05) between different cell types and growth conditions. Post hoc comparisons using a Tukey HSD test indicate that at all four enhancers, growth in the presence of tamoxifen for 6 h resulted in statistically significant binding of PUER when compared with no tamoxifen, and at Il1a and Peli1, differences were already statistically significant after 1 h. *, p < 0.01. D, a BAF155 ChIP was performed with cells as in C, and a statistical analysis confirmed the significance of the differences in BAF155 recruitment as described for PU.1/PUER binding in C. *, p < 0.01.
BAF/PBAF Recruitment Is a Consequence of PUER Translocation to the Nucleus
To determine how BAF/PBAF is recruited to macrophage-specific enhancers, we turned to the PUER-expressing cell line that we had previously used to determine the effects of PU.1 binding on nucleosome occupancy (8). This cell line was derived from hematopoietic progenitors of the fetal liver of a PU.1−/− mouse and expresses the pioneer TF PU.1 as an estrogen receptor fusion (PUER). Growth for prolonged times (i.e. 4 days) in the presence of tamoxifen leads to differentiation of these cells into macrophage-like cells (24). Alternatively, they can be differentiated into mast cells or erythrocyte precursors, indicating that they are multipotent progenitors. We and others previously showed that when these cells were grown in the presence of tamoxifen, PUER bound to the enhancer of Il1a and other inducible genes, which led to reduced nucleosome binding at these sites (6, 8). We had also shown that PUER did not bind to the enhancer of Il12b and several other inducible macrophage-specific enhancers that are bound by PU.1 in BMDMs, consistent with published results (6). Instead this subset of inducible genes became associated with the polycomb repressive complex PRC2 (i.e. Suz12) and acquired repressive histone marks (i.e. H3K27me3) when the cells were differentiated into macrophage-like cells, indicating that facultative heterochromatin was formed at these sites in the absence of PU.1 binding. To determine whether PUER recruited BAF/PBAF to macrophage-specific enhancers that could bind the pioneer TF in this system, we performed a ChIP experiment probing for the BAF155 subunit and for PU.1 and found that recruitment of BAF/PBAF indeed correlated with PUER binding to the enhancers of Il1a, Peli1, Il6, and Ccl5. Statistically significant BAF155 recruitment and PUER binding was detected as early as 1 h after the addition of tamoxifen at Il1a and Peli1 (Fig. 1, C and D, orange bars) and further increased with prolonged growth in the presence of tamoxifen to reach significant levels at all four enhancers after 6 h (red bars). We had shown previously that at this time the cells still resemble progenitors and that the associated genes are not induced and signal-induced TFs are not bound (8). The rapid appearance of BAF155 binding after tamoxifen addition suggests that remodeler recruitment is a direct consequence of PUER translocation to the nucleus. We speculate that PU.1 may directly recruit BAF/PBAF to these enhancers, although further experiments will have to be performed to confirm this conclusion. We also demonstrated that in primary HSPCs from bone marrow, where BAF/PBAF was not recruited to the enhancers (Fig. 1, A, B, and D, cyan bars), PU.1 was absent as well (Fig. 1C, cyan bars), further supporting the idea that BAF/PBAF recruitment is a consequence of PU.1 binding in primary macrophages. Together our results suggest that up-regulation of PU.1 expression during macrophage differentiation (25) induces PU.1 binding and concomitant recruitment of BAF/PBAF to enhancers of macrophage-specific genes, which primes these genes for induction in mature macrophages.
BAF/PBAF Is Required for Il12b and Il1a Induction in BMDMs
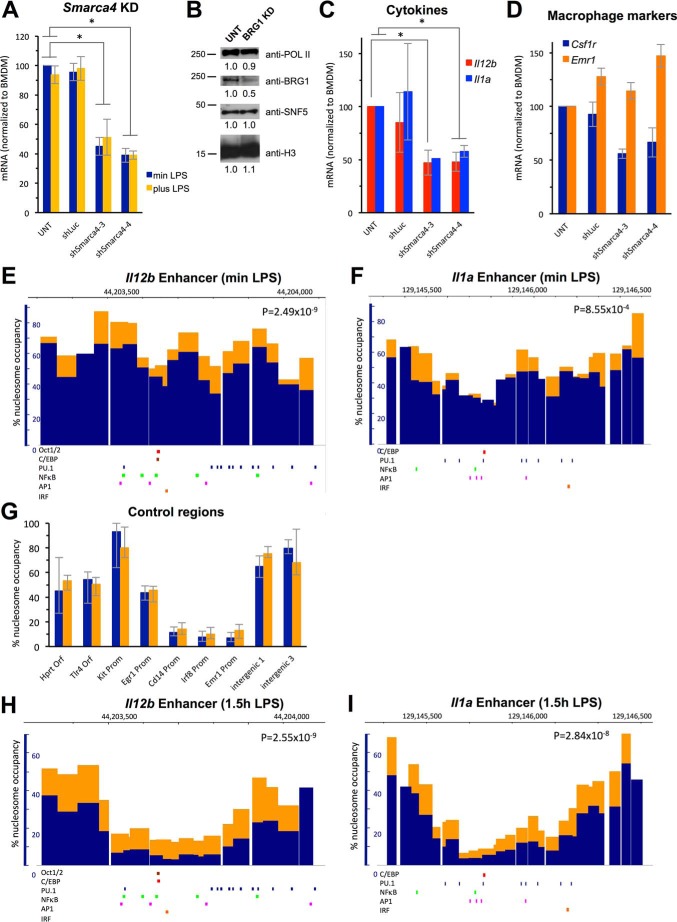
To determine whether recruitment of the BAF/PBAF complex rendered the enhancers of Il12b and Il1a accessible during macrophage differentiation, we used a lentiviral shRNA-mediated knockdown approach. For these experiments, bone marrow cells were transduced with lentivirus containing shRNAs targeting BRG1, encoded by the Smarca4 gene, or with control shRNA targeting firefly luciferase (shLuc). The effect of BRG1 KD was then analyzed in transduced cells that had been differentiated into macrophages in the presence of M-CSF for 9 days. We identified two shRNAs from a pool of shRNAs pre-validated by the Broad Consortium (shSmarca4-3 and shSmarca4-4) that yielded 50–60% knockdown of Smarca4 as determined by mRNA analysis (Fig. 2A) and resulted in reduction of chromatin-associated BRG1 protein by 50% (Fig. 2B). This level of knockdown reduced Il12b and Il1a expression 1.5 h after LPS addition by 50% (Fig. 2C). Previous studies in the macrophage cell line J774 had shown that Il12b expression was dependent on BRG1, but these investigators had classified Il1a as a BAF/PBAF-independent gene, although a small decrease in Il1a expression was reported (23). We believe that the more pronounced effect of our BRG1 KD on Il1a induction may be due to differences between the macrophage cell line J774 and primary BMDMs. The cells differentiated under these conditions still resembled macrophages and expressed the macrophage marker F4/80 (i.e. Emr1, orange bars in Fig. 2D). However, we found that other macrophage-specific, constitutively expressed genes were expressed at lower levels in BRG1 KD cells (i.e. Csf1r, blue bars in Fig. 2D).
FIGURE 2.
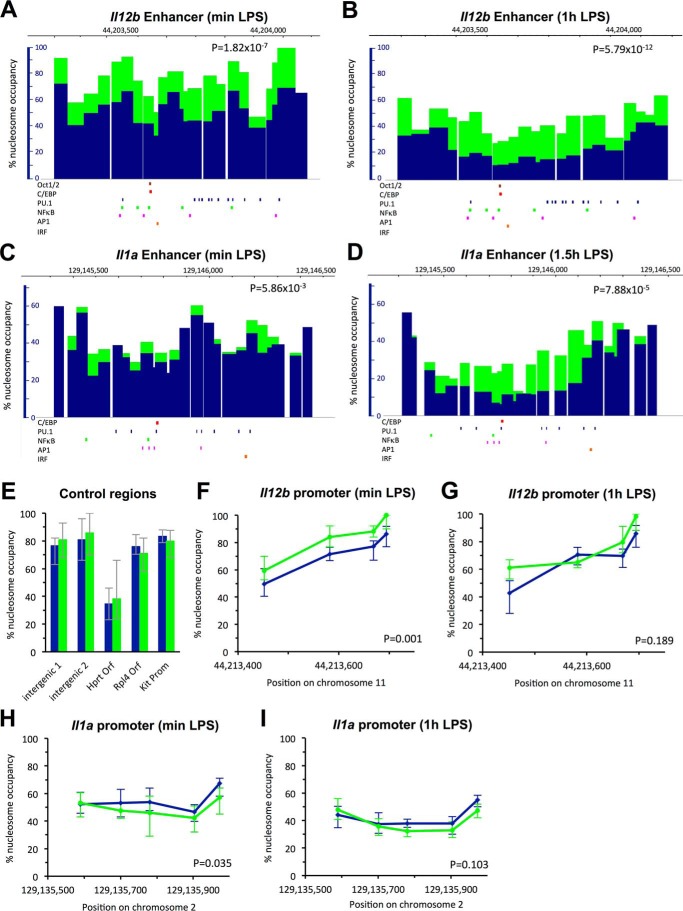
KD of the catalytic BAF/PBAF subunit BRG1. A, BRG1 was knocked down in hematopoietic progenitors using two shRNAs (shSmarca4-3 and shSmarca4-4), and cells were differentiated into BMDMs as described under “Experimental Procedures.” Cells transduced with control shLuc are also shown. mRNA levels of the Smarca4 gene were analyzed in untreated (UNT) BMDMs, as well as in cells transduced with control and specific shRNAs as indicated. Cells were either grown without LPS (blue, min LPS) or with LPS (yellow, plus LPS) for 1.5 h. Data were normalized to mRNA levels found in untreated BMDMs grown in the absence of LPS; experiments were performed at least four times, and S.E. are indicated by the error bars. One-way ANOVA shows statistical significance between differently treated cells (p < 0.05), and a post hoc Tukey HSD test confirms statistical significance between untreated (or shLuc-treated) and shSmarca4-treated cells. *, p < 0.01. B, BRG1 protein abundance was determined by Western analysis in the chromatin fraction of untreated BMDMs, as well as in that of cells transduced with either of the BRG1-specific shRNAs identified in A and pooled before fractionation. SNF5, RNA polymerase II (POLII), and histone H3 levels are shown as controls. Relative abundance of proteins when compared with untreated BMDMs is indicated. C, mRNA of Il12b (red) and Il1a (blue) in cells as described in A and grown in the presence of LPS for 1.5 h. One-way ANOVA shows statistical significance between differently treated cells (p < 0.05), and a post hoc Tukey HSD test confirms statistical significance between untreated and shSmarca4-3- or shSmarc4-treated cells for Il12b, and shSmarca4-4-treated cells for Il1a induction. Induction data for shLuc-treated cells showed higher variability, but were not statistically significantly different from untreated cells. *, p < 0.01. D, mRNA of the macrophage markers Csf1r (blue) and Emr1 (orange) is shown in cells as in A grown in the absence of LPS. E–I, untreated BMDMs (blue) and BRG1 KD cells (orange) were obtained as described in B. E, nucleosome occupancy at the Il12b enhancer in cells grown without LPS is shown as a bar graph with the width of each bar corresponding to the size of each amplicon. p value of a Student's t test shows the significance of the differences between untreated and BRG1 KD cells. IRF, interferon response factor. F, nucleosome occupancy at the Il1a enhancer in cells grown without LPS. G, nucleosome occupancy at control regions in cells grown in the absence of LPS. H, nucleosome occupancy at the Il12b enhancer in cells grown in the presence of LPS for 1.5 h. I, nucleosome occupancy at the Il1a enhancer in cells grown in the presence of LPS for 1.5 h.
BRG1 KD Affects Nucleosome Occupancy and Eviction at the Il12b and Il1a Enhancers
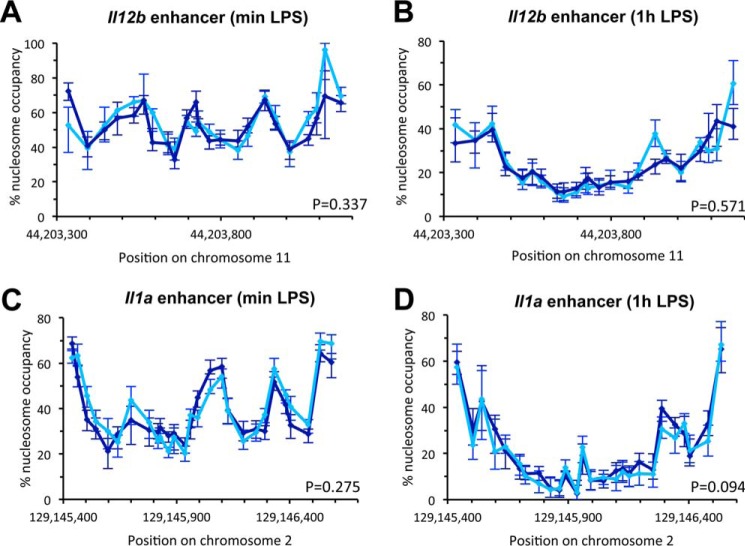
To analyze the effect of knocking down BRG1 on nucleosome occupancy at enhancers, we pooled cells transduced with lentivirus containing either of the two BRG1-specific shRNAs we had identified, and then performed the quantitative nucleosome occupancy assay. We found that nucleosome occupancy over the whole Il12b enhancer was higher in BRG1 KD when compared with untreated control cells (Fig. 2E). Nucleosome occupancy at preferred positions increased by 10–25%, resulting in peak occupancies of 75–90%. Positioning of nucleosomes was largely unaffected, suggesting that other factors determine nucleosome positioning in the Il12b enhancer. Knockdown of BRG1 in hematopoietic progenitors also led to increased nucleosome occupancy at the Il1a enhancer, although the effect was less pronounced than at Il12b (Fig. 2F). p values of Student's t tests showed that the differences found over the whole enhancer region between BRG1 KD and control cells were statistically significant. Control regions were not affected by BRG1 KD (Fig. 2G). Analysis of nucleosome occupancy 1.5 h after LPS addition showed less nucleosome eviction at both enhancers in BRG1 KD when compared with untreated cells (Fig. 2, H and I). For example, occupancy at positions in the Il12b enhancer that are completely cleared of nucleosomes in response to LPS in untreated cells (<5%) remained associated with nucleosomes in 15–20% of the population when BRG1 was knocked down. Control experiments showed that transduction with shLuc had no effect on occupancy before or upon LPS induction (Fig. 3, A–D). Together our results indicate that recruited BAF/PBAF prevents high levels of nucleosome binding at the Il12b and Il1a enhancers in resting macrophages and stimulates nucleosome eviction from the enhancers upon LPS induction. However, nucleosomes were still partially evicted in the absence of BRG1, suggesting that a BRM containing BAF complex may partially compensate for the loss of BRG1.
FIGURE 3.
Nucleosome occupancy in shLuc-treated and untreated control cells. A, nucleosome occupancy at the Il12b enhancer in BMDMs (dark blue) and cells transduced with shLuc as described under “Experimental Procedures” grown without LPS (sky blue, min LPS). Data are shown as line graphs with each point representing the midpoint of a single amplicon, and error bars indicate the confidence interval derived from curve fitting. B, nucleosome occupancy at the Il12b enhancer in cells as in A grown with LPS for 1 h (1h LPS). C, nucleosome occupancy at the Il1a enhancer in cells grown without LPS. D, nucleosome occupancy at the Il1a enhancer grown in the presence of LPS for 1 h. p values of Student's t tests indicate that differences between untreated and shLuc-transduced cells are not statistically significant.
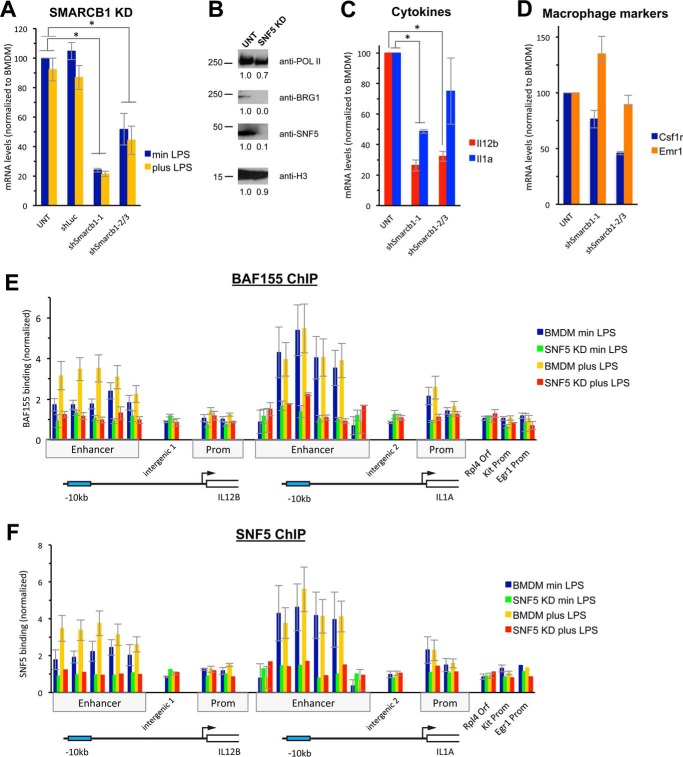
Knockdown of SNF5 Abolishes BAF/PBAF Binding at the Il12b and Il1a Enhancers
To determine whether inactivation of both BAF and PBAF has a stronger effect on nucleosome occupancy at the enhancers, we knocked down the shared core subunit SNF5 in hematopoietic progenitors using the same lentiviral approach. As shown in Fig. 4A, we identified three shRNAs (shSmarcb1-1, shSmarcb1-2, and shSmarcb1-3) that knocked down Smarcb1 (the gene encoding SNF5). shSmarcb1-1 yielded better knockdown (∼80%) than either of the other two shRNAs (shown as average), and we therefore selected shSmarcb1-1 for further analysis. Western blotting confirmed that KD by shSmarcb1-1 reduced the levels of chromatin-associated SNF5 protein by about 90% (Fig. 4B). Moreover, the catalytic subunit BRG1 was no longer detectable in the chromatin-bound fraction when SNF5 was knocked down (Fig. 4B). Under these conditions, Il12b induction was reduced by about 75% 1.5 h after LPS addition and Il1a induction was reduced by about 50% (Fig. 4C). Similar to our findings in BRG1 KD cells, we found that SNF5 KD cells still resembled macrophages and expressed macrophage markers (Fig. 4D). However, we noted that many cells died during the time course of differentiation when we knocked down SNF5, suggesting that loss of SNF5 impairs differentiation and that a minimal amount of SNF5 may be necessary for cells to differentiate into macrophages. Cell survival was also impaired upon BRG1 KD, but to a lesser extent. When we analyzed recruitment of the BAF/PBAF complex to the Il12b and Il1a enhancers by ChIP, we found that recruitment of BAF155, both before and upon LPS induction, was strongly reduced in the SNF5 KD (Fig. 4E); as expected, SNF5 was no longer detected at the enhancers under these conditions (Fig. 4F). This result suggests that the SNF5 subunit is required for either recruitment of BAF/PBAF to the Il12b and Il1a enhancers or formation of a stable complex. Previous results indicated that a BAF/PBAF complex is still formed in the absence of SNF5 in rhabdoid tumor cell lines (26), but our attempts to determine whether BAF/PBAF stability was affected when we knocked down SNF5 in BMDMs were unsuccessful, because low abundance of the complex in whole cell lysates of primary BMDMs made detection of the complex difficult.4
FIGURE 4.
KD of the shared BAF/PBAF core subunit SNF5. SNF5 was knocked down in hematopoietic progenitors and cells were differentiated into BMDMs as described under “Experimental Procedures.” A, mRNA levels of the Smarcb1 gene were analyzed in untreated BMDMs, as well as in cells transduced with control and specific shRNAs, as indicated. Results in cells transduced with shSmarcb1-2 and shSmarcb1-3 are shown as an average. Cells were either grown without LPS (blue, min LPS) or with LPS (yellow, plus LPS) for 1.5 h, and data were normalized to uninduced BMDMs. One-way ANOVA shows statistical significance between differently treated cells (p < 0.05), and a post hoc Tukey HSD test confirms statistical significance between untreated (UNT) (or shLuc-treated) and shSmarcb1-1- and shSmarcb1-2- or shSmarcb1-3-treated cells. *, p < 0.01. B, SNF5 protein was analyzed in the chromatin fractions of untreated BMDMs and of cells transduced with shSmarcb1-1. Western analysis shows loss of SNF5 and BRG1 in the SNF5 KD. RNA polymerase II (POLII) and histone H3 are shown as controls. Relative abundance of proteins when compared with untreated BMDMs is indicated. C, mRNA of Il12b (red) and Il1a (blue) in cells as described in A and grown in the presence of LPS for 1.5 h. One-way ANOVA shows statistical significance between differently treated cells (p < 0.05), and a post hoc Tukey HSD test confirms statistical significance between untreated and shSmarcb1-1- or shSmarcb2/3-treated cells for Il12b and for shSmarcb1-1-treated cells for Il1a. *, p < 0.01. D, mRNA of the macrophage markers Csf1r (blue) and Emr1 (orange) is shown in cells as in A grown in the absence of LPS. E and F, a BAF155 ChIP (E) and a SNF5 ChIP (F) were performed in untreated BMDMs grown in the absence (blue) or presence (yellow) of LPS for 1.5 h or in cells knocked down for SNF5 (shSmarcb1-1) and grown in the absence (green) or presence (red) of LPS. BAF155 binding to Il12b, Il1a, and control regions is shown as described in the legend for Fig. 1A. One-way ANOVA shows that differences between BMDMs and SNF5 KD cells are statistically significant (p < 0.05). A post hoc Fisher LSD test confirms that differences at the enhancers are statistically significant, whereas differences at control regions are not. Prom, promoter.
Nucleosome Occupancy at the Il12b and Il1a Enhancers Increases in the Absence of BAF/PBAF Recruitment
We analyzed nucleosome occupancy in BMDMs that had been transduced with shSmarcb1-1-expressing lentivirus and found increased nucleosome occupancy at the Il12b and Il1a enhancers, both before and upon LPS induction (Fig. 5, A–D). The increase in nucleosome occupancy at Il12b was even more pronounced than in the BRG1 KD and resulted in occupancies at preferred nucleosomal positions at about 85–100% before LPS induction (Fig. 5A), whereas control regions were not affected (Fig. 5E). 1 h after LPS addition, nucleosomes remained associated with the Il12b enhancer in 40–50% of the population (Fig. 5B) and occupancy did not decrease further with prolonged LPS induction for 1.5 h (see Fig. 6C). This result is consistent with the more pronounced effect of SNF5 KD on Il12b expression when compared with KD of BRG1 (compare Fig. 2C with Fig. 4C). We also found increased nucleosome occupancy at the Il1a enhancer both before and upon LPS induction (Fig. 5, C and D). The increase in occupancy at the Il1a enhancer before induction was similar to what we had found in the BRG1 KD, whereas nucleosome eviction at Il1a upon LPS induction was more strongly affected by SNF5 KD. Nevertheless, we note that some level of nucleosome eviction was still seen at both enhancers in the SNF5 KD, and nucleosome occupancy at the Il1a enhancer before induction was only moderately affected. Whether this is due to the activity of residual SNF5 under the conditions of our KD, or whether other remodelers play a role in addition to BAF/PBAF at these as well as at other enhancers that may regulate these genes, remains to be determined.
FIGURE 5.
Nucleosome occupancy in SNF5 KD cells. A, nucleosome occupancy at the Il12b enhancer is shown in untreated BMDMs (blue) and SNF5 KD (green) cells grown in the absence of LPS (min LPS). p values of Student's t tests indicate the significance of differences. IRF, interferon response factor. B, nucleosome occupancy at the Il12b enhancer in cells grown for 1 h in the presence of LPS (1h LPS). C, nucleosome occupancy at the Il1a enhancer in cells grown in the absence of LPS. D, nucleosome occupancy at the Il1a enhancer in cells grown for 1.5 h in the presence of LPS (1.5h LPS). E, nucleosome occupancy at control regions indicated in cells grown in the absence of LPS. Prom, promoter. F and G, nucleosome occupancy at the Il12b promoter in cells grown in the absence of LPS (F) and in the presence of LPS (G) for 1 h. H and I, nucleosome occupancy at the Il1a promoter in cells grown in the absence of LPS (H) and in the presence of LPS (I) for 1 h.
FIGURE 6.
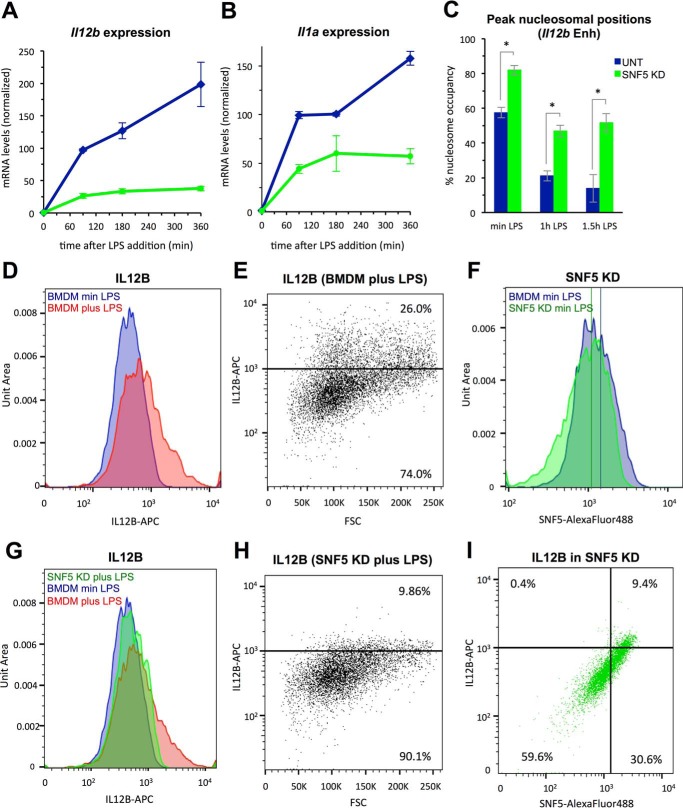
Cytokine expression in SNF5 KD cells. A, mRNA levels of Il12b in control BMDMs (blue) and SNF5 KD cells (green). Cells were grown in the absence of LPS, or for increasing times in the presence of LPS as indicated. mRNA levels after 1.5 h were set to 100%. Error bars represent the S.E. of at least two measurements. B, mRNA levels of Il1a in cells as in A. C, the average occupancy at the three peak nucleosomal positions in the Il12b enhancer (Il12b Enh) is shown in cells as in A grown without or with LPS for 1 h and 1.5 h. One-way ANOVA followed by a post hoc Tukey HSD test (p < 0.05) shows that differences between control and SNF5 KD cells are statistically significant, whereas occupancy levels in SNF5 KD cells after 1 h and 1.5 h are indistinguishable. UNT, untreated. *, p < 0.01. D–I, flow cytometry was performed as described under “Experimental Procedures.” D, IL12B protein accumulation in control BMDMs grown in the absence of LPS (blue, min LPS) or presence of LPS (red, plus LPS) for 3 h was measured by staining with anti-IL12B-APC. Normalized cell counts are displayed as Unit Area. E, scatterplot representation of the data from the experiment described in D. A threshold was set with unstained control BMDMs. FSC, forward scatter. F, SNF5 protein levels in control BMDMs (blue) and SNF5 KD cells (green) were measured by staining with anti-SNF5-Alexa Fluor 488. Mean fluorescence intensities of each population are indicated by lines of the respective color. G, IL12B accumulation in control BMDMs grown in the absence (blue) or presence of LPS for 3 h (red), as well as in SNF5 KD cells grown in the presence of LPS for 3 h (green), was measured by staining with anti-IL12B-APC. Note that data for BMDMs are the same as in D. H, scatterplot representation of the SNF5 KD data from the experiment described in G. I, correlation between IL12B and SNF5 protein levels in SNF5 KD cells grown in the presence of LPS for 3 h was measured by double-staining with anti-IL12B-APC and anti-SNF5-Alexa Fluor 488. Quartile thresholds were set by analysis of unstained control BMDMs.
We also analyzed nucleosome occupancy at the Il12b and Il1a promoters in the SNF5 KD, but effects on nucleosome occupancy both before and upon LPS induction at promoters were small when compared with those detected at the enhancers (Fig. 5, F–I). Together our results indicate that BAF/PBAF regulates nucleosome occupancy at the enhancers of Il12b and Il1a and less so at their promoters. This finding is consistent with our previous data showing that nucleosomes at the promoters of Il12b and Il1a were not stably evicted under inducing conditions (4), which may contribute to the highly stochastic expression of these genes (27, 28).
FACS Analysis Reveals Effects of SNF5 KD on Cytokine Expression in Single Cells
To determine whether knockdown of SNF5 merely slowed down the rate of mRNA production in the whole macrophage population or also affected the final levels of cytokine expression, we performed a time-course study of LPS induction in SNF5 KD cells. In untreated macrophages, Il12b and Il1a mRNA levels increased during the whole 6-h time course of LPS induction as we had shown previously (4)(Fig. 6, A and B, blue lines). In contrast, when SNF5 was knocked down, Il1a and Il12b mRNA levels reached steady state after 90–180 min and did not increase further (green lines). Moreover, as mentioned above, we found that nucleosome eviction at the Il12b enhancer did not increase further with prolonged LPS induction and that nucleosome levels 1.5 h after LPS addition were similar to levels seen after 1 h (Fig. 6C). These results suggested that a fraction of cells may not express Il12b or Il1a when levels of SNF5 are limiting. To further address this question, we analyzed Il12b expression in single cells by FACS. We used accumulation of newly synthesized intracellular IL12B protein in cells that had been treated with the Golgi inhibitor brefeldin A to prevent protein secretion to assess Il12b expression as described (27). In control macrophages, induction of Il12b by LPS for 3 h led to accumulation of significant levels of IL12B protein in about 26% of the cells (compare red areas with blue areas in Fig. 6D, and see scatterplot in Fig. 6E), consistent with results by others (27). When we knocked down SNF5 and monitored intracellular SNF5 protein levels, we found that KD reduced mean SNF5 levels in the population (indicated by the vertical lines in Fig. 6F). More significantly, the fraction of cells with high levels of SNF5 protein was reduced (compare blue shoulder areas with green shoulder areas in Fig. 6F). When we analyzed Il12b expression in SNF5 KD cells, we found that the fraction of cells accumulating IL12B protein was dramatically reduced to about 9% (compare green area with red area in Fig. 6G and see scatterplot in Fig. 6H). Furthermore, we found that cells that expressed Il12b in the SNF5 KD population expressed only low levels of Il12b and accumulated less IL12B protein than control macrophages (compare the magnitude of the anti-IL12B-APC fluorescence intensity signal in Fig. 6, E and H, on the y axis). As shown in Fig. 6I, we found that IL12B protein accumulation correlated with residual levels of SNF5 protein present in SNF5 KD cells, further demonstrating that the remodeler is required for Il12b expression.
Discussion
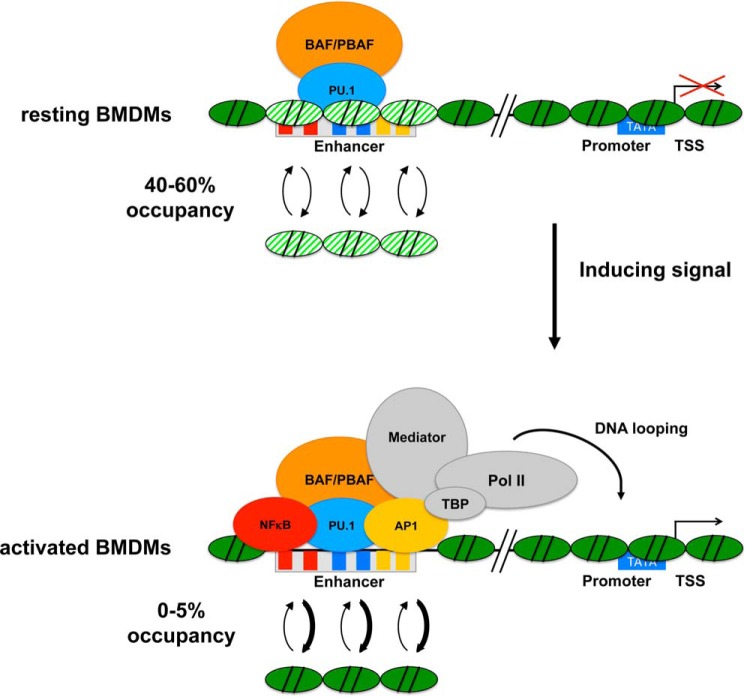
Our results suggest that BAF/PBAF is recruited to macrophage-specific enhancers in response to PUER translocation to the nucleus (Fig. 1), and we speculate that PU.1 recruits the remodeler to these sites. Whether PU.1 directly interacts with BAF/PBAF subunits or whether the interaction is mediated by another factor remains to be determined. We and others showed previously that PU.1 binds to many enhancers together with C/EBPβ, the other macrophage-lineage determining pioneer TF, and C/EBPβ has been shown to directly interact with BAF/PBAF and to mediate its recruitment in other myeloid cells, suggesting that C/EBPβ may recruit BAF/PBAF together with PU.1 in macrophages (6, 8, 17). The absence of PU.1 and BAF/PBAF at macrophage-specific enhancers in HSPCs suggests that binding of the pioneer TF and recruitment of the remodeler occurs at some time during macrophage differentiation. Whether the presence of the remodeler in turn stabilizes PU.1 binding to enhancers remains to be determined. If BAF/PBAF is already recruited by PU.1 to some extent prior to gene induction in resting macrophages (Fig. 1), how might complete nucleosome eviction be accomplished at enhancers under inducing conditions? We propose that recruited BAF/PBAF increases nucleosome turnover (Fig. 7), so that fractional occupancies of enhancer nucleosomes are about 40–60% in a population of resting BMDMs (Figs. 2 and 5). Upon induction by LPS, signal-induced TFs such as NFκB and AP1 are activated and compete with nucleosomes for binding to their sites in the enhancers. This shifts the equilibrium toward nucleosome removal (0–5%). We call this model remodeler-assisted competition between TFs and nucleosomes for binding to enhancers. In the absence of BAF/PBAF, enhancers become more highly occupied by nucleosomes, which impairs gene expression in mature cells in response to an appropriate stimulus (Figs. 2 and 4). Our model predicts that in the absence of BAF/PBAF, nucleosome turnover is low, and signal-induced TFs and the transcriptional machinery are recruited only infrequently, because nucleosome formation is favored over TF binding. This prediction is borne out by our experiments in single cells, where we found that the fraction of cells expressing Il12b was reduced in the SNF5 KD (Fig. 6, G and H). The model further predicts that in the absence of BAF/PBAF, competing nucleosomes reduce the residence times of signal-induced TFs at enhancers, which in turn may decrease the stability of a transcription complex and therefore the transcriptional output from that promoter. Our findings in single cells support this notion, because we found that the levels of IL12B protein that accumulated in individual cells were higher when BAF/PBAF was present at the Il12b enhancer than in its absence in the SNF5 KD (compare the magnitude of the IL12B-APC signal in Fig. 6E versus Fig. 6H). This finding suggests that in the absence of SNF5, a transcription complex at a promoter may only fire once before it falls apart, whereas in the presence of SNF5, such a complex may be stable for several rounds of transcription. Previous studies at various genes have suggested that enhancers can function either by increasing the probability that a competent transcription complex is formed at a promoter or by increasing the probability that another round of transcription is initiated from the same promoter (for a review, see Ref. 29). Our results indicate that the distal enhancer of Il12b may play a role in both initiation and re-initiation and that remodeler-assisted competition facilitates TF over nucleosome binding to the enhancer to stimulate both processes.
FIGURE 7.
Remodeler-assisted competition favors TF over nucleosome binding to sites in enhancers. Our model proposes that recruitment of BAF/PBAF to the distal enhancers of Il12b and Il1a by PU.1 during macrophage differentiation increases turnover of nucleosomes to prevent high occupancy in fully differentiated BMDMs. This results in fractional occupancies of 40–60% for enhancer nucleosomes in the cell population. Under inducing conditions, the equilibrium is shifted toward nucleosome removal as signal-induced TFs (e.g. NFκB and AP1) bind to their sites in the enhancers. Note that increased BAF/PBAF recruitment under inducing conditions (at some enhancers) may further shift the equilibrium toward nucleosome removal. Subsequent steps that result in assembly of a pre-initiation complex at the promoter are not shown.
Experimental Procedures
Cell Isolation and Culture
Bone marrow cells and splenic B-cells were isolated as described from 6–8-week-old C57BL/6 female mice (NCI, Charles River) with Institutional Animal Care and Use Committee (IACUC) oversight (4). To obtain BMDMs, cells were differentiated into macrophages by growth in the presence of M-CSF as described (30). BMDMs were induced with LPS as described (4). The PU.1−/− and PUER cells were grown as described previously (8). HSPCs were isolated using the EasySepTM Mouse Hematopoietic Progenitor Cell Isolation Kit (Stemcell Technologies) per the manufacturer's instructions, with an additional red blood lysis step prior to progenitor isolation. Briefly, 2–3 × 107 cells were resuspended in 1 ml of red cell lysis buffer (Sigma) and mixed gently for 2 min before the addition of 9 ml of Iscove's modified Dulbecco's medium (Gibco) and centrifugation at 400 × g for 5 min. The resulting cell pellet was used for HSPC isolation.
shRNA-mediated Knockdown of BRG1 and SNF5
Lentiviral transductions were performed essentially as described (8). Briefly, lentiviral particles containing shRNAs targeting either BRG1 (Smarca4) or SNF5 (Smarcb1) selected from a pool that had been pre-validated by the Broad Consortium (TRC collection MISSION shRNA library, Sigma) or control shRNA targeting firefly luciferase were produced in HEK293T cells. For lentiviral transductions, bone marrow cells from the femur and tibia of 6–8-week-old C57BL/6 female mice were infected with lentivirus after they had been grown for 48 h in BMDM medium containing L929 cell supernatant as a source of M-CSF. 4 h after viral infection, the medium was replaced to remove the virus and cells were grown for 48 h. Transduced cells were then selected by growth in the presence of 5 μg/ml puromycin for 5 days. Cells were harvested for various experiments as described (4).
Quantitative Nucleosome Occupancy Assay
The assay was performed essentially as described in Ref. 4 except that cross-linked chromatin from 0.5 to 1 × 107 cells was used per experiment and the micrococcal nuclease (New England Biolabs) concentrations were adjusted to a range from 0.0027 units to 13.3 units. Bar graphs and overlays were generated using the Integrated Genome Browser (IGB). Primer pairs for the amplicons used can be given upon request.
Chromatin Immunoprecipitation
ChIP experiments were performed essentially as described (4) except that sonicated chromatin from 1.5 to 2.5 × 106 cells per antibody was diluted 2.5-fold with low salt ChIP buffer (20 mm Tris-HCl, pH 8, 200 mm NaCl, 0.5% Triton X-100, 2 mm EDTA, HaltTM Protease Inhibitor (Thermo Scientific)) to a total volume of 375 μl and incubated overnight at 4 °C with either 5 μl of anti-SNF5 (ab126734; Abcam), 5 μl of anti-BAF155 (D7F8S; Cell Signaling Technology), or 2 μl of anti-PU.1 (sc-352; SCBT). Immunoprecipitated DNA was quantified on a LightCycler 480 (Roche Applied Science). LPS-inducible enhancers measured were identified by Ghisletti et al. (5) and have the following genomic locations: 44 kb upstream of Peli1, 64 kb upstream of IL6, and 3.9 kb upstream of Ccl5 (5). Intergenic region 1 is located 7 kb upstream of the transcription start site of Il12b, intergenic region 2 is located 25 kb upstream of the transcription start site of Il1a, and intergenic region 3 is located is in the HOX cluster between Hoxd11 and Hoxd10. Primer sequences can be given upon request. ChIP data are displayed as the -fold binding over average binding at control regions (i.e. the Kit promoter, Rpl4 ORF, and intergenic region 1).
mRNA Determination
RNA isolation and cDNA synthesis were performed as described (4). cDNA was analyzed by quantitative RT-PCR on a LightCycler 480 (Roche Applied Science) using gene-specific primer pairs. Primer sequences can be given upon request.
Chromatin Fractionation and Western Blotting
Chromatin fractionation was performed essentially as described using the high salt extraction protocol of Ref. 31. Briefly, from 1.5 to 2 × 106 cells that had been transduced with lentivirus bearing specific shRNAs or untreated control BMDMs were resuspended in 400 μl of extraction buffer (10 mm HEPES, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 0.34 m sucrose, 10% glycerol, HaltTM Protease Inhibitor (Thermo Scientific)), which contained 0.2% Nonidet P-40 but no sodium butyrate. The solution was centrifuged for 5 min at 6,500 × g. The nuclear pellet was washed in 400 μl of extraction buffer without Nonidet P-40 or sodium butyrate and then centrifuged again for 5 min at 6,500 × g. Nuclei were resuspended by vortexing in 400 μl of no-salt buffer (10 mm HEPES, pH 7.9, 3 mm EDTA, 0.2 mm EGTA). The solution was placed on a rotator at 4 °C for 30 min and then spun at 6,500 × g for 5 min. The pellet containing chromatin was resuspended in 160 μl of high salt solubilization buffer (50 mm Tris-Cl, pH 8.0, 2.5 m NaCl, 0.05% Nonidet P-40), vortexed, and incubated on a rotator at 4 °C for 30 min. The samples were then centrifuged for 10 min at 16,000 × g. The supernatant containing solubilized proteins was collected as the chromatin-associated fraction. TCA was added at a final concentration of 10%, and samples were incubated for 15 min and then centrifuged at 21,000 × g for 15 min. The resulting pellet was washed with 500 μl of acetone and resuspended in 40 μl of LDS sample buffer (106 mm Tris-HCl, 141 mm Tris base, 2% LDS, glycerol 10%, 0.51 mm EDTA, pH 8.5). To 30 μl of each sample, 3 μl of 0.1% Coomassie Blue, 2 μl of 1 m DTT, and 5 μl of 2× LDS sample buffer were added, and then samples were incubated at 75 °C for 10 min, and each fraction was analyzed by SDS-PAGE on a 4–12% Bis-Tris Plus gel (Novex, Life Technologies). Western analysis was performed after protein transfer for 2 h at 90 V onto a nitrocellulose membrane and quantification of total protein was performed by Ponceau Red staining, using antibodies against RNA polymerase II (sc-56767; SCBT), BRG1 (sc-10768; SCBT), SNF5 (ab12167; Abcam), and histone H3 (ab1791; Abcam). Chemiluminescent signal after incubation with appropriate secondary antibodies was quantified on a ChemiDoc MP Imaging System (Bio-Rad) or using ImageJ.
Flow Cytometry
Analysis was performed on a BD Biosciences LSR II flow cytometer. 1 × 105 cells were used per antibody. To determine IL12B production, Golgi inhibitor GolgiPlugTM (BD Biosciences) was added to prevent cytokine secretion as described (27). Briefly, 1 μl/ml GolgiPlugTM was added in medium without FBS and cells were incubated for 1 h at 37 °C. Then 1 μg/ml LPS from Escherichia coli strain EH100 (Ra mutant)(Sigma) was added and cells were grown for 3 h. Cells were collected using Versene (Life Technologies) treatment and washed with PBS once. Cells were fixed with 1% formaldehyde for 10 min and washed with PBS once. To block nonspecific Fc receptor binding, cells were incubated with 2.42G supernatant for 10 min, followed by a wash with PBS. Staining was performed in permeabilization buffer (PBS, 5% FBS, 0.1% sodium azide, 0.5% Triton X-100) for 30 min in the dark with anti-IL12B-APC (554480; BD Pharmingen) and anti-SNF5-Alexa Fluor 488 (bs-6109R; Bioss), and cells were subsequently washed twice in flow wash buffer (PBS, 5% FBS, 0.1% sodium azide). Due to differences in total count after size gating, fluorescence histograms were normalized, and unit areas are shown in overlays instead of absolute cell counts.
Isolation of Lin− cells was confirmed by flow cytometry using the lineage antibody mixture provided in the EasySepTM Hematopoietic Progenitor Cell Isolation Kit probing for CD5, CD11b, CD19, CD45R/B220, Ly6G/C(Gr-1), TER119, and 7-4 (19856; Stemcell Technologies) followed by secondary incubation with streptavidin-phycoerythrin (Life Technologies), as well as with anti-CD117/KIT (60025; Stemcell Technologies) and anti-SCA1 (60032; Stemcell Technologies) followed by secondary incubation with anti-mouse-FITC (55499; MP Biomedicals). BMDMs were also analyzed using anti-F4/80-APC (eBioscience 17-4801) and anti-CD11b-FITC (eBioscience 10-0112) antibodies.
Statistical Analysis
Knockdown experiments were performed at least four times for each shRNA, and mRNA results for target genes and cytokine induction are shown as the average of all experiments. The statistical significance of differences was determined by one-way ANOVA analysis and confirmed by a post hoc Tukey HSD test. BAF155, SNF5, and PU.1 ChIP experiments were performed at least twice. The statistical significance of the observed differences was determined by one-way ANOVA and confirmed by post hoc Tukey HSD or Fisher LSD tests. To determine binding to the enhancers, all the data from the different enhancer amplicons tested were analyzed together for statistical significance and compared with all the control amplicons. Nucleosome occupancy experiments were performed twice for each knockdown, and a full analysis including all the amplicons in each enhancer was performed once for the BRG1 KD and twice for the SNF5 KD. The error bars represent the confidence intervals of the curve-fitting analysis for a representative experiment. p values in the figures indicate the statistical The significance of differences between different conditions as determined by paired, two-tailed Student's t tests.
Author Contributions
M. F. conceptualized the study; A. G., M. F., M. J. M., and T. M. developed the methodology; A. G., M. F., and M. J. M. performed validation and formal analysis; A. G., M. F., M. J. M., M. T., and T. M. performed the investigation; M. F. wrote the original draft; M. F. and M. J. M. reviewed and edited the study; M. F. was responsible for supervision, resources, and funding acquisition.
Acknowledgments
We thank Amy Ralston for helpful discussions; David Arnosti, Jason Knott, Min-Hao Kuo, and Erik Martinez-Hackert for careful reading of the manuscript; Louis King, Nara Parameswaran, and Michael Steury for help with FACS; and Steve Suhr and John LaPres for help with lentiviral transductions.
Note Added in Proof
The shLuc control data in Fig. 4A were inadvertently duplicated from Fig. 2A in the version of this manuscript that was published as a Paper in Press on July 5, 2016. This error has now been corrected. In addition, the shLuc controls in Fig. 4, C and D, have been removed as they are identical to the data shown in Fig. 2, C and D.
This work was supported by a Scientist-Development-Grant from the American Heart Association (to M. F.) (13SDG17260004). The authors declare that they have no conflicts of interest with the contents of this article.
This article was selected as a Paper of the Week.
M. Floer and A. Gjidoda, unpublished data.
- TF
- transcription factor
- BMDM
- bone marrow-derived macrophage
- HSPC
- hematopoietic stem and progenitor cell
- KD
- knockdown
- APC
- allophycocyanin
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol
- ANOVA
- analysis of variance
- HSD
- honest significant difference
- C/EBPβ
- CCAAT/enhancer-binding protein β.
References
- 1. Zaret K. S., and Carroll J. S. (2011) Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25, 2227–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mouse ENCODE Consortium, Stamatoyannopoulos J. A., Snyder M., Hardison R., Ren B., Gingeras T., Gilbert D. M., Groudine M., Bender M., Kaul R., Canfield T., Giste E., Johnson A., Zhang M., Balasundaram G., Byron R., et al. (2012) An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol. 13, 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Teif V. B., Vainshtein Y., Caudron-Herger M., Mallm J. P., Marth C., Höfer T., and Rippe K. (2012) Genome-wide nucleosome positioning during embryonic stem cell development. Nat. Struct. Mol. Biol. 19, 1185–1192 [DOI] [PubMed] [Google Scholar]
- 4. Gjidoda A., Tagore M., McAndrew M. J., Woods A., and Floer M. (2014) Nucleosomes are stably evicted from enhancers but not promoters upon induction of certain pro-inflammatory genes in mouse macrophages. PLoS ONE 9, e93971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ghisletti S., Barozzi I., Mietton F., Polletti S., De Santa F., Venturini E., Gregory L., Lonie L., Chew A., Wei C. L., Ragoussis J., and Natoli G. (2010) Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32, 317–328 [DOI] [PubMed] [Google Scholar]
- 6. Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., and Glass C. K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. cell 38, 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barozzi I., Simonatto M., Bonifacio S., Yang L., Rohs R., Ghisletti S., and Natoli G. (2014) Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol. Cell 54, 844–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tagore M., McAndrew M. J., Gjidoda A., and Floer M. (2015) The lineage-specific transcription factor PU.1 prevents polycomb-mediated heterochromatin formation at macrophage-specific genes. Mol. Cell. Biol. 35, 2610–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bryant G. O., Prabhu V., Floer M., Wang X., Spagna D., Schreiber D., and Ptashne M. (2008) Activator control of nucleosome occupancy in activation and repression of transcription. PLoS Biol. 6, 2928–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reinke H., and Hörz W. (2003) Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11, 1599–1607 [DOI] [PubMed] [Google Scholar]
- 11. Floer M., Wang X., Prabhu V., Berrozpe G., Narayan S., Spagna D., Alvarez D., Kendall J., Krasnitz A., Stepansky A., Hicks J., Bryant G. O., and Ptashne M. (2010) A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell 141, 407–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Badis G., Chan E. T., van Bakel H., Pena-Castillo L., Tillo D., Tsui K., Carlson C. D., Gossett A. J., Hasinoff M. J., Warren C. L., Gebbia M., Talukder S., Yang A., Mnaimneh S., Terterov D., et al. (2008) A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell 32, 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hartley P. D., and Madhani H. D. (2009) Mechanisms that specify promoter nucleosome location and identity. Cell 137, 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bultman S., Gebuhr T., Yee D., La Mantia C., Nicholson J., Gilliam A., Randazzo F., Metzger D., Chambon P., Crabtree G., and Magnuson T. (2000) A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6, 1287–1295 [DOI] [PubMed] [Google Scholar]
- 15. Reyes J. C., Barra J., Muchardt C., Camus A., Babinet C., and Yaniv M. (1998) Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α). EMBO J. 17, 6979–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Choi J., Ko M., Jeon S., Jeon Y., Park K., Lee C., Lee H., and Seong R. H. (2012) The SWI/SNF-like BAF complex is essential for early B cell development. J. Immunol. 188, 3791–3803 [DOI] [PubMed] [Google Scholar]
- 17. Kowenz-Leutz E., and Leutz A. (1999) A C/EBPβ isoform recruits the SWI/SNF complex to activate myeloid genes. Mol. Cell 4, 735–743 [DOI] [PubMed] [Google Scholar]
- 18. Hu G., Schones D. E., Cui K., Ybarra R., Northrup D., Tang Q., Gattinoni L., Restifo N. P., Huang S., and Zhao K. (2011) Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res. 21, 1650–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shain A. H., and Pollack J. R. (2013) The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS ONE 8, e55119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Klochendler-Yeivin A., Fiette L., Barra J., Muchardt C., Babinet C., and Yaniv M. (2000) The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 1, 500–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roberts C. W., Galusha S. A., McMenamin M. E., Fletcher C. D., and Orkin S. H. (2000) Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc. Natl. Acad. Sci. U.S.A. 97, 13796–13800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang X., Sansam C. G., Thom C. S., Metzger D., Evans J. A., Nguyen P. T., and Roberts C. W. (2009) Oncogenesis caused by loss of the SNF5 tumor suppressor is dependent on activity of BRG1, the ATPase of the SWI/SNF chromatin remodeling complex. Cancer Res. 69, 8094–8101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ramirez-Carrozzi V. R., Braas D., Bhatt D. M., Cheng C. S., Hong C., Doty K. R., Black J. C., Hoffmann A., Carey M., and Smale S. T. (2009) A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 138, 114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Walsh J. C., DeKoter R. P., Lee H. J., Smith E. D., Lancki D. W., Gurish M. F., Friend D. S., Stevens R. L., Anastasi J., and Singh H. (2002) Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity 17, 665–676 [DOI] [PubMed] [Google Scholar]
- 25. Laiosa C. V., Stadtfeld M., and Graf T. (2006) Determinants of lymphoid-myeloid lineage diversification. Annu. Rev. Immunol. 24, 705–738 [DOI] [PubMed] [Google Scholar]
- 26. Doan D. N., Veal T. M., Yan Z., Wang W., Jones S. N., and Imbalzano A. N. (2004) Loss of the INI1 tumor suppressor does not impair the expression of multiple BRG1-dependent genes or the assembly of SWI/SNF enzymes. Oncogene 23, 3462–3473 [DOI] [PubMed] [Google Scholar]
- 27. Weinmann A. S., Plevy S. E., and Smale S. T. (1999) Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity 11, 665–675 [DOI] [PubMed] [Google Scholar]
- 28. Shalek A. K., Satija R., Adiconis X., Gertner R. S., Gaublomme J. T., Raychowdhury R., Schwartz S., Yosef N., Malboeuf C., Lu D., Trombetta J. J., Gennert D., Gnirke A., Goren A., Hacohen N., et al. (2013) Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 498, 236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blackwood E. M., and Kadonaga J. T. (1998) Going the distance: a current view of enhancer action. Science 281, 60–63 [DOI] [PubMed] [Google Scholar]
- 30. Nociari M., Ocheretina O., Schoggins J. W., and Falck-Pedersen E. (2007) Sensing infection by adenovirus: Toll-like receptor-independent viral DNA recognition signals activation of the interferon regulatory factor 3 master regulator. J. Virol. 81, 4145–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shechter D., Dormann H. L., Allis C. D., and Hake S. B. (2007) Extraction, purification and analysis of histones. Nat. Protoc. 2, 1445–1457 [DOI] [PubMed] [Google Scholar]