Abstract
PgpB belongs to the lipid phosphate phosphatase protein family and is one of three bacterial integral membrane phosphatases catalyzing dephosphorylation of phosphatidylglycerol phosphate (PGP) to generate phosphatidylglycerol. Although the structure of its apo form became recently available, the mechanisms of PgpB substrate binding and catalysis are still unclear. We found that PgpB was inhibited by phosphatidylethanolamine (PE) in a competitive mode in vitro. Here we report the crystal structure of the lipid-bound form of PgpB. The structure shows that a PE molecule is stabilized in a membrane-embedded tunnel formed by TM3 and the “PSGH” fingerprint peptide near the catalytic site, providing structural insight into PgpB substrate binding mechanism. Noteworthy, in silico docking of varied lipid phosphates exhibited similar substrate binding modes to that of PE, and the residues in the lipid tunnel appear to be important for PgpB catalysis. The catalytic triad in the active site is essential for dephosphorylating substrates lysophosphatidic acid, phosphatidic acid, or sphingosine-1-phosphate but surprisingly not for the native substrate PGP. Remarkably, residue His-207 alone is sufficient to hydrolyze PGP, indicating a specific catalytic mechanism for PgpB in PG biosynthesis. We also identified two novel sensor residues, Lys-93 and Lys-97, on TM3. Our data show that Lys-97 is essential for the recognition of lyso-form substrates. Modification at the Lys-93 position may alter substrate specificity of lipid phosphate phosphatase proteins in prokaryotes versus eukaryotes. These studies reveal new mechanisms of lipid substrate selection and catalysis by PgpB and suggest that the enzyme rests in a PE-stabilized state in the bilayer.
Keywords: phosphatase, phosphatidylglycerol, phospholipid metabolism, protein-lipid interaction, X-ray crystallography, lipid phosphate dephosphorylation, substrate selectivity
Introduction
Phosphatidylglycerol (PG)2 is one of the major phospholipids in the bacterial inner membrane, accounting for ∼20% of total lipid composition. PG is generated by dephosphorylation of phosphatidylglycerol-phosphate (PGP) catalyzed by three membrane-associated phosphatidylglycerol phosphatases, PgpA, PgpB, and PgpC (1, 2). These phosphatidylglycerol phosphatases exhibit no sequence homology with each other, and their active sites are predicted to face different sides of the cytoplasmic membrane. PgpB is the only phosphatidylglycerol phosphatase protein containing multiple (six) transmembrane helices (TM) (3). Raetz and coworkers (1) have shown that PgpB alone is sufficient to maintain PG at the wild type level in Escherichia coli, although the specific role of each phosphatidylglycerol phosphatase is still unclear. In addition to PGP, in vitro studies showed that PgpB was able to dephosphorylate other lipid phosphate substrates, including lysophosphatidic acid (LPA), phosphatidic acid (PA), diacylglycerol pyrophosphate (DGPP), and undecaprenyl phosphate (2–4). Any involvement of PgpB in these bacterial phospholipid catalyzes will require experimental verification in vivo, although substrate diversity has been reported in PgpB's eukaryotic homologs.
PgpB is a member of the lipid phosphate phosphatase protein family (LPP), which is universally found in all kingdoms of life (5). Numerous studies have illustrated their important roles in lipid signaling processes in attenuating a variety of lipid phosphate levels in eukaryotic cells (6). For instance, mammalian LPP1 hydrolyzes LPA to reduce cell proliferation and increase apoptosis (7). Both LPP3 and SPP, a sphingosine-1-phosphate (S1P) phosphatase, were found to be associated with embryonic development and cell death (8, 9). In Arabidopsis, LPPs modulate the stress response via PA and DGPP-mediated lipid signaling (10). This substrate versatility may be advantageous for LPPs to participate in diverse signaling networks simultaneously. It has long been proposed that LPPs including PgpB utilize a common acid phosphatase mechanism mediated by three essential catalytic motifs (C1–C3) to catalyze dephosphorylation of structurally different lipid phosphate substrates (11).
Recently, the structure of the apo form of PgpB from E. coli has been reported. The structure showed that the three catalytic motifs form a catalytic site on the extracellular surface in a conformation found in many globular acidic phosphatase proteins (12). Fan et al. (12) also used mutations to prove that all six conserved residues in the catalytic motifs are essential for PgpB catalysis of LPA, PA, and DGPP. They proposed a substrate lateral diffusion model for substrate access from the bilayer. Unfortunately, a native substrate for PGP was not tested. Thus, important questions remain unanswered. 1) How does PgpB recognize its substrates? The substrate binding mode in the membrane bilayer was not deduced from the apo form structure. 2) Does PgpB utilize a common LPP mechanism for PG production? 3) How do LPPs including PgpB distinguish among structurally different lipid substrates, lyso or diacyl forms, to perform efficient dephosphorylation reactions? Answering these questions may help to understand the LPPs' role as a regulator of multiple signaling pathways.
To address these fundamental questions, here we report the crystal structure of the PE-bound state of PgpB. The PE binding conformation provides a structural basis to help understand the substrate binding mechanism of PgpB. Our data also reveal a novel PgpB dephosphorylation mechanism specific for PGP. We also identified a pair of lysine residues on TM3 that enable PgpB to distinguish the lyso form from diacyl-form substrates based on their acyl chain conformations. In addition, a novel regulatory effect of PE lipid on PgpB catalysis is discussed.
Results
Functional Characterization of PgpB in Vitro
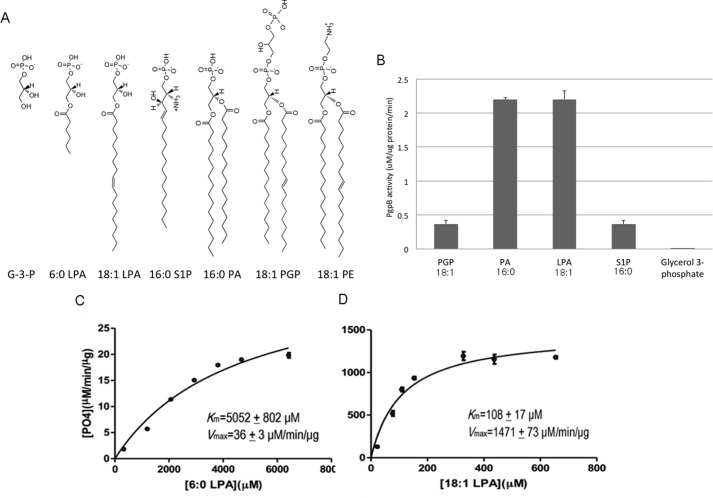
The PgpB protein fused with a His6 tag at its C terminus was expressed and purified using the detergent n-dodecyl β-D-maltoside (DDM). The activity of the purified protein was measured using a standard colorimetric phosphatase assay in vitro. Although PGP is the only PgpB substrate that has been characterized in vivo (1), PGP catalysis has not yet been analyzed in vitro. To compare PGP with other LPP substrates, we also chose two lyso forms (LPA and S1P) and the diacyl form PA (Fig. 1A). Both PGP and PA have similar chemical structures except that PGP has an additional phosphoglycerol moiety on the phosphate group. PgpB apparently dephosphorylates all four substrates in vitro; however, the activity for LPA or PA was substantially (∼6×) higher than for S1P or PGP (Fig. 1B).
FIGURE 1.
In vitro functional characterization of PgpB. A, chemical structure of lipid phosphate substrates and inhibitors of PgpB. B, each reaction of 100 μl contains 2 μg of PgpB protein and 5 μg of each substrate (30 mm for glycerol 3-phosphate) mixed for 40 min at room temperature and then characterized by colorimetric phosphatase assay. The activity was normalized with an individual background control. C and D, Michaelis-Menten kinetic assays using 6:0 LPA or 18:1 LPA as substrates plotted using GraphPad PrismTM software. G-3-P, glycerol-3-phosphate.
All PgpB substrates tested above contain a long (>C16) acyl chain(s) despite their different head group structures. To examine whether the acyl chain is involved in substrate binding, we tested PgpB using three LPA analogs with different acyl chain lengths: 18:1, 6:0, and acyl-less glycerol 3-phosphate. Michaelis-Menten kinetic analysis yielded a Km = 108 ± 17 μm and Vmax = 1471 ± 73 μm/μg/min for 18:1 LPA (Fig. 1C). PgpB activity showed a strong dependence on substrate acyl chain length. As seen in Fig. 1D, the activity of 6:0 LPA was dramatically reduced, resulting in a Km = 5052 ± 802 μm and Vmax = 36 ± 3 μm/min/μg for 6:0 LPA. No activity was detected with 30 mm glycerol 3-phosphate in the reaction (Fig. 1B). These data showed that PgpB exclusively catalyzes lipid substrates. The reduction (>40×) in catalytic kinetics, in both Km and Vmax, indicated that a long acyl chain is important not only for substrate binding but also for enzyme catalysis.
PE Inhibition of PgpB in Vitro
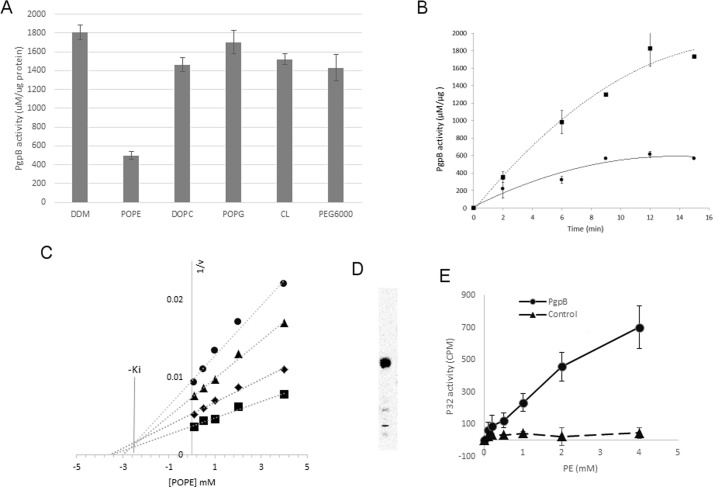
Orthovanadate is the only identified inhibitor for the LPP proteins. Interestingly, we also found that the activity of PgpB was inhibited by 70% after adding 3 mm POPE (the concentration in the crystallization buffer; see below) in the reaction (Fig. 2A). No apparent inhibition was observed in the presence of POPG or cardiolipin (CL) at the same concentration. Fig. 2B shows a time course for PgpB-catalyzed LPA hydrolysis in the presence of POPE. To further characterize this novel PE inhibition, we generated a Dixon plot by measuring PgpB activity at varied substrate concentrations as a function of inhibitor concentration. The Dixon plot in Fig. 2C is characteristic of competitive inhibition. The Ki value was estimated to be 2.7 mm based on the intersection of linear regressions at varied LPA concentrations. The lack of inhibition by PG suggested the ethanolamine head group is required for PE inhibition. Phosphocholine shares a zwitterionic lipid conformation similar to PE but has an additional trimethyl group. To probe the role of inhibitor conformation, we tested DOPC. However, the addition of DOPC to the reaction had no noticeable effect (Fig. 2A), indicating the specificity of PE inhibition.
FIGURE 2.
In vitro inhibition of PgpB by PE. A, 2 μg of PgpB protein was premixed with 3 mm POPE, DOPC, POPG, or CL, 0.5% DDM, or 10% PEG 6000 for 10 min at room temperature before adding 18:1 LPA for 30 min. The remaining PgpB activity was monitored using a colorimetric phosphatase assay. B, a time course of LPA reaction of PgpB (squares) or in the presence of 3 mm POPE (circles). C, Dixon plot of reciprocal rates of phosphate release (1/v) as a function of POPE concentration. Each line represents linear regression analysis of the reciprocal of average simulated rates for different substrate concentrations as a function of inhibitor concentration. Substrate concentrations: 10 μm (circles), 50 μm (triangles), 100 μm (diamonds), 250 μm (squares). −Ki value of inhibition was estimated based on the intersection of linear regression. D, TLC analysis of 32P-labeled total lipids from E. coli UE54 strain, visualized by a phosphorimaging system. E, [32P]PE pulldown assay using the PgpB protein (circles) on a Ni-NTA resin as a function of POPE concentration. Empty resin (triangles) was used as control. The bound PE ligand was quantified by a scintillation counter.
To directly measure PE interaction, we also performed a ligand pulldown assay with the purified PgpB protein anchored on Ni2+-NTA resin. We generated 32P-labeled PE from E. coli strain UE54 (13) (Fig. 2C). Based on radioactivity, the binding of PgpB with PE increased as a function of lipid concentration (Fig. 2D). In the presence of 4 mm PE, 7% (∼700 cpm) of the added [32P]PE (total ∼10,000 cpm) was associated with the PgpB protein compared with only 0.6% (∼60 cpm) bound to the control empty resin lacking PgpB, indicating specific interaction of PE with PgpB and also lending support to the evidence for PE as an in vitro PgpB inhibitor.
Structural Determination of the PgpB-PE Complex
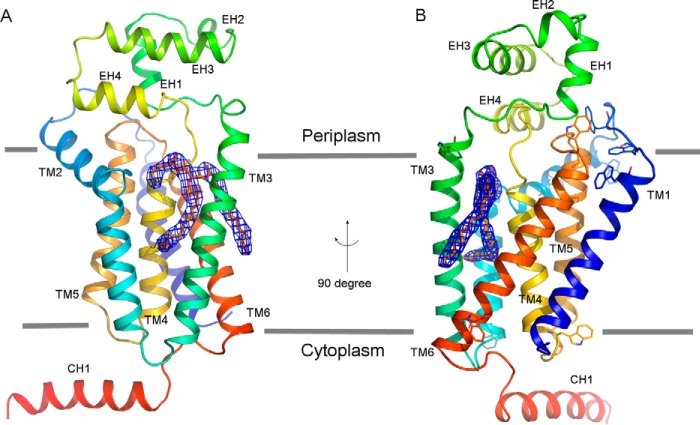
Identification of PE as a specific in vitro inhibitor provided an opportunity to study the lipid substrate binding mechanism of PgpB. Attempts to crystallize PgpB in the presence of synthetic POPE were unsuccessful. We were able to crystallize the PgpB protein using the vapor diffusion method in the presence of 5 mm E. coli total lipid extract, which contained ∼3 mm PE. The structure was determined at 3.2 Å resolution using the SAD (single wavelength anomalous dispersion) method (Table 1). Although the PgpB lipid complex was crystallized under different conditions from that used for the previous apo form (12), both structures exhibited nearly identical overall conformations with an root mean square deviation value of 0.9 Å. However, in the PgpB lipid complex, a strong and lipid-like electron density was observed in the central cavity assembled by six TM helices (see below) in the Fo − Fc difference map. A PE molecule (1,2-dilauroyl-sn-glucero-3-phosphoethanolamine) fits well in the density region. We excluded other possibilities such as PG, cardiolipin, DDM or PEG6000, which were also present in the crystallization solution due to the following facts: 1) their headgroups did not fit in the density and 2) no noticeable inhibition by these additives was observed under their crystallization conditions (Fig. 2A). The electron density shown in Fig. 3, A and B, represents the final PE conformation after structural refinement using Refmac. Both the PE headgroup and Sn2 tail exhibit similar thermal B values to those of their neighboring peptides (∼90), whereas the B value of the Sn1 tail is 20% higher, perhaps due to its localization on the protein surface (see Fig. 5A and also see below). The final structural model contains the full-length sequence (amino acid residues 1–254) together with the C-terminal His6 tag and a PE molecule.
TABLE 1.
Statistics of data collection and structural refinement
| Data collection | |
| Wavelength (Å) | 0.9794 |
| Resolution range (Å) | 132.1-3.2 (3.314-3.2) |
| Space group | P 21 21 21 |
| Unit cell | 39.5, 73.6, 132.1 |
| Total reflections | 95,776 (9,665) |
| Unique reflections | 6,800 (664) |
| Multiplicity | 14.1 (14.6) |
| Completeness (%) | 99.9 (100.0) |
| Mean I/σ(I) | 27.8 (4.5) |
| Wilson B-factor | 89.1 |
| R-merge | 0.102 (0.778) |
| R-measured | 0.109 |
| Structural refinement | |
| R-work | 0.267 (0.344) |
| R-free | 0.305 (0.376) |
| Number of non-hydrogen atoms | 2,149 |
| Macromolecules | 2,104 |
| Ligands | 43 |
| Water | 2 |
| Root mean square (bonds) | 0.008 |
| Root mean square (angles) | 1.28 |
| Ramachandran favored (%) | 93 |
| Ramachandran allowed (%) | 7 |
| Average B-factor | 94 |
| Macromolecules | 94 |
| Ligands | 95.5 |
| Solvent | 52.2 |
FIGURE 3.
Overall structure of the PgpB-PE complex. A, front view in the membrane plane. B, side view rotated in a 90° direction. The PgpB protein drawn as rainbow schematic contains a TM domain formed by TMs 1–6, an extracellular domain formed by EHs 1–4, and an intracellular helix CH1. PE lipid depicted as yellow sticks is superimposed with the 2Fo − Fc electron density map (blue meshes) contoured at 1.0 σ. The membrane boundaries (gray lines) are estimated from the positions of the multiple tryptophan residues (sticks) on both membrane surfaces.
FIGURE 5.
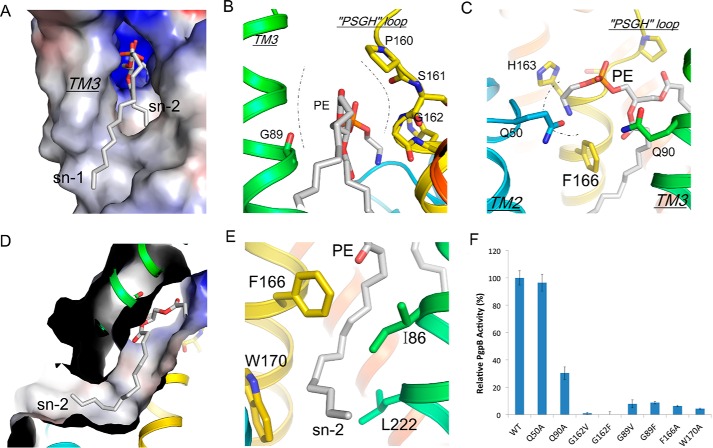
PE conformation in the TM3 tunnel. A, electrostatic surface representation of the TM3 tunnel with PE (sticks). B, TM3 tunnel is constructed by TM3 (green) and PSGH sequence (yellow). The tunnel neck is drawn as dashed lines. C, the headgroup conformation of bound PE in the front of the catalytic site. D, a cut-through view of the electrostatic surface shows the Sn2 tail of PE inside the acyl chain pore. E, the conformation of the pore-lining residues stabilizes the PE Sn2 tail (gray). F, characterization of PgpB mutations in the TM3 tunnel or the front V-shaped cavity using 18:1 LPA as substrate. All activities were normalized to that of wild type.
Overall Conformation of PgpB
Monomeric PgpB contains a transmembrane domain, an extracellular domain, and an intracellular helix CH1 (Fig. 3A). Six TM helices (TMs 1–6) assemble the integral membrane domain with both N and C termini in the cytoplasm. The extracellular domain formed by four short helices (EHs 1–4) between TMs 3 and 4 of the PgpB molecule tilts by 65° in the bilayer based on the membrane boundaries determined by the positions of 10 tryptophan residues from different TMs (Fig. 3B).
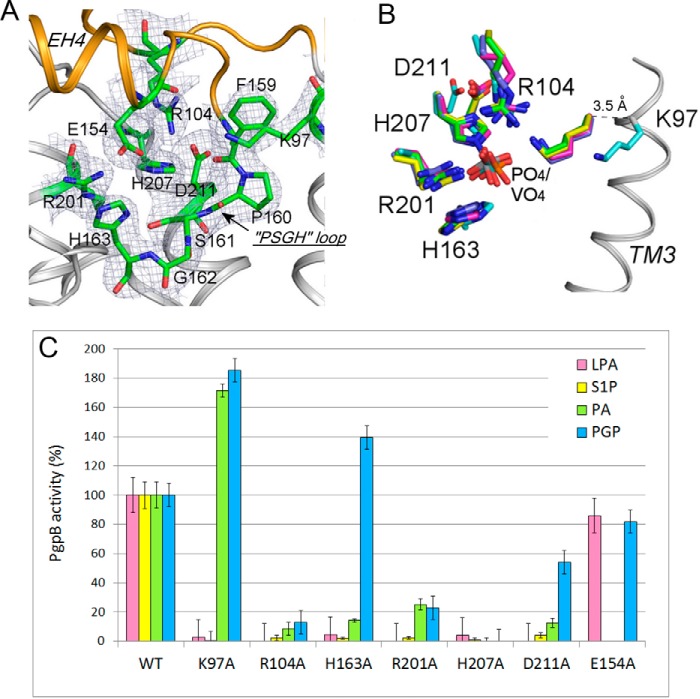
The catalytic site is located in a positively charged cavity on the interface between the membrane domain and the periplasmic domain (Fig. 3B). Six conserved residues in the C1-C3 motifs essential for LPPs' activity are assembled together to form a catalytic site (Fig. 4A). Three conserved residues, Lys-97, Arg-104, and Arg-201, in a triangular orientation, create a phosphate binding site. His-163, His-207, and Asp-211 form the catalytic triad aligning across the phosphate binding site.
FIGURE 4.
Conformation of the PgpB catalytic site. A, the catalytic site of PgpB on the interface between the extracellular domain (orange) and the TM domain (gray). The residues in the catalytic site shown as green sticks are superimposed with the electron density map (2Fo − Fc contoured at 1σ). B, structural similarity of the catalytic motifs. The PgpB structure (cyan) was superimposed with those of soluble phosphatase proteins bound with phosphate or vanadate: vanadium chloroperoxidase from Curvularia inaequalis (green), vanadium-dependent bromoperoxidase from Ascophyllum nodosum (yellow), nonspecific acid phosphatase from Salmonella typhimurium (blue), acid phosphatase from Escherichia blattae (magenta). C, characterization of PgpB mutations in the catalytic site using four different substrates, LPA, S1P, PA, and PGP (except Glu-154 tested with LPA and PGP only). The activity was represented as a percentage compared with the wild type.
PE Binding Conformation
In both the apo and PE-bound forms, TM3 is angled away from the other five TM helices, creating a V-shaped groove beneath the active site and a membrane-embedded tunnel (referred to as the TM3 tunnel) (Fig. 3B and 5A) (12). The tunnel is narrow, ∼4 Å in diameter, estimated by MOLE and is mainly constrained by a loop consisting of four residues (PSGH) that comprise the LPP fingerprint sequence (Fig. 5B).
In the lipid-bound structure, PE was found in the tunnel, which was sandwiched between TM3 and TMs 4 and 6 (Figs. 3B and 5A). The ethanolamine headgroup was stabilized in a three-walled cage formed by His-163, Gln-50, and Phe-166 in front of the catalytic site via cation-π interactions (Fig. 5C). A similar PE interaction was previously reported in the structure of cytochrome c oxidase (14). The H-bond interaction between the amine group of PE and the side-chain oxygen atom of Gln-50 stabilized the whole Gln-50 residue, which was invisible in the apo form structure (an alanine was assigned at this position instead) (12). In the tunnel, the PE glycerol group is constrained by two glycine residues, Gly-89 from TM3 and Gly-162 from the PSGH loop (Fig. 5B). The PE Sn2 tail deeply inserts into a narrow pore (referred to as the acyl pore) with its tip reaching out of the front TM2–3 groove (Fig. 5D). The acyl pore is formed by several hydrophobic residues including Leu-82, Ile-86 from TM3, Phe-166 and Trp-170 from TM4, and Leu-222 from TM6 (Fig. 5E). It should be noted that the PE molecule and its adjacent structural components are not involved in crystal packing.
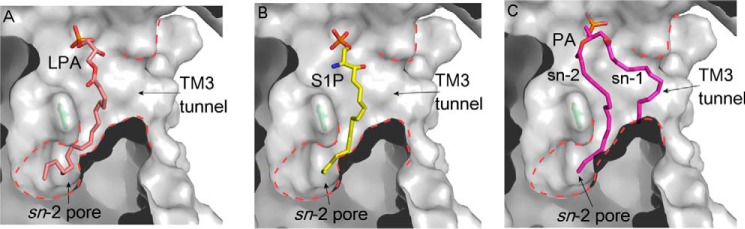
Docked Substrate Conformations
The PE conformation provides insight into the PgpB substrate binding mechanism. As a further test, we examined substrate docking using the AutoDock program (15). As seen in Fig. 6, LPA, S1P, and PA are stabilized in the TM3 tunnel in a conformation strikingly similar to that of PE observed in the structure. Interestingly, in all three docking models, one fatty acyl chain, either the Sn1 tail of LPA or S1P or the Sn2 tail of PA, was found in the acyl pore, suggesting that the pore is energetically favorable for fatty acyl chain binding. This finding also explained results from the PgpB kinetic study showing that a long fatty acyl chain is important for substrate binding (Fig. 1, C and D). In contrast to PE, these lipid phosphates positioned their phosphor headgroups toward the catalytic center in proximity to the predicted phosphate binding position.
FIGURE 6.
Substrate docking conformations. A, LPA (pink sticks). B, S1P (yellow sticks). C, PA (magenta sticks). Surface cut-through views show substrate conformations inside the TM3 tunnel, and the acyl pore is highlighted using red dashed lines.
The TM3 Tunnel Is Important for PgpB Catalysis
To gain further insight into the substrate binding mechanism in the TM3 tunnel, we first tested the effect of blocking substrate tunnel access by placing bulky residues, either valine or phenylalanine, at the Gly-89 or Gly-162 positions in the tunnel neck. As expected, all four mutations (G89V, G89F, G162V and G162F) completely abrogated PgpB activity. In addition, acyl chain binding in the acyl pore appeared to be important for substrate binding. We tested this by mutating two aromatic pore-lining residues. Phe-166 interacted with the C1-C6 atoms, whereas Trp-170 reached the C10-C12 atoms at the front exit (Fig. 5E). Both alanine substitutions at these two positions resulted in a dramatic reduction (>90%) of C18-LPA catalysis (Fig. 4F), supporting our hypothesis.
In addition to the TM3 tunnel, the structure suggested that the active site may be accessed from the front V-shaped groove (Fig. 3A), although the tilting orientation of PgpB would argue against substrate lateral diffusion from the membrane (Fig. 3B). Previously, Fan et al. (12) showed that mutations on the groove surface largely preserved PgpB activity. To carefully exclude this access possibility, we mutated two glutamine residues, Gln-50 from TM2 and Gln-90 from TM3, at the entrance of the groove (Fig. 5C). Q50A was as fully active as the wild type, and Q90A maintained >30% activity (Fig. 4F). Taken together, these results do not support this alternative substrate accessing route.
The Catalytic Triad Is Not Required for PGP Activity
PgpB was thought to use a common acidic phosphatase mechanism requiring all six residues in the active site for hydrolysis of different lipid substrates. This hypothesis was tested using substrates LPA, PA, and DGPP (12). To study the PGP catalytic mechanism and substrate specificity, we generated six single alanine mutants in the active site and tested them with a new set of four lipid phosphates including LPA, S1P, PA, and PGP (Fig. 4C). Our results using LPA, S1P, and PA are consistent findings from the previous study by Fan et al. (12) that the entire catalytic triad (His-163, His-207, and Asp-211) are crucial for dephosphorylation of these substrates. Surprisingly, this was not the case for PGP. Results shown in Fig. 4C show that His-207 appears to be the only residue essential for hydrolysis of PGP. In fact, mutation H163A increased PGP hydrolysis by 50%, and mutant D211A retained 50% activity. Therefore, PgpB may utilize a unique catalytic mechanism for PGP dephosphorylation.
In many histidine phosphatases, the dephosphorylation reaction is mediated by a catalytic dyad composed of a histidine as an H+ acceptor and another carboxylate residue as an H+ donor (16). On the periplasmic EH4 helix, the carboxylate group of Glu-154 is closed (4.5 Å) to the imidazole ring of His-163. However, mutation of Glu-154 to alanine retained ∼80% activity for both LPA and PGP, excluding it as an alternative H+ donor in the PGP reaction.
Selectivity for Lyso Versus Diacyl Substrates
Substrate discrimination was also observed among the conserved triple Lys/Arg residues in the active site. Mutation of R104A or R201A inside the active site was lethal for all four substrates. But Lys-97 located at the cavity entry was only required for the lyso forms LPA and S1P. In fact, K97A enhanced the activity for diacyl PA (instead of the complete activity loss observed in the Fan et al. (12) study) and for PGP by >70%, indicating its distinctive role in the catalysis of lyso versus diacyl substrates.
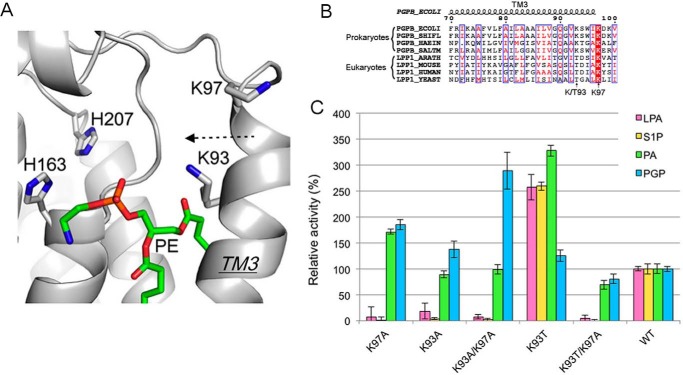
The novel acyl chain selectivity of K97A drew our further attention to the TM3 helix. Adjacent to Lys-97, another lysine residue, Lys-93, is also located near the active site (Fig. 7A). Interestingly, similar to K97A, the mutant K93A also showed complete activity loss for LPA and S1P, whereas it retained full activity for PA and gained 40% more activity toward PGP compared with wild type (Fig. 7C). To probe any functional cross-talk between these two adjacent lysine residues, we generated a double mutant, K93A/K97A. The double mutant exhibited a similar selective activity change as seen with single mutants K93A and K97A. Furthermore, the double alanine mutant further enhanced PGP catalysis by nearly 3-fold (Fig. 7C). Those results indicated that both lysine residues are absolutely required for lyso form substrates, whereas they do not appear to be involved in catalysis of diacyl form substrates. Given that these substrate selectivity properties occur at two positions in TM3, these results further support our structural observation that TM3 is important for substrate binding.
FIGURE 7.
Role of Lys-93 and Lys-97 on substrate selectivity. A, the conformation of Lys-93 and Lys-97 on TM3. PgpB is shown as a gray schematic. PE is depicted as green sticks. B, sequence alignment of TM3 from prokaryotic PgpBs (E. coli, Shigella flexneri, Haemophilus influenza, and Salmonella) and eukaryotic LPP1s (Arabidopsis thaliana, Mus musculus, Homo sapiens, and Saccharomyces cerevisiae). Conserved amino acids are white in red-filled rectangles. Identical residues are in red surrounded by blue lines. C, characterization of PgpB mutations at the Lys-93 and Lys-97 positions using four types of lipid phosphates. Activities were normalized to that of wild type.
Selectivity for Prokaryotic Versus Eukaryotic Substrates
Lys-97 is highly conserved in the LPP family, but evolutionary divergence clearly appears at the Lys-93 position (Fig. 7B). Lys-93 is conserved in prokaryotes, but a threonine residue was found at the analogous position in all eukaryotic LPPs. To further explore the specific role of Lys-93, we mutated Lys-93 to threonine (Fig. 7C). Strikingly, K93T significantly improved activity for LPA, S1P, and PA by 2.5- or 3.4-fold but not for PGP. LPA, S1P, and PA are the major substrates of eukaryotic LPPs. In eukaryotic cells, PGP dephosphorylation is actually catalyzed by a different phosphatase enzyme Gep4 localized in mitochondria (17). Because K93T boosted overall PgpB activity, we questioned if K93T alone is sufficient to maintain PgpB activity. The double mutant K93T/K97A lost all activity for LPA or S1P but retained 50% activity for PA or PGP. These results further confirmed the critical role of Lys-97 for lyso substrates and suggested that the residue at Lys-93 serves as a tuner to adjust LPP activity for individual substrates.
Discussion
PgpB is an intramembrane lipid phosphatase protein promoting PG biosynthesis in bacteria. We identified a novel in vitro PE inhibition and determined the structure of the PgpB PE-bound form. The PE binding conformation together with substrate in silico docking and mutagenesis studies provides new insights into the mechanisms of lipid substrate recognition and catalysis of prokaryotic PgpB and other LPPs in eukaryotes.
Potential Role of PE on PgpB
The in vitro inhibitory effect of PE also raised a fundamental question about its function in vivo as PgpB is an integral membrane protein embedded in the inner membrane, which contains 70% PE. In the apo form structure, the hydrophobic TM3 tunnel is largely open toward the membrane bilayer. Whether PgpB rests as a lipid-occluded state in the membrane is unknown as all in vitro studies were carried out using detergent-delipidized protein.
Lipid is important for the stability of the PgpB protein. Fan et al. (12) has shown that adding lipid substrates with a long-chain (C18) but not a short-chain (C8) enhanced thermal stability of PgpB. In addition, PgpB could be stabilized substantially by adding MoO42− in the protein solution (12). It was expected that MoO42− binding at the active site draws TM3 to close the TM3 tunnel. These data collectively implied that a lipid-occluded TM3 tunnel is energetically unfavorable and may thus require lipid association in the bilayer. Zwitterionic PE usually increases the stability of the folded state of membrane proteins (18). Conversely, an excess of negatively charged lipids such as PG could severely destabilize a transmembrane helix bundle structure (19). Therefore, the PE-bound form demonstrated in this work may represent the resting state of PgpB stabilized in the cell membrane in vivo.
Several lines of evidences have suggested that accommodation of a defined lipid species within a membrane protein might allow the activity of the protein to be controlled by altering its stability (20, 21). In the case of PgpB, the substrate binding affinity is significantly higher (∼20-fold) than the Ki value for PE that shows only partial inhibition even at high PE concentrations (Figs. 1D and 2B). The modest inhibition is perhaps due to the fact that the bulky phosphoethanolamine headgroup cannot enter the phosphate binding site (Figs. 5C and 6). A resting conformation would allow efficient replacement of PE with lipid phosphate substrates in the active state. By balancing stability and activity, the optimal existence and functioning of PgpB may be reached in the lipid environment.
Although the physiological importance of the anionic lipids PG and CL has been recognized, the regulation of a key enzyme for their biosynthesis, the phosphatase that converts PGP to PG, remained obscure. PE regulation may allow PgpB activity to remain at a physiologically required threshold. PE can dissociate as soon as its concentration drops. The level of PE can be altered at different growth phases, and its accumulation could shift depending on the stage of the cell cycle (22). These events could contribute to regulation of anionic and zwitterionic glycerophospholipid synthesis during cell growth. The elevation of PE levels would lead to an increased level of PGP coincident with its use as a phosphorylated precursor in the synthesis of PG and CL (1). Thus, PE could be crucial for controlling the activity of PgpB via its dependence on the presence of an amount of PE required for stabilizing this protein. Changes in PE concentration can also be produced by the action and regulation of several lipid enzymes (phosphatidylserine synthase, phosphatidylserine decarboxylase, and phospholipase A) that are accordingly predicted to affect the activity of PgpB during different growth phases via the maintenance of appropriate PG, CL, and PE levels.
Substrate Binding Mechanism: Lyso Versus Diacyl Lipids
Our structural and functional analyses suggested that the TM3 tunnel is the major lipid substrate binding site. Lateral diffusion into the tunnel from the membrane entry between TMs 3 and 6 provides a convenient access to the catalytic site (Fig. 3B). Stabilizing a long acyl chain(s) in the tunnel is crucial not only for lipid substrate binding (Km) but also for the catalytic rate (Vmax) as seen in the kinetic analyses of 18:1 versus 6:0 LPA (Fig. 1, C and D). Based on the structure and docking model, 6:0 LPA might not be able to be stabilized in the tunnel due to its short length. Instead, phosphor binding in the active site would pull its short acyl chain out of the tunnel to a position in the front of the catalytic cavity. As expected, a flexible glycerol-acyl chain would unavoidably interfere with the His-163 side chain at the entry, affecting proton transfer by the catalytic triad evidenced by the dramatic drop in the rate. Most natural bacterial lipids have a C16-C18 acyl chain. By stabilizing the long acyl chain in the tunnel, an optimal lipid catalysis can be achieved. Our data also revealed an important role of Lys-97 for lyso substrates (Fig. 7). In the LPP catalytic mechanism, Lys-97 was expected to stabilize the phosphor headgroup (5). As seen in many acid phosphatase and haloperoxidase structures (23–25), the analogous residues of Lys-97, Arg-104, and Arg-201 coordinate a phosphate/vanadate ion in the catalytic site (Fig. 4B). In contrast to other residues found in nearly identical positions, Lys-97 is 3.5 Å away from the ligand-bound state of its counterparts. To form proper phosphor coordination, Lys-97 may need to move toward the catalytic center. Phosphor binding would inevitably draw TM3 toward the PSGH loop to constrain the TM3 tunnel, in turn stabilizing the slim lyso substrate in the tunnel (Fig. 6, A and B). This conformational change may require a joint effort with Lys-93 by interacting with the glycerol backbone of LPA or the sphingoid base of S1P to enhance the stability of the lysolipid in the tunnel.
However, this motion may not be required or may even be unfeasible for diacyl substrates due to the fact that their two acyl chains simultaneously enter into the TM3 tunnel (Fig. 6C). In the PE complex structure, the glycerol backbone of PE actually blocked the tunnel neck (Fig. 4B). The full occupancy is probably sufficient to stabilize the substrate in the tunnel, as indicated by the dispensability of Lys-97 for diacyl substrates. Such a conformational adaptation induced by substrate binding allows both substrate diversity and optimal catalysis to be achieved. Determination of any protein motion with lyso versus diacyl lipids would provide a test for the induced-fit mechanism.
PGP-specific Catalytic Mechanism
Both Fan et al. (12) and our studies support a common LPP catalytic mechanism (11) for PgpB catalysis of LPA, S1P, PA, or DGPP. The catalytic triad Asp-211, His-207, and His-163 establishes a charge relay system; 1) D201 stabilizes His-207 for a nucleophilic attack to form a phosphohistidine intermediate with the substrate; 2) His-163 at the tunnel entry facilitates hydrolysis of the intermediate to release the phosphate ion to the periplasm and the dephosphorylated lipid back to the membrane layer.
However, this mechanism may be inapplicable to PGP as both Asp-211 and His-163 are not required for its dephosphorylation (Fig. 4C). But the essential role of His-207, which is located deeply inside the active site, indicates that the PGP reaction takes place in the same catalytic site. PGP has an additional phosphoglycerol moiety compared with PA (Fig. 1A). Attempts to dock PGP into the structure did not lead to any meaningful binding information, arguing that the protein may undergo a conformational change to accommodate the large PGP headgroup. Based on the structure, this could be achieved by moving the imidazole ring of His-163 away from the catalytic site entry (Fig. 4A). The fact that the H163A mutation improved PGP hydrolysis supports this idea (Fig. 4C). Depositioning the side chain of His-163 would probably disrupt the existing charge-relay wire. Therefore, an alternative mechanism has to be established for PGP hydrolysis.
The search for an alternative proton transfer route on the protein was not successful. Specific PGP binding conformations may allocate a water molecule as a H+ donor to initialize hydrophilic attack given the surface location of the active site. Whether PGP itself is involved in its own catalysis is unknown. In the mechanism of phosphoglycerate mutase, the phosphor-isomerization reaction is mediated by a single histidine residue and the substrate itself (26). PGP is significantly different from other lipid phosphate substrates. A bulky phosphoglycerol moiety as a replacement for His-163 at the catalytic entry may serve as an alternative H+ donor to deliver a H+ to the terminal phosphor group given their adjacent positions (Fig. 1A). A self-catalysis mechanism might explain the specificity of the PGP reaction due to its unique chemical structure.
Experimental Procedures
Materials
Phospholipids used in the PgpB assay including 18:1 LPA (1-(9Z-octadecenoyl)-sn-glycero-3-phosphate), 6:0 LPA (1-hexanoyl-sn-glycero-3-phosphate), S1P ((2S,3R,4E)-2-aminooctadec-4-ene-1,3-diol-1-phosphate), PA (1,2-dihexadecanoyl-sn-glycero-3-phosphate), POPE (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine), POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol), DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine) CL, and glycerol 3-phosphate were purchased from Cayman, Avanti Polar Lipids, or Sigma. PGP (1,2-dipalmitoryl-sn-glycero-3-[phospho-rac-(1′-(3′-phospho)glycerol)]) was custom-synthesized by Avanti Polar Lipids. DDM is a product of Anatrace. [32P]Orthophosphate was purchased from MP Biomedicals.
Protein Expression and Purification
The E. coli pgpB gene was cloned into the pET22b vector (Novagen) to generate the C-terminal His6-tagged protein. Mutants were generated using standard site-directed mutagenesis. The PgpB wild type and mutant proteins were expressed in E. coli strain C41(DE3) grown in autoinduction medium (27) at 37 °C for 2 h followed by a 20-h incubation at 25 °C. To express the selenomethionine derivative, autoinduction minimal medium (28) was used. Harvested cells were suspended in lysis buffer containing 50 mm Tris-HCl, pH 7.5, 500 mm NaCl, and 10% glycerol and then ruptured using an EmulsiFlex-C3 homogenizer (Avestin) at 15,000 p.s.i. Cell debris was removed by centrifugation. The supernatant was ultracentrifuged at 150,000 × g for 1 h. Membrane fractions were suspended in the lysis buffer and then incubated with 1% (w/v) DDM for 1.5 h at 4 °C. After another ultracentrifugation at 150,000 × g for 30 min, the supernatant was loaded on a Ni2+-NTA affinity column (GE Healthcare). The resin was washed with lysis buffer containing 40 mm imidazole. The protein was eluted with a buffer containing 50 mm Tris-HCl, pH 7.5, 500 mm NaCl, 400 mm imidazole, and 0.05% DDM. The protein was further purified using a size exclusion Superdex-200 GL column (GE Healthcare) equilibrated in a buffer of 20 mm Tris-HCl, pH 7.5, 200 mm NaCl, and 0.03% DDM. The peak fraction was directly used for in vitro assay or concentrated to 8 mg ml−1 for crystallization.
In Vitro Colorimetric Assay
Lipid phosphatase assay was carried out in vitro using a standard phosphate colorimetric approach (29). Briefly, the colorimetric reagent was prepared by mixing 34 mm ammonium molybdate in 5 m hydrochloric acid with 2.16 mm malachite green oxalate in a ratio of 1:3 (v/v) at room temperature and filtered before use. All substrates were dissolved in the reaction buffer containing 20 mm Tris-HCl, pH 7.5, 200 mm NaCl, 4% glycerol, and 0.5% DDM before use.
Each assay was carried out in a 100-μl reaction mixture containing the appropriate amount of purified PgpB proteins at room temperature so that no more than 30% of the substrates was hydrolyzed during the reaction. The reaction was terminated by the addition of 200 μl of the colorimetric reagent. 30 μl of 1% polyvinyl alcohol was then added to the reaction to stabilize the phosphate complex. The reaction mixture was measured spectroscopically at 660 nm. The free phosphate concentration was calculated based on a standard curve measured with potassium phosphate.
Preparation of 32P-Labeled PE
Radioactive PE was made biosynthetically in E. coli strain UE54 (MG1655 lpp-2Δara714 rcsF::mini-Tn10 cam pgsA::FRT-kan-FRT) in LB medium containing 5 μCi ml−1 [32P]orthophosphate. This strain lacks the major anionic phospholipids PG and CL, accumulating PE to 95% of total phospholipids in deep stationary grown cells (13). The cells from 50 ml of culture were harvested, and the lipids were extracted as described. The purity of 32P-labeled PE was verified by TLC on Silica Gel G thin-layer plates developed with chloroform/methanol/ammonia/H2O (60:37.5:1:3 v/v) and then visualized by a phosphorimaging system.
In Vitro PE Binding Assay
250 μg of PgpB protein was incubated with 50 μl of Ni2+-NTA affinity resin for 1 h. Unbound proteins were removed by washing with 20 mm Tris-HCl, pH 7.5, buffer containing 100 mm NaCl, 0.03% DDM, and 40 mm imidazole. 32P-Labeled PE prepared above was dried, solubilized in 0.3% DDM, and then diluted 10× as in binding assay. About 10,000 cpm of 32P-labeled PE premixed with the indicated concentrations of POPE was incubated with the resin for 10 min. The resulting resin was washed with 500 μl of the washing buffer 3 times and then eluted with a buffer supplemented with 400 mm imidazole. Radioactivity of the eluate was measured by liquid scintillation spectrometry to assess lipid-protein interaction. A resin without the PgpB protein was used as the control.
Crystallization of the PgpB-Lipid Complex
An E. coli total lipid extract (Avanti Polar Lipids) in chloroform was dried in an argon gas flow and resuspended in 1.25% DDM at a concentration of ∼30 mm. The selenomethionine-substituted PgpB protein premixed with 5 mm (final concentration) of the lipid extract was crystallized using the sitting-drop vapor diffusion method at 20 °C. The crystallization buffer contained 0.1 m N-carbamoylmethyl iminodiacetic acid, pH 6.5, and 10% polyethylene glycol 6000. Crystals were soaked in a cryo-protectant solution supplemented with 30% glycerol and then directly flash-frozen under liquid nitrogen. Note that crystallization and structural determination were done before the apo form structure was published; therefore, selenomethionine-derivative protein was used in the crystallization.
Structural Determination
Crystal diffraction data were collected at 100 K at the Swiss Light Source beamline X06SA (SLS, Villigen, Switzerland). Data processing was carried out using the programs XDS (30) and SCALA (31). The phases were obtained using the single wavelength anomalous dispersion (SAD) method by Autosol in the PHENIX software suite (32). The PgpB model was initially generated by the PHENIX Autosolution program and then completed manually in Coot (33). The PE ligand was carefully built based on the electron density map. The structure refinement was performed using Refmac (34, 35). All residues fell into the favorable and allowable region of the Ramachandran plot. The statistics of data collection and refinement are shown in Table 1. The atomic coordinates of the PgpB-PE complex structure has been deposited in the Protein Data Bank (entry code 5JWY). All structural figures were prepared using graphic programs PyMOL (35).
Author Contributions
S. T., Y. L., S. L., M. B., M. W., and L. Z. designed the study and performed the experiments. S. T., M. B., and L. Z. wrote the paper. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank William Dowhan for providing the UE54 strain, the staff of the Swiss Light Source for diffraction data collection and Julia Lever for comments on manuscript.
This work was supported by National Institute of General Medical Sciences Grants R01GM098572 and R01GM097290 (to L. Z.). This work was also supported by American Heart Association postdoctoral fellowships (to S. T.) The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The atomic coordinates and structure factors (code 5JWY) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- PG
- phosphatidylglycerol
- PGP
- 1,2-dipalmitoryl-sn-glycero-3-[phospho-rac-(1′-(3′-phospho)glycerol)]
- PgpB
- phosphatidylglycerol phosphatase
- LPP
- lipid phosphate phosphatase
- TM
- transmembrane helix
- PE
- phosphatidylethanolamine
- POPE
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- POPG
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine
- CL
- cardiolipin
- 18:1 LPA
- 1-(9Z-octadecenoyl)-sn-glycero-3-phosphate
- 6:0 LPA
- 1-hexanoyl-sn-glycero-3-phosphate
- S1P
- (2S,3R,4E)-2-aminooctadec-4-ene-1,3-diol-1-phosphate
- PA
- 1,2-dihexadecanoyl-sn-glycero-3-phosphate
- DGPP
- diacylglycerol pyrophosphate
- DDM
- n-dodecyl β-d-maltoside
- Ni2+-NTA
- nickel-nitrilotriacetic acid.
References
- 1. Lu Y. H., Guan Z., Zhao J., and Raetz C. R. (2011)Three phosphatidylglycerol-phosphate phosphatases in the inner membrane of Escherichia coli. J. Biol. Chem. 286, 5506–5518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Icho T., and Raetz C. R. (1983) Multiple genes for membrane-bound phosphatases in Escherichia coli and their action on phospholipid precursors. J Bacteriol, 153, 722–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Touzé T., Blanot D., and Mengin-Lecreulx D. (2008) Substrate specificity and membrane topology of Escherichia coli PgpB, an undecaprenyl pyrophosphate phosphatase. J. Biol. Chem. 283, 16573–16583 [DOI] [PubMed] [Google Scholar]
- 4. Dillon D. A. (1996) The Escherichia coli pgpB gene encodes for a diacylglycerol pyrophosphate phosphatase activity. J. Biol. Chem. 271, 30548–30553 [DOI] [PubMed] [Google Scholar]
- 5. Sigal Y. J., McDermott M. I., and Morris A. J. (2005) Integral membrane lipid phosphatases/phosphotransferases: common structure and diverse functions. Biochem. J. 387, 281–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brindley D. N., and Pilquil C. (2009) Lipid phosphate phosphatases and signaling. J. Lipid Res. 50, S225–S230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tanyi J. L., Hasegawa Y., Lapushin R., Morris A. J., Wolf J. K., Berchuck A., Lu K., Smith D. I., Kalli K., Hartmann L. C., McCune K., Fishman D., Broaddus R., Cheng K. W., and Atkinson E. N. (2003) Role of decreased levels of lipid phosphate phosphatase-1 in accumulation of lysophosphatidic acid in ovarian cancer. Clin Cancer Res. 9, 3534–3545 [PubMed] [Google Scholar]
- 8. Escalante-Alcalde D., Hernandez L., Le Stunff H., Maeda R., Lee H. S., Jr-Gang-Cheng, Sciorra V. A., Daar I., Spiegel S., Morris A. J., and Stewart C. L. (2003) The lipid phosphatase LPP3 regulates extra-embryonic vasculogenesis and axis patterning. Development 130, 4623–4637 [DOI] [PubMed] [Google Scholar]
- 9. Mandala S. M., Thornton R., Galve-Roperh I., Poulton S., Peterson C., Olivera A., Bergstrom J., Kurtz M. B., and Spiegel S. (2000) Molecular cloning and characterization of a lipid phosphohydrolase that degrades sphingosine-1-phosphate and induces cell death. Proc. Natl. Acad. Sci. U.S.A. 97, 7859–7864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pierrugues O. (2001) Lipid phosphate phosphatases in Arabidopsis: regulation of the AtLPP1 gene in response to stress. J. Biol. Chem. 276, 20300–20308 [DOI] [PubMed] [Google Scholar]
- 11. Stukey J., and Carman G. M. (1997) Identification of a novel phosphatase sequence motif. Protein Sci. 6, 469–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fan J. (2014) Crystal structure of lipid phosphatase Escherichia coli phosphatidylglycerophosphate phosphatase B. Proc. Natl. Acad. Sci. U.S.A. 111, 7636–7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shiba Y., Yokoyama Y., Aono Y., Kiuchi T., Kusaka J., Matsumoto K., and Hara H. (2004) Activation of the Rcs signal transduction system is responsible for the thermosensitive growth defect of an Escherichia coli mutant lacking phosphatidylglycerol and cardiolipin. J. Bacteriol. 186, 6526–6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shinzawa-Itoh K., Aoyama H., Muramoto K., Terada H., Kurauchi T., Tadehara Y., Yamasaki A., Sugimura T., Kurono S., Tsujimoto K., Mizushima T., Yamashita E., Tsukihara T., and Yoshikawa S. (2007) Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 26, 1713–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Trott O., and Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rigden D. J. (2008) The histidine phosphatase superfamily: structure and function. Biochem. J. 409, 333–348 [DOI] [PubMed] [Google Scholar]
- 17. Osman C., Haag M., Wieland F. T., Brügger B., and Langer T. (2010) A mitochondrial phosphatase required for cardiolipin biosynthesis: the PGP phosphatase Gep4. EMBO J. 29, 1976–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meijberg W., and Booth P. J. (2002) The activation energy for insertion of transmembrane α-helices is dependent on membrane composition. J. Mol. Biol. 319, 839–853 [DOI] [PubMed] [Google Scholar]
- 19. Hong H., and Bowie J. U. (2011) Dramatic destabilization of transmembrane helix interactions by features of natural membrane environments. J. Am. Chem. Soc. 133, 11389–11398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Valiyaveetil F. I., Zhou Y., and MacKinnon R. (2002) Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry 41, 10771–10777 [DOI] [PubMed] [Google Scholar]
- 21. Laganowsky A., Reading E., Allison T. M., Ulmschneider M. B., Degiacomi M. T., Baldwin A. J., and Robinson C. V. (2014) Membrane proteins bind lipids selectively to modulate their structure and function. Nature 510, 172–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mozharov A. D., Shchipakin V. N., Fishov I. L., and Evtodienko Y. uV. (1985) Changes in the composition of membrane phospholipids during the cell cycle of Escherichia coli. FEBS Lett. 186, 103–106 [DOI] [PubMed] [Google Scholar]
- 23. Messerschmidt A., and Wever R. (1996) X-ray structure of a vanadium-containing enzyme: chloroperoxidase from the fungus Curvularia inaequalis. Proc. Natl. Acad. Sci. U.S.A. 93, 392–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weyand M. (1999) X-ray structure determination of a vanadium-dependent haloperoxidase from Ascophyllum nodosum at 2.0 Å resolution. J. Mol. Biol. 293, 595–611 [DOI] [PubMed] [Google Scholar]
- 25. Makde R. D., Mahajan S. K., and Kumar V. (2007) Structure and mutational analysis of the PhoN protein of Salmonella typhimurium provide insight into mechanistic details. Biochemistry 46, 2079–2090 [DOI] [PubMed] [Google Scholar]
- 26. Winn S. I. (1981) Structure and activity of phosphoglycerate mutase. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 293, 121–130 [DOI] [PubMed] [Google Scholar]
- 27. Studier F. W. (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 28. Sreenath H. K., Bingman C. A., Buchan B. W., Seder K.D., Burns B. T., Geetha H. V., Jeon W. B., Vojtik F. C., Aceti D. J., Frederick R. O., Phillips G. N. Jr., and Fox B. G. (2005) Protocols for production of selenomethionine-labeled proteins in 2-L polyethylene terephthalate bottles using auto-induction medium. Protein Expr. Purif. 40, 256–267 [DOI] [PubMed] [Google Scholar]
- 29. Havriluk T., Lozy F., Siniossoglou S., and Carman G. M. (2008) Colorimetric determination of pure Mg-dependent phosphatidate phosphatase activity. Anal. Biochem. 373, 392–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 32. Adams P. D. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34. Murshudov G. N., Vagin A. A., and Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 35. DeLano W. L. (2012) The PyMOL Molecular Graphics System. Schrodinger, LLC, New York [Google Scholar]