Abstract
Dysfunction of cilia is associated with common genetic disorders termed ciliopathies. Knowledge on the interaction networks of ciliary proteins is therefore key for understanding the processes that are underlying these severe diseases and the mechanisms of ciliogenesis in general. Cep104 has recently been identified as a key player in the regulation of cilia formation. Using a combination of sequence analysis, biophysics, and x-ray crystallography, we obtained new insights into the domain architecture and interaction network of the Cep104 protein. We solved the crystal structure of the tumor overexpressed gene (TOG) domain, identified Cep104 as a novel tubulin-binding protein, and biophysically characterized the interaction of Cep104 with CP110, Cep97, end-binding (EB) protein, and tubulin. Our results represent a solid platform for the further investigation of the microtubule-EB-Cep104-tubulin-CP110-Cep97 network of proteins. Ultimately, such studies should be of importance for understanding the process of cilia formation and the mechanisms underlying different ciliopathies.
Keywords: biophysics, centriole, cilia, circular dichroism (CD), x-ray crystallography, Cep104, TOG domain
Introduction
Cilia and flagella are evolutionary conserved organelles that contain a microtubule (MT)3-based cytoskeleton called the axoneme. They are crucial for motility, fluid flow, and coordination of many signaling pathways during cell growth, development, cell mobility, and tissue homeostasis (1, 2). The majority of mammalian cells have the capacity to form cilia. Conversely, most cancer cells lack cilia (3). Moreover, defects in ciliogenesis have been implicated in a wide range of other human disorders, including polycystic kidney disease, obesity, mental retardation, blindness, as well as various other developmental malformations (4). As a result, there has been great interest in understanding the regulation of both ciliary assembly/disassembly and the control of cilia length.
Ciliary assembly and disassembly is precisely coordinated during cell cycle progression. Cilia are formed from the distal end of the mother centriole upon exit from the cell cycle and entry into the G0 phase, become shorter as cells progress from the G1 to the S phase and disassemble in mitosis (5). The process of cilia formation is tightly controlled through a delicate balance of positive and negative regulators. Cep104 has recently been identified as a key player in the regulation of cilia formation (6).
Cep104 localizes to the distal ends of both centrioles throughout the whole cell cycle. At the G0 phase of the cell cycle, the mother centriole converts to a basal body that gives rise to a cilium. During ciliogenesis Cep104 shifts from the mother centriole to the tip of the elongating cilium. It has been shown that the Cep104 protein and its Chlamydomonas orthologue, FAP256, are essential for cilia formation in a high percentage of cells and in cases where, after depletion of Cep104/FAP256, cilia could still form, the ciliary tips displayed major structural deformation (6). Mutations in the Cep104 gene have been directly linked to Joubert syndrome, which is a genetically heterogeneous ciliopathy characterized by a distinctive mid-hindbrain and cerebellar malformation (7).
Cep104 has recently been identified as an EB protein-dependent MT plus-end tracking protein (+TIP). Based on immunoprecipitation studies with cell lysates, Cep104 was found to interact with CP110 and Cep97, two proteins that affect the length of the centriole and that interact with each other (8). Depletion of either CP110 or Cep97 causes abnormal elongation of centrioles and enhances primary cilia formation in growing cells (9, 10). Based on these observations, it was proposed that CEP104 regulates the function of the CP110-Cep97 complex when cilia outgrowth is initiated by mediating the connection between the CP110-Cep97 complex and dynamic MT ends (6).
Cep104 is crucial for the conversion of the mother centriole into the ciliary basal body (6). The exact role of Cep104 in this process and how it collaborates with its binding partners in the regulation of ciliogenesis is, however, not fully understood. Moreover, a biophysical or structural characterization of the Cep104 protein and its interaction network is currently missing. To better understand the role of the Cep104 protein in ciliogenesis, we analyzed the protein sequence in detail and revealed the presence of four conserved functional domains in the Cep104 molecule (Fig. 1A). Based on these results, we carried out a detailed biophysical characterization of the individual domains and their interactions with the binding partners Cep97, CP110, and EB. Moreover, we solved the crystal structure of the Cep104 TOG domain and identified Cep104 as a novel tubulin-binding protein.
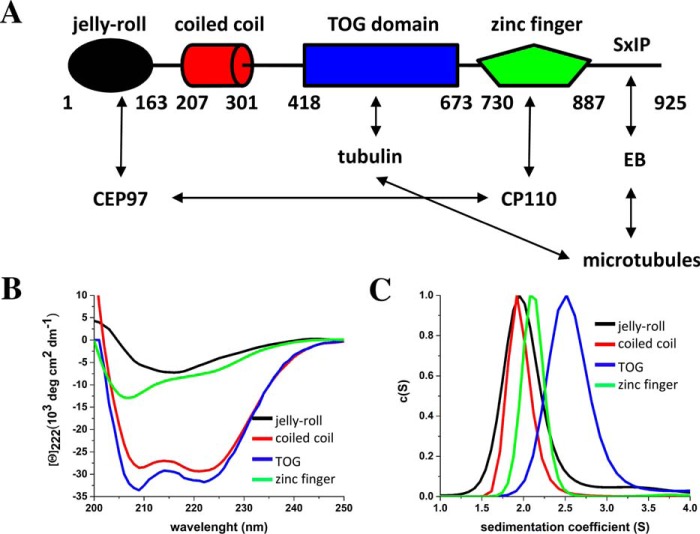
FIGURE 1.
Biophysical characterization of Cep104 domains. A, schematic representation of the Cep104 interaction network. Domains are shown as cylinders, jelly-roll in black, coiled coil in red, TOG in blue, and zinc finger in green. Interactions are denoted by arrows; B, CD spectra of Cep104 domains. The spectra were recorded using a protein concentration of 5 μm. C, c(S) distributions show a single peak for all Cep104 domains. The jelly-roll, TOG, and zinc-finger domains are monomeric, whereas the coiled coil is dimeric.
Results
Biophysical Characterization of the Cep104 Domains
To better understand the role of Cep104 in ciliogenesis, we first analyzed its amino acid sequence by secondary structure prediction and protein threading that revealed the presence of four conserved functional domains (Fig. 1A). The first 163 residues fold into an N-terminal domain that is predicted to adopt a jelly-roll-fold similar to IFT25 (11). The jelly-roll domain consists of nine β-strands that are organized into two antiparallel sheets stacked on top of each other. Previously, this domain has been identified as a binding site for Cep97 (8). The N-terminal domain is followed by a predicted coiled coil region (residues 207–301). The α-helical coiled coil is one of the principal subunit oligomerization motifs in proteins (12) and based on the MultiCoil algorithm (13) the Cep104 coiled coil is predicted to be responsible for dimerization of the protein. Following the coiled-coil region, the Phyre2 fold recognition server (14) identified a putative TOG domain (residues 418–673). TOG domains are present in proteins that regulate MT dynamics (15). These proteins either increase MT polymerization rates by recruiting soluble tubulin via their conserved TOG domains to polymerizing MT plus ends or decrease MT catastrophe and activate MT rescue by recruiting soluble tubulin to depolymerizing MT plus ends (15). The C-terminal part of Cep104 (residues 730–887) is predicted to fold into a zinc-finger domain. Previously, this domain has been identified as a binding site for the CP110 protein (8). Furthermore, an SXIP motif is present at the very C terminus of the Cep104 protein. The SXIP motif is a short linear motif that is recognized by EB proteins that then target the proteins to growing MT ends (16) (Fig. 1A).
To characterize the identified domains, we first cloned and expressed them individually in Escherichia coli. The TOG domain and the C-terminal zinc-finger domain expressed well as N-terminally His6-tagged variants. In contrast, expression of the N-terminal jelly-roll domain and the coiled-coil region required the insertion of a SUMO tag and a thioredoxin tag, respectively, between the His6 tag and the target proteins. All the His6-tagged fusion proteins were cleaved off by proteases before performing biophysical analysis (for details, see “Experimental Procedures”). All proteins migrated at the expected positions on SDS-PAGE gels and no degradation products were observed.
We characterized the recombinant proteins by CD spectroscopy and sedimentation velocity (SV) analytical ultracentrifugation (AUC) to confirm their correct folding based on their secondary structure content (Fig. 1B) and assess their oligomerization state, respectively (Fig. 1C). The N-terminal jelly-roll domain revealed a CD spectrum rich in β-sheet structure, which correlates well with the secondary structure prediction and the Phyre2 model based on the similarity with IFT25 structure (11). c(S) distribution shows a single peak with a sw value of 2.0. This corresponds to a Smax/S value of 1.29 and therefore suggests a monomeric globular shape of the domain (theoretical molecular mass of monomer: 19 kDa) that is consistent with the Phyre2 prediction. The CD spectrum of the coiled-coil domain revealed a significant amount of α-helical structure with distinct minima near 208 and 222 nm. The stability of the protein was assessed by thermal unfolding monitored by CD at 222 nm, which revealed a sigmoidal-shaped unfolding profile with a Tm of 55 °C (Fig. 4B). Characteristic of a coiled-coil structure, the unfolding was reversible. c(S) distribution analysis revealed that the coiled coil sediments with a sw value of 1.9 S. This yields a Smax/S of 1 for a monomer and a Smax/S of 1.57 for a dimer. As a Smax/S value of 1 is not possible for proteins, we conclude that the coiled-coil domain forms a dimer in solution (theoretical molecular mass of monomer 12 kDa). This is in good agreement with the theoretical prediction by MultiCoil (13). Furthermore, a Smax/S value of 1.57 is consistent with an elongated shape of the domain, which is characteristic of coiled coils. CD spectroscopy of the putative TOG domain showed a significant amount of α-helical structure that is typical for this type of fold. c(S) distribution revealed a single peak with a sw value of 2.5 that amounts to a Smax/S of 1.36 (theoretical molecular mass of monomer: 29 kDa). This result suggests a moderately elongated monomeric structure in solution, which is characteristic of TOG domains.
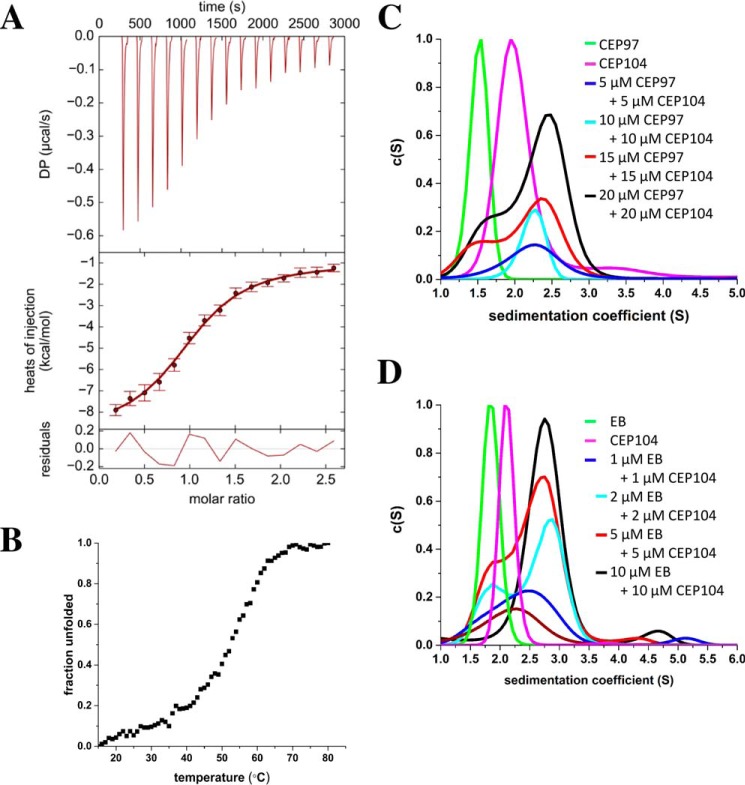
FIGURE 4.
Cep104 forms a dimer in solution that directly interacts with CP110, Cep97, and EB. A, ITC measurement of the interaction between the C-terminal part of Cep104 (residues 730–925) and the synthetic CP110 peptide (residues 907–936). The CP110 peptide interacts with Cep104 with an apparent dissociation constant of 4 μm and in a 1:1 stoichiometry; B, normalized thermal unfolding profile and of the Cep104 coiled-coil region (residues 207–301); C, SV AUC profiles showing the interaction between the N-terminal part of Cep104 (residues 1–163) and central part of Cep97 (residues 310–480). Green, Cep97; magenta, Cep104; blue, mixture of 5 μm Cep97 and 5 μm Cep104; cyan, mixture of 10 μm Cep97 and 10 μm Cep104; red, mixture of 15 μm Cep97 and 15 μm Cep104; black, mixture of 20 μm Cep97 and 20 μm Cep104; D, SV AUC profiles showing the interaction between the C-terminal part of Cep104 (residues 730–925) and EB1 (residues 191–267). Green, EB1; magenta, Cep104; brown, mixture of 1 μm EB and 1 μm Cep104; blue, mixture of 2 μm EB and 2 μm Cep104; cyan, mixture of 5 μm EB and 5 μm Cep104; red, mixture of 10 μm EB and 10 μm Cep104; black, mixture of 15 μm EB and 15 μm Cep104.
The CD spectrum of the C-terminal zinc-finger domain confirmed the folded state of the purified protein and revealed a mixture of α-helixes, β-sheets, and random coils. c(S) distribution showed a single peak with a sw value of 1.9 S. This corresponds to a Smax/S of 1.49, suggesting a slightly elongated shape and monomeric state of the protein in solution (theoretical molecular mass of monomer: 22 kDa). Taken together, our bioinformatics analysis and biophysical characterization revealed the presence of four conserved functional domains in the Cep104 molecule (Fig. 1A).
Cep104 Is a Novel Tubulin-binding Protein
We identified a putative TOG domain in the central part of the Cep104 molecule. To assess the functional role of this domain, we performed a detailed biophysical and structural characterization. We used SV AUC to test tubulin binding. Tubulin forms dimers in solution that are prone to polymerization. To avoid formation of higher oligomers, we kept the concentration of tubulin constant at 1 μm. At that concentration, the c(S) distribution showed a single peak corresponding to tubulin dimers (sw 5.8, Smax/S for dimer: 1.34, theoretical molecular mass of monomer: 50 kDa) without signs of aggregation (Fig. 2B). To study its binding to tubulin, different concentrations of the putative TOG domain were added to 1 μm tubulin. sw isotherm analysis revealed that the TOG domain of Cep104 directly binds to tubulin dimers with a 1:1 stoichiometry and an apparent dissociation constant of 1 μm (Fig. 2, A and B).
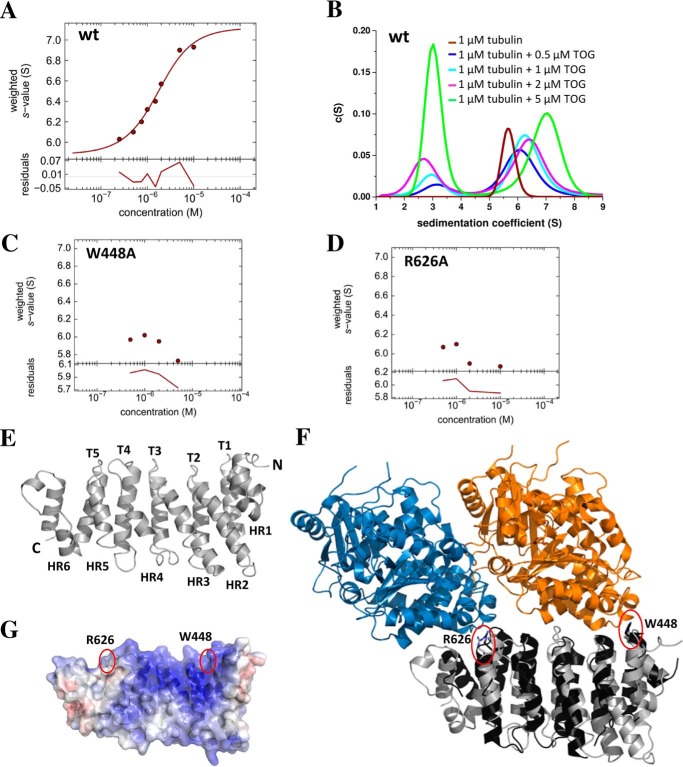
FIGURE 2.
Cep104 is a novel tubulin binding protein. A, sw isotherm showing the interaction between the TOG domain of Cep104 and tubulin. B, SV AUC profiles showing the interaction between the TOG domain of Cep104 (residues 418–673) and tubulin. Brown curve, tubulin; blue curve, mixture of 1 μm tubulin and 0.5 μm TOG domain; cyan curve, mixture of 1 μm tubulin and 1 μm TOG domain; magenta, mixture of 1 μm tubulin and 2 μm TOG domain; green curve, mixture of 1 μm tubulin and 5 μm TOG domain; C and D, sw isotherms showing that mutation of Trp-448 or Arg-626 completely abolishes binding to tubulin; E, schematic representation of x-ray structure of the Cep104 TOG domain comprising 6 HEAT repeats (HR1–6) and 5 loop regions (T1-T5). The loop regions form the tubulin-binding site; F, model of a complex between the TOG domain of Cep104 and an unpolymerized tubulin dimer in the curved conformation that was generated based on the x-ray crystal structure of the TOG1 domain of the Stu2 complexed with yeast tubulin dimer (PDB code 4FFB). The model was generated by superimposition of the Cep104 TOG domain with the TOG1 domain of the Stu2-TOG1-tubulin complex. Residues important for the interaction with tubulin, Trp-448 and Arg-626 in Cep104 TOG, and Trp-23 and Arg-200 in Stu2-TOG1, are highlighted with a red circle. Blue, α-tubulin; orange, β-tubulin; gray, Cep104 TOG domain; black, Stu2-TOG1; G, surface charge representation of the x-ray crystal structure of the Cep104 TOG domain colored according to its electrostatic potential (±8 kT/e, where k is the Boltzmann constant, T is temperature, and e is the elementary charge): blue denotes basic residues, red denotes acidic residues, and white denotes hydrophobic residues. Residues important for the interaction with tubulin are labeled and highlighted with a red circle.
To further study the Cep104 TOG domain and its interaction with tubulin, we decided to characterize it by x-ray crystallography. Unfortunately, the human TOG domain did not yield any crystals; therefore we performed a screen of a limited number of Cep104 TOG domains from different species, including Mus musculus, Gallus gallus, Xenopus tropicalis, Drosophila melanogaster, Danio rerio, Tetrahymena thermophile, and Chlamydomonas reinhardtii. The only crystals were obtained from the chicken TOG domain, which possesses 73% amino acid sequence identity to its human homolog. The structure was solved at 1.4-Å resolution by using selenomethionine-labeled protein for obtaining experimental phases. The overall structure resembles a canonical TOG-domain fold comprising six HEAT repeats (HR1–6) forming flat paddles with wide thin edges (Fig. 2E).
Although the overall architecture of TOG domains structures is the same, small structural variations that have been linked to diversified interactions with tubulin have been described. The PDBefold identified the crescerin TOG2 domain (PDB code 5DN7) as the most similar structure (Q-score (quality score): 0.31). Although both domains share a sequence identity of only 14%, their structures can be superimposed with a root mean square deviation of 2.94 Å. Both TOG domains have the tubulin binding intra-HEAT loop surface in a straight conformation that resembles the Stu2 TOG1 structure and contrasts with the bent architectures observed in ch-TOG TOG4, MAST TOG1, and hCLASP1 TOG2 (17–21). The straight conformation and the similarity to Stu2-TOG1 complement our finding that the Cep104 TOG domain is involved in binding of free tubulin and suggest a possible role of Cep104 in regulation of MT dynamics.
Although the sequence identity among different TOG domains is generally very low (typically below 20%), they share two highly conserved features: hydrophobic residues buried between neighboring HEAT repeat α-helices that stabilize the overall structure of the domain and five short intra-HEAT repeat loops (T1-T5) on one narrow edge creating a binding site for tubulin. Single amino acid mutations in this highly conserved loop region often completely abolishes binding to tubulin dimer (15). The TOG domain of Cep104 possesses two highly conserved residues, Trp-448 in the HEAT repeat loop T1 and Arg-626 in the HEAT repeat loop T5, amino acids that have previously been shown to play an important role in tubulin binding. Additionally, a basic overall electrostatic net charge distribution is seen across the first five intra-HEAT loops that in other TOG domains was shown to complement the acidic nature of the TOG domain binding site in tubulin (Fig. 2G). A hypothetical model of a complex between the TOG domain of Cep104 and an unpolymerized tubulin dimer (Fig. 2F) that was generated based on the x-ray structure of the TOG1 domain of Stu2 and yeast tubulin dimer (PDB code 4FFB) indeed suggests that both Cep104 residues Trp-448 and Arg-626 make very similar contacts with tubulin as their equivalents in Stu2-TOG1 (Trp-23 and Arg-200, respectively). We used site-directed mutagenesis and SV AUC to probe the importance of Trp-448 and Arg-626 for the TOG/tubulin interaction. As expected, both mutants display substantially reduced binding to tubulin dimers (Fig. 2, C and D).
Taken together, we identified Cep104 as a novel tubulin-binding protein. Tubulin therefore adds to the complexity of Cep104 interaction network, which comprises CP110, Cep97, and EB proteins.
Cep104 Directly Interacts with CP110 and Cep97
Jiang et al. (8) identified CP110 and Cep97 as novel Cep104 binding partners. They mapped the binding sites and found that the C-terminal part of CP110 (residues 791–991) interacts with the zinc-finger domain of Cep104 (residues 730–887) and the central part of Cep97 (residues 310–480) binds to the jelly-roll domain of Cep104 (residues 1–163) (8). Because the authors used an immunoprecipitation assay with cell extracts to map the binding sites, indirect binding could not be excluded. To test whether the interaction with the binding partners involves direct binding, we performed SV AUC experiments.
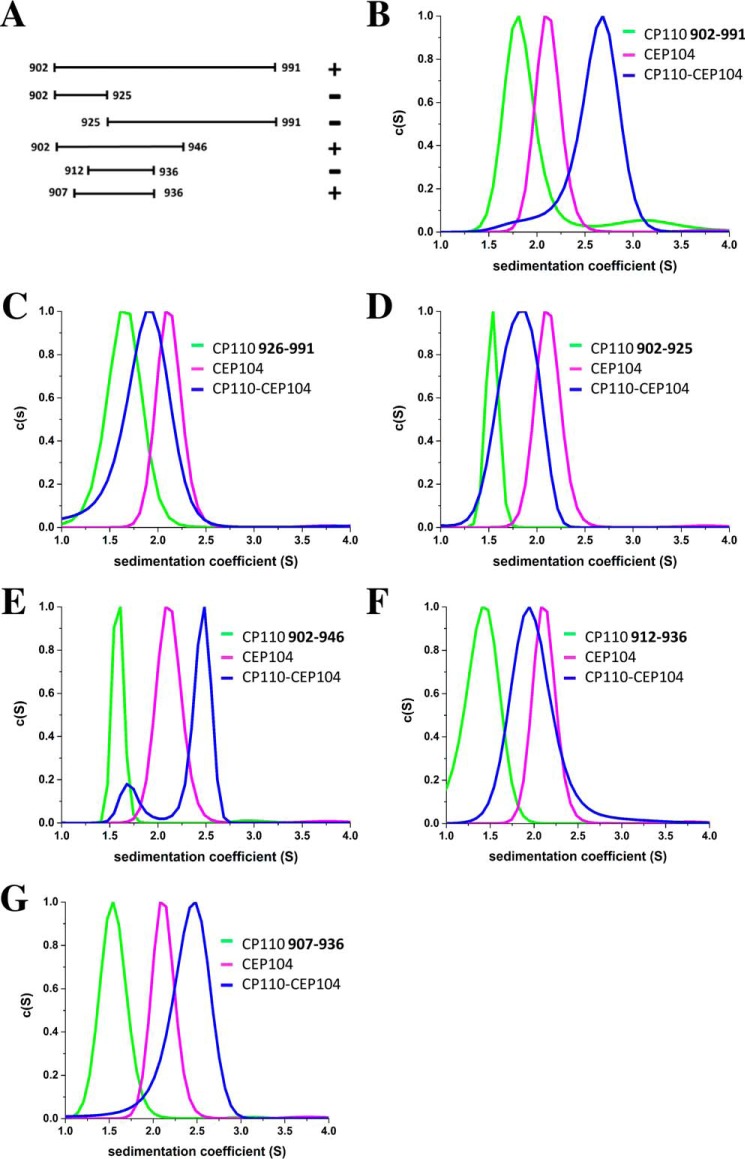
The CP110 791–991 region is predicted to be unstructured and when expressed in E. coli it forms mostly insoluble inclusion bodies; therefore the boundaries were first narrowed down to residues 902–991 using pull-down experiments with cell lysates. This region of CP110 can be expressed when fused to thioredoxin, and the uncleaved fusion protein sediments with a sw value of 1.8. This corresponds to a Smax/S ratio of 1.67, which suggests an asymmetric shape of the molecule (theoretical molecular mass of monomer: 24 kDa). Based on the fusion of the compact thioredoxin fold with the unstructured CP110 moiety, an asymmetric shape is expected. c(S) distribution clearly shows that the fusion protein directly interacts with zinc-finger domain of Cep104 with a 1:1 stoichiometry (Fig. 3B). Using N- and C-terminal truncations, we were able to map the Cep104 binding region even more precisely (Fig. 3, A-G). The smallest region identified contains only 30 amino acids (residues 907–936) (Fig. 3G). For screening, we used recombinantly expressed peptides fused to thioredoxin. However, for the final affinity measurements by isothermal titration calorimetry (ITC) we used a synthetic peptide. The ITC results revealed that the CP110 peptide (residues 907–936) interacts with Cep104 with an apparent dissociation constant of 4 μm and in a 1:1 stoichiometry (Fig. 4A).
FIGURE 3.
Mapping of the interaction between the C-terminal part of Cep104 (residues 730–925) and different CP110 polypeptide chain fragments. A, schematic representation of CP110 polypeptide chain fragments used in this study; B-G, SV AUC profiles of the C-terminal part of Cep104 (magenta), different uncleaved thioredoxin-CP110 fusion proteins (residues 902–991, 926–991, 902–946, 912–936, 902–925, 907–936 in green) and their mixtures (blue). Only CP110 polypeptide chain fragments containing residues 902–991, 902–946, and 907–936 have the ability to interact with Cep104. The experiments were performed using a protein concentration of 30 μm.
The central part of Cep97 (residues 310–480) is also predicted to be unstructured in solution but can be expressed in the cytosol of E. coli as a His6-tagged version. c(S) distribution showed a single peak with a sw of 1.5 and a Smax/S of 1.7, which is consistent with a monomeric state of the protein (theoretical molecular mass of monomer: 19 kDa). The Smax/S of 1.7 indicates an asymmetric shape of the polypeptide chain fragment and corresponds well with the unstructured nature of the protein. By measuring different ratios in SV AUC experiments, we found that the jelly-roll domain of Cep104 directly interacts with the central part of Cep97 with an apparent dissociation Kd of 3 μm and in a 1:1 stoichiometry (Fig. 4C).
Cep104 Directly Interacts with EB Proteins
Cep104 has been recently identified as a MT plus-end tracking protein (+TIP) (8). Like many other +TIPs it binds to EB proteins through intrinsically disordered basic and serine-rich polypeptide chain regions containing a C-terminal core SXIP motif (S, serine; X, any amino acid; I, isoleucine; P, proline). The conserved SXIP peptide motif binds to the hydrophobic groove formed by the C-terminal helix bundle of the EB homology domain dimer (16). To confirm a direct interaction between the C-terminal part of the Cep104 and the EB homology domain of EB1 proteins and to assess the binding affinities, we performed SV AUC measurements. For these experiments, we used the C-terminal part of Cep104 that consists of the zinc-finger domain followed by a 40-amino acid long unstructured region. The SXIP motif is located within this unstructured part. In agreement with the literature, the EB homology domain forms tight dimers. c(S) distribution shows a single peak with a sw value of 1.6 S. A calculated Smax/S value of 1.56 is consistent with a dimeric-fold of the EB homology domain (theoretical molecular mass of monomer: 9 kDa). By measuring different protein concentration ratios using SV AUC, we determined that the C-terminal part of Cep104 directly interacts with the EB homology domain dimers with an apparent dissociation Kd of 2 μm (Fig. 4D). The strength of the interaction is comparable with those of other known EB-binding proteins.
Discussion
Dysfunction of cilia is associated with common genetic disorders called ciliopathies. Understanding the structure, function, and interaction networks of ciliary proteins is therefore key for a better understanding of these severe diseases and ciliogenesis in general. In this study, we characterized in detail the domain organization and interaction network of Cep104, a protein that plays a crucial role in ciliogenesis.
We found that Cep104 is a multidomain protein that interacts with a number of other proteins: CP110, Cep97, tubulin, and EB. This suggests that the function of Cep104 is complex and tightly regulated through various interactions. In this study, we showed that the Cep104 protein directly interacts with tubulin through a conserved TOG domain. The TOG domain array-containing proteins ch-TOG and CLASP are key regulators of cytoplasmic MTs (15). The cytoplasmic TOG array proteins have been extensively studied, however, the first TOG array protein that regulates ciliary MT, crescerin, has been reported only very recently (22). The number of TOG domains in each array differs. Higher eukaryotic ch-TOG family members use a pentameric TOG domain array to promote MT polymerization, whereas CLASP members use a trimeric TOG domain array to stabilize MTs (15). Members of the newly described crescerin family contain four TOG domains and have been suggested to promote MT polymerization in vitro (22). Cep104 contains only one TOG domain, but the whole protein is dimeric in solution due to the presence of a coiled-coil region. It has been shown previously, i.e. for Stu2 TOG domains, that two TOG domains of the same type are sufficient for altering the MT dynamics (23). MT binding and plus end localization of Cep104 is also provided by the interaction with EB proteins (8). The Cep104 protein localizes to the tips of growing cilia and possesses 2 TOG domains per Cep104 dimer and EB-dependent plus end tracking activity suggests that Cep104 could be a novel MT dynamics regulator and therefore complement the function of crescerin in cilia.
It was suggested that CP110 and Cep97 proteins act as a “cap” at the distal end of the centriole and collaborate to limit centriolar length and block the ciliogenic branch of centriole function (6). An attractive hypothesis is that in the regulation of ciliogenesis Cep104 mediates the connection between the CP110-Cep97 complex and dynamic MT ends because neither CP110 nor Cep97 have been reported to directly interact with MTs. Via the EB interaction, Cep104 could potentially recruit the Cep97-CP110 complex to the growing MT tips. The CP110-Cep97 complex at the distal end of the centriole blocks cilium elongation, whereas Cep104, based on its domain organization, could potentially be involved in the initiation of cilia formation. However, the initiation of ciliogenesis by Cep104 can probably only start, once the CP110-Cep97 inhibitory complex is released from the distal end of the centriole. The observation that depletion of either CP110 or Cep97 enhances primary cilia formation in growing cells is consistent with this hypothesis (9, 10). The permanent presence of Cep104 at the tip of dynamic MTs and its necessity for ciliogenesis (6) support our suggestion of Cep104 being novel MT regulator.
Our study provides new insights into Cep104 domain organization and interaction network. However, further work will be necessary to fully understand Cep104 function in cilia and at the distal end of the centriole. Further experiments will examine the effect of ablating the Cep104 protein in human cells on cilia formation. A dissection of the contribution of the different domains to the function of the Cep104 protein (i.e. deleting the TOG domain and/or introducing a point mutation (W448A or R626A) could help to answer whether Cep104 plays a role in MT dynamics. It will also be important to understand how the different Cep104 binding partners Cep97, CP110, EB, and tubulin, regulate its function. This work provides a solid platform for performing such experiments.
Experimental Procedures
DNA Constructs and Protein Production
Human CEP104 cDNA fragments encoding the TOG domain (residues 418–673) and the C-terminal zinc-finger domain (residues 730–925) were ligated into the bacterial expression vector pET-15b. The N-terminal jelly-roll domain (residues 1–163) was cloned into a modified version of pET-15b vector that includes a SUMO fusion protein after the His6 tag and the coiled-coil domain (residues 207–301) into a modified version of the pET-15b vector, pHisTrx2, which includes a thioredoxin fusion protein after the His6 tag. A synthetic, codon-optimized gene fragment encoding the chicken TOG domain (residues 428–686) used for crystallization and the human Cep97 (residues 310–480) were cloned into pET-15b. cDNAs encoding and all CP110 polypeptide chain fragments (residues 902–991, 926–991, 902–946, 912–936, 902–925, 907–936) were cloned into pHisTrx2. All mutants were generated using the QuikChange approach.
Recombinant proteins were produced in E. coli BL21(DE3) cells. Bacterial cultures were induced at OD 0.7 with 0.5 mm isopropyl d-thiogalactopyranoside and grown for an additional 16 h at 20 °C. Recombinant proteins were purified using immobilized-metal affinity chromatography according to a standard protocol. Depending on the experiment, the His6-tagged fusion proteins were cleaved by incubation with thrombin or sumoprotease. Size exclusion chromatography on Superdex 75 or 200 columns (GE Healthcare) was used as a final purification step or for tag removal (20 mm HEPES (pH 7.5), 150 mm NaCl and 2 mm β-mercaptoethanol). The EB1 protein fragment (residue 191–267) was purified as reported previously (16). Bovine brain tubulin was prepared according to well established protocols (24). The CP110 peptide (residue 907–936) was assembled on an automated continuous flow synthesizer employing standard methods.
Analytical Ultracentrifugation
SV experiments were performed at 20 °C and 42,000 rpm in a Beckman Coulter ProteomeLab XL-I analytical ultracentrifuge using standard protocols (25, 26); a 400-μl sample at different concentrations in 20 mm HEPES (pH 7.5), 150 mm NaCl, 2 mm β-mercapthoethanol was filled in 12-mm charcoal-filled Epon double-sector centerpieces and matched with an equal volume of buffer in the reference sector.
Sedimentation data were collected with absorbance (230, 250, or 280 nm) or interference optical systems. Protein partial specific volumes, solution density, and viscosity were calculated in SEDNTERP. Data were analyzed in terms of a continuous c(s) distribution of Lamm equation solutions with the software SEDFIT (27). Scan file time stamps were corrected (28) and good fits were obtained with root mean square deviation values corresponding to typical instrument noise values. Sedimentation coefficients were corrected to standard conditions, s20,w. The shapes and oligomerization states of the proteins were determined on the basis of the Smax/S ratio. Smax was calculated by the following equation: Smax = 0.00361 × M2/3, where M is molecular weight of the protein (29). The Gilbert sw fast isotherm was determined by integration of the c(S) peaks of the fast boundary component in c(S); integration was done from 4.5 to 9 S. The isotherm was created by plotting the weighted average sedimentation coefficients sw as a function of TOG domain concentration (27, 30). The fitting was done with the software SEDPHAT (31). Isotherm figures were prepared using the GUSSI software package.
Circular Dichroism (CD)
CD spectra measurements were carried out on a Chirascan-Plus instrument (Applied Photophysics Ltd.) equipped with a computer-controlled Peltier element. All experiments were performed in PBS. The CD spectra were obtained at 5 °C by scanning wavelengths from 200 to 260 nm in 1-nm steps using a protein concentration of 5 μm. A ramping rate of 1 °C per min was used to record the thermal unfolding profiles. Midpoints of the transitions, Tm, were taken at the maximum of the derivative d[θ]222/dT.
Isothermal Titration Calorimetry
ITC measurements were performed using an ITC200 instrument (MicroCal). The CP110 peptide and the C-terminal Cep104 polypeptide chain fragment (residues 730–925) were dialyzed against 20 mm Tris-HCl (pH 7.5), 150 mm NaCl prior to analysis. Titrations were performed at 0.2-min intervals at a stirring speed of 1000 s−1. The resulting heats were integrated using NITPIC software (32) and isotherms were fitted using a 1:1 bimolecular interaction model in program SEDPHAT (31). All final figures were prepared using the GUSSI software package.
Crystallization and X-ray Structure Determination
The recombinant TOG domain (from G. gallus) was concentrated to 20 mg/ml. Crystals suitable for structure determination were grown at 20 °C in sitting drops composed of a 1:1 mixture of the protein solution and a well solution consisting of 25% PEG 1500 and 0.1 m MMT buffer (pH 5.0). For cryo-protection, the reservoir solution was supplemented with 20% ethylene glycol. Native and selenomethionine SAD data were acquired at 1-Å wavelength (0.96 Å for SAD) and 100 K at the X06SA beamline of the Swiss Light Source (Paul Scherrer Institute) to a resolution of 1.4 and 2.10 Å, respectively. The acquired datasets were reduced, scaled, and merged using XDS, XSCALE, and XDSCONV (33, 34). The structure was solved in the space group P22121 using AUTOSOL (35). The initial model building was done via AUTOBUILD and was used as a molecular replacement search model for phasing the native dataset by PHASER (36). Model building and refinement were performed via COOT (37) and phenix.refine from the PHENIX suite (38). Crystallographic data collection and refinement statistics are summarized in Table 1.
TABLE 1.
Crystallographic data collection and refinement statistics table
| TOG selenomethionine dataset | TOG native dataset | |
|---|---|---|
| Data collection | ||
| Wavelength | 0.96 Å | 1.0 Å |
| Resolution range | 42.23–2.1 (2.175–2.1) | 50–1.4 (1.44–1.4) |
| Space group | P2 21 21 | P 2 21 21 |
| Unit cell | 46.15 56.21 104.72 90 90 90 | 48.36 57.22 102.96 90 90 90 |
| Total reflections | 858,343 (88163) | 914,221 (68112) |
| Unique reflections | 16,513 (1607) | 55,668 (4135) |
| Multiplicity | 52.0 (54.8) | 16.4 (16.5) |
| Completeness | 1.00 (1.00) | 0.97 (1.0) |
| Mean I/σ(I) | 33.54 (2.30) | 19.02 (1.05) |
| Wilson B-factor | 42.43 | 21.4 |
| Rmerge | 0.1347 (2.472) | 0.065 (2.76) |
| Rmeas | 0.136 (2.494) | 0.067 (2.852) |
| CC1/2 | 1 (0.754) | 1 (0.445) |
| Data refinement | ||
| Reflections used in refinement | 55,550 (3847) | |
| Reflections used for R-free | 1982 (142) | |
| R-work | 0.2186 (0.4068) | |
| R-free | 0.2383 (0.4180) | |
| CC (work) | 0.963 | |
| CC (free) | 0.926 | |
| Number of non-hydrogen atoms | 2,321 | |
| Number of protein atoms | 2,073 | |
| Protein residues | 256 | |
| Root mean square deviation (bonds) | 0.006 | |
| Root mean square deviation (angles) | 1.00 | |
| Ramachandran favored (%) | 99 | |
| Ramachandran allowed (%) | 0.77 | |
| Ramachandran outliers (%) | 0 | |
| Rotamer outliers (%) | 1.3 | |
| Clashscore | 2.82 | |
| Average B-factor | 28.49 | |
| Protein | 27.49 | |
| Solvent | 36.77 | |
Author Contributions
L. R., A. A., M. O. S., and R. A. K. designed the research, L. R. carried out the research, L. R. and S. H. W. K analyzed the data, L. R. and R. A. K. wrote the manuscript with input from the other authors.
Acknowledgments
We thank beamline scientists at beamline X06SA of the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland) for excellent technical assistance with the collection of x-ray data.
The work was supported in part by Swiss National Science Foundation Grants 310030B_138659 (to M. O. S.) and 31003A_163449 (to R. A. K.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 5JO8) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- MT
- microtubule
- EB
- end-binding
- TOG
- tumor overexpressed gene
- ITC
- isothermal titration calorimetry
- AUC
- analytical ultracentrifugation
- SV
- sedimentation velocity.
References
- 1. Mahjoub M. R. (2013) The importance of a single primary cilium. Organogenesis 9, 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nachury M. V. (2014) How do cilia organize signalling cascades? Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, pii. 20130465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Plotnikova O. V., Golemis E. A., and Pugacheva E. N. (2008) Cell cycle-dependent ciliogenesis and cancer. Cancer Res. 68, 2058–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nigg E. A., and Raff J. W. (2009) Centrioles, centrosomes, and cilia in health and disease. Cell 139, 663–678 [DOI] [PubMed] [Google Scholar]
- 5. Izawa I., Goto H., Kasahara K., and Inagaki M. (2015) Current topics of functional links between primary cilia and cell cycle. Cilia 4, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Satish Tammana T. V., Tammana D., Diener D. R., and Rosenbaum J. (2013) Centrosomal protein CEP104 (Chlamydomonas FAP256) moves to the ciliary tip during ciliary assembly. J. Cell Sci. 126, 5018–5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Srour M., Hamdan F. F., Mcknight D., Davis E., Mandel H., Schwartzentruber J., Martin B., Patry L., Nassif C., Dionne-laporte A., Ospina L. H., Lemyre E., Massicotte C., Laframboise R., Maranda B., et al. (2015) Joubert syndrome in French Canadians and identification of mutations in CEP104. 97, 744–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jiang K., Toedt G., Montenegro Gouveia S., Davey N. E., Hua S., van der Vaart B., Grigoriev I., Larsen J., Pedersen L. B., Bezstarosti K., Lince-Faria M., Demmers J., Steinmetz M. O., Gibson T. J., and Akhmanova A. (2012) A proteome-wide screen for mammalian SxIP motif-containing microtubule plus-end tracking proteins. Curr. Biol. 22, 1800–1807 [DOI] [PubMed] [Google Scholar]
- 9. Spektor A., Tsang W. Y., Khoo D., and Dynlacht B. D. (2007) Cep97 and CP110 suppress a cilia assembly program. Cell. 130, 678–690 [DOI] [PubMed] [Google Scholar]
- 10. Schmidt T. I., Kleylein-Sohn J., Westendorf J., Le Clech M., Lavoie S. B., Stierhof Y. D., and Nigg E. A. (2009) Control of centriole length by CPAP and CP110. Curr. Biol. 19, 1005–1011 [DOI] [PubMed] [Google Scholar]
- 11. Bhogaraju S., Taschner M., Morawetz M., Basquin C., and Lorentzen E. (2011) Crystal structure of the intraflagellar transport complex 25/27. EMBO J. 30, 1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Burkhard P., Stetefeld J., and Strelkov S. V (2001) Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 11, 82–88 [DOI] [PubMed] [Google Scholar]
- 13. Wolf E., Kim P. S., and Berger B. (1997) MultiCoil: a program for predicting two-and three-stranded coiled coils. Protein Sci. 6, 1179–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Slep K. C. (2009) The role of TOG domains in microtubule plus end dynamics. Biochem. Soc. Trans. 37, 1002–1006 [DOI] [PubMed] [Google Scholar]
- 16. Honnappa S., Gouveia S. M., Weisbrich A., Damberger F. F., Bhavesh N. S., Jawhari H., Grigoriev I., van Rijssel F. J., Buey R. M., Lawera A., Jelesarov I., Winkler F. K., Wüthrich K., Akhmanova A., and Steinmetz M. O. (2009) An EB1-binding motif acts as a microtubule tip localization signal. Cell 138, 366–376 [DOI] [PubMed] [Google Scholar]
- 17. Slep K. C., and Vale R. D. (2007) Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol. Cell. 27, 976–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ayaz P., Ye X., Huddleston P., Brautigam C. A., and Rice L. M. (2012) A TOG: αβ-tubulin complex structure. Science 337, 857–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. De la Mora-Rey T., Guenther B. D., and Finzel B. C. (2013) The structure of the TOG-like domain of Drosophila melanogaster Mast/Orbit. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 69, 723–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leano J. B., Rogers S. L., and Slep K. C. (2013) A cryptic TOG domain with a distinct architecture underlies CLASP-dependent bipolar spindle formation. Structure 21, 939–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fox J. C., Howard A. E., Currie J. D., Rogers S. L., and Slep K. C. (2014) The XMAP215 family drives microtubule polymerization using a structurally diverse TOG array. Mol. Biol. Cell 25, 2375–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Das A., Dickinson D. J., Wood C. C., Goldstein B., and Slep K. C. (2015) Crescerin uses a TOG domain array to regulate microtubules in the primary cilium. Mol. Biol. Cell 26, 4248–4264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ayaz P., Munyoki S., Geyer E. A., Piedra F.-A., Vu E. S., Bromberg R., Otwinowski Z., Grishin N. V., Brautigam C. A., and Rice L. M. (2014) A tethered delivery mechanism explains the catalytic action of a microtubule polymerase. Elife 3, e03069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Andreu J. M. (2007) Large scale purification of brain tubulin with the modified Weisenberg procedure. Methods Mol. Med. 137, 17–28 [DOI] [PubMed] [Google Scholar]
- 25. Zhao H., Brautigam C. A., Ghirlando R., and Schuck P. (2013) Overview of current methods in sedimentation velocity and sedimentation equilibrium analytical ultracentrifugation. Curr. Protoc. Protein Sci. Chapter 20, unit 20.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brown P. H., Balbo A., and Schuck P. (2008) Characterizing protein-protein interactions by sedimentation velocity analytical ultracentrifugation. Curr. Protoc. Immunol. Chapter 18, unit 18.15 [DOI] [PubMed] [Google Scholar]
- 27. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao H., Ghirlando R., Piszczek G., Curth U., Brautigam C. A., and Schuck P. (2013) Recorded scan times can limit the accuracy of sedimentation coefficients in analytical ultracentrifugation. Anal. Biochem. 437, 104–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Erickson H. P. (2009) Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol. Proced. Online 11, 32–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dam J., Velikovsky C. A., Mariuzza R. A., Urbanke C., and Schuck P. (2005) Sedimentation velocity analysis of heterogeneous protein-protein interactions: Lamm equation modeling and sedimentation coefficient distributions c(s). Biophys. J. 89, 619–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schuck P. (2003) On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal. Biochem. 320, 104–124 [DOI] [PubMed] [Google Scholar]
- 32. Keller S., Vargas C., Zhao H., Piszczek G., Brautigam C. A., and Schuck P. (2012) High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal. Chem. 84, 5066–5073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kabsch W. (2010) Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Terwilliger T. C., Adams P. D., Read R. J., McCoy A. J., Moriarty N. W., Grosse-Kunstleve R. W., Afonine P. V., Zwart P. H., and Hung L. W. (2009) Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 65, 582–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson J. S., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]