Abstract
The coagulation factor prothrombin has a complex spatial organization of its modular assembly that comprises the N-terminal Gla domain, kringle-1, kringle-2, and the C-terminal protease domain connected by three intervening linkers. Here we use single molecule Förster resonance energy transfer to access the conformational landscape of prothrombin in solution and uncover structural features of functional significance that extend recent x-ray crystallographic analysis. Prothrombin exists in equilibrium between two alternative conformations, open and closed. The closed conformation predominates (70%) and features an unanticipated intramolecular collapse of Tyr93 in kringle-1 onto Trp547 in the protease domain that obliterates access to the active site and protects the zymogen from autoproteolytic conversion to thrombin. The open conformation (30%) is more susceptible to chymotrypsin digestion and autoactivation, and features a shape consistent with recent x-ray crystal structures. Small angle x-ray scattering measurements of prothrombin wild type stabilized 70% in the closed conformation and of the mutant Y93A stabilized 80% in the open conformation directly document two envelopes that differ 50 Å in length. These findings reveal important new details on the conformational plasticity of prothrombin in solution and the drastic structural difference between its alternative conformations. Prothrombin uses the intramolecular collapse of kringle-1 onto the active site in the closed form to prevent autoactivation. The open-closed equilibrium also defines a new structural framework for the mechanism of activation of prothrombin by prothrombinase.
Keywords: enzyme kinetics, prothrombin, single-molecule biophysics, structure-function, thrombin
Introduction
Key biological processes such as blood coagulation and immune response depend on trypsin-like proteases generated from cascades of sequential zymogen activation. Factors involved in these cascades share common ancestry (1) and a modular assembly where the protease domain is coupled to auxiliary components that regulate the proteolytic conversion of zymogen to active enzyme (2). Resolving the spatial organization of these zymogens has long posed a challenge for structural biology. Successes have been few but highly significant (3–7). In some cases, multiple relative arrangements of individual domains have been detected from x-ray analysis, underscoring both the plasticity of the fold and the need for validation from solution studies where the protein is unconstrained by crystal packing. The zymogen prothrombin offers a biologically relevant example as the key player of the coagulation cascade and one of the most abundant proteins circulating in the blood (8).
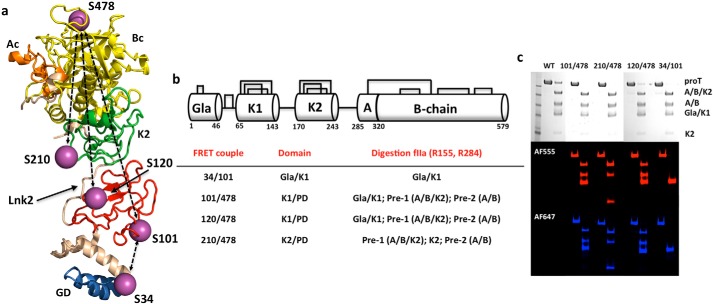
The modular assembly of prothrombin comprises the Gla domain (residues 1–46), kringle-1 (residues 65–143), kringle-2 (residues 170–248), and the protease domain (residues 285–579) connected by three intervening linkers (see Fig. 1, a and b). The linker connecting the two kringles (Lnk2) spans residues 144–169 and is highly flexible (7, 9). Crystallization of prothrombin has succeeded only recently and required deletion of the Gla domain (10) or significant portions of the flexible Lnk2 (7, 9). The molecular picture emerging from these structures is that prothrombin is organized as two rigid ends, the N-terminal Gla domain/kringle-1 pair and the C-terminal kringle-2/protease domain pair, capable of different relative arrangements mediated by Lnk2 (see Fig. 1a). Such plasticity has physiological significance: once prothrombin is anchored to the membrane via its Gla domain, Lnk2 pivots the C-terminal kringle-2/protease domain end over the N-terminal Gla domain/kringle-1 end and regulates how the sites of cleavage at Arg271 and Arg320 are presented to prothrombinase to promote conversion to the mature enzyme thrombin (9). The plasticity of prothrombin supported by recent crystal structures is compelling, but requires validation from independent measurements in solution where the conformational landscape of the protein is accessible and unconstrained by crystal packing. Similar considerations apply quite generally to other zymogens with modular assembly involved in blood coagulation, immune response, and fibrinolysis.
FIGURE 1.
Prothrombin structure and smFRET. a, high resolution structure of prothrombin devoid of residues 154–167 in Lnk2 (9) (PDB ID 5EDM) showing the overall arrangement of the Gla domain (GD, marine), kringle-1 (K1, red), Lnk2 (wheat), kringle-2 (K2, green), and protease domain comprising the A chain (Ac, orange) and catalytic B chain (Bc, yellow). Ser residues mutated to Cys for conjugation with the thiol-reactive dyes AF555 and AF647 used in smFRET measurements are indicated by purple spheres and labeled. The four FRET couples 34/101, 101/478, 120/478, and 210/478 used in the study (see also panel b) are indicated by dotted lines. b, schematic representation of the modular assembly of prothrombin with the Gla domain (Gla), two kringles (K1 and K2), and protease domain (PD) containing the A and B chains connected by a disulfide bond. Three intervening linkers connect the Gla domain to kringle-1, the two kringles (Lnk2), and kringle-2 to the protease domain. Prothrombin has 24 Cys residues paired into 12 disulfide bonds. FRET couples are listed with their respective domains and the products of digestion of each construct with thrombin (fIIa). Pre-1, prethrombin-1; Pre-2, prethrombin-2. c, incorporation of the probes was checked by limited proteolysis with thrombin. After the addition of 10 μl of loading buffer, proteins were loaded into a gradient 4–12% polyacrylamide gel in the presence of SDS and visualized by Coomassie Brilliant Blue R-250 (black and white) or fluorescence intensity by exciting donor at 532 nm (red panel) and acceptor at 640 nm (blue panel). Prothrombin (proT) wild type shows no detectable fluorescence after being treated under the same conditions. Thrombin cleaves prothrombin at Arg155 and generates prethrombin-1 and the Gla domain/kringle-1 pair containing residues 101 and 120. A second cleavage at Arg284 produces prethrombin-2 and kringle-2 containing residue 210. The band corresponding to prethrombin-2 appears in the 101/478, 120/478, and 210/478 couples because residue 478 is in the B chain of the protease domain.
Single molecule Förster resonance energy transfer (smFRET)3 is an essential tool to probe molecular interactions, folding pathways, and conformational changes of freely diffusing single molecules in solution (11–15). Advancements in experimental setup and data analysis ensure greater insights into the properties of macromolecular systems, especially when used in conjunction with structural information (16–19). Here we present smFRET measurements of full-length prothrombin in solution and unravel new features not detected by recent crystallographic analysis. The results broaden our understanding of this important coagulation factor in ways that benefit the study of other zymogens with modular assembly.
Results
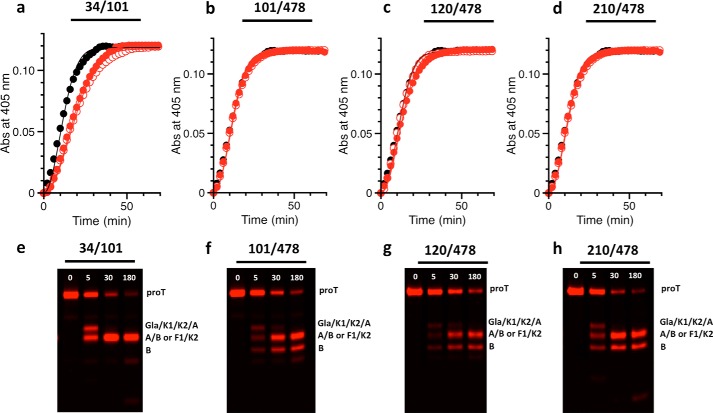
smFRET analysis of protein conformational equilibria requires preparation of labeled constructs and appropriate controls. The conformation of full-length prothrombin in solution was investigated with FRET couples positioned in the N-terminal Gla domain/kringle-1 pair (34/101), the C-terminal kringle-2/protease domain pair (210/478), or across the flexible Lnk2 (101/478 and 120/478) (Fig. 1a). Limited proteolysis of these constructs with thrombin revealed incorporation of the probes at the expected positions within each domain (Fig. 1c). Thrombin cleaves prothrombin at Arg155 and generates prethrombin-1 and the Gla domain/kringle-1 pair containing residues 101 and 120. A second cleavage at Arg284 produces prethrombin-2 and kringle-2 containing residue 210. The band corresponding to prethrombin-2 appears only in the 101/478, 120/478, and 210/478 couples because residue 478 is in the B chain of the protease domain (Fig. 1c). Prothrombin wild type shows no detectable fluorescence after being treated under the same conditions (Fig. 1c), as expected. The functional properties of all constructs were tested by activation with the prothrombinase complex through a continuous enzymatic assay utilizing a chromogenic substrate (9) (Fig. 2, a–d) or standard SDS-PAGE analysis (Fig. 2, e–h). No differences were detected in either the rate or the pathway of activation between labeled or unlabeled constructs when compared with wild type.
FIGURE 2.
Activation of prothrombin constructs by prothrombinase. a–d, all prothrombin mutants labeled for smFRET measurements (filled red circles) are activated by prothrombinase at a rate similar to that of wild type (black circles) or of the unlabeled constructs (open red circles). Measurements were carried out by a continuous assay of substrate hydrolysis (9) using 1 μm prothrombin, 1 pm factor Xa, 20 μm phospholipids, 10 nm cofactor Va, and the chromogenic substrate H-d-Phe-Pro-Phe-p-nitroanilide under experimental conditions: 150 mm NaCl, 5 mm CaCl2, 0.1% PEG 8000, 20 mm Tris, pH 7.4, at 25 °C. Abs, absorbance. e–h, SDS-PAGE analysis of the conversion of prothrombin (proT, 1.4 μm) to thrombin by prothrombinase (0.2 nm factor Xa, 20 μm phospholipids, 10 nm cofactor Va), that occurs with similar efficiency for all constructs within 180 min. The lack of B chain in the 34/101 construct (e) confirms selective incorporation of the probes in the Gla domain/kringle-1 pair.
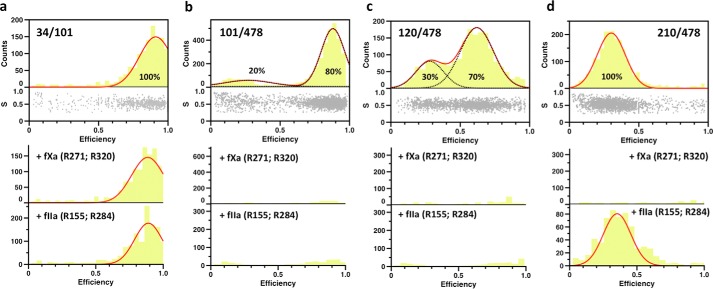
The FRET couple 210/478 positioned above Lnk2 reports a single population of interprobe distances (Fig. 3d). The signal is retained upon cleavage by thrombin at Arg155 (7) in Lnk2, but disappears upon cleavage by factor Xa at Arg271. Likewise, the FRET couple 34/101 below Lnk2 reports a single population (Fig. 3a) with a signal retained upon incubation with thrombin or factor Xa, as expected. The results are consistent with all current crystal structures of prothrombin (7, 9, 10). Assuming a Förster distance Ro = 51 Å for the AF555/AF647 FRET couple, the calculated interprobe distances of 34/101 and 210/478 are 34 ± 2 and 58 ± 4 Å, respectively. The latter distance is practically identical to the Cα-Cα distance between residues Ser210 and Ser478 in structures of prothrombin lacking the Gla domain (10) or portions of Lnk2 (7, 9). The former distance agrees with the Cα-Cα distance between residues Ser34 and Ser101 in three structures of prothrombin with portions of Lnk2 deleted (7, 9). In both cases, a single population of conformers validates the rigidity of the two ends of the zymogen caused by extensive intramolecular contacts between the Gla domain and kringle-1 or kringle-2 and the protease domain (7, 9, 10, 20).
FIGURE 3.
smFRET measurements of prothrombin in solution. Shown are histograms of the four FRET couples probing the conformation of prothrombin in solution. The bottom section of the top graph of each construct depicts the stoichiometry, S, versus FRET efficiency for each diffusing molecule that contains both AF555 and AF647 fluorophores. The upper section shows the one-dimensional efficiency histogram of the molecules in the bottom section. Populations were fit to a single (34/101 and 210/478 FRET couples) or double (101/478 and 120/478 FRET couples) Gaussian distribution (red lines). The percentage of each population is indicated. The bottom graphs depict the results of incubation with factor Xa (fXa) or thrombin (fIIa) for 3 h at room temperature. a–d, proteolytic digestion by factor Xa or thrombin minimally affects the 34/101 couple (a), but abrogates the FRET signal for the 101/478 and 120/478 couples (b and c), as expected. The FRET signal for the 210/478 couple (d) disappears upon digestion with factor Xa due to cleavage at Arg271. Cleavage at Arg320 by factor Xa does not separate the A and B chains that remain connected through the disulfide bond between Cys293 and Cys439. Cleavage at Arg284 by thrombin drastically reduces the signal, as evidenced by the lower counts, but it is not as efficient as the one at R155.
The FRET couples 101/478 and 120/478 positioned across Lnk2 report two distinct populations of interprobe distances (Fig. 3, b and c). The FRET signals disappear in all cases upon cleavage by thrombin at Arg155 or factor Xa at Arg271, as expected. However, analysis of the interprobe distances associated with these FRET couples reveals significant variations with Cα-Cα distances reported in existing crystal structures of prothrombin (7, 9, 10). The flexibility of Lnk2 produces relative arrangements of the rigid ends of the zymogen in solution not captured by x-ray structural analysis of constructs devoid of Gla domain (10) or several portions of Lnk2 (7, 9). The distances associated with the low FRET population tend to be shorter than the corresponding Cα-Cα distances in the crystals by 6–20 Å. The difference is even more significant for the high FRET population. The value measured for the 101/478 couple is 37 ± 2 Å, or >30 Å shorter than that documented in all existing crystal structures. Notably, the value is 21 Å shorter than the interprobe distance reported by the 210/478 couple within the rigid kringle-2/protease domain pair, suggesting that kringle-1 and the protease domain are close to each other rather than being widely separated along a vertical axis. The structural architecture of prothrombin detected by the high FRET signal may be even more contorted than that reported by structures of the zymogen with deletions of Lnk2 (7, 9). Further support to this conclusion comes from the interprobe distances associated with the high FRET population reported by the 120/478 couple. The 47 ± 3 Å distance is >20 Å shorter than that reported in all but one of the existing crystal structures. The data in Fig. 3 support a pre-existing equilibrium between two main conformations that interconvert over a time scale faster than the time of diffusion of prothrombin through the confocal volume, estimated to be 0.3 ms. Optimal resolution of the two conformations is achieved with the 120/478 couple whose interprobe distances are close to the Förster distance Ro = 51 Å for the AF555/AF647 FRET couple. The 120/478 FRET couple was therefore selected for further analysis.
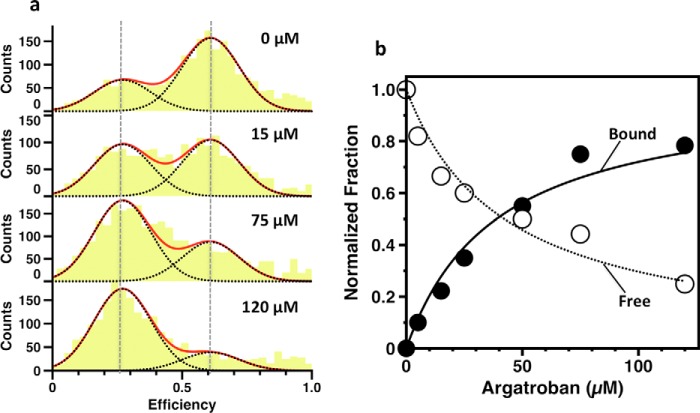
Prethrombin-2 is the zymogen intermediate generated when prothrombinase cleaves prothrombin at Arg271 and sheds the Gla domain and two kringles (21). The conformation of prethrombin-2 (22) is almost identical to the protease domain in all documented structures of prothrombin (7, 9, 10) and enables binding of the inhibitor argatroban at the active site (23). Consistent with these previous findings, argatroban binds to prothrombin with a Kd = 42 μm (Fig. 4b) comparable with that of prethrombin-2 (23). Interestingly, the binding causes the smFRET profile of the 120/478 couple to shift in favor of the low FRET population without affecting the position of the peaks of the two distributions (Fig. 4a). The effect provides direct evidence of ligand binding to only one of two pre-existing conformations according to a mechanism of conformational selection (24, 25). It is unlikely that the two populations detected by smFRET reflect the pre-existing E*-E equilibrium of the trypsin fold where access to the active site is open (E) or closed (E*) by residues within the substrate binding site (26, 27). The concerted displacement of these residues is <3 Å, and the time scale for the E*-E interconversion in a zymogen such as prethrombin-2 is 5 ms (28), or >10-fold slower than the transition between the two populations detected by smFRET. Occlusion of the active site as prothrombin transitions from the open (low FRET) to closed (high FRET) population has a different structural origin that is detected by FRET probes positioned >300 residues apart and across Lnk2.
FIGURE 4.
Binding of the active site inhibitor argatroban monitored by smFRET. a, efficiency histograms of the 120/478 couple as a function of argatroban concentration (0–120 μm). Vertical dashed lines indicate the mean efficiency value of free (E = 0.62) and bound (E = 0.27) forms that dominate in the absence or nearly saturating (120 μm) concentrations of argatroban. Histograms were fitted to a double Gaussian distribution (red lines) with well defined single components (black dotted lines). b, the individual areas of the low (bound) and high (free) FRET populations calculated at each concentration of argatroban, normalized by the total change observed upon saturation, show a hyperbolic dependence on the inhibitor concentration. Values of Kd obtained from independent fit of the two curves are 42 ± 5 μm (open circles) and 45 ± 5 μm (closed circles). Data points are averages of triplicate measurements with standard errors within 3%.
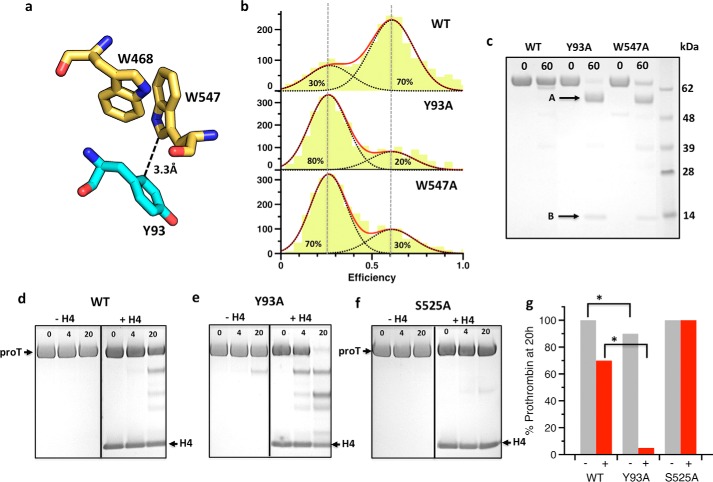
Insight into the nature of the high FRET population comes from a crystal packing interaction documented in all current structures of prothrombin (7, 9, 10): Tyr93 in kringle-1 penetrates the active site of a symmetry-related molecule and makes direct contact with Trp547 (Fig. 5a), a residue critical to substrate recognition in the mature enzyme (29, 30). We therefore tested the hypothesis that such an interaction would occur in solution within the same molecule of prothrombin and result in steric blockage of the active site. The mutations Y93A and W547A introduced in the S120C/S478C background produce a remarkably similar smFRET profile with two populations peaked at the same positions as wild type but shifted in favor of the open form (Fig. 5b). Replacement of two residues widely separated in space, according to existing crystal structures of prothrombin (7, 9, 10), perturbs the pre-existing open-closed equilibrium in a similar fashion and points to a common structural origin of the effect. The Y93A and W547A mutants represent key reagents to interrogate the structural and functional properties of prothrombin in the open form, and offer a relevant comparison with wild type existing predominantly in the closed form. The mutants are completely cleaved by chymotrypsin at residue Trp468 in the autolysis loop within 60 min, but the wild type is not (Fig. 5c), suggesting that this residue may not be accessible to proteolysis in the closed form (Fig. 5a). Prothrombin has been shown to spontaneously convert to thrombin upon mutations that promote exposure of the site of activation at Arg320 (22, 23). The reaction is started by prothrombin itself and therefore requires the catalytic Ser525, but is then propagated by the mature enzyme. Autoactivation is also triggered in the wild type under physiological conditions upon interaction with histone H4 (31). The reaction is slow, but within the half-life (60 h) of prothrombin in the blood. When compared with wild type (Fig. 5d), the mutant Y93A shows increased propensity to autoactivation with and without histone H4 (Fig. 5e). In the presence of histone H4, conversion of the mutant Y93A to the mature enzyme thrombin starts within 4 h and is completed by 20 h (Fig. 5g). No autoactivation is observed under the same conditions for the S525A mutant where the catalytic Ser is replaced by Ala (Fig. 5, f and g). Hence, a potentially important role for the closed conformation of prothrombin is to protect the zymogen from autoactivation to thrombin that is either spontaneous or induced by external cofactors such as histone H4 released in the blood upon cellular damage.
FIGURE 5.
The Tyr93–Trp547 intramolecular interaction stabilizes the closed form of prothrombin. a, a crystal packing interaction present in all current structures of prothrombin brings Tyr93 from kringle-1 (cyan sticks) in contact with Trp547 in the active site of a symmetry-related molecule (yellow sticks). The side chain of Trp547 is also stabilized by Trp468 in the neighbor autolysis loop of the protease domain. The Tyr93-Trp547 interaction occurs in solution within the same prothrombin molecule and occludes the active site in the closed form. b, mutation of Tyr93 or Trp547 destabilizes the high FRET closed form and shifts the pre-existing equilibrium of prothrombin in favor of the low FRET open form. c, the conformational difference between open and closed forms is directly documented by proteolytic digestion with chymotrypsin, which readily cleaves the Y93A and W547A mutants within 60 min, but leaves the wild type intact over the same time frame (band at 72 kDa in the acrylamide gel). N-terminal sequencing of the fragments confirms cleavage at Trp468 in the mutants (A: 1ANTLF5; B: 469TANVG473). This residue is inaccessible to proteolysis in the wild type due to interaction with Trp547 (a). d–g, the open and closed forms of prothrombin also differ in their propensity to autoactivate to thrombin (d–f), with (+, red bar) and without (−, grey bar) histone H4 (g), as monitored by SDS-PAGE. The mutant Y93A autoactivates significantly faster than wild type, especially in the presence of histone H4. Replacement of the active site Ser525 with Ala serves as control. Cleavage at Trp155 is confirmed by the appearance of an intense band that migrates at ∼50 kDa corresponding to prethrombin-1. Errors in panel g are within 2%.
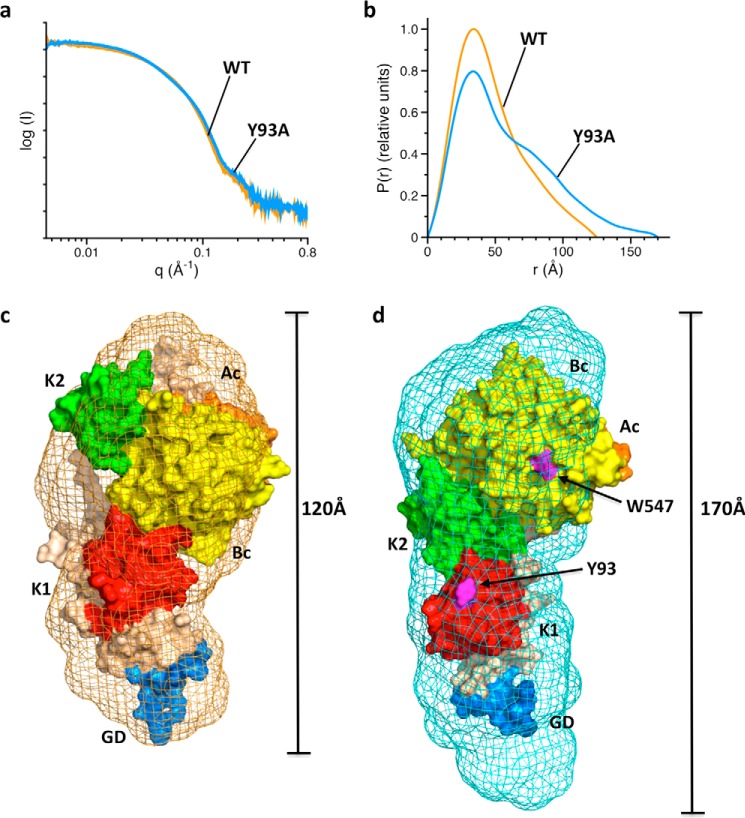
Comparison between wild type and mutant Y93A also offers critical insight into the structural differences between the open and closed conformations of prothrombin in solution. Small angle X-ray scattering (SAXS) measurements (Fig. 6) show a compact envelope for wild type that becomes significantly (50 Å) more elongated for the Y93A mutant. Using information on the rigidity of the N-terminal Gla domain/kringle-1 pair and C-terminal kringle-2/protease domain pair from existing crystal structures of prothrombin (7, 9, 10) and the interprobe distances measured by smFRET (Fig. 3), we built a model of the closed conformation of prothrombin in solution that is consistent with the compact envelope revealed by SAXS (Fig. 6c). The distinguishing feature of this conformation, not captured so far by x-ray structural biology, is the intramolecular collapse of kringle-1 onto the protease domain mediated by the Tyr93-Trp547 interaction. These two residues become separated in the open conformation (Fig. 6d) whose SAXS envelope, revealed by the mutant Y93A, accommodates the existing high resolution structure of prothrombin devoid of residues 154–167 of Lnk2 (9). The bigger SAXS envelope around this structure is explained by the missing portion of Lnk2 and the more compact conformation of the Gla domain bound to Mg2+.
FIGURE 6.
Closed and open conformations of prothrombin revealed by SAXS. a and b, scattering profiles (a) and pair distance distribution functions (b) for prothrombin wild type (orange) and mutant Y93A (blue). c and d, ab initio envelopes calculated from scattering profiles for wild type (c, orange mesh) and mutant Y93A (d, blue mesh) superimposed on an atomic model of the closed conformation (c) or the crystal structure of prothrombin lacking residues 154–167 in Lnk2 (PDB ID 5EDM) (9) (d). The envelope of wild type spans 120 Å and is 50 Å shorter than that of the mutant Y93A. The arrangement of domains in the structure 5EDM is consistent with the elongated SAXS envelope, whose extra volume can be accounted for by the missing portion of Lnk2 (residues 154–167) and the more compact conformation of the Gla domain (GD, marine) in the presence of Mg2+ used in the crystallization buffer. Note how transition from the closed to open conformation separates Tyr93 and Trp547 (magenta). Wheat, Lnk2; red, kringle-1 (K1); green, kringle-2 (K2); orange, A chain (Ac); yellow, B chain (Bc).
Discussion
The results reported in this study show how smFRET measurements combined with structural information afford an effective strategy to unravel the conformational complexity of proteins with modular assembly. FRET probes positioned with the help of available crystal structures reveal conformational transitions affecting distinct domains of the molecule and point to validation by site-directed mutagenesis and solution methods such as SAXS. Using this experimental strategy with the coagulation factor prothrombin, a biologically relevant zymogen with modular assembly, we have identified a pre-existing equilibrium between open and closed forms in solution whose structural underpinnings have been resolved only partially by previous crystallographic analysis. The closed form is dominant (70%) and features an unanticipated intramolecular collapse of Tyr93 in kringle-1 onto Trp547 in the protease domain that obliterates access to the active site and protects prothrombin from autoproteolytic attack, leading to conversion to the mature enzyme thrombin. The open conformation (30%) has a more elongated shape where Tyr93 and Trp547 separate and make the zymogen available to proteolytic digestion by chymotrypsin at Trp468 and more prone to autoactivation. Importantly, smFRET measurements of argatroban binding to prothrombin (Fig. 4a) also provide new and direct evidence that prothrombin functions according to conformational selection (24, 25), a general feature of the trypsin fold so far supported by rapid kinetic studies (24, 28) and observational evidence from the structural biology database (26, 27). Of the two pre-existing forms of prothrombin in equilibrium, only one has the active site open and is populated at saturation with ligand. Furthermore, binding to the open form does not change the properties of this pre-existing conformation. In the alternative mechanism of induced fit, binding produces new conformations that do not exist in the absence of ligand (32), a scenario that finds no support from smFRET measurements of argatroban binding to prothrombin (Fig. 4a).
The smFRET and SAXS measurements reported in this study refer to prothrombin free in solution and are relevant to the function of the zymogen before it binds to membranes and becomes a substrate of prothrombinase in the penultimate step of the coagulation cascade. It should be stressed that prothrombin is synthesized in the liver as other vitamin K-dependent coagulation factors, but its auxiliary domains and codon usage of key Ser residues (33) support a more ancestral evolutionary origin and biological roles that may extend beyond the coagulation cascade (1, 2, 33, 34). Knock-out of the prothrombin gene in the mouse results in embryonic and neonatal lethality (35, 36), a wastage more consistent with a deficit of growth factor function than loss of procoagulant/prothrombotic function that leads to less severe phenotypes (37, 38). Prothrombin is also expressed in the developing and adult rat brains (39), and becomes elevated in the cerebrospinal fluid of patients suffering from progressive neurodegenerative diseases (40). The open and closed conformations of prothrombin likely have distinct physiological roles, and the availability of reagents such as Y93A will be key to future studies that may address this issue in vivo as recently done with other prothrombin mutants (41). The closed form protects the zymogen from autoactivation when it circulates in the blood at high concentration (0.1 mg/ml) and over a long half-life (60 h). This is an important physiological function because factors that promote rapid autoactivation may trigger microvascular thrombosis (42). Protection of the zymogen form from autoproteolytic or proteolytic cleavage is a common theme among proteins with modular assembly. Plasminogen assumes a closed form stabilized by intramolecular interaction of the protease domain with kringles that keeps the zymogen in an activation-resistant conformation (6). Binding of kringles to fibrin clots and cell surface receptors is assumed to induce a transition to an open form that can be cleaved and converted to plasmin by physiological activators. The metalloprotease ADAMTS13 has its distal domains closed on the catalytic site, and the autoinhibition is removed upon binding of its physiological substrate von Willebrand factor (43).
Whether the open and closed forms play a role in the activation of prothrombin by prothrombinase remains to be investigated further. Activation of Y93A and W547A by prothrombinase occurs with the same catalytic rate and along the same pathway as wild type (data not shown). Plausible explanations include equally accessible sites of cleavage at Arg271 and Arg320 in the open and closed forms, or stabilization of a single prothrombin conformation upon binding to prothrombinase. The possibility that multiple conformations of prothrombinase differentially interact with different conformations of prothrombin should also be considered. smFRET measurements of prothrombin in the presence of prothrombinase will address these possibilities directly.
Materials and Methods
Cys Substitutions in Prothrombin
Prothrombin cDNA wild type (residues 1–579) modified to include an epitope for the HPC4 antibody at the C-terminal was cloned into a pDEST40 expression vector using the Gateway cloning technology (Life Technologies). To investigate the architecture of the modular assembly in the full-length zymogen, we expressed and labeled several constructs for smFRET studies with probes located below (S34C/S101C), above (S210C/S478C), or across Lnk2 (S101C/S478C, S120C/S478C, S120C/S478C/Y93A, and S120C/S478C/W547A). Probes were attached to exposed Ser residues mutated to Cys. The mutants were generated using the QuikChange Lighting kit (Agilent, Santa Clara, CA) and appropriate primers (Integrated DNA Technologies, Coralville, IA). After sequencing, the proteins were expressed in baby hamster kidney cells and purified by affinity chromatography, ion exchange chromatography, and size exclusion chromatography as described previously (10, 23, 26). SDS-PAGE and N-terminal sequencing verified homogeneity and chemical identity of final preparations.
Protein Labeling for Single Molecule Detection
Because prothrombin has a total of 24 Cys residues arranged into 12 disulfide bonds, great care was taken to reduce Cys residues introduced in the constructs for smFRET measurements without altering the overall architecture of the zymogen. To this end, we tested different combinations of DTT, tris(2-carboxyethyl)phosphine (TCEP), pH, and temperature. Selective reaction of the unpaired Cys residues was achieved by reducing prothrombin (12–14 μm) in the presence of 350 mm NaCl, 20 mm Tris, pH 7.4 at room temperature for 1 h in the dark, in the presence of DTT at a molar ratio of [-SH]:[DTT] = 1:1.4. Constructs were then labeled with an equal molar mixture of the thiol-reactive dyes, using Alexa Fluor 555-C2-maleimide (AF555) as donor and Alexa Fluor 647-C2-maleimide (AF647) as acceptor (Thermo Fisher Scientific). The labeling reaction was carried out for 2 h at room temperature in the dark. The monomeric protein free of unreacted dyes was purified on an analytical Superdex 200 column (GE Healthcare), and the efficiency of derivatization was assessed by UV-visible measurements.
Analysis of Labeled Prothrombin
Specific incorporation of fluorophores was probed by limited proteolysis with thrombin. Prothrombin wild type and double Cys mutants were dissolved in 20 mm Tris, 145 mm NaCl, 10 mm CaCl2, pH 7.4, at 0.1 mg/ml. Following the addition of thrombin (300 nm) for 3 h at room temperature, aliquots (4 μg or 10 ng) were quenched with 10 μl of NuPAGE lithium dodecyl sulfate (LDS) buffer containing β-mercaptoethanol as reducing agent. Samples were processed by SDS-PAGE. Gels were stained with Coomassie Brilliant Blue R-250 or imaged using a Typhoon 9410 (GE Healthcare). Prothrombin constructs were tested by activation with the prothrombinase complex using a continuous assay as reported elsewhere (9). Briefly, prothrombin (1.4 μm) activation was measured in the presence of 1 pm factor Xa, 20 μm phospholipids, and 10 nm cofactor Va, using 24 μm chromogenic substrate H-d-Phe-Pro-Phe-p-nitroanilide. Data were collected on a SpectraMax i3x Multi-Mode Detection Platform and analyzed with Origin 2015 (OriginLab Corp., Northampton, MA). Prothrombin activation was also monitored by SDS-PAGE. In this case, prothrombin (1.4 μm) dissolved in 150 mm NaCl, 20 mm Tris, 5 mm CaCl2 was activated with 0.2 nm factor Xa, 20 μm phospholipids, and 30 nm cofactor Va in the presence 60 μm dansylarginine-N-(3-ethyl-1,5-pentanediyl)amine (DAPA). Following the addition of the prothrombinase complex, samples (5 μl) were quenched at different time intervals with 10 μl of NuPAGE LDS buffer containing β-mercaptoethanol as the reducing agent and 20 mm EDTA. Samples were loaded into 12% SDS-polyacrylamide gels or processed by NuPAGE Novex 4–12% Bis-Tris protein gels run with MES buffer. Gels were imaged using a Typhoon 9410 (GE Healthcare).
Single Molecule PIE-FRET Measurements
FRET measurements of freely diffusing single molecules were performed with a confocal microscope MicroTime 200 (PicoQuant, Berlin, Germany). Experiments were carried out with pulsed interleaved excitation (PIE), which reports the status of both donor and acceptor fluorophores by sorting molecules on the basis of relative donor:acceptor stoichiometry (S) and apparent FRET efficiency (E). This technique overcomes the limitations of ensemble studies and greatly simplifies the experimental design by avoiding purification of the incomplete labeled species, a task that we could not achieve with prothrombin using standard chromatographic methods based on hydrophobic, affinity, and ionic interaction. In a typical smFRET experiment, thanks to the PIE technology, we were able to isolate and analyze as low as 6–12% of the labeled protein that had incorporated the fluorophores with the correct stoichiometry. The donor and acceptor dyes were excited with a ps pulsed diode laser at 532 and 638 nm, respectively. To achieve pulsed interleaved excitation (19), the 532 nm laser was electronically delayed 25 ns relative to the 638 nm laser (44, 45). A dual band dichroic mirror reflecting 532 and 638 nm guided the light to a high numerical aperture apochromatic objective (60×, N.A. 1.2, water immersion, Olympus) that focused the light to a confocal volume of 1.0 fl for excitation at 532 nm and detection at 575 nm. Fluorescence from excited molecules was collected with the same objective and focused onto a 50-μm diameter pinhole. The donor and acceptor emissions were separated via a dichroic long pass filter with a dividing edge at 620 nm. Suited bandpass filters were inserted to eliminate the respective excitation wavelength and minimize spectral crosstalk. The fluorescence was detected with two avalanche photodiodes using Time-correlated Single Photon Counting with the TimeHarp 200 board. Data were stored in the Time-tagged Time-resolved Mode. Measurements were performed 25 μm deep in the solution with a total acquisition time of 1 h and repeated fresh up to four times on each protein sample (50–80 pm) in 20 mm Tris, 145 mm NaCl, 5 mm CaCl2 0.01% Tween 20, pH 7.4. Signals from single molecules were observed as bursts of fluorescence. Bursts with more than 35 counts were searched with the all photon burst search (APBS) algorithm while integration time was set to 0.5 ms (46). This binning ensured the resolution of two distinct populations for prothrombin (see “Results”). Appropriate correction for direct excitation of the acceptor at the donor excitation wavelength, leakage of the donor in the acceptor channel, and the instrumental γ factor was calculated using a mixture of double-stranded DNA models with known FRET efficiency and stoichiometry labeled with dyes AF555 and AF647 (47). Only molecules with a stoichiometry in the range S = 0.25–0.75 were considered in the final analysis, and their distribution was fit to Gaussian curves using Origin 2015 (OriginLab). The number of independent Gaussians was determined according to the corrected Akaike information criterion (AICc). Data recording and initial data analysis were performed using the SymphoTime Software 6.4 (PicoQuant, Berlin). Further analysis was carried out with MATLAB routines kindly provided by Dr. Don C. Lamb (Munich, Germany).
Binding of Argatroban
Aliquots of a solution (0.1–5 mm) of the active site inhibitor argatroban (Sigma-Aldrich) were added to 200 μl of protein sample (50–80 pm) in 20 mm Tris, 145 mm NaCl, 5 mm CaCl2, 0.01% Tween 20, pH 7.4. At each concentration, measurements were performed 25 μm deep in the solution with a total acquisition time of 1 h. The peak positions of the free and bound forms of prothrombin were obtained by fitting the E histograms obtained at zero and saturating concentration of inhibitor to a double Gaussian function. The high and low FRET peaks were then used as constraints while globally fitting the entire dataset (up to 7 concentrations). The dissociation constant Kd was derived from a fit of the area of the high FRET distribution as a function of inhibitor concentration according to a single site model.
Digestion with Chymotrypsin
Prothrombin wild type and mutants Y93A and W547A were dissolved in 20 mm Tris, 145 mm NaCl, 10 mm CaCl2, pH 7.4, at 0.1 mg/ml. Following the addition of sequencing grade chymotrypsin (Promega) at 1:180 (w/w) ratio, aliquots (40 μl) were quenched at different time intervals with 10 μl of NuPAGE LDS buffer. Samples were loaded into 12% SDS-polyacrylamide gels and processed by NuPAGE Novex 4–12% Bis-Tris protein gels run with MES buffer. Gels were stained with Coomassie Brilliant Blue R-250 and analyzed by quantitative densitometry.
Autoactivation of Prothrombin
Autoactivation was monitored as described previously (31). Briefly, physiological concentrations (0.1 mg/ml) of prothrombin wild type, Y93A, and S525A were incubated at 37 °C in the absence and presence of H4 (4 μm) in buffer containing 150 mm NaCl, 5 mm CaCl2, 20 mm Tris, pH 7.4. Prothrombin autoactivation was followed for up to 40 h. The reaction leading to generation of mature enzyme and depletion of zymogen was quenched at different times with 15 μl of NuPAGE LDS buffer containing β-mercaptoethanol as reducing agent. Samples were processed by SDS-PAGE. Gels were stained with Coomassie Brilliant Blue R-250 and analyzed by quantitative densitometry.
All-atom Three-dimensional Model of the Closed Form of Prothrombin
A model of the closed form of prothrombin was developed from the high resolution structure devoid of residues 154–167 in Lnk2 (Protein Data Bank (PDB) ID 5EDM) (9) by constraining the relative orientation of the rigid N-terminal Gla domain/kringle-1 pair and C-terminal kringle-2/protease domain pair on the interprobe distances measured by smFRET and the packing observed in the structure devoid of Gla domain (PDB ID 4HZH) (10), where kringle-1 comes in contact with the protease domain of a symmetry-related molecule. The portion of Lnk2 missing in the structure 5EDM, with sequence 154PRSEGSSVNLSPPL167, was built manually using COOT. Agreement between the model and distances calculated from the high FRET population was checked with the FRET-restrained positioning and screening (FPS) software (18) in which suitable dyes were attached to residues 101, 120, and 478. Docking simulations of isolated fragment-1 (residues 1–155, comprising the rigid Gla domain/kringle pair) onto prethrombin-1 (residues 156–569, comprising the rigid kringle-2/protease domain pair) constrained by interprobe distances from smFRET measurements returned a bundle of solutions that included a model similar to the closed form of prothrombin. Independent measurements of fragment-1 binding to prethrombin-2 by surface plasmon resonance supported a weak but specific interaction between the two domains (data not shown). Importantly, the intramolecular collapse of kringle-1 onto the protease domain in the model of the closed form was also obtained as an alternative solution of the original data of the structure of prothrombin devoid of Gla domain (PDB ID 4HZH).
SAXS
SAXS data were collected at the beamline 12-ID-B of the Advanced Photon Source at Argonne National Laboratory (Argonne, IL) on prothrombin wild type and mutant Y93A to detect differences between the closed and open conformations. In both cases, constructs carried the S525A substitution to prevent autoproteolytic digestion and were used at concentrations of 1, 2, and 5 mg/ml. Scattered x-rays at 14 keV radiation energy were measured using a Pilatus 2 M detector with a sample-to-detector distance of 2 m. A flow cell was used to reduce radiation damage. Thirty images were collected for each sample and buffer blank. The scattering vector q = 4πsin(θ/2)/λ is the momentum transfer defined by the scattering angle θ and x-ray wavelength λ. The isotropic two-dimensional images were converted to one-dimensional SAXS profiles, i.e. intensity versus q, followed by averaging and background subtraction using software packages at the beamline. The radius of gyration, Rg, was determined using the Guinier approximation in the low q region (qRg<1.3), and its linearity served as an initial assessment of data and sample quality. Maximum particle dimension, Dmax, and distance distribution function, P(r), were calculated using GNOM (48). The low resolution envelopes were produced using both GASBOR (48) (q up to 0.8 Å−1) and DAMMIN (49) (q up to 0.3 Å−1) by directly fitting the reciprocal space scattering profile. Twenty models were generated for every calculation and then aligned and averaged using DAMAVER (49). The results of GASBOR and DAMMIN were very similar, but only GASBOR results were reported here. Structural figures were prepared using PyMOL.
Author Contributions
N. P. and E. D. C. designed experiments; N. P. set up the confocal microscope and developed the workflow for data collection and analysis; N. P. and D. B. expressed proteins; N. P., D. B., and X. Z. collected data; N. P., X. Z., and E. D. C. analyzed the results; and N. P. and E. D. C. wrote the manuscript.
Acknowledgments
We gratefully acknowledge Dr. David Gohara and Zhiwei Chen for their assistance with the model of the closed conformation of prothrombin, Daniel Shropshire and Leslie Pelc for expression and labeling of several constructs for smFRET measurements, and Dr. Tomasz Heyduk and Rachel Hickey for providing double-stranded DNA constructs used for calibration of the MicroTime instrument and control experiments. This research used resources of the Advanced Photon Source, a U. S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357.
This work was supported in part by American Heart Association Grant 15SDG25550094 (to N. P.), a grant from the Wroclaw Centre of Biotechnology the Leading National Research Centre (KNOW) program (2014–2018) (to D. B.), and National Institutes of Health Research Grants HL049413, HL073813, and HL112303 (to E. D. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- smFRET
- single molecule Förster resonance energy transfer
- PIE
- pulsed interleaved excitation
- SAXS
- small angle x-ray scattering
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. Krem M. M., and Di Cera E. (2002) Evolution of enzyme cascades from embryonic development to blood coagulation. Trends Biochem. Sci. 27, 67–74 [DOI] [PubMed] [Google Scholar]
- 2. Patthy L. (1985) Evolution of the proteases of blood coagulation and fibrinolysis by assembly from modules. Cell 41, 657–663 [DOI] [PubMed] [Google Scholar]
- 3. Papagrigoriou E., McEwan P. A., Walsh P. N., and Emsley J. (2006) Crystal structure of the factor XI zymogen reveals a pathway for transactivation. Nat. Struct. Mol. Biol. 13, 557–558 [DOI] [PubMed] [Google Scholar]
- 4. Forneris F., Ricklin D., Wu J., Tzekou A., Wallace R. S., Lambris J. D., and Gros P. (2010) Structures of C3b in complex with factors B and D give insight into complement convertase formation. Science 330, 1816–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Banner D. W., D'Arcy A., Chène C., Winkler F. K., Guha A., Konigsberg W. H., Nemerson Y., and Kirchhofer D. (1996) The crystal structure of the complex of blood coagulation factor VIIa with soluble tissue factor. Nature 380, 41–46 [DOI] [PubMed] [Google Scholar]
- 6. Law R. H., Caradoc-Davies T., Cowieson N., Horvath A. J., Quek A. J., Encarnacao J. A., Steer D., Cowan A., Zhang Q., Lu B. G., Pike R. N., Smith A. I., Coughlin P. B., and Whisstock J. C. (2012) The x-ray crystal structure of full-length human plasminogen. Cell Rep. 1, 185–190 [DOI] [PubMed] [Google Scholar]
- 7. Pozzi N., Chen Z., Pelc L. A., Shropshire D. B., and Di Cera E. (2014) The linker connecting the two kringles plays a key role in prothrombin activation. Proc. Natl. Acad. Sci. U.S.A. 111, 7630–7635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Degen S. J., and Davie E. W. (1987) Nucleotide sequence of the gene for human prothrombin. Biochemistry 26, 6165–6177 [DOI] [PubMed] [Google Scholar]
- 9. Pozzi N., Chen Z., and Di Cera E. (2016) How the linker connecting the two kringles influences activation and conformational plasticity of prothrombin. J. Biol. Chem. 291, 6071–6082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pozzi N., Chen Z., Gohara D. W., Niu W., Heyduk T., and Di Cera E. (2013) Crystal structure of prothrombin reveals conformational flexibility and mechanism of activation. J. Biol. Chem. 288, 22734–22744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Juette M. F., Terry D. S., Wasserman M. R., Zhou Z., Altman R. B., Zheng Q., and Blanchard S. C. (2014) The bright future of single-molecule fluorescence imaging. Curr Opin Chem. Biol. 20, 103–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Santoso Y., Joyce C. M., Potapova O., Le Reste L., Hohlbein J., Torella J. P., Grindley N. D., and Kapanidis A. N. (2010) Conformational transitions in DNA polymerase I revealed by single-molecule FRET. Proc. Natl. Acad. Sci. U.S.A. 107, 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tomescu A. I., Robb N. C., Hengrung N., Fodor E., and Kapanidis A. N. (2014) Single-molecule FRET reveals a corkscrew RNA structure for the polymerase-bound influenza virus promoter. Proc. Natl. Acad. Sci. U.S.A. 111, E3335–3342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang S., Vafabakhsh R., Borschel W. F., Ha T., and Nichols C. G. (2016) Structural dynamics of potassium-channel gating revealed by single-molecule FRET. Nat. Struct. Mol. Biol. 23, 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chung H. S., Cellmer T., Louis J. M., and Eaton W. A. (2013) Measuring ultrafast protein folding rates from photon-by-photon analysis of single molecule fluorescence trajectories. Chem. Phys. 422, 229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roy R., Hohng S., and Ha T. (2008) A practical guide to single-molecule FRET. Nat. Methods 5, 507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kudryavtsev V., Sikor M., Kalinin S., Mokranjac D., Seidel C. A., and Lamb D. C. (2012) Combining MFD and PIE for accurate single-pair Forster resonance energy transfer measurements. Chemphyschem 13, 1060–1078 [DOI] [PubMed] [Google Scholar]
- 18. Kalinin S., Peulen T., Sindbert S., Rothwell P. J., Berger S., Restle T., Goody R. S., Gohlke H., and Seidel C. A. (2012) A toolkit and benchmark study for FRET-restrained high-precision structural modeling. Nat. Methods 9, 1218–1225 [DOI] [PubMed] [Google Scholar]
- 19. Müller B. K., Zaychikov E., Bräuchle C., and Lamb D. C. (2005) Pulsed interleaved excitation. Biophys. J. 89, 3508–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen Z., Pelc L. A., and Di Cera E. (2010) Crystal structure of prethrombin-1. Proc. Natl. Acad. Sci. U.S.A. 107, 19278–19283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rosing J., Tans G., Govers-Riemslag J. W., Zwaal R. F., and Hemker H. C. (1980) The role of phospholipids and factor Va in the prothrombinase complex. J. Biol. Chem. 255, 274–283 [PubMed] [Google Scholar]
- 22. Pozzi N., Chen Z., Zapata F., Pelc L. A., Barranco-Medina S., and Di Cera E. (2011) Crystal structures of prethrombin-2 reveal alternative conformations under identical solution conditions and the mechanism of zymogen activation. Biochemistry 50, 10195–10202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pozzi N., Chen Z., Zapata F., Niu W., Barranco-Medina S., Pelc L. A., and Di Cera E. (2013) Autoactivation of thrombin precursors. J. Biol. Chem. 288, 11601–11610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vogt A. D., and Di Cera E. (2012) Conformational selection or induced fit? A critical appraisal of the kinetic mechanism. Biochemistry 51, 5894–5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Boehr D. D., Nussinov R., and Wright P. E. (2009) The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 5, 789–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pozzi N., Vogt A. D., Gohara D. W., and Di Cera E. (2012) Conformational selection in trypsin-like proteases. Curr. Opin. Struct. Biol. 22, 421–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gohara D. W., and Di Cera E. (2011) Allostery in trypsin-like proteases suggests new therapeutic strategies. Trends Biotechnol. 29, 577–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vogt A. D., Chakraborty P., and Di Cera E. (2015) Kinetic dissection of the pre-existing conformational equilibrium in the trypsin fold. J. Biol. Chem. 290, 22435–22445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bode W., Turk D., and Karshikov A. (1992) The refined 1.9-Å x-ray crystal structure of d-Phe-Pro-Arg chloromethylketone-inhibited human α-thrombin: structure analysis, overall structure, electrostatic properties, detailed active-site geometry, and structure-function relationships. Protein Sci. 1, 426–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arosio D., Ayala Y. M., and Di Cera E. (2000) Mutation of W215 compromises thrombin cleavage of fibrinogen, but not of PAR-1 or protein C. Biochemistry 39, 8095–8101 [DOI] [PubMed] [Google Scholar]
- 31. Barranco-Medina S., Pozzi N., Vogt A. D., and Di Cera E. (2013) Histone H4 promotes prothrombin autoactivation. J. Biol. Chem. 288, 35749–35757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tang C., Schwieters C. D., and Clore G. M. (2007) Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR. Nature 449, 1078–1082 [DOI] [PubMed] [Google Scholar]
- 33. Krem M. M., and Di Cera E. (2001) Molecular markers of serine protease evolution. EMBO J. 20, 3036–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Patthy L. (1999) Genome evolution and the evolution of exon-shuffling: a review. Gene 238, 103–114 [DOI] [PubMed] [Google Scholar]
- 35. Sun W. Y., Witte D. P., Degen J. L., Colbert M. C., Burkart M. C., Holmbäck K., Xiao Q., Bugge T. H., and Degen S. J. (1998) Prothrombin deficiency results in embryonic and neonatal lethality in mice. Proc. Natl. Acad. Sci. U.S.A. 95, 7597–7602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xue J., Wu Q., Westfield L. A., Tuley E. A., Lu D., Zhang Q., Shim K., Zheng X., and Sadler J. E. (1998) Incomplete embryonic lethality and fatal neonatal hemorrhage caused by prothrombin deficiency in mice. Proc. Natl. Acad. Sci. U.S.A. 95, 7603–7607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Coughlin S. R. (2000) Thrombin signalling and protease-activated receptors. Nature 407, 258–264 [DOI] [PubMed] [Google Scholar]
- 38. Suh T. T., Holmbäck K., Jensen N. J., Daugherty C. C., Small K., Simon D. I., Potter S., and Degen J. L. (1995) Resolution of spontaneous bleeding events but failure of pregnancy in fibrinogen-deficient mice. Genes Dev. 9, 2020–2033 [DOI] [PubMed] [Google Scholar]
- 39. Dihanich M., Kaser M., Reinhard E., Cunningham D., and Monard D. (1991) Prothrombin mRNA is expressed by cells of the nervous system. Neuron 6, 575–581 [DOI] [PubMed] [Google Scholar]
- 40. Huang Y. C., Wu Y. R., Tseng M. Y., Chen Y. C., Hsieh S. Y., and Chen C. M. (2011) Increased prothrombin, apolipoprotein A-IV, and haptoglobin in the cerebrospinal fluid of patients with Huntington's disease. PLoS ONE 6, e15809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Flick M. J., Chauhan A. K., Frederick M., Talmage K. E., Kombrinck K. W., Miller W., Mullins E. S., Palumbo J. S., Zheng X., Esmon N. L., Esmon C. T., Thornton S., Becker A., Pelc L. A., Di Cera E., et al. (2011) The development of inflammatory joint disease is attenuated in mice expressing the anticoagulant prothrombin mutant W215A/E217A. Blood 117, 6326–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gould T. J., Lysov Z., and Liaw P. C. (2015) Extracellular DNA and histones: double-edged swords in immunothrombosis. J. Thromb. Haemost. 13, Suppl. 1, S82–S91 [DOI] [PubMed] [Google Scholar]
- 43. Muia J., Zhu J., Gupta G., Haberichter S. L., Friedman K. D., Feys H. B., Deforche L., Vanhoorelbeke K., Westfield L. A., Roth R., Tolia N. H., Heuser J. E., and Sadler J. E. (2014) Allosteric activation of ADAMTS13 by von Willebrand factor. Proc. Natl. Acad. Sci. U.S.A. 111, 18584–18589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gopich I. V., and Szabo A. (2007) Single-molecule FRET with diffusion and conformational dynamics. J. Phys. Chem. B 111, 12925–12932 [DOI] [PubMed] [Google Scholar]
- 45. Kapanidis A. N., Lee N. K., Laurence T. A., Doose S., Margeat E., and Weiss S. (2004) Fluorescence-aided molecule sorting: analysis of structure and interactions by alternating-laser excitation of single molecules. Proc. Natl. Acad. Sci. U.S.A. 101, 8936–8941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nir E., Michalet X., Hamadani K. M., Laurence T. A., Neuhauser D., Kovchegov Y., and Weiss S. (2006) Shot-noise limited single-molecule FRET histograms: comparison between theory and experiments. J. Phys. Chem. B 110, 22103–22124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee N. K., Kapanidis A. N., Wang Y., Michalet X., Mukhopadhyay J., Ebright R. H., and Weiss S. (2005) Accurate FRET measurements within single diffusing biomolecules using alternating-laser excitation. Biophys. J. 88, 2939–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Svergun D. I., Petoukhov M. V., and Koch M. H. (2001) Determination of domain structure of proteins from x-ray solution scattering. Biophys. J. 80, 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Svergun D. I. (1999) Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]