Abstract
Regenerative medicine holds great promise for the treatment of degenerative retinal disorders. Krüppel-like factors (KLFs) are transcription factors that have recently emerged as key tools in regenerative medicine because some of them can function as epigenetic reprogrammers in stem cell biology. Here, we show that KLF16, one of the least understood members of this family, is a POU4F2 independent transcription factor in retinal ganglion cells (RGCs) as early as embryonic day 15. When overexpressed, KLF16 inhibits RGC neurite outgrowth and enhances RGC growth cone collapse in response to exogenous ephrinA5 ligands. Ephrin/EPH signaling regulates RGC connectivity. The EphA5 promoter contains multiple GC- and GT-rich KLF-binding sites, which, as shown by ChIP-assays, bind KLF16 in vivo. In electrophoretic mobility shift assays, KLF16 binds specifically to a single KLF site near the EphA5 transcription start site that is required for KLF16 transactivation. Interestingly, methylation of only six of 98 CpG dinucleotides within the EphA5 promoter blocks its transactivation by KLF16 but enables transactivation by KLF2 and KLF15. These data demonstrate a role for KLF16 in regulation of RGC neurite outgrowth and as a methylation-sensitive transcriptional regulator of EphA5 expression. Together, these data identify differential low level methylation as a novel mechanism for regulating KLF16-mediated EphA5 expression across the retina. Because of the critical role of ephrin/EPH signaling in patterning RGC connectivity, understanding the role of KLFs in regulating neurite outgrowth and Eph receptor expression will be vital for successful restoration of functional vision through optic nerve regenerative therapies.
Keywords: chromatin immunoprecipitation (ChIP), DNA methylation, gene regulation, Kruppel-like factor (KLF), mouse, neurite outgrowth, neurodevelopment, promoter, protein expression, retina
Introduction
Progressive loss of retinal ganglion cells (RGCs)2 leads to blindness in diseases such as glaucoma, the leading cause of irreversible bilateral blindness worldwide (1). Current treatments can slow progression but cannot restore lost vision. Recent advances in generating retinal cells from embryonic and induced pluripotent stem cells have stimulated interest in the potential for neuronal regeneration to treat blindness (2–4). For successful optic nerve regeneration, newly regenerated RGCs will need to extend axons considerable distances to reach the visual centers of the brain, where they must make the correct patterns of termination on post-synaptic neurons to reestablish retinotopic maps. Thus, it is critical to understand the mechanisms that regulate expression of molecules involved in axonal growth and connectivity.
Krüppel-like factors (KLFs) are members of the SP/KLF family of transcription factors that are characterized by three zinc finger DNA binding domains near their C terminus (5). KLFs bind to GT- or GC-rich sequences and can function as either transcriptional activators or repressors, depending on the promoter context (6). At least 15 of the 17 members of the KLF transcription factor family are expressed in RGCs, and overexpression of different KLF transcription factors in RGCs, hippocampal neurons, or cortical neurons can enhance or inhibit neurite outgrowth (7). Such differential promotion and inhibition of growth are unusual among transcriptional regulators of axon growth. KLF16 and three other members of the basic transcription element binding protein (BTEB) subfamily reduce neurite outgrowth in cortical neurons (7). However, the roles of KLF16 in the retina have not yet been determined.
The neural retina is part of the central nervous system and offers an accessible model for studying cellular processes related to neurogenesis, degenerative neuropathies, and therapeutic interventions. RGCs, the retina's projection neurons, convey visual information from the retina to post-synaptic targets in the brain. Like other sensory and motor systems, the visual system is organized topographically, such that neighboring RGCs terminate on neighboring post-synaptic cells in visual centers of the brain. This retinotopic mapping of RGC terminals maintains the spatial organization of visual stimuli at every level of visual processing, beginning in the retina (8, 9). A key mechanism regulating the formation of retinotopic maps during development is signaling between ephrin receptors (EPH) and ephrin ligands (EFN) (10, 11).
EPH receptors are the largest identified family of tyrosine kinase receptors, and the 13 family members present in mammals are classified into EPHA and EPHB subclasses, based on their structure and ligand binding affinities (12). Ligand binding activates bidirectional intracellular signaling through both the receptor and ligand, and it mediates a wide range of cellular activities (13–16). In projection neurons such as RGCs, activation of EPH/EFN signaling reduces cell adhesion and leads to growth cone collapse (17). Eph receptor and Efn ligand mRNA expression patterns in the retina are complex and vary between species, depending on the size of ipsilateral projections, but the overall mechanisms underlying retinotopic mapping are highly conserved (18). In the mouse, gradients of Eph mRNA expression across the nasal/temporal (EphA) and dorsal/ventral (EphB) axes of the retina result in different levels of EPH receptors on RGC growth cones. The EPH receptors interface with counter-gradients of cognate ligands in the superior colliculus to drive initial patterns of axon connectivity (19–21).
Despite the extensive characterization of expression patterns and functions of EPH/EFNs in RGC connectivity, the mechanisms that establish and maintain the spatial patterns of Eph expression in the retina are not well understood. There is evidence for silencing of EphA5 expression by hypermethylation in cancer (22–24). We previously showed that total CpG methylation abolishes activity of mEphA5 promoter-reporter constructs in R28 retinal progenitor cells and that transcriptional silencing of the endogenous EphA5 gene in a mouse retinal cell line (ImM10) is reversed by genome-wide demethylation (25). Intriguingly, our prior analysis also showed that in the nasal retina, where EphA5 expression is lowest, CpG methylation is increased specifically in a cluster of CpG dinucleotides immediately downstream of the EphA5 transcription start site. This region contains putative KLF-binding sites, suggesting a role for KLFs and DNA methylation in regulation of EphA5 in the retina. These studies were designed to test the hypothesis that KLF16 regulates neurite outgrowth and is a methylation-sensitive transcriptional regulator of EphA5 expression in RGCs.
Results
KLF16 Expression Is Enriched in the Developing Retina
Previous RT-PCR analysis showed KLF16 expression in the developing retina (7). To determine KLF16 cellular expression, we used immunofluorescence on frozen sections of mouse retina, cut along the nasal/temporal axis. At E15.5, a time point when RGC axons have reached the optic tract (26), KLF16 was observed in cells in the nascent GCL (Fig. 1, A–C). To confirm the RGC identity of the KLF16-positive cells, we performed co-localization studies using antibodies against POU4F2 (BRN3b), a transcription factor that is expressed in ∼80% of RGCs, beginning when they exit the cell cycle (27). As expected, POU4F2-positive nuclei of new RGCs were present near the ventricular zone at E15.5, and post-mitotic RGCs continued to express POU4F2 as they migrated to the retinal GCL (Fig. 1B). The newly post-mitotic RGCs near the ventricular zone were KLF16-negative. However, POU4F2 and KLF16 were co-localized in the nuclei of RGCs within the nascent GCL (Fig. 1C). Thus, KLF16 is expressed at stages of RGC development consistent with a role in regulation of EphA5 expression and neurite out-growth.
FIGURE 1.
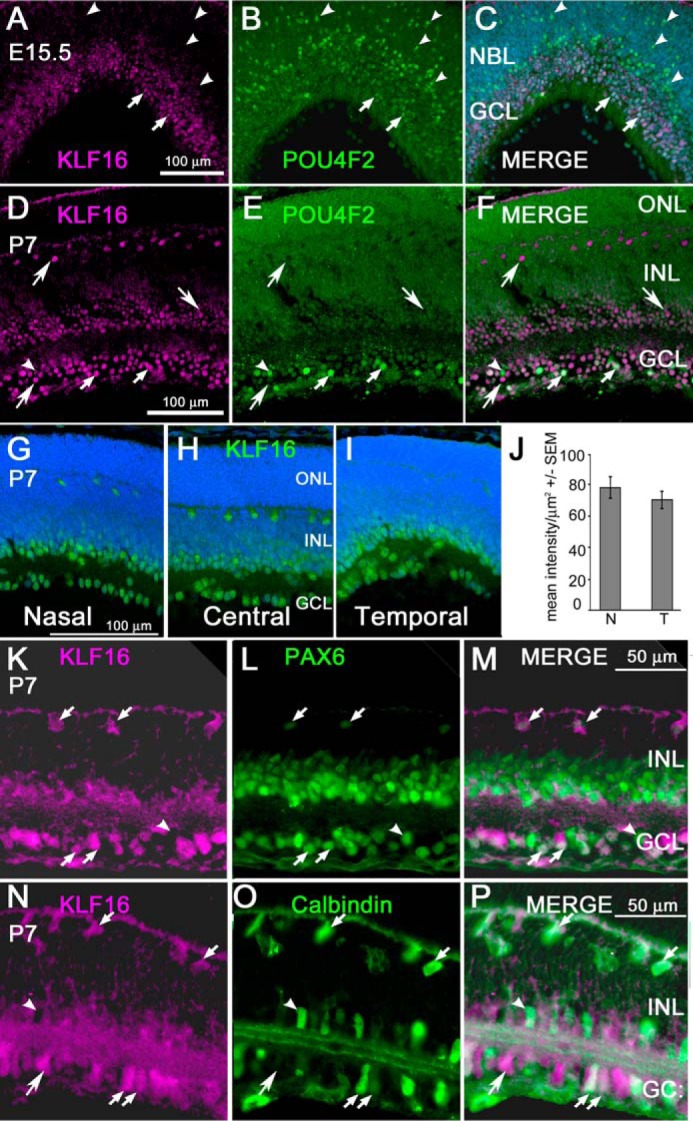
KLF16 protein expression in retina during development. A–C, retinal section from an E15.5 mouse showing immunoreactivity for KLF16 (A) and POU4F2 (B) in nascent retinal ganglion cells in the central retina. D–F, retinal section from postnatal mouse (P7) showing immunoreactivity for KLF16 (D) and POU4F2 (E); F, overlay of D and E. A–F, small arrows indicate nuclei immunopositive for both KLF16 and POU4F2; arrowheads indicate nuclei immunopositive for POU4F2 only; large arrows indicate nuclei immunopositive for KLF16 only. G–I, representative images showing immunostaining for KLF16 (green) and DAPI (blue) in the nasal, central, and temporal retinas of a single section cut along the nasal/temporal axis from P7 mice. G–I, contrast and brightness were adjusted simultaneously. J, semi-quantitative analysis of KLF16 immunostaining intensity in nasal versus temporal ganglion cell layer in P0 retinas. Bars show mean pixel intensity/mm2 ± S.E. K–M, retinal section double-stained with antibodies against KLF16 (K) and PAX6 (L), with merged image (M) showing co-localization in P7 the mouse retina. N–P, retinal section double-stained with antibodies against KLF16 (N) and calbindin (O), with merged image showing co-localization (P). In all panels, large arrows indicate cells immunopositive for KLF16 only; small arrows indicate nuclei immunopositive for both proteins; arrowheads indicate nuclei immunonegative for KLF16. NBL, neuroblastic layer; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bars, 100 μm in A–I and 50 μm in K–P.
At P7, KLF16 was present in nuclei in the GCL, in the inner portion of the inner nuclear layer (INL), and in scattered INL cells that stratified immediately adjacent to the outer plexiform layer (Fig. 1, D–F). In the GCL, KLF16 and POU4F2 immunoreactivity co-localized in some nuclei, but there were also KLF16-positive nuclei negative for POU4F2 at both E15.5 (Fig. 1C) and at P7 (Fig. 1F). KLF16+/POU4F2 negative cells in the GCL likely include at least some of the 20% of RGCs that do not express POU4F2 (28, 29). However, other differentiated cell types are present the INL and GCL at this developmental stage. Therefore, additional double immunostaining was used to further characterize the KLF16-expressing cells in the postnatal retina.
PAX6 is expressed in RGCs, amacrine cells, and horizontal cells in the mature retina, in a pattern similar to that observed for KLF16 (Fig. 1, K–M) (30). There was co-localization of PAX6 and KLF16 in the nuclei of most, but not all, cells in the GCL. In the inner nuclear layer, KLF16 immunoreactivity was closely associated with PAX6-positive nuclei of amacrine and horizontal cells. At P7, KLF16 immunoreactivity appeared to be both nuclear and cytoplasmic in some cells, and KLF16 staining was present in horizontal cell processes (Fig. 1K), and the inner plexiform layer (Fig. 1M). The non-nuclear staining in RGCs and horizontal cells likely reflects trafficking of KLF16 between the nuclear and cytoplasmic compartments, as we reported previously in uterine cells (31).
In the mouse retina, calbindin is a marker of horizontal cells, a subset of amacrine cells in the inner nuclear layer and displaced amacrine cells in the GCL (32). Consistent with the PAX6 double-staining, horizontal and amacrine cells in the inner nuclear layer were double labeled for KLF16 and calbindin (Fig. 1, N–P). We also observed double-labeled cells in the GCL with robust processes extending to the inner plexiform layer, identifying them as displaced amacrine cells. There were additional cells in the GCL that were KLF16-positive and calbindin-negative, consistent with a ganglion cell identity.
KLF16 Inhibits Neurite Outgrowth in RGCs
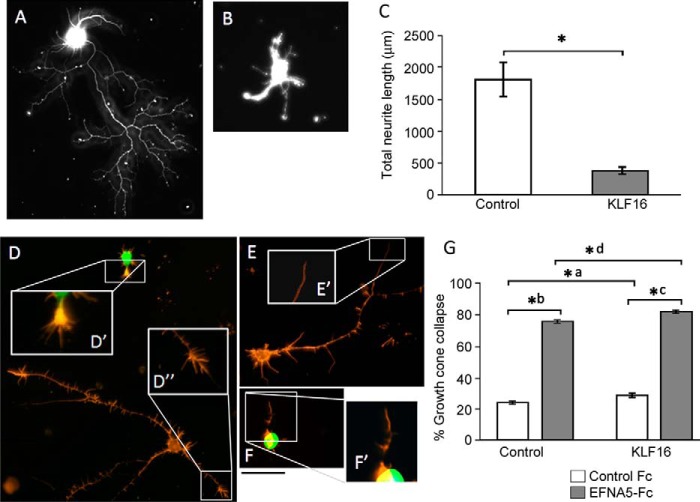
Our prior results showed that KLF16 reduces neurite outgrowth of cortical neurons. KLF16 expression in RGCs prompted us to test whether KLF16 also modulates neurite outgrowth in RGCs. To test the biological effect of exogenous KLF16, expression plasmids were electroporated into primary RGCs in vitro. Consistent with our previous findings in cortical neurons (7), KLF16 overexpression decreased neurite growth by 77.5% (p = 0.0001) compared with control-electroporated RGCs (Fig. 2, A–C; Table 1).
FIGURE 2.
Effects of KLF16 overexpression and EphrinA5 ligands on growth cone collapse in RGCs. A and B, representative images of purified P5 RGCs transduced with mCherry (A) or KLF-16 (B) and immunostained against β-III tubulin to highlight neurites. C, quantification of neurite growth in RGCs (n = 4 with at least n = 50 cells per condition; error bars are standard error of the mean (S.E.); *, p < 0.0001 by Student's t test). D–F, representative images of RGCs treated with cross-linked EphrinA5 (EFNA5-Fc) or control anti-human Fc antibody for 30 min and then labeled with Alexa Fluor 555-phalloidin (F-actin-rich neurites and growth cones, orange); GFP is detected in KLF16-transduced cells (green). Phalloidin staining highlights example spread (D′ and D″) and collapsed (E′ and F′) growth cones. G, EFNA5-Fc promotes growth cone collapse in both control transduced (*a, p < 0.0001) and KLF16-transduced RGCs (*d, p < 0.0001). KLF-16 overexpression resulted in a statistically significant but marginal increase in collapse both in control-Fc (*b, p < 0.0001) and EFNA5-Fc-treated RGCs (*c, p < 0.0001) At least 100 growth cones were scored per condition. Statistics calculated by two-way analysis of variance. Error bars show standard error of the mean (S.E.). Scale bar, 50 μm (A, B, and D–F) and 20 μm (D′, D″, E′, and F′).
TABLE 1.
Effects of KLF16 overexpression on total neurite length in rat retinal ganglion cells
| Plasmid | Mean length/cell | S.E. | No. of cells analyzed |
|---|---|---|---|
| μm | |||
| KLF16 | 39 | 44.8 | 57 |
| mCherry | 1800 | 287.7 | 74 |
KLF16 Is a Novel Activator of EphA5, a Critical Regulator of RGC Connectivity in the Visual System
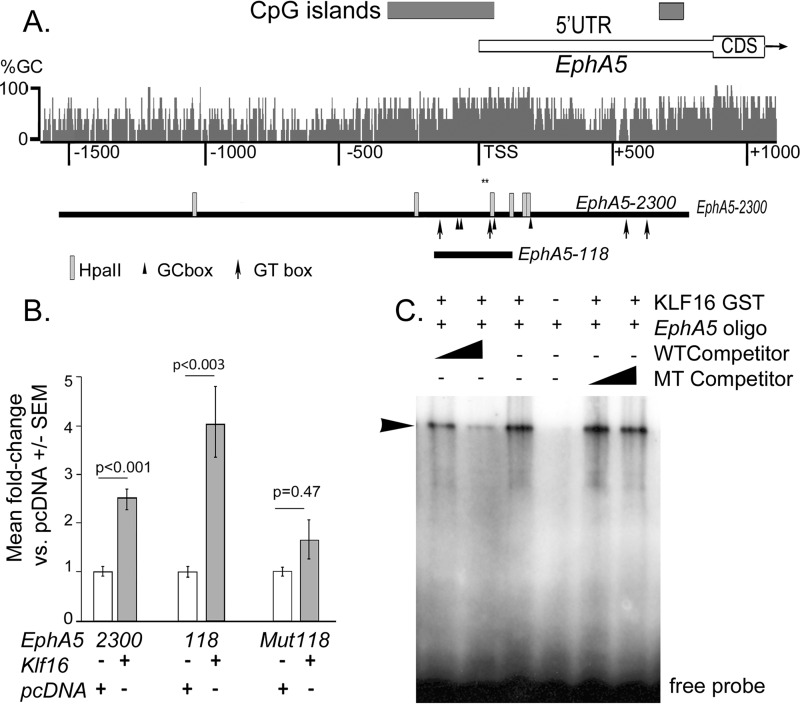
EphA5 is one of two EPHA receptors that regulate positional mapping of RGC axons along the nasal temporal axis of the retina, and targeted EphA5 KO results in ectopic RGC terminations in the superior colliculus (33). The known pattern of EphA5 expression in RGCs and inner retinal neurons (33) is similar to KLF16, suggesting a potential regulatory relationship. Bioinformatics analysis of the mouse EphA5 proximal promoter identified multiple core binding sites for KLF family transcription factors: GT boxes (GGGTG/CACCC) and GC boxes (GGGCG/CGCCC) (Fig. 3A). Therefore, we next asked whether EphA5 was a regulatory target of KLF16.
FIGURE 3.
KLF16 activation and binding to EphA5 proximal promoter in vitro. A, schematic diagram showing EphA5 exon 1 (white box) with the 5′-UTR and coding region (CDS), GC percent (histogram), and CpG islands (dark gray boxes). Numbers indicate distance in base pairs from the transcription start site (TSS). Regions cloned for analysis are indicated by black bars below, with predicted KLF-binding sites (GC boxes, arrowheads; GT boxes, arrows) and HpaII methylation sites (light rectangles). Asterisks show region containing six CpG dinucleotides previously shown to have increased methylation in nasal retina (25). B, effects of Klf16 overexpression on mEphA5 promoter deletion constructs; Mut118 construct is identical to EphA5-118 but with GT box at +47 to +51 mutated (GGGTG to TTTTG). Bars show mean fold-change ± standard error of the mean for indicated EphA5 promoter constructs, co-transfected with Klf16 (gray) or empty vector pcDNA3.1 (white). Statistical comparisons used ANOVA, with post hoc t tests versus control and Holm-Bonferroni correction for multiple comparisons. p ≤ 0.05 was considered statistically significant. C, electrophoretic mobility shift assays of KLF16 binding to oligonucleotide containing GT box located in the EphA5-118 promoter (see also Fig. 1). Arrowhead shows shifted DNA-protein complexes. Competitor contained the native (WT) or mutated (Mut) GT box (5th and 6th lanes). Representative gel shown from three independent experiments.
We found that KLF16 overexpression transactivated the EphA5-2300 promoter construct, as well as the deletion constructs (Fig. 3B). Compared with pcDNA controls, KLF16 transactivated the EphA5-2300 promoter by 2.51 ± 0.23-fold (p < 0.001). Promoter transactivation was greater when KLF16 was co-transfected with the EphA5-118 promoter compared with pcDNA control (4.09 ± 0.69-fold, p < 0.003). These results are consistent with the presence of a positive regulatory element within the region contained within EphA5-118.
Regulation of the EphA5 Promoter Requires KLF16 Binding to a Distinct Cis-regulatory Element Adjacent to the Transcriptional Initiation Site
The EphA5-118 promoter construct extends from −118 to +123 bp relative to the transcription start site and contains one GT box (GGGTG, at +47 to +51) and three GC boxes (GGGCG, at −71 to −75, −56 to −60 and +64 to +68) (Fig. 3A). Interestingly, the GT box is positioned immediately 3′ to a cluster of CpG dinucleotides that we previously found to have increased methylation in genomic DNA from the nasal but not temporal retina (25). Therefore, subsequent analysis focused on this site. Mutation of the GT box from GGGTG to TTTTG (mutEphA5-118) was sufficient to reduce KLF16 transactivation of the shortest promoter construct to near control levels (1.6 ± 0.37-fold, p = 0.47 versus pcDNA) (Fig. 3B).
In EMSA, KLF16 protein bound to oligonucleotides containing the GT box (Fig. 3C), consistent with the GT box functioning as the KLF16-binding site. Unlabeled oligonucleotides containing the native sequence competed with radiolabeled probes containing the native sequence for binding with the KLF16 protein. In contrast, unlabeled oligonucleotides containing the mutated site did not compete. Thus, decreased KLF16 binding at this GT box was associated with and likely explained the loss of KLF16 transactivation of the mutEphA5-118 promoter construct.
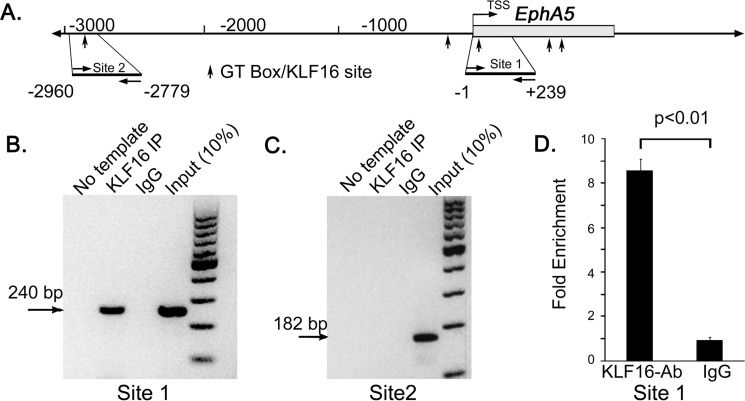
KLF16 Binds to the EphA5 Promoter in Retinal Cells in Vivo
If KLF16 functions as an in vivo regulator of EphA5, it should occupy the EphA5 promoter in retinal cells. To test this, we performed chromatin immunoprecipitation (ChIP) using KLF16 antibodies and chromatin isolated from retinas of P7 mice. The genomic region containing the GT box/KLF16-binding site required for KLF16 activation of the EphA5-118 promoter was amplified from KLF16 ChIP'ed chromatin (Fig. 4, A, B, and D), but an upstream GT box (−2960 to −2779 bp, Fig. 4C) was not. Both sites were amplified from input chromatin, but not from IgG ChIP'ed chromatin. This localizes the in vivo KLF16-binding site to the region of EphA5 containing the GT box required for KLF16 transactivation and binding in vitro. Although we cannot attribute the KLF16 binding specifically to RGCs, our ChIP results are consistent with KLF16 occupying the EphA5 promoter in co-expressing cells that are present in the INL and GCL.
FIGURE 4.
Chromatin immunoprecipitation (ChIP) analysis of KLF16 binding to EphA5 promoter in the mouse retina. A, schematic diagram of EphA5 promoter (solid line) and exon 1 (open box) showing the 5′-untranslated region, coding region, and location of predicted KLF16-binding sites/GT boxes (arrows). Amplicons for sites 1 and 2 are indicated, with position relative to transcription start site. B and C, representative agarose gels of PCR amplicons from ChIP analysis of site 1 containing the GT box analyzed in mutational and EMSA analysis (A) and site 2 (B) containing an upstream GT box. D, real time PCR analysis of site 1, showing fold-enrichment in KLF16 ChIP reactions versus rabbit IgG. Bars show mean fold change ± S.E. from three independent experiments.
KLF16 Regulates Cone Collapse in RGCs
If KLF16 is a transcriptional regulator of EphA5, we predicted that overexpression would alter RGC response to exogenous EFNA5 in a growth cone collapse assay. Postnatal RGCs in culture normally exhibit growth cones with a spread morphology containing a central domain and actin-rich filopodia (Fig. 2D), but a fraction will also exhibit collapsed growth cones (Fig. 2, E and F) (34, 35). In transfected RGCs treated with control Fc antibodies, overexpression of KLF16 resulted in a small, but statistically significant, increase in the number of collapsed growth cones (1.2%, p = 0.0001) compared with control transfected RGCs (*a in Fig. 2G; Table 2). EFNA5-Fc elicited significant increases in growth cone collapse in control transfected RGCs (3.2-fold versus control Fc; *b in Fig. 2G) and KLF16-transfected RGCs (2.9-fold versus control Fc; *c in Fig. 2G) compared with RGCs treated with control Fc antibodies (p = 0.0001 for both comparisons). The mean number of collapsed growth cones per cell in EFNA5-Fc-treated cells was marginally higher in KLF16-overexpressing RGCs compared with control transfected RGCs (76.2 ± 3.9 for control versus 81.5 ± 7.8 for KLF16-transfected RGCs, p = 0.0001) (*d in Fig. 2G). Thus, exogenous KLF16 expression suppresses RGC axon growth but only modestly increases the overall RGC response to EFNA5 at the level of acute growth cone collapse.
TABLE 2.
Effects of KLF16 overexpression on ephrinA5-mediated growth cone collapse in rat retinal ganglion cells
| Plasmid | Ligand | Mean no. of collapsed/cells | S.D. | No. of cells analyzed |
|---|---|---|---|---|
| KLF16 | Control Fc | 28.5 | 1.7 | 27 |
| KLF16 | EphrinA5-Fc | 81.5 | 7.8 | 35 |
| mCherry | Control Fc | 23.6 | 1.73 | 25 |
| mCherry | EphrinA5-Fc | 76.2 | 3.9 | 36 |
KLF16-related Transcription Factors Contribute to the Regulation of the EphA5 Promoter
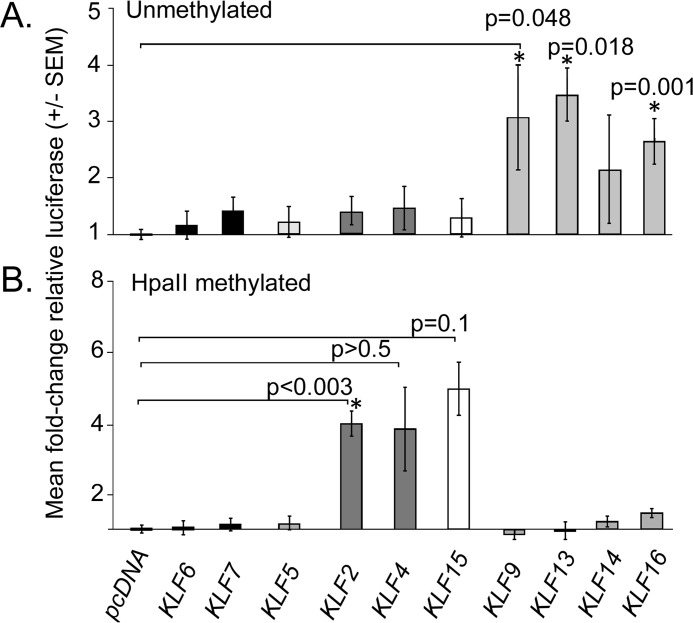
As other KLFs can reduce or promote neurite outgrowth, we asked whether other KLF factors can function as transcriptional regulators of EphA5. Eukaryotic expression plasmids containing Klf cDNAs were co-transfected with the EphA5-2300-luc promoter construct and analyzed for promoter activation in luciferase assays (Fig. 5A). KLF9, KLF13, and KLF16, which are all BTEB class KLFs in subgroup III (36), transactivated the EphA5 promoter, as measured by increased expression of the luciferase reporter. KLF13 and KLF16 up-regulated the EphA5-2300 promoter construct most robustly, increasing reporter expression by 3.47 ± 0.41-fold (p = 0.018) and 2.64 ± 0.4-fold (p < 0.001), respectively, compared with pcDNA controls. KLF9 activation of EphA5-2300 was more variable, increasing reporter expression by 3.07 ± 0.93-fold when compared with pcDNA controls (p = 0.048). Other KLFs tested did not transactivate the EphA5-2300 promoter. Thus, three of the four BTEB class KLF transcription factors transactivate of the EphA5 promoter.
FIGURE 5.
Activation of unmethylated and methylated EphA5 promoter by KLF transcription factors. A, effects of Klf overexpression on EphA5-2300-luc promoter activity in HEK293FT cells. KLFs are grouped and shaded by phylogenetic relationships (36). B, effects Klf overexpression on the activity of HpaII-methylated pCpGL-EphA5-luc promoter constructs in HEK293FT cells. Bars show mean fold-change in relative luciferase versus controls ± standard error of the mean (S.E.) for empty vector pcDNA3.1+EphA5-2300-luc (A) and empty vector pcDNA3.1+HpaII methylated pCpGL-EphA5-2300-luc) (B). For each condition, at least 3 independent experiments were analyzed using ANOVA, with post-hoc, t-tests versus control and Holm-Bonferroni correction for multiple comparisons. *, p ≤ 0.05 was considered statistically significant.
KLF16-dependent Transactivation of mEphA5 Is Methylation-sensitive
The GT box required for KLF16 binding and transactivation of the EphA5 promoter is located near CpG dinucleotides with increased methylation in nasal retina. To test whether methylation altered the ability of KLF16 to transactivate the EphA5 promoter, the EphA5-2300 promoter was subcloned into the CpG-free luciferase reporter construct (25, 37), and the promoter was methylated with HpaII methyltransferase. HpaII methyltransferase targets the internal cytosines on both strands of a CCGG consensus sequence, and therefore it methylates only six of the 98 CpG dinucleotides in the mEphA5-2300 promoter. HpaII methylation effectively blocked activation of the mEphA5-2300 promoter by KLF9, KLF13, and KLF16 (Fig. 5B). Interestingly, KLF2, which had no effect on the unmethylated EphA5 promoter, did transactivate HpaII methylated pCpG-EphA5-2300 (4.01 ± 0.36-fold, p < 0.003) compared with pcDNA controls (Fig. 5B). KLF15 also activated the HpaII-methylated pCpG-EphA5-2300 promoter, although it only reached the 90% confidence level after correcting for multiple comparisons. Thus, different KLF proteins transactivate the EphA5 promoter, depending on the methylation status of only six CpG dinucleotides in the promoter.
Nasal/Temporal Expression of KLF16 and EphA5 in the Retina Is Independent of Pou4f2
Pou4f2 is an important transcription factor for RGC differentiation and maturation, and both KLF16 and EphA5 are expressed in RGCs. Therefore, we asked whether expression of either of these two genes depends on Pou4f2. RNA isolated from both nasal and temporal thirds of retinas from Pou4f2 homozygous KO mice and wildtype (WT) littermates at P0 to P2 was analyzed by RT-qPCR. This age was selected for analysis because the retinas are large enough to reliably isolate nasal from temporal retina thirds, and sufficient numbers of RGCs are still present in the Pou4f2-KO retina to permit analysis (27, 29).
We first asked whether Klf16 transcripts were differentially expressed in the nasal versus temporal retina in WT-Pou4f2 mice. This was not the case, and we detected no difference in Klf16 mRNA expression in the temporal retina versus nasal retina (0.85 ± 0.19-fold; p = 0.226) (Fig. 6). We confirmed this at the protein level on KLF16 immunostained retinas using image analysis and found no differences in KLF16 immunoreactivity between nasal and temporal retina (Fig. 1J).
FIGURE 6.
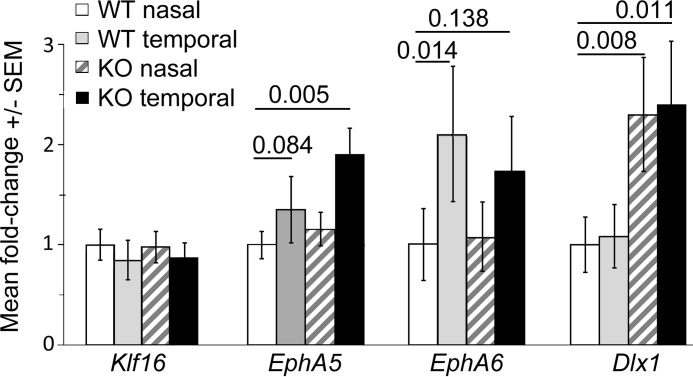
Quantitative RT-PCR analysis of mRNA expression in nasal versus temporal retina from Pou4f2+/+ (WT) and Pou4f2−/− (KO) mice. Bars show mean fold-change in mRNA expression versus WT nasal retina, normalized to Rplp0, Sdha, and Srp14. Horizontal lines show p values versus WT nasal retina, as determined using the relative expression software tool (75). Error bars, standard error of the mean (S.E.).
We next analyzed expression of EphA5 and EphA6 in the same RNA samples. Both of these genes are expressed in temporal high/nasal low gradients across the retina at this stage (33), and our results (Fig. 6) were consistent with previous reports. Both EphA5 and EphA6 had higher expression in the temporal retina of WT littermates mice, with EphA5 increased 1.35 ± 0.33-fold (p = 0.084) and EphA6 increased 2.1 ± 0.67-fold (p = 0.014) in temporal versus nasal retina. Although the small increase in EphA5 did not achieve statistical significance after normalization, the difference is similar to the 1.5-fold increase in EphA5 we previously reported in temporal versus nasal retinas of C57BL/6J mice (25). These data are consistent with the known spatial differences in EphA5 and EphA6 mRNA expression across the retina, validating the nasal versus temporal origins of the samples and supporting our finding that Klf16 is not differentially expressed between nasal and temporal retina.
We then asked whether Klf16 expression was dependent on Pou4f2 by analyzing expression in the retinas of the Pou4f2-KO mice. Klf16 mRNA expression levels in the retinas of Pou4f2-KO mice were no different from WT controls and again showed no apparent nasal/temporal differences (Fig. 6). Expression of EphA5 and EphA6 was also largely unaffected by the absence of Pou4f2, and their overall temporal high/nasal low expression patterns persisted (Fig. 6).
Dlx1 mRNA Expression Increases in the Retina Homozygous Pou4f2
KO mice were compared with wild type and hemizygous controls (38, 39). Therefore, we analyzed Dlx1 expression in the same samples to further verify the genotyping of the homozygous Pou4f2-KO mice. As expected, Dlx1 expression was increased in both nasal and temporal retinas of Pou4f2-KO mice by 2.30 ± 0.57-fold (p = 0.008) and 2.40 ± 0.63-fold (p = 0.011), respectively (Fig. 6), with no differences in Dlx1 expression between temporal and nasal retinas in either WT or KO Pou4f2 mice. Taken together, the RT-qPCR data are consistent with the notion that Klf16, EphA5, and EphA6 expression does not depend on Pou4f2.
Discussion
KLF16 Inhibition of RGC Neurite Outgrowth and Effects on EFNA5-induced Growth Cone Collapse
KLF16 is one of several KLFs that inhibit neurite outgrowth of cortical neurons in vitro (7). Here, we show that overexpression of KLF16 also inhibits neurite outgrowth of RGCs. KLF16 is expressed in RGCs at the appropriate developmental time to be an in vivo transcriptional regulator of EphA5, and using ChIP, KLF16 occupies the proximal promoter region of EphA5 in the retina. It is well known that activation of EPHA5 signaling by EFNA ligands triggers growth cone collapse (40–42) and that the extent of EFN-mediated growth cone collapse reflects the relative abundance of the receptor present in the growth cones (43). Thus, if overexpression of KLF16 increases EphA5 expression, we predicted it would also enhance growth cone collapse in RGCs treated with EFNA5 ligands. Compared with control Fc-treated cells, EFNA5 treatment resulted in robust increases in growth cone collapse in both KLF16- and control-transfected cells. Unexpectedly, the extent of EFNA5-induced growth cone collapse in KLF16-overexpressing RGCs was only slightly greater than in control transfected RGCs. One possible explanation is that the overall reduction in neurite outgrowth resulting from KLF16 overexpression itself is so robust that it renders the assay relatively insensitive for detecting changes resulting from EPH/EFN-mediated growth cone collapse.
Another possibility is that endogenous EphA5 expression in cultured RGCs is already near maximal levels, and overexpression of KLF16 only modestly increases the overall levels of EphA5 expression. For example, forskolin, used in the culture media to maintain RGCs in vitro, up-regulates EphA5 transcription in immortalized rat ovarian granulosa (GFSHR-17) cells (44) and human neuroepithelioma cells (SK-N-MC) (45). Forskolin and the resultant increase in cAMP alters RGC axon outgrowth and regeneration and influences retinotectal map formation (46–51). Although the forskolin concentration used in our experiments (5 μm) was lower than that used in several other studies (10 μm), both of these concentrations result in similar plateau elevations of cAMP levels (52–54). Therefore, the presence of forskolin in the culture media has the potential to increase basal levels of EphA5 expression, contributing to the unexpectedly modest effects of KLF16 overexpression on EFNA5-induced growth cone collapse. Future experiments characterizing EphA5 expression in RGCs in vitro and in vivo in response to KLF16 overexpression and knockdown could address this hypothesis.
Regulation of EphA5 Expression by KLFs and DNA Methylation
These experiments analyzed regulation of EphA5 promoter activation by DNA methylation and KLF transcription factors. The rationale was based on evidence that overexpression of different KLF transcription factors promote or inhibit neurite outgrowth of RGCs and other neurons (7), the role of EPHA5 in patterning RGC axon connectivity during early visual system development (33), and our previous identification of putative KLF-binding sites associated with differential CpG methylation within the EphA5 promoter in the nasal versus temporal retina (25). DNA hypermethylation is a well known epigenetic mechanism for gene silencing, and hypermethylation of the EPHA5 promoter (>70%) has been detected in prostate (24), colorectal (23), and breast cancers (22) and is highly correlated with transcriptional silencing of EPHA5. We previously showed that EphA5 promoter transactivation in luciferase assays is blocked by SssI methylation of all CpG dinucleotides and that transcription of the endogenous EphA5 gene in a mouse Müller glial cell line is silenced by hypermethylation (25). This study, however, focused on the consequences of low level methylation on transcriptional activation of the EphA5 promoter. A key finding in this study was that methylation of only six out of 98 CpG dinucleotides by HpaII methyltransferase changed the ability of different subclasses of KLF transcription factors to transactivate the EphA5 promoter.
Methylation and KLF Binding Specificity
KLF transcription factors have phylogenetically conserved C-terminal DNA binding domains that consist of three zinc fingers, which recognize a 9-bp target site (55). Previous studies showed that the second zinc finger recognizes the GTG or GCG triplet in the core-binding site, whereas the third zinc finger plays an important role in overall binding site preferences (56, 57). Among the KLFs analyzed in this study, the third zinc finger shows the greatest variation in amino acid sequence. This is consistent with our finding that only a subset of KLFs function as positive regulators of EphA5 and likely reflects the differences in binding site preferences of each factor.
Despite the effects of HpaII methylation on KLF transactivation of the EphA5 promoter, none of the six HpaII methylation sites were located in the critical GT box (Fig. 3A). Therefore, sequences outside of this KLF16-binding site must mediate methylation sensitivity. DNA binding by the structurally related SP1 and SP3 transcription factors is unaffected by methylation of the CpG dinucleotides in the GC box itself, but it is inhibited by methylation of CpG dinucleotides located 6–12 bp outside of the central GC box within the SP1-binding site (58). Consistent with this, one of the HpaII sites analyzed is located 7–10 bp from the core GT box that is required for KLF16 to bind and transactivate the EphA5 promoter. In addition, the CpG dinucleotides with increased methylation in the nasal retina are located between 8 and 32 bp from the KLF16-binding site in the EphA5 promoter (25). Thus, CpG dinucleotides in the EphA5 promoter that preferentially are methylated in nasal compared with temporal retina are appropriately positioned to modulate binding specificity of KLF transcription factors based on methylation status.
Taken in conjunction with our previously published studies (25), our data are consistent with low level CpG methylation serving as a mechanism by which a uniformly expressed transcription factor, such as KLF16, can regulate spatially restricted patterns of gene expression in the retina (Fig. 7). Additional research is required to determine whether similar mechanisms regulate the spatial expression patterns of EphA5 and other Eph receptors or Efn ligands throughout the central nervous system.
FIGURE 7.
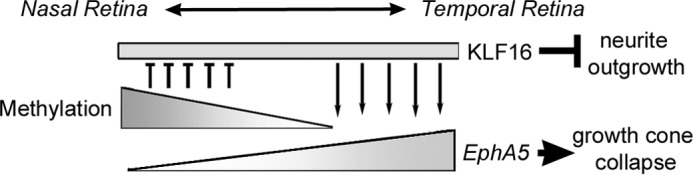
Proposed model for methylation-sensitive regulation of EphA5 expression by KLF16 in the retina. In the nasal retina, DNA methylation near the transcription start site of the EphA5 promoter (upper triangle) blocks promoter transactivation by KLF16 (rectangle), which is uniformly expressed along the nasal/temporal axis, resulting in low EphA5 mRNA levels. In the temporal retina, the EphA5 promoter is unmethylated, and KLF16 increases EphA5 transcription (lower triangle).
KLF16 Regulation of EphA5 in Retinal Development
A key question raised by these data is which of the KLFs are likely to regulate EphA5 expression in vivo. EphA5 expression begins during embryogenesis, and the nasal/temporal gradient is detected as early as E9 (59). In the post-natal retina, EphA5 is expressed in both the GCL and inner nuclear layer, and the nasal/temporal differences persist in the adult, albeit with a slightly flattened gradient (60). Of the KLF transcription factors analyzed, the temporal pattern of KLF16 expression is most consistent with regulating EphA5 in RGCs. KLF16 is expressed in nascent RGCs as early as E15.5 (Fig. 1A) and continues to be expressed in both RGCs and inner retinal neurons in the mature retina. In contrast, our prior RT-PCR analysis showed that Klf9 mRNA is not detected in RGCs until postnatal day 8; Klf13 mRNA expression is much lower levels than Klf16 in RGCs from both embryonic and mature retina (7). KLF16 is expressed in newly generated RGCs, but not until they reach the nascent GCL (Fig. 1C). Interestingly, KLF16 expression begins after POU4F2, yet expression of Klf16 and EphA5 mRNA (Fig. 7) was essentially unchanged in the retinas of Pou4f2-KO mice. We interpret these results to show that their expression is independent of POU4F2.
Despite our evidence supporting a role for KLF16 as a transcriptional regulator of EphA5, there was not complete concordance of expression in early retinal development. KLF16 was not detected in retinal progenitors, although in situ hybridization and LacZ transgenic reporters have shown EphA5 expression in the neuroblastic layer as early as E9 (59). Therefore, KLF16 cannot be responsible for initiating EphA5 expression in the retina. Rather, it is more likely to play a role in sustaining EphA5 expression and in modulating the nasal/temporal differences in EphA5 expression via differential activation of the methylated versus unmethylated promoter (Fig. 7). An interesting but unanswered question is whether this type of regulatory relationship exists in other tissues. Based on the distribution of ESTs in Unigene (www.ncbi.nlm.nih.gov), Klf16 is more broadly expressed than EphA5 in both mouse and humans. However, expression of both Klf16 and EphA5 has been reported in the cortex, spinal cord, tectum, thalamus, and hippocampus (61–64). Thus, there is potential for KLF16 to function as a transcriptional regulator of EphA5 in multiple regions of the brain.
Experimental Procedures
Animals
All studies were performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals and the Association for Research in Vision and Ophthalmology Guidelines for the Use of Animals in Vision Research. All animal use was approved by Institutional Animal Care and Use Committees of the University of Houston (mice) and the University of California at San Diego (rats). C57BL/6J (The Jackson Laboratory), Pou4f2+/− mice on a C57BL/6J;129 mixed genetic background (gift from Dr. W. H. Kline; M.D. Anderson Cancer Center, Houston, TX), and Sprague-Dawley rats (Harlan Laboratories; Placentia, CA) were housed with a 12-h light/12-h dark light cycle and fed standard rodent diet ad libitum.
Immunostaining
Embryos from timed pregnancies or whole eyes from post-natal C57BL/6J mice were fixed in 4% paraformaldehyde (2 h), cryoprotected in sucrose, and flash-frozen in Optimal Cutting Temperature (OCT, Sakura). Cryosections (10 μm) were immunostained, as described previously (65), using 1% SDS in PBS to permeabilize and 10% goat serum or 2% donkey serum in PBS for blocking. Primary antibodies were rabbit polyclonal anti-KLF16 (1:100; H-46; Santa Cruz Biotechnology), goat polyclonal anti-POU4F2 (1:100; H-18, Santa Cruz Biotechnology) (66), mouse monoclonal anti-calbindin (1:400; catalog no. 300, Swant) (67), and mouse monoclonal anti-PAX6 (1:400; Developmental Studies Hybridoma Bank) (68). Secondary antibodies were diluted 1:400 (Alexa Fluor® 488 or Alexa Fluor® 555 conjugates; Molecular Probes, ThermoFisher). Slides were coverslipped using VECTASHIELD® containing DAPI (Vector Laboratories). Images were captured using a Nikon ECLIPSE Ti microscope and NIS Elements software, pseudocolored, assembled into montages, and adjusted for brightness and contrast (Photoshop Elements 11).
Image Analysis
Images of KLF16 immunostaining in P0 retinas were captured on an inverted microscope (IX71; Olympus) with a monochrome, cooled CCD digital camera (Rolera-XR; Q-Imaging) and Q-Capture software using constant exposure times. With orientation masked to the observer, the GCL, defined as the band of DAPI-stained nuclei located between the vitreous cavity and the innermost boundary of the neuroblastic layer, was digitally isolated. The mean pixel intensity of the peripheral 500 μm of the nasal and temporal GCL was measured (ImageJ) and averaged across two non-adjacent sections (n = 3 eyes). Nasal versus temporal intensity was compared using paired t tests, with p < 0.05 considered statistically significant.
Retinal Ganglion Cell Purification
RGC cultures were prepared as described previously (69, 70) Briefly, the retinas were dissected from 3- to 5-day-old Sprague-Dawley rat pups in Ca2+- and Mg2+-free Dulbecco's PBS (ThermoFisher). Papain-digested retinas were immunopanned using antibodies against THY1 (T11D7, ATCC), as described (70). Immediately after purification, expression plasmids (mCherry control or KLF16) were electroporated into RGCs using the rat neuron Nucleofector kit (Amaxa, Lonza). Cells were plated at low density on poly-d-lysine- (10 mg/ml) and laminin (1 mg/ml)-coated tissue culture plates (Falcon) and cultured for 3 days in Neurobasal A media (ThermoFisher) containing sodium pyruvate, N-acetylcysteine, l-glutamine, Sato supplement, insulin, GS21, brain-derived neurotrophic factor (BDNF), ciliary neurotrophic factor (CNTF), and forskolin at 37 °C with 10% CO2 (71).
Axonal Length Quantification
At day 3 in culture, RGCs were fixed in 4% paraformaldehyde and immunostained against rabbit β3-tubulin (D71G9) (1:300; ab5568, Cell Signaling) overnight at 4 °C. Secondary antibodies, goat anti-rabbit 546 (Alexa Fluor® 546; ThermoFisher), were applied (2 h 22 °C). Images were captured using a fluorescent microscope (Zeiss) with treatment condition masked to the experimenter. Neurite length of RGCs was traced manually for at least 50 neurons per condition using NeuronJ plug-in for Fiji-ImageJ. Four independent experiments were conducted for each condition.
Growth Cone Collapse Assay
EFNA5 (10 μg/ml; RD Biosciences) was cross-linked in the presence of goat anti-human Fc antibody (0.45 mg/ml; The Jackson Laboratory) for 45 min at 37 °C in Dulbecco's PBS (ThermoFisher), as described previously (42). Cross-linked EFNA5-Fc or control anti-human Fc was added to the culture for 30 min. Cells were fixed in pre-warmed 4% paraformaldehyde (15 min), permeabilized with 0.3% Triton X-100 (20 min 22 °C), and labeled with Alexa Fluor 555-conjugated phalloidin (2 units/ml, Molecular Probes). Growth cones on at least 25 cells per condition were scored for growth cone collapse by a masked observer.
Cloning
Full-length cDNAs for KLF2 (hKLF2), hKLF4, hKLF9, mouse Klf5 (mKlf5), mKlf6, mKlf7, mKlf13, mKlf15, and mKlf16 were subcloned in pcDNA3.1 + HisC (ThermoFisher) and sequence-verified. All Klf cDNA inserts have been previously described (7), with the exception of the human KLF9 cDNA insert (GenBankTM accession number BC074880), which was purchased (GE Healthcare). The mouse EphA5-2300 promoter clones in pGl2-basic and the CpG-free, pCpGL-basic vectors have been previously described (25, 37). EphA5-118 was generated by PCR amplification (Platinum Taq, ThermoFisher) of the EphA5-2300 clone using primers modified to include an upstream KpnI restriction site (in boldface): EphA5-118: forward 5′-ACGTTGGTACCACACTTACAGTAGCTTGTCAC-3′, and reverse 5′-CCTGTACCTTCTGGCTCTCGAC-3′). Gel-purified KpnI- and XhoI-digested fragments were directionally cloned into pGL2-basic (Promega) or pCpGL-basic (37). The GT box in EphA5-118-luc was mutated (GGGTG to TTTTG) by site-directed mutagenesis (QuikChange site-directed mutagenesis kit; Agilent Technologies). All constructs were confirmed by DNA sequencing. DNA-modifying enzymes were obtained from Promega or New England Biolabs unless otherwise specified.
Luciferase Assays
HEK293FT cells (72, 73) used for luciferase analysis were purchased directly from ThermoScientific (P/N51-0035, lot no. 1705662), and cell identity was confirmed following the experiments by STR DNA fingerprinting (Promega 16 High Sensitivity STR kit DC2100). The STR profiles were compared with on-line search databases (DSMZ/ATCC/JCRB/RIKEN) of 2455 known profiles, along with the MD Anderson Characterized Cell Line Core (CCLC) database of 2556 known profiles. Cells were maintained in DMEM (4500 mg/liter glucose; Sigma) with 10% heat-inactivated FBS (Atlanta Biologicals) and penicillin/streptomycin (Gibco). DNA for transfection experiments was purified using endotoxin-free DNA affinity columns (Qiagen). Transfection reactions containing 5 μl of lipid-based transfection reagent (FuGENE 6; Promega) diluted in 300 μl of serum-free medium with 2.5 μg of the promoter-reporter plasmid, 1.25 μg of the expression plasmid, and 0.125 μg of pCMV-LacZ plasmid (Clontech) were divided between two plates of suspended cells. Control transfections included promoterless pGl2-basic or pCpGL-basic + empty pcDNA3.1 plasmids, with the total amount of plasmid DNA held constant within experiments by addition the corresponding empty vector controls.
Luciferase expression was assayed in cell lysates at 48 h post-transfection (Reporter Lysis Buffer; Dual-Luciferase Reporter System, Promega) and luciferase measured using a luminometer (20/20, Promega). β-Gal was measured using a chemiluminescence reporter system (TROPIX® Galacto-Light Plus; ThermoFisher). Relative luciferase activity for each sample was calculated as the luciferase/β-gal ratio, and samples with a luciferase/β-gal ratio greater than three standard deviations from the overall mean were designated as outliers and excluded from analysis. Relative luciferase for each sample was normalized to the mean relative luciferase of control transfections and compared across all conditions using analysis of variance with post hoc t tests (Microsoft Excel) and the Holm-Bonferroni correction for multiple comparisons. p ≤ 0.05 was considered statistically significant.
EMSA
Isolation of GST-tagged, KLF16 zinc finger fusion protein and EMSAs followed previously described protocols (31, 74), with 10 mm ZnCl2 added to the media and protein isolation buffers. EMSA used dsDNA probes containing the GT box located at +47 to +51 of mEphA5 (wild-type GT box, 5′-AGCGCCGCGCGGGTGCAGTCGCCG-3′; mutated GT box: 5′-AGCGCCGCGCTTTTGCAGTCGCCGG-3′) (IDT), end-labeled with [γ-32P]ATP, incubated with 1 μg of GST-KLF16-zinc finger fusion protein in 5× binding buffer supplemented with 50 μm ZnCl2 (30 min 22 °C). In some reactions, excess unlabeled WT or mutant GT box probes were added as competitors. EMSAs were replicated at least three times.
Chromatin Isolation
Chromatin was isolated from retinas of seven C57BL/6J mice at postnatal day 7 (P7) using Tru ChIP chromatin shearing kit according to the manufacturer's instructions (Covaris) with modifications. Briefly, retinas were cross-linked in fixing buffer with 1% formaldehyde (10 min 22 °C), quenched, washed in PBS (4 °C), pelleted by centrifugation, flash frozen in liquid nitrogen, and stored at −80 °C for no more than 3 days. Cross-linked retinas were thawed in lysis buffer, incubated on ice (20 min), and lysed using a handheld Dounce homogenizer. Pelleted nuclei were resuspended in SDS shearing buffer and sonicated (12 min at a peak intensity of 105 watts, 2% duty cycle (factor), 200 cycles/burst) in an S220 Focused Ultrasonicator (Covaris), running SonolabTM software (version 7.1.4). Insoluble components were removed by centrifugation, and chromatin was stored at 4 °C for no more than 2 days. For each chromatin preparation, ∼4% of the sonicated chromatin was analyzed by agarose gel electrophoresis to verify chromatin fragmentation to 200 to 1500 bp.
Chromatin Immunoprecipitation (ChIP)
ChIP used the ChIP-IT® express kit (Active Motif) as per the manufacturer's directions with protein A magnetic beads (Pierce) and rabbit anti-KLF16 (H46) or rabbit IgG antibodies (Santa Cruz Biotechnology). Protein A beads were blocked in PBS with 0.05% Tween 20, BSA (10 mg/ml), and herring sperm genomic DNA (10 mg/ml) overnight at 4 °C. Chromatin was pre-cleared by incubation with blocked protein A beads (15 min 4 °C). For each experiment, 10% of the pre-cleared chromatin was reserved for the “input” positive control, and the remaining chromatin was diluted in ChIP buffer (Active Motif) and divided between the ChIP reactions as follows: 1) chromatin + anti-KLF16; 2) chromatin + rabbit IgG (negative control), and 3) KLF16 antibody only (no chromatin, negative control). Following overnight incubation (4 °C with rotation), blocked protein A beads were added (2 h, 22 °C). Cross-links were reversed by incubation in ChIP buffer with 3.5 m NaCl (15 min 95 °C), followed by RNaseA and proteinase K digestion, phenol/chloroform extraction, and ethanol precipitation. DNA was resuspended in 10 mm Tris-HCl, pH 8.5, and analyzed by PCR.
Primers for ChIP analysis were as follows: site 1 forward, 5′-CAGACACAGCAGGAGCGAGC-3′, and reverse, 5′-AACTGGAGAGGCTGGCAGAG-3′; site 2 forward, 5′-AACTTATTCATATATCCAC-3′, reverse, 5′-TCAAGATGAGAATGTCTCTG-3′, with a 60 °C annealing temperature. Experimental, negative control, and chromatin input samples were amplified in parallel, using 5% of the purified DNA in each reaction. Standard PCR used GoTaq polymerase (Promega) with 30 cycles, and quantitative PCR used SYBR Green chemistry (Brilliant II SYBR, Low Rox; Agilent). Quantitative PCRs were performed in triplicate using an MX3005P thermocycler (Agilent) with relative quantification (KLF16 versus IgG) calculated as ΔCt = (Ct-KLF16-IP) − (Ct-IgG-IP), and fold enrichment was calculated by using formula 2−ΔCt. The entire experiment was repeated with three independent chromatin preparations.
In Vitro DNA Methylation
Plasmid DNA was isolated from transformed Escherichia coli (DH5-α), a strain that does not methylate CpG dinucleotides. Thus, all purified plasmid DNA used in transfections was unmethylated, unless subsequently methylated in vitro using HpaII methyltransferase. The mouse EphA5 promoter, subcloned into the CpG-free pCpGL-basic vector (pCpGL-EphA5–2.3), was methylated with 3 units of HpaII methyltransferase and 160 μm S-adenosylmethionine per μg of DNA (4 h 37 °C). Mock methylated DNA samples were prepared in parallel, without addition of HpaII methyltransferase. Methylation was verified by restriction enzyme digestion using HpaII restriction endonuclease, which cuts the consensus CCGG site only when the internal CpGs are unmethylated. Methylated plasmids used in luciferase assays remained uncut following HpaII restriction digestion.
RT-qPCR
For RNA isolation, retinal tissue was collected from neonatal (P0 to P2), homozygous Pou4f2 KO mice (gift of W. H. Klein), and non-transgenic littermates as identified by PCR genotyping as described previously (29). Orientation marks were added prior to enucleation, and retinas were dissected in cold diethyl pyrocarbonate-treated PBS. The nasal and temporal thirds of the retinas from each mouse were isolated, transferred to lysis buffer (RLT, Qiagen) with 1% β-mercaptoethanol (Sigma), and stored at −80 °C. Retinal samples from 2 to 3 pups of the same genotype, from 1 or more litters, were pooled prior to RNA purification (RNeasy Mini or RNeasy Micro; Qiagen) and quantification (Nanodrop). All samples had A260/A280 ratios between 1.96 and 2.11 and RNA integrity numbers of 6.0 or higher (RNA Nano Kit, Bioanalyzer 2100; Agilent). For each genotype, three independent samples from nasal and three from temporal retina were analyzed.
RNA was reverse-transcribed (0.25 μg/reaction) with oligo(dT) primers (AffinityScript, Agilent), and quantitative PCR was performed in triplicate 25-μl reactions containing 2% of each RT reaction and 250 nm primers with SYBR chemistry (Brilliant II, Agilent) (95 °C 30 s, 60 °C 30 s, and 72 °C 30 s for 40 cycles), with dissociation curves (Mx3005p, Stratagene) and gel electrophoresis to confirm product size. For Dlx1 primers, the annealing temperature was 55 °C, and the amplification read was at 86 °C, to avoid detection of a minor nonspecific band with a melting temperature (Tm) of 83 °C. All primer pairs (available on request) spanned an intron, with the exception of Klf16. Amplification efficiency for each primer pair was determined by standard curves spanning a 4–5-log range. Controls using RT reactions without reverse transcriptase were performed for all samples and primer pairs and showed no amplification.
RT-qPCR Analysis
Data analysis used Relative Expression Software Tool version 2 (75), a method that includes efficiency corrections and uses mathematical modeling to test expression ratios for significance using a pairwise fixed reallocation randomization test (2000 iterations). Three normalizing genes were used, Rplp0, Sdha, and Srp14 (76, 77). A fourth normalizing gene, B2m, was tested, but it showed consistently increased expression in samples from Pou4f2-KO mice and was excluded from normalization. p < 0.05 was considered statistically significant.
Author Contributions
J. W. conducted experiments, analyzed data, prepared figures, and edited the manuscript. J. G. performed the axon outgrowth/growth cone collapse assays, analyzed data, prepared figures, and edited the manuscript. K. M. B. performed the RT-qPCR and image analysis experiments, analyzed data, and prepared figures. W. L. assisted with confocal image capture and analysis. R. A. U. advised and provided critical reagents for experiments, contributed to data interpretation, critical reading, and revising the manuscript for important intellectual content. J. L. G. contributed to experimental design and analysis of axon outgrowth assays, writing, and editing the manuscript. D. C. O. conceived the overall experimental question and approach, analyzed and interpreted data, prepared figures and tables, and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgments
STR DNA fingerprinting was done by the Cancer Center Support Grant-funded Characterized Cell Line Core (National Institutes of Health NCI Grant P30 CA016672). We thank Dr. William Klein (M. D. Anderson Cancer Center, Houston TX) for the Pou4f2 knock-out mice, Dr. Robert Schwartz (University of Houston, Houston TX) for use of the imaging system, and Micah Mesko-Smith for expert technical assistance.
Note Added in Proof
Fig. 4 was inadvertently omitted in the version of this article that was published as a Paper in Press on July 11, 2016. The figure is now shown.
This work was supported in whole or part by National Institutes of Health Grants R01-EY021792 (to D. C. O.), P30-EY007551 (University of Houston, Core), R01-EY020913 (to J. L. G.), P30-EY022589 (University of California at San Diego), R01-DK052913 (to R. A. U.), P30-DK084567 (Mayo Clinic, Core), P50-CA102701 (Mayo Clinic, SPORE), and Research to Prevent Blindness, Inc. (University of California at San Diego). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- RGC
- retinal ganglion cell
- KLF
- Krüppel-like factor
- qPCR
- quantitative PCR
- EPH
- ephrin receptor
- EPN
- ephrin ligand
- GCL
- ganglion cell layer
- INL
- inner nuclear layer.
References
- 1. Tham Y. C., Li X., Wong T. Y., Quigley H. A., Aung T., and Cheng C. Y. (2014) Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology 121, 2081–2090 [DOI] [PubMed] [Google Scholar]
- 2. Meyer J. S., Howden S. E., Wallace K. A., Verhoeven A. D., Wright L. S., Capowski E. E., Pinilla I., Martin J. M., Tian S., Stewart R., Pattnaik B., Thomson J. A., and Gamm D. M. (2011) Optic vesicle-like structures derived from human pluripotent stem cells facilitate a customized approach to retinal disease treatment. Stem Cells 29, 1206–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Meng F., Wang X., Gu P., Wang Z., and Guo W. (2013) Induction of retinal ganglion-like cells from fibroblasts by adenoviral gene delivery. Neuroscience 250, 381–393 [DOI] [PubMed] [Google Scholar]
- 4. Xie B. B., Zhang X. M., Hashimoto T., Tien A. H., Chen A., Ge J., and Yang X. J. (2014) Differentiation of retinal ganglion cells and photoreceptor precursors from mouse induced pluripotent stem cells carrying an Atoh7/Math5 lineage reporter. PLoS ONE 9, e112175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Philipsen S., and Suske G. (1999) A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 27, 2991–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pearson R., Fleetwood J., Eaton S., Crossley M., and Bao S. (2008) Kruppel-like transcription factors: a functional family. Int. J. Biochem. Cell Biol. 40, 1996–2001 [DOI] [PubMed] [Google Scholar]
- 7. Moore D. L., Blackmore M. G., Hu Y., Kaestner K. H., Bixby J. L., Lemmon V. P., and Goldberg J. L. (2009) KLF family members regulate intrinsic axon regeneration ability. Science 326, 298–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Simon D. K., and O'Leary D. D. (1992) Development of topographic order in the mammalian retinocollicular projection. J. Neurosci. 12, 1212–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tootell R. B., Silverman M. S., Switkes E., and De Valois R. L. (1982) Deoxyglucose analysis of retinotopic organization in primate striate cortex. Science 218, 902–904 [DOI] [PubMed] [Google Scholar]
- 10. Feldheim D. A., and O'Leary D. D. (2010) Visual map development: bidirectional signaling, bifunctional guidance molecules, and competition. Cold Spring Harb. Perspect. Biol. 2, a001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Triplett J. W., and Feldheim D. A. (2012) Eph and ephrin signaling in the formation of topographic maps. Semin Cell Dev. Biol. 23, 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grassot J., Mouchiroud G., and Perrière G. (2003) RTKdb: database of receptor tyrosine kinase. Nucleic Acids Res. 31, 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Funk S. D., and Orr A. W. (2013) Ephs and ephrins resurface in inflammation, immunity, and atherosclerosis. Pharmacol. Res. 67, 42–52 [DOI] [PubMed] [Google Scholar]
- 14. Perez White B. E., and Getsios S. (2014) Eph receptor and ephrin function in breast, gut, and skin epithelia. Cell Adh. Migr. 8, 327–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen J., Song W., and Amato K. (2015) Eph receptor tyrosine kinases in cancer stem cells. Cytokine Growth Factor Rev. 26, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Park I., and Lee H. S. (2015) EphB/ephrinB signaling in cell adhesion and migration. Mol. Cells 38, 14–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Drescher U., Kremoser C., Handwerker C., Löschinger J., Noda M., and Bonhoeffer F. (1995) In vitro guidance of retinal ganglion cell axons by RAGS, a 25-kDa tectal protein related to ligands for Eph receptor tyrosine kinases. Cell 82, 359–370 [DOI] [PubMed] [Google Scholar]
- 18. Otteson D. C., and Petkova T. D. (2014) in Ganglion Cells: Morphology, Functional Development and Role in Disease (Vlastimil L., ed) pp. 1–70, Nova Biomedical, New York [Google Scholar]
- 19. Marcus R. C., Gale N. W., Morrison M. E., Mason C. A., and Yancopoulos G. D. (1996) Eph family receptors and their ligands distribute in opposing gradients in the developing mouse retina. Dev. Biol. 180, 786–789 [DOI] [PubMed] [Google Scholar]
- 20. Feldheim D. A., Vanderhaeghen P., Hansen M. J., Frisén J., Lu Q., Barbacid M., and Flanagan J. G. (1998) Topographic guidance labels in a sensory projection to the forebrain. Neuron 21, 1303–1313 [DOI] [PubMed] [Google Scholar]
- 21. Thakar S., Chenaux G., and Henkemeyer M. (2011) Critical roles for EphB and ephrin-B bidirectional signalling in retinocollicular mapping. Nat. Commun. 2, 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fu D. Y., Wang Z. M., Wang B. L., Chen L., Yang W. T., Shen Z. Z., Huang W., and Shao Z. M. (2010) Frequent epigenetic inactivation of the receptor tyrosine kinase EphA5 by promoter methylation in human breast cancer. Hum. Pathol. 41, 48–58 [DOI] [PubMed] [Google Scholar]
- 23. Kober P., Bujko M., Oledzki J., Tysarowski A., and Siedlecki J. A. (2011) Methyl-CpG binding column-based identification of nine genes hypermethylated in colorectal cancer. Mol. Carcinog. 50, 846–856 [DOI] [PubMed] [Google Scholar]
- 24. Li S., Zhu Y., Ma C., Qiu Z., Zhang X., Kang Z., Wu Z., Wang H., Xu X., Zhang H., Ren G., Tang J., Li X., and Guan M. (2015) Downregulation of EphA5 by promoter methylation in human prostate cancer. BMC Cancer 15, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Petkova T. D., Seigel G. M., and Otteson D. C. (2011) A role for DNA methylation in regulation of EphA5 receptor expression in the mouse retina. Vis. Res. 51, 260–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sretavan D. W. (1990) Specific routing of retinal ganglion cell axons at the mammalian optic chiasm during embryonic development. J. Neurosci. 10, 1995–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gan L., Xiang M., Zhou L., Wagner D. S., Klein W. H., and Nathans J. (1996) POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc. Natl. Acad. Sci. U.S.A. 93, 3920–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moshiri A., Gonzalez E., Tagawa K., Maeda H., Wang M., Frishman L. J., and Wang S. W. (2008) Near complete loss of retinal ganglion cells in the math5/brn3b double knockout elicits severe reductions of other cell types during retinal development. Dev. Biol. 316, 214–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang S. W., Mu X., Bowers W. J., Kim D. S., Plas D. J., Crair M. C., Federoff H. J., Gan L., and Klein W. H. (2002) Brn3b/Brn3c double knockout mice reveal an unsuspected role for Brn3c in retinal ganglion cell axon outgrowth. Development 129, 467–477 [DOI] [PubMed] [Google Scholar]
- 30. de Melo J., Qiu X., Du G., Cristante L., and Eisenstat D. D. (2003) Dlx1, Dlx2, Pax6, Brn3b, and Chx10 homeobox gene expression defines the retinal ganglion and inner nuclear layers of the developing and adult mouse retina. J. Comp. Neurol. 461, 187–204 [DOI] [PubMed] [Google Scholar]
- 31. Daftary G. S., Lomberk G. A., Buttar N. S., Allen T. W., Grzenda A., Zhang J., Zheng Y., Mathison A. J., Gada R. P., Calvo E., Iovanna J. L., Billadeau D. D., Prendergast F. G., and Urrutia R. (2012) Detailed structural-functional analysis of the Kruppel-like factor 16 (KLF16) transcription factor reveals novel mechanisms for silencing Sp/KLF sites involved in metabolism and endocrinology. J. Biol. Chem. 287, 7010–7025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mojumder D. K., Wensel T. G., and Frishman L. J. (2008) Subcellular compartmentalization of two calcium binding proteins, calretinin and calbindin-28 kDa, in ganglion and amacrine cells of the rat retina. Mol. Vis. 14, 1600–1613 [PMC free article] [PubMed] [Google Scholar]
- 33. Feldheim D. A., Nakamoto M., Osterfield M., Gale N. W., DeChiara T. M., Rohatgi R., Yancopoulos G. D., and Flanagan J. G. (2004) Loss-of-function analysis of EphA receptors in retinotectal mapping. J. Neurosci. 24, 2542–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Steketee M. B., Oboudiyat C., Daneman R., Trakhtenberg E., Lamoureux P., Weinstein J. E., Heidemann S., Barres B. A., and Goldberg J. L. (2014) Regulation of intrinsic axon growth ability at retinal ganglion cell growth cones. Invest. Ophthalmol. Vis. Sci. 55, 4369–4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Steketee M. B., Moysidis S. N., Jin X. L., Weinstein J. E., Pita-Thomas W., Raju H. B., Iqbal S., and Goldberg J. L. (2011) Nanoparticle-mediated signaling endosome localization regulates growth cone motility and neurite growth. Proc. Natl. Acad. Sci. U.S.A. 108, 19042–19047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kaczynski J., Cook T., and Urrutia R. (2003) Sp1- and Kruppel-like transcription factors. Genome Biol. 4, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Klug M., and Rehli M. (2006) Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector. Epigenetics 1, 127–130 [DOI] [PubMed] [Google Scholar]
- 38. Mu X., Beremand P. D., Zhao S., Pershad R., Sun H., Scarpa A., Liang S., Thomas T. L., and Klein W. H. (2004) Discrete gene sets depend on POU domain transcription factor Brn3b/Brn-3.2/POU4f2 for their expression in the mouse embryonic retina. Development 131, 1197–1210 [DOI] [PubMed] [Google Scholar]
- 39. Qiu F., Jiang H., and Xiang M. (2008) A comprehensive negative regulatory program controlled by Brn3b to ensure ganglion cell specification from multipotential retinal precursors. J. Neurosci. 28, 3392–3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gao P. P., Sun C. H., Zhou X. F., DiCicco-Bloom E., and Zhou R. (2000) Ephrins stimulate or inhibit neurite outgrowth and survival as a function of neuronal cell type. J. Neurosci. Res. 60, 427–436 [DOI] [PubMed] [Google Scholar]
- 41. Wahl S., Barth H., Ciossek T., Aktories K., and Mueller B. K. (2000) Ephrin-A5 induces collapse of growth cones by activating Rho and Rho kinase. J. Cell Biol. 149, 263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sahin M., Greer P. L., Lin M. Z., Poucher H., Eberhart J., Schmidt S., Wright T. M., Shamah S. M., O'connell S., Cowan C. W., Hu L., Goldberg J. L., Debant A., Corfas G., Krull C. E., and Greenberg M. E. (2005) Eph-dependent tyrosine phosphorylation of ephexin1 modulates growth cone collapse. Neuron 46, 191–204 [DOI] [PubMed] [Google Scholar]
- 43. Brown A., Yates P. A., Burrola P., Ortuño D., Vaidya A., Jessell T. M., Pfaff S. L., O'Leary D. D., and Lemke G. (2000) Topographic mapping from the retina to the midbrain is controlled by relative but not absolute levels of EphA receptor signaling. Cell 102, 77–88 [DOI] [PubMed] [Google Scholar]
- 44. Buensuceso A. V., and Deroo B. J. (2013) The ephrin signaling pathway regulates morphology and adhesion of mouse granulosa cells in vitro. Biol. Reprod. 88, 25. [DOI] [PubMed] [Google Scholar]
- 45. Jassen A. K., Yang H., Miller G. M., Calder E., and Madras B. K. (2006) Receptor regulation of gene expression of axon guidance molecules: implications for adaptation. Mol. Pharmacol. 70, 71–77 [DOI] [PubMed] [Google Scholar]
- 46. Nicol X., Voyatzis S., Muzerelle A., Narboux-Nême N., Südhof T. C., Miles R., and Gaspar P. (2007) cAMP oscillations and retinal activity are permissive for ephrin signaling during the establishment of the retinotopic map. Nat. Neurosci. 10, 340–347 [DOI] [PubMed] [Google Scholar]
- 47. Cai D., Qiu J., Cao Z., McAtee M., Bregman B. S., and Filbin M. T. (2001) Neuronal cyclic AMP controls the developmental loss in ability of axons to regenerate. J. Neurosci. 21, 4731–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Monsul N. T., Geisendorfer A. R., Han P. J., Banik R., Pease M. E., Skolasky R. L. Jr., and Hoffman P. N. (2004) Intraocular injection of dibutyryl cyclic AMP promotes axon regeneration in rat optic nerve. Exp. Neurol. 186, 124–133 [DOI] [PubMed] [Google Scholar]
- 49. Chierzi S., Ratto G. M., Verma P., and Fawcett J. W. (2005) The ability of axons to regenerate their growth cones depends on axonal type and age, and is regulated by calcium, cAMP and ERK. Eur. J. Neurosci. 21, 2051–2062 [DOI] [PubMed] [Google Scholar]
- 50. Hu Y., Cui Q., and Harvey A. R. (2007) Interactive effects of C3, cyclic AMP and ciliary neurotrophic factor on adult retinal ganglion cell survival and axonal regeneration. Mol. Cell. Neurosci. 34, 88–98 [DOI] [PubMed] [Google Scholar]
- 51. Dhande O. S., Bhatt S., Anishchenko A., Elstrott J., Iwasato T., Swindell E. C., Xu H. P., Jamrich M., Itohara S., Feller M. B., and Crair M. C. (2012) Role of adenylate cyclase 1 in retinofugal map development. J. Comp. Neurol. 520, 1562–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Alewijnse A. E., Smit M. J., Rodriguez Pena M. S., Verzijl D., Timmerman H., and Leurs R. (1997) Modulation of forskolin-mediated adenylyl cyclase activation by constitutively active G(S)-coupled receptors. FEBS Lett. 419, 171–174 [DOI] [PubMed] [Google Scholar]
- 53. Clancy J. P., Ruiz F. E., and Sorscher E. J. (1999) Adenosine and its nucleotides activate wild-type and R117H CFTR through an A2B receptor-coupled pathway. Am. J. Physiol. 276, C361–C369 [DOI] [PubMed] [Google Scholar]
- 54. Goldberg J. L., Espinosa J. S., Xu Y., Davidson N., Kovacs G. T., and Barres B. A. (2002) Retinal ganglion cells do not extend axons by default: promotion by neurotrophic signaling and electrical activity. Neuron 33, 689–702 [DOI] [PubMed] [Google Scholar]
- 55. Polozov R. V., Sivozhelezov V. S., Chirgadze Y. N., and Ivanov V. V. (2015) Recognition rules for binding of Zn-Cys2His2 transcription factors to operator DNA. J. Biomol. Struct. Dyn. 33, 253–266 [DOI] [PubMed] [Google Scholar]
- 56. Singleton B. K., Lau W., Fairweather V. S., Burton N. M., Wilson M. C., Parsons S. F., Richardson B. M., Trakarnsanga K., Brady R. L., Anstee D. J., and Frayne J. (2011) Mutations in the second zinc finger of human EKLF reduce promoter affinity but give rise to benign and disease phenotypes. Blood 118, 3137–3145 [DOI] [PubMed] [Google Scholar]
- 57. Schuetz A., Nana D., Rose C., Zocher G., Milanovic M., Koenigsmann J., Blasig R., Heinemann U., and Carstanjen D. (2011) The structure of the Klf4 DNA-binding domain links to self-renewal and macrophage differentiation. Cell. Mol. Life Sci. 68, 3121–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhu K., Hamilton A. D., and Sebti S. M. (2003) Farnesyltransferase inhibitors as anticancer agents: current status. Curr. Opin. Invest. Drugs 4, 1428–1435 [PubMed] [Google Scholar]
- 59. Cooper M. A., Crockett D. P., Nowakowski R. S., Gale N. W., and Zhou R. (2009) Distribution of EphA5 receptor protein in the developing and adult mouse nervous system. J. Comp. Neurol. 514, 310–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rodger J., Lindsey K. A., Leaver S. G., King C. E., Dunlop S. A., and Beazley L. D. (2001) Expression of ephrin-A2 in the superior colliculus and EphA5 in the retina following optic nerve section in adult rat. Eur. J. Neurosci. 14, 1929–1936 [DOI] [PubMed] [Google Scholar]
- 61. Hwang C. K., D'Souza U. M., Eisch A. J., Yajima S., Lammers C. H., Yang Y., Lee S. H., Kim Y. M., Nestler E. J., and Mouradian M. M. (2001) Dopamine receptor regulating factor, DRRF: a zinc finger transcription factor. Proc. Natl. Acad. Sci. U.S.A. 98, 7558–7563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. D'Souza U. M., Lammers C. H., Hwang C. K., Yajima S., and Mouradian M. M. (2002) Developmental expression of the zinc finger transcription factor DRRF (dopamine receptor regulating factor). Mech. Dev. 110, 197–201 [DOI] [PubMed] [Google Scholar]
- 63. Washburn C. P., Cooper M. A., and Zhou R. (2007) Expression of the tyrosine kinase receptor EphA5 and its ligand ephrin-A5 during mouse spinal cord development. Neurosci. Bull. 23, 249–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gamble J. A., Karunadasa D. K., Pape J. R., Skynner M. J., Todman M. G., Bicknell R. J., Allen J. P., and Herbison A. E. (2005) Disruption of ephrin signaling associates with disordered axophilic migration of the gonadotropin-releasing hormone neurons. J. Neurosci. 25, 3142–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wilson D. M. 3rd, and Bianchi C. (1999) Improved immunodetection of nuclear antigens after sodium dodecyl sulfate treatment of formaldehyde-fixed cells. J. Histochem. Cytochem. 47, 1095–1100 [DOI] [PubMed] [Google Scholar]
- 66. Fu X., Kiyama T., Li R., Russell M., Klein W. H., and Mu X. (2009) Epitope-tagging Math5 and Pou4f2: new tools to study retinal ganglion cell development in the mouse. Dev. Dyn. 238, 2309–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Haverkamp S., and Wässle H. (2000) Immunocytochemical analysis of the mouse retina. J. Comp. Neurol. 424, 1–23 [PubMed] [Google Scholar]
- 68. Ericson J., Rashbass P., Schedl A., Brenner-Morton S., Kawakami A., van Heyningen V., Jessell T. M., and Briscoe J. (1997) Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell 90, 169–180 [DOI] [PubMed] [Google Scholar]
- 69. Barres B. A., Silverstein B. E., Corey D. P., and Chun L. L. (1988) Immunological, morphological, and electrophysiological variation among retinal ganglion cells purified by panning. Neuron 1, 791–803 [DOI] [PubMed] [Google Scholar]
- 70. Meyer-Franke A., Kaplan M. R., Pfrieger F. W., and Barres B. A. (1995) Characterization of the signaling interactions that promote the survival and growth of developing retinal ganglion cells in culture. Neuron 15, 805–819 [DOI] [PubMed] [Google Scholar]
- 71. Corredor R. G., Trakhtenberg E. F., Pita-Thomas W., Jin X., Hu Y., and Goldberg J. L. (2012) Soluble adenylyl cyclase activity is necessary for retinal ganglion cell survival and axon growth. J. Neurosci. 32, 7734–7744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Graham F. L., Smiley J., Russell W. C., and Nairn R. (1977) Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36, 59–74 [DOI] [PubMed] [Google Scholar]
- 73. Harrison T., Graham F., and Williams J. (1977) Host-range mutants of adenovirus type 5 defective for growth in HeLa cells. Virology 77, 319–329 [DOI] [PubMed] [Google Scholar]
- 74. Kaczynski J. A., Conley A. A., Fernandez Zapico M., Delgado S. M., Zhang J. S., and Urrutia R. (2002) Functional analysis of basic transcription element (BTE)-binding protein (BTEB) 3 and BTEB4, a novel Sp1-like protein, reveals a subfamily of transcriptional repressors for the BTE site of the cytochrome P4501A1 gene promoter. Biochem. J. 366, 873–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pfaffl M. W., Horgan G. W., and Dempfle L. (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tiwari S., Dharmarajan S., Shivanna M., Otteson D. C., and Belecky-Adams T. L. (2014) Histone deacetylase expression patterns in developing murine optic nerve. BMC Dev. Biol. 14, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Simpson D. A., Feeney S., Boyle C., and Stitt A. W. (2000) Retinal VEGF mRNA measured by SYBR green I fluorescence: A versatile approach to quantitative PCR. Mol. Vis. 6, 178–183 [PubMed] [Google Scholar]