Abstract
Collagen is post-translationally modified by prolyl and lysyl hydroxylation and subsequently by glycosylation of hydroxylysine. Despite the widespread occurrence of the glycan structure Glc(α1–2)Gal linked to hydroxylysine in animals, the functional significance of collagen glycosylation remains elusive. To address the role of glycosylation in collagen expression, folding, and secretion, we used the CRISPR/Cas9 system to inactivate the collagen galactosyltransferase GLT25D1 and GLT25D2 genes in osteosarcoma cells. Loss of GLT25D1 led to increased expression and intracellular accumulation of collagen type I, whereas loss of GLT25D2 had no effect on collagen secretion. Inactivation of the GLT25D1 gene resulted in a compensatory induction of GLT25D2 expression. Loss of GLT25D1 decreased collagen glycosylation by up to 60% but did not alter collagen folding and thermal stability. Whereas cells harboring individually inactivated GLT25D1 and GLT25D2 genes could be recovered and maintained in culture, cell clones with simultaneously inactive GLT25D1 and GLT25D2 genes could be not grown and studied, suggesting that a complete loss of collagen glycosylation impairs osteosarcoma cell proliferation and viability.
Keywords: collagen, CRISPR/Cas, endoplasmic reticulum (ER), glycosylation, osteosarcoma, protein trafficking (Golgi)
Introduction
Collagens, the most abundant animal proteins, are essential components of the extracellular matrix of various tissues and organs. Collagens are mainly located in connective tissue and regulate a variety of biological processes such as cell attachment, migration, proliferation, and differentiation (1). Collagens feature specific domains composed of Gly-X-Y repeats with proline and lysine often occupying the X and Y positions. Nascent procollagen chains are modified co-translationally by prolyl 4-hydroxylation (2), prolyl 3-hydroxylation (3), lysyl hydroxylation (4), and glycosylation of selected hydroxylysine (Hyl)2 residues (5). Collagen modifications take place in the endoplasmic reticulum (ER) before completion of triple helix assembly (6). The importance of collagen hydroxylation is underlined by various diseases linked to defective collagen modifications. For example, prolyl 3-hydroxylase 1 deficiency causes osteogenesis imperfecta with severe skeletal deformation (7), and mutation in the lysyl hydroxylase 1 PLOD1 gene causes Ehlers-Danlos syndrome type VI (8). Mutations in the lysyl hydroxylase 2 PLOD2 gene cause Bruck syndrome (9), and mutations in the lysyl hydroxylase 3 PLOD3 gene cause connective tissue defects typical of collagen disorders (10). A recently identified mutation of the prolyl 4-hydroxylase β-subunit protein disulfide isomerase causes Cole-Carpenter syndrome (11). By contrast, the biological significance of collagen glycosylation remains elusive because no disease has been associated with the process, and no model organism harboring defective collagen glycosylation has been described to date.
The GLT25D1 and GLT25D2 genes encode Hyl-specific galactosyltransferase enzymes, which initiate collagen glycosylation (12, 13). The gene encoding the α1–2 glucosyltransferase enzyme, which adds Glc to Gal, has not been identified yet, although the lysyl hydroxylase 3 enzyme has been claimed to also act as a collagen glucosyltransferase (14). The resulting Glc(α1–2)Gal disaccharide is strongly conserved in all animal collagens, from sponges up to mammals (14–17). Whereas GLT25D1 is broadly expressed across tissues, GLT25D2 is mainly expressed in brain tissue and at low levels in skeletal muscle (13). The GLT25D1 and GLT25D2 galactosyltransferases share identical enzymatic activities and substrate specificities, because they are able to glycosylate various types of collagen to similar levels (13).
Despite the recent identification of the GLT25D1 and GLT25D2 galactosyltransferases, little is known about the functional role of collagen glycosylation. Glycosylation has been shown to affect the binding of the urokinase-type plasminogen activator receptor associated protein (uPARAP) to collagen type IV, thereby implying collagen glycosylation in receptor-mediated matrix remodeling (18). Also integrins appear to be sensitive to collagen glycosylation, because decreased integrin-mediated cell adhesion was measured on galactosylated collagen peptides compared with unmodified peptides (19, 20). Glycosylation of Hyl is notably not confined to collagens. The collagen domains of multimeric proteins such as adiponectin and mannose-binding lectin also carry glycosylated Hyl, where lysyl hydroxylation and glycosylation influence protein oligomerization (21–23). Considering the possible functional involvement of glycosylation in collagen folding and intracellular trafficking, we investigated collagen properties after inactivation of the GLT25D1 and GLT25D2 galactosyltransferase genes in osteosarcoma cells, which produce large amounts of fibrillar collagens, including collagen types I and V and minor amounts of collagen type III.
Results
GLT25D1 and GLT25D2 Inactivation in Osteosarcoma Cells
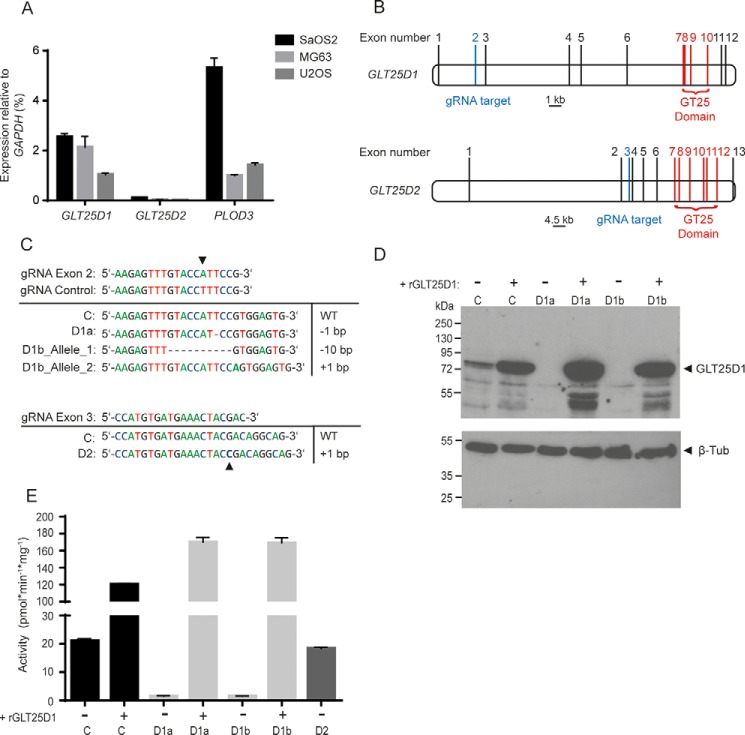
Collagen glycosylation is initiated by the transfer of Gal to Hyl catalyzed by GLT25D1 and GLT25D2 galactosyltransferase enzymes. To identify osteosarcoma cell lines generating high amounts of collagen galactosylation, we first analyzed GLT25D1 and GLT25D2 gene expression in the three collagen-producing osteosarcoma cell lines SaOS-2, MG63, and U2OS (24). As expected, GLT25D1 was the main collagen galactosyltransferase isoform expressed in osteosarcoma cells considering the restricted expression of GLT25D2 in brain and skeletal muscle (13). The transcript levels of GLT25D2 represented only 1–4% of GLT25D1 levels in the three cell lines investigated (Fig. 1A). As a comparison, the PLOD3 gene encoding the lysyl hydroxylase 3 enzyme was expressed between 0.5- and 2-fold the levels of GLT25D1 transcripts (Fig. 1A). The three collagen-modifying genes were expressed at the highest levels in SaOS-2 cells. Considering the strong expression of collagen galactosyltransferases and lysyl hydroxylase 3 genes in SaOS-2 cells, as well as the prominent production of collagen type I and highly glycosylated collagen type V in these cells (25), we primarily investigated the role of collagen glycosylation in SaOS-2 cells.
FIGURE 1.
Characterization of GLT25D1 and GLT25D2 inactivation in osteosarcoma cell lines. A, real time PCR analysis of GLT25D1, GLT25D2, and PLOD3 relative to GAPDH expression levels in SaOS-2, MG63, and U2OS cells (means ± S.D., n = 3 independent experiments). B, representation of GLT25D1 and GLT25D2 gene structure. Lines mark exons, blue lines mark the guide RNA (gRNA) target region, and red lines mark the glycosyltransferase coding region. C, sequences of gRNAs targeting GLT25D1 and GLT25D2 and sequences of the targeted segment in cell clones transfected with control gRNA (row C), with gRNA targeting GLT25D1 (row D1a, clone 1; row D1b, clone 2), and GLT25D2 (row D2). D, Western blotting of GLT25D1 in GLT25D1-null cells with and without overexpression of GLT25D1 cDNA (rGLT25D1). One representative experiment is shown (total n = 3 independent experiments). E, galactosyltransferase activity in lysates of control (column C), GLT25D1-null (columns D1a and D1b), and GLT25D2-null (column D2) cells with and without GLT25D1 cDNA (rGLT25D1) overexpression (mean ± S.D., n = 3 independent experiments).
The GLT25D1 and GLT25D2 galactosyltransferase genes were inactivated in SaOS-2 cells using the CRISPR/Cas9 system (26). Exons 7–10 of GLT25D1 and exons 7–12 of GLT25D2 encode the GT25 domain required for galactosyltransferase activity (Fig. 1B). We therefore targeted exon 2 to disrupt GLT25D1 and exon 3 to disrupt GLT25D2. To control for annealing specificity, we used a guide RNA (gRNA) sequence nearly identical to the GLT25D1-targeting gRNA by including a single base mismatch (Fig. 1C). We obtained 2 cell clones with null mutations at the expected location of GLT25D1 after screening 10 clones and 1 clone of 16 with a null mutation in GLT25D2. Sanger sequencing of the targeted GLT25D1 exon 2 revealed a homozygous mutation in the first cell clone and compound heterozygous mutations in the second cell clone. Sequencing of GLT25D2 exon 3 confirmed a homozygous point mutation at the expected genomic location (Fig. 1C). The frameshift mutations detected in the GLT25D1 clones yielded truncated open reading frames that lack the catalytic GT25 domain. To obtain cell clones containing both GLT25D1-null and GLT25D2-null genes, we transfected GLT25D1-null cells with the CRISPR/Cas9 construct targeting the GLT25D2 exon 3, which was successfully applied to disrupt GLT25D2. After screening 100 cell clones, we identified 14 clones carrying a single mutated GLT25D2 allele, but none that carried GLT25D1 and GLT25D2 genes inactivated on both alleles. SaOS-2 cells transfected with the mismatch control gRNA construct did not yield any genomic mutation in the GLT25D1 targeted region after screening 10 cell clones. These 10 cell clones were pooled and used as control cells in subsequent experiments. The loss of GLT25D1 protein expression in SaOS-2 cells bearing inactivating mutations was confirmed by Western blotting (Fig. 1D). Changes in GLT25D2 protein levels could not be assessed because GLT25D2 remained undetectable by Western blotting analysis in the three osteosarcoma cell lines tested. The inactivation of the GLT25D1 gene yielded a strong decrease of collagen galactosyltransferase activity down to 3–7% of the reference activity in native SaOS-2 cells and in the transfection control (Fig. 1E). Stable overexpression of a GLT25D1 cDNA transgene in GLT25D1-null cells increased collagen galactosyltransferase by 6–8-fold. By contrast, the inactivation of GLT25D2 in SaOS-2 cells only marginally decreased collagen galactosyltransferase activity to 92% of control values (Fig. 1E), thereby confirming GLT25D1 as the main enzyme mediating collagen galactosylation in SaOS-2. Taken together, these data confirmed the successful functional inactivating role of the introduced mutations.
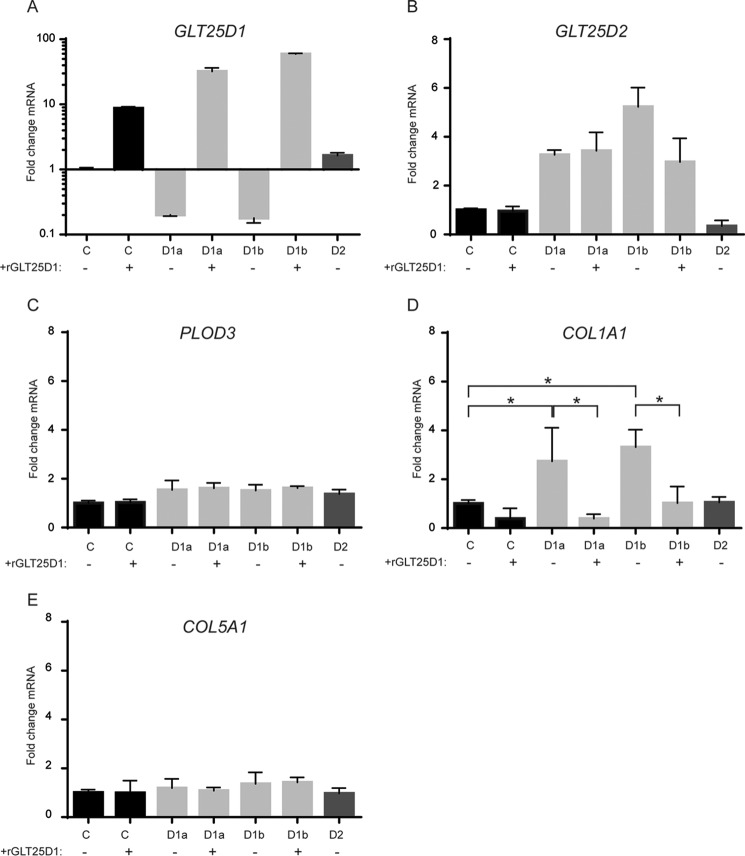
GLT25D1 Inactivation Up-regulates GLT25D2 and COL1A1 mRNA Levels
GLT25D1 mRNA levels were significantly reduced in GLT25D1-null cells, indicating induction of mRNA nonsense-mediated decay (27) (Fig. 2A). GLT25D1 mRNA levels were increased by 25% in GLT25D2-null cells, suggesting a possible compensatory effect by GLT25D1 upon inactivation of GL525D2. This increase of GLT25D1 transcript levels, however, did yield a corresponding increase of collagen galactosyltransferase activity as observed in Fig. 1E. Correspondingly, GLT25D2 was up-regulated 3–5-fold in GLT25D1-null cells (Fig. 2B). Interestingly, GLT25D2 expression was also increased in GLT25D1-null cells overexpressing a GLT25D1 cDNA transgene. As documented in a recent study, functional inactivation of single genes often results in compensatory overexpression by paralogous genes (28). Whereas GLT25D2 expression showed such a compensatory pattern, PLOD3 expression remained unchanged upon GLT25D1 and GLT25D2 inactivation (Fig. 2C), indicating that loss of collagen galactosyltransferase activity did not lead to increased lysyl hydroxylase gene expression. Surprisingly, GLT25D1 inactivation induced a strong expression of the COL1A1 gene encoding a collagen type I polypeptide (Fig. 2D). Normal COL1A1 expression was restored to control levels in GLT25D1-null cells upon overexpression of a GLT25D1 cDNA transgene, indicating that collagen galactosyltransferase activity was critical in regulating COL1A1 expression. Accordingly, COL1A1 mRNA levels were normal in GLT25D2-null cells (Fig. 2D). The impact of collagen galactosyltransferase activity on collagen expression was specific to COL1A1 because COL5A1, encoding a collagen type V polypeptide, was insensitive to GLT25D1 and GLT25D2 alterations (Fig. 2E).
FIGURE 2.
Transcription analysis of collagen and collagen modifying enzymes. Real time PCR analysis was performed on GLT25D1-null (bars D1a and D1b), GLT25D2-null (bars D2), and control (bars C) cell lines with and without GLT25D1 (rGLT25D1) overexpression. Primers specific for GLT25D1 (A), GLT25D2 (B), PLOD3 (C), COL1A1 (D), and Col5A1 (E) were used. Statistically significant differences as determined by two-tailed Student's t test (p < 0.05) are marked by asterisks (n = 3 independent experiments).
Collagen Glycosylation in GLT25D1-null Cells Is Partially Restored by GLT25D2
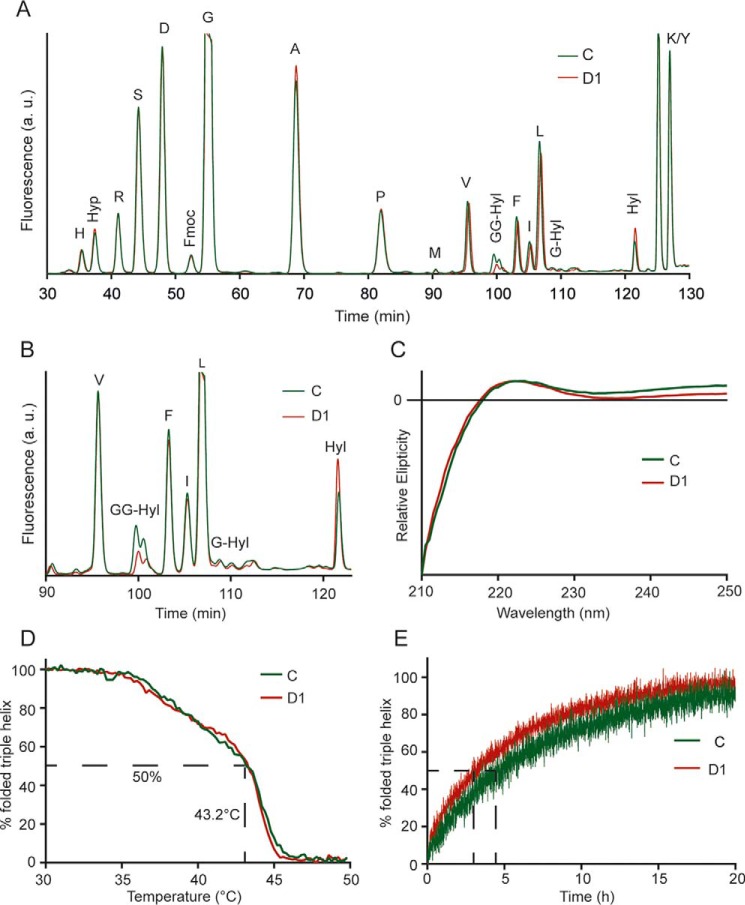
To quantify the compensatory effect of GLT25D2 on collagen glycosylation in GLT25D1-null cells, we determined collagen post-translational modifications in endogenously produced collagen by amino acid analysis. Whereas the overall levels of modified lysine remained unchanged, the amount of glycosylated Hyl carrying the Glc(α1–2)Gal disaccharide decreased in GLT25D1-null cells by 42 to 60%, and the amounts of free Hyl increased correspondingly (Fig. 3, A and B). Alterations of collagen folding result in overmodification of collagen because of extended exposure of collagen substrates to prolyl hydroxylases in the endoplasmic reticulum (ER) compartment (7, 29). Here, we did not detect any difference in hydroxyproline levels in GLT25D1-null collagen compared with collagen from control cells, suggesting a normal rate of collagen folding in GLT25D1-null cells.
FIGURE 3.
Analysis of collagen post-translational modifications and triple helical stability. A, amino acid analysis of collagens extracted from control (line C) and GLT25D1-null cells (line D1). Collagens were alkaline-hydrolyzed and Fmoc (N-(9-fluorenyl)methoxycarbonyl)-labeled before separation by HPLC. Hyp, hydroxyproline; GG-Hyl, glucosylgalactosyl-hydroxylysine; Hyl, hydroxylysine. B, zoom of region containing glycosylated Hyl. C, circular dichroism of collagens extracted from control (line C) and GLT25D1-null (line D1) cell lines. Spectra were recorded at 10 °C between 210 and 250 nm in a spectropolarimeter. D, thermal transition of control (line C) and GLT25D1-null (line D1) collagens in 0.1 m acetic acid. Temperature was raised from 30 to 50 °C with 0.5 °C/min. Tm value was calculated at 50% triple helical signal. One representative experiment is shown (total n = 3 independent experiments). E, refolding of control (line C) and GLT25D1-null (line D1) collagens in PBS. Collagens were denatured for 5 min at 50 °C prior to refolding at 20 °C.
Defects of triple helix formation because of altered post-translational modifications of collagen often impair the trafficking of collagens (7, 30). Under physiological conditions, only triple helical collagen is secreted, indicating that collagen secretion is directly proportional to triple helix formation (31). We assessed the triple helical conformation of endogenous collagen in control and GLT25D1-null cells using circular dichroism. The ellipticity spectra showed the typical peak at 222 nm, as well as a minimum under 200 nm in both control and GLT25D1-null cells (Fig. 3C). Thermal stability was determined by monitoring changes in ellipticity at 222 nm during heating collagen from 30 to 50 °C. The heating curve consisted of two phases, possibly reflecting the mixed composition consisting of collagen type I and collagen type V. Both collagens extracted from GLT25D1-null and control cell lines exhibited similar thermal properties with a normal melting temperature of 43.2 °C (Fig. 3D). By contrast, denatured collagen derived from GLT25D1-null cells refolded 40% faster than collagen from control cells (Fig. 3E). These results indicated that decreased collagen glycosylation affected the kinetic of triple helix formation but not triple helix thermal stability.
ER Accumulation of Collagen Type I in GLT25D1-null Cells
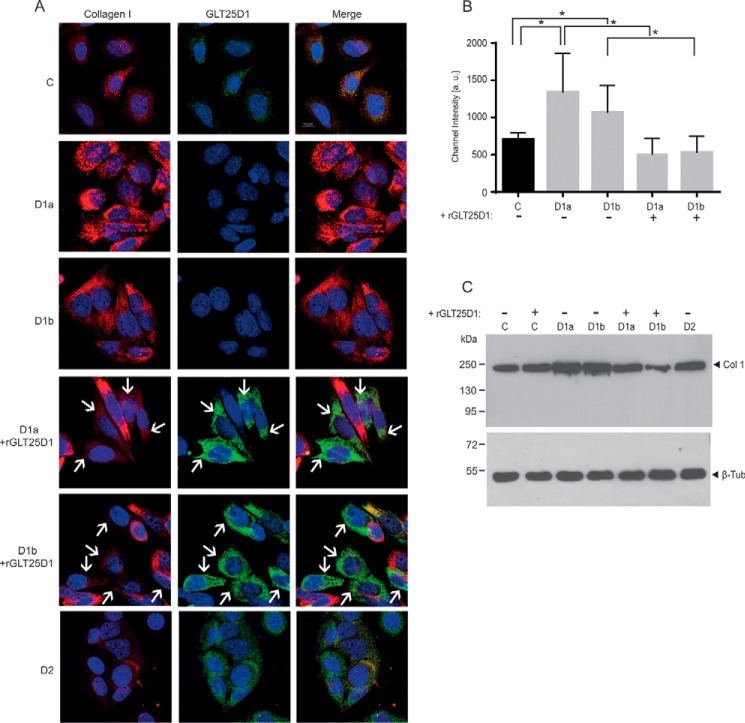
To confirm the increased collagen type I production in GLT25D1-null cells at the protein level, we first analyzed intracellular collagen amounts and distribution in control, GLT25D1-null, and GLT25D2-null SaOS-2 cells. In GLT25D1-null cells, collagen type I levels reached 150–190% of control values as measured by immunofluorescent channel intensity (Fig. 4, A and B). Collagen type I levels normalized upon GLT25D1 cDNA overexpression in GLT25D1-null cells. As noted for COL1A1 mRNA levels, inactivation of GLT25D2 did not alter intracellular collagen type I amounts. The increased collagen type I levels detected by immunofluorescence were further confirmed by Western blotting analysis (Fig. 4C). Collagen expression in GLT25D2-inactivated cells was equal to control cells and to GLT25D1 cDNA-overexpressing GLT25D1-null cells (Fig. 4A), indicating a minor impact of GLT25D2 inactivation on collagen expression.
FIGURE 4.
Immunofluorescent analysis. A, collagen type I and GLT25D1 staining in control (row C), GLT25D1-null (rows D1a and D1b), and GLT25D2-null (row D2) cells with and without overexpression of GLT25D1 cDNA (+GLT25D1). White arrows mark cells expressing the transfected GLT25D1 cDNA. Scale bar equals 10 μm. B, quantification of collagen type I channel intensity based on 50 cells. Asterisks above bars indicate statistically significant differences based on two-tailed paired t test (p < 0.05). C, Western blotting of collagen type I in GLT25D1-null (columns D1a and D1b), GLT25D2-null (column D2), control (column C), and GLT25D1 cDNA (+rGLT25D1) overexpressing cells. One representative experiment is shown (total n = 3 independent experiments).
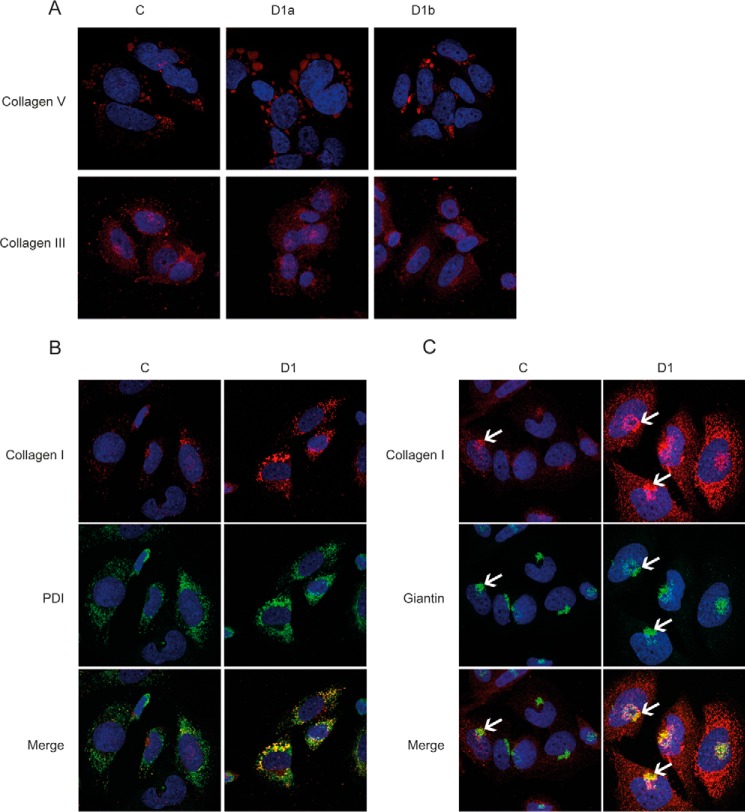
Even though collagen type V contains 10 times more glycosylated Hyl residues than collagen type I (32), increased cellular amounts were only visible for collagen type I. Immunofluorescent staining for collagens type III and V remained unchanged in GLT25D1-null cells (Fig. 5A). Inactivation of GLT25D1 may only affect collagen type I levels because this type of collagen is by far produced at highest amounts in SaOS-2 cells. To locate the site of collagen accumulation, we co-localized collagen type I with the ER marker protein disulfide-isomerase. Elevated collagen type I levels were clearly associated with the ER compartment as shown by co-localization with protein disulfide-isomerase (Fig. 5B). Staining of SaOS-2 cells with the Golgi marker giantin showed a dilatation of Golgi stacks and co-localization with collagen type I, thus indicating the translocation of collagen type I from the ER to the Golgi (Fig. 5C).
FIGURE 5.
Immunofluorescent analysis of collagen type III and V and co-localization of collagen type I with ER and Golgi. A, immunofluorescent staining of control (column C) and GLT25D1-null (column D1) cells with anti-collagen type V and anti-collagen type III antibodies (red). B, co-localization of collagen type I (red) and the ER marker protein disulfide-isomerase (PDI, green). C, co-localization of collagen type I (red) and the Golgi marker giantin (green). White arrows point to Golgi and collagen type I-positive region.
Increased Collagen Type I Does Not Induce ER Stress
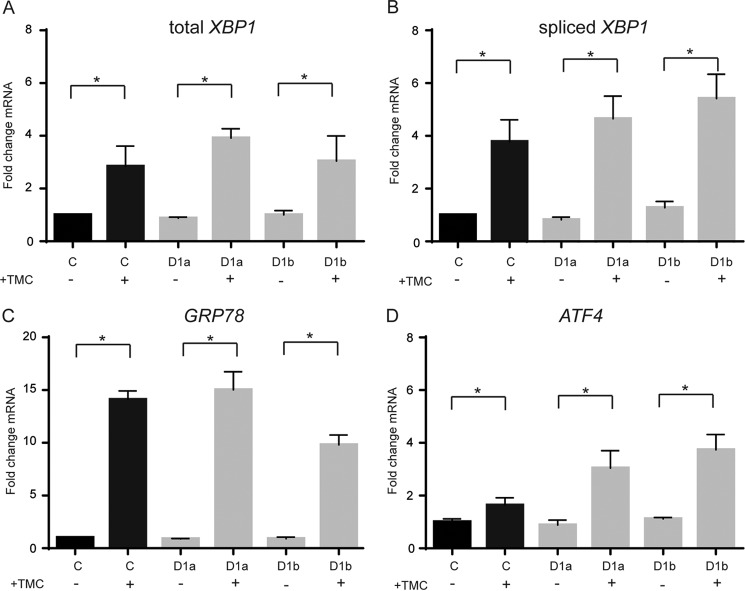
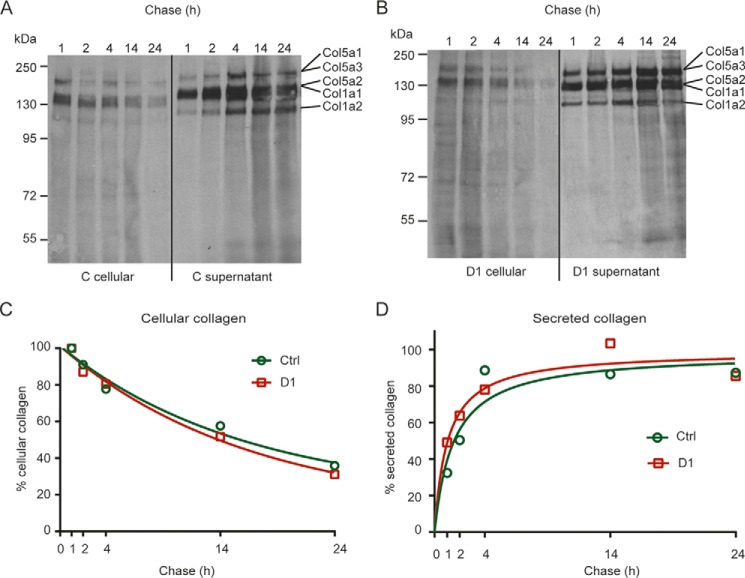
The intracellular accumulation of collagen type I and the occurrence of possibly improperly folded collagens could induce ER stress and thereby an unfolded protein response (33) in GLT25D1-null cells. To assess the possible induction of an unfolded protein response, we measured splicing of XBP1 (Fig. 6, A and B), the mRNA levels of GRP78 (Fig. 6C) and ATF4 (Fig. 6D) as markers of the unfolded protein response. Tunicamycin was used as a positive control to induce the unfolded protein response (34). Whereas tunicamycin treatment induced a robust unfolded protein response in control and GLT25D1-null cells, the three markers investigated remained unchanged in GLT25D1-null cells under conditions of collagen type I ER accumulation (Fig. 6, A–D). The absence of unfolded protein response in GLT25D1-null cells indicated that the accumulation of collagen type I was not related to impaired collagen folding in the ER and delayed transfer to the Golgi apparatus. Because the accumulation of collagen type I could be related to delayed collagen secretion, we quantified collagen production and secretion. The main types of collagen secreted by SaOS-2 cells are collagen type I and the highly glycosylated collagen type V (24). Both control and GLT25D1-null cells produced and secreted these two types of collagens normally as assessed by pulse-chase experiments (Fig. 7, A and B). Even tough total collagen secretion was increased in GLT25D1-null cells, the rate of collagen degradation (Fig. 7C) and secretion (Fig. 7D) remained largely unaffected in GLT25D1-null cells. The amount of collagen secreted measured after 1 and 2 h of chase was slightly higher in GLT25D1-null cells, but no difference was noticeable after 10 h, thus showing that loss of GLT25D1 maintained a normal rate collagen secretion.
FIGURE 6.
Analysis of the unfolded protein response. A–D, real time PCR analysis of RNA extracted from control (bars C) and GLT25D1-null (bars D1) cells. Specific primers for XBP1 (A), spliced XPB1 (B), GRP78 (C), and ATF4 (D) were used with or without induction of the unfolded protein response using tunicamycin (TMC). Statistically significant differences as determined by two-tailed Student's t test (p < 0.05) are marked by asterisks (n = 3 independent experiments).
FIGURE 7.
Analysis of collagen secretion. A–D, pulse-chase analysis of collagens from control (A) and GLT25D1-null (B) cells after pulse period of 4 h. C and D, collagen bands were quantified for cellular collagen (C) and for secreted collagens (D) using ImageJ. One representative experiment is shown (total n = 3 independent experiments).
Discussion
The inactivation of collagen galactosyltransferase GLT25D1 and GLT25D2 genes in osteosarcoma cells delineated the role of glycosylation in collagen expression and intracellular trafficking. Defective glycosylation had no impact on the rate of collagen secretion and triple helix thermal stability. By contrast, the loss of the main collagen galactosyltransferase isoform GLT25D1 led to ER accumulation of collagen type I in SaOS-2 cells. The intracellular accumulation of collagen type I resulted from increased collagen type I gene expression induced by low collagen galactosyltransferase activity. In addition, GLT25D1 inactivation was compensated at the transcriptional level by induction of GLT25D2, which is normally hardly expressed in osteosarcoma cells.
Genetic compensation consecutive to gene inactivation is an effect that has previously been described in animals and plants such as Arabidopsis (28). Accordingly, compensation by induction of paralogous genes often occurs in parallel to lethal mutations (35). Our results on the induction of GLT25D2 in GLT25D1-null cells are in agreement with such a compensatory reaction and underline the necessity to inactivate both collagen galactosyltransferase isoforms to obtain a complete loss of collagen glycosylation. Our failure to isolate cell clones harboring both inactive GLT25D1 and GLT25D2 genes supports the notion that collagen glycosylation could be essential for osteosarcoma cell viability and that the induction of GLT25D2 is required to prevent the complete loss of collagen galactosyltransferase activity when the main GLT25D1 isoform is disrupted.
The inactivation of GLT25D1 led to the up-regulation of collagen type I expression, which is the main type of collagen produced in SaOS-2 cells. Up-regulation of collagen type I in response to mutation of the post-translational machinery is also seen in fibroblasts from Bruck syndrome type 2 patients, in which mutations in PLOD2 encoding the lysyl hydroxylase 2 isoform decrease the level of telopeptide lysyl hydroxylation and thereby the formation of telopeptide-based intermolecular cross-links (9). By contrast, hypermodification of collagen, as observed in osteogenesis imperfecta caused by delayed collagen folding, does not increase collagen type I production (36, 37). The capping of Hyl by glycosylation may act as a signal for the packaging of folded collagen into vesicular structures (38) to be transported to Golgi cisternae. Other classes of glycosylation, such as N-linked glycosylation, also convey signals for folded glycoproteins to depart the ER compartment (39). Carbohydrate-binding proteins, such as ERGIC53 (40), recognize glycan chains on glycoproteins and mediate their transfer to the cis-Golgi compartment.
Collagen type I is regulated by various factors including interleukins, insulin-like growth factor-1, and transforming growth factor-β (41). Mice deficient for β1-integrin lack feedback regulation of collagen synthesis (42). Because glycosylation was shown to impact collagen-integrin interaction, integrins may be involved in the collagen type I up-regulation seen in GLT25D1-null cells by modulated binding of collagen to integrin and subsequent alteration of transforming growth factor-β signaling. Another collagen-interacting receptor, uPARAP, also depends on collagen glycosylation for matrix remodeling. uPARAP mediates the preferential endocytosis of glycosylated collagen IV and V (18, 43). uPARAP itself has no direct regulatory function in collagen expression because uPARAP−/− mice show normal collagen production. Nevertheless, uPARAP could be regulating collagen homeostasis by altered collagen uptake, thereby changing the intracellular collagen pool and making it available for intracellular feedback regulation.
Collagen glycosylation is conserved in the animal kingdom from sponges to humans (14–17), suggesting an important contribution of glycosylation to collagen properties in the extracellular matrix. In addition to the intracellular functions investigated in the present study of osteosarcoma cells, glycosylation may be involved in the organization of collagens in the extracellular space. Glycosylation has been suggested to regulate cross-link formation in fibrillar collagens (44). Also, the glycosylation of collagenous domains of adiponectin, serum mannose-binding lectin, and complement factor C1q has been involved in the control of subunit oligomerization (45, 46). Whereas the present study uncovered an unexpected link between collagen glycosylation and collagen type I expression in osteosarcoma cells, the inactivation of GLT25D1 and GLT25D2 in model organisms will be required to uncover the role of collagen glycosylation in shaping the extracellular matrix.
Materials and Methods
Cell Lines and Culture Conditions
SaOS-2 cells (ATCC: HTB-85), U2OS cells (ATCC: HTB-96), and MG63 cells (ATCC: CRL-1427) were provided by Dr. Roman Muff (Sarkomzentrum Zürich, University of Zurich). All cell lines were authenticated (Microsynth, Balgach, Switzerland) and negatively tested for mycoplasma contamination. The cells were grown in McCoy's 5A (modified) medium (Thermo Fisher, Waltham, MA) containing 15% fetal bovine serum (Biochrom, Berlin, Germany) at 37 °C in 5% CO2.
Cloning and Transfection of CRISPR/Cas9 Vectors
The gRNA sequences for GLT25D1 exon 2 (forward: 5′-CACCGGAAGAGTTTGTACCATTCCG-3′, reverse: 5′-AAACCGGAATGGTACAAACTCTTC-3′), for GLT25D2 exon 3 (forward: 5′-CACCGCCATGTGATGAAACTACGAC-3′, reverse: 5′-AAACGTCGTAGTTTC-ATCACATGGC-3′), and for the control construct (forward: 5′-CACCGGAAGAGTTTGTACCTTTCC G-3′, reverse: 5′-AAACCGGAAAGGTACAAACTCTTC-3′) were ligated into the BbsI sites of pSpCas9(BB) (gift from Dr. Feng Zhang, Addgene plasmid no. 48139). Osteosarcoma cells were transfected using the AMAXA nucleofector kit (Lonza, Basel, Switzerland) according to the manufacturer protocol. The cells were selected for positive transfection with 0.5 μg/ml of puromycin (Santa Cruz Biotechnology Inc., Dallas, TX). Single clones were isolated and analyzed for mutations in the targeted genes using the Surveyor nuclease assay (Integrated DNA Technologies, Coralville, IA) (47) and the primers 5′-GGAGAAGTGTCCTGTCCAGGGATAC-3′ and 5′-ACAGGGAACGGCTTGGGCAAAGGTC-3′ for GLT25D1 exon 2 and 5′-CCCTGATGAAATTGGACCAAAGC-3′ and 5′-TGCCTTTCTTAAAAAGTGGGGG-3′ for GLT25D2 exon 3. Mutations were confirmed by Sanger sequencing (Microsynth).
Cloning and Transfection of GLT25D1 Overexpression Vector
GLT25D1 cDNA was cloned from the pFastBacI baculovirus transfer vector from Ref. 13 in pcDNA3.1(+) (Thermo Fisher) using the NotI and XbaI restriction sites. SaOS-2 cells were transfected using AMAXA nucleofector kit (Lonza). The cells were selected 48 h after positive transfection using 2.5 μg/ml of Geneticin (Thermo Fisher) for 10 days.
Collagen Galactosyltransferase Assay
107 cells were lysed in 200 μl of Tris-buffered saline, 1% Triton X-100, pH 7.4, for 15 min on ice. Nuclei and debris were removed by spinning at 13,000 × g for 10 min at 4 °C, and supernatants were used as enzyme source. Bovine collagen type I was heat-denatured for 10 min at 60 °C and kept on ice until use. Activity assays included 10 μl of cell lysate, 0.5 mg/ml denatured collagen acceptor, 60 μm UDP-Gal, 50,000 cpm of UDP-[14C]Gal (GE Healthcare), 10 mm MnCl2, 20 mm NaCl, 50 mm MOPS (pH 7.4) and 1 mm 1,4-dithiothreitol. The reactions were incubated for 3 h at 37 °C and stopped by the addition of 500 μl of 5% trichloroacetic acid 5% phosphotungstic acid for 30 min on ice. Precipitated proteins were recovered on filters using a vacuum manifold and washed with 15 ml of 50% ethanol, and radioactivity was measured in a Tri-Carb 2900TR scintillation counter (PerkinElmer Life Sciences).
Quantitative PCR Analysis
Total RNA was extracted from 5 × 106 cells using TRIzol reagent (Life Technologies, Inc.) according to the manufacturer protocol. First strand cDNA was produced using 2 μg of total RNA and RevertAid reverse transcriptase (Life Technologies, Inc.). 10 μl of 2× SsoAdvanced universal SYBR Green Supermix (Bio-Rad) were mixed with 1 μl of translated cDNA and 1 μl of 10 μm diluted primer pair (Table 1) in a 20-μl reaction. Specific real time PCR primers (Table 1) for the unfolded protein response were selected from Oslowski and Urano (34). Primers for collagen type I COL1A1 and collagen type V COL5A1 mRNA quantification were designed using the primer-BLAST software (48) and encompassed exon sequences flanking at least one intron.
TABLE 1.
Primer pairs used for amplification in real time PCRs
| Gene | Forward primer | Reverse primer |
|---|---|---|
| COL1A1 | 5′-GCTCGTGGAAATGATGGTGC-3′ | 5′-ACCCTGGGGACCTTCAGAG-3′ |
| COL5A1 | 5′-CTTGGCCCAAAGAAAACCCG-3′ | 5′-TAGGAGAGCAGTTTCCCACG-3′ |
| GAPDH | 5′-CGCTCTCTGCTCCTCCTGTT-3′ | 5′-CCATGGTGTCTGAGCGATGT-3′ |
| Spliced XBP1 | 5′-TGCTGAGTCCGCAGCAGGTG-3′ | 5′-ATCCATGGGGAGATGTTCTGG-3′ |
| Unspliced XBP1 | 5′-CAGCACTCAGACTACGTGCA-3′ | 5′-TGGCCGGGTCTGCTGAGTCCG-3′ |
| ATF4 | 5′-GTTCTCCAGCGACAAGGCTA-3 | 5′-ATCCTCCTTGCTGTTGTTGG-3′ |
| GRP78 | 5′-TGTTCAACCAATTATCAGCAAACTC-3′ | 5′-TTCTGCTGTATCCTCTTCACCAGT-3′ |
| GLT25D1 | 5′-ACTCACGCTACGAGCATGTC-3′ | 5′-GTGTCAGGGTTGAGGATCAG-3′ |
| GLT25D2 | 5′-ACTATGGCTACCTGCCCATC-3′ | 5′-GGGACAACTGAGACATACTG-3′ |
| PLOD3 | 5′-AGAACCTCAACGGGGCTTTA-3′ | 5′-CTTAGTGGGACCGTTTCCAT-3′ |
Collagen Extraction
Osteosarcoma cells were grown to 70% confluency in DMEM containing 2% fetal calf serum, 50 μg/ml ascorbate, and 50 μg/ml catalase. Ascorbate and catalase were replenished every 48 h for 8 days. Medium and cells were collected and digested at 10 μg/ml pepsin in 1 m HCl for 6 h at 22 °C. Neutral pH was restored by addition of 1 m NaOH, and collagens were precipitated in 50% ethanol at −20 °C overnight. Collagens were resuspended in 10 ml of 0.1 m acetic acid at 4 °C for 48 h and then concentrated and purified using 100-kDa cutoff Amicon ultra centrifugal filter units (Sigma-Aldrich). Collagens were diluted to 0.1 mg/ml and stored at 4 °C for 72 h prior to analysis.
HPLC Amino Acid Analysis
Purified collagens (10 μg) were hydrolyzed in 500 μl of 4 m KOH at 105 °C for 20 h. Hydrolysates were neutralized using perchloric acid. Salt precipitates were removed, and supernatant was dried down under nitrogen, washed twice with 500 μl of water, and then dried down again. Samples were resuspended in 100 μl of water and derivatized using 9-fluorenylmethoxycarbonyl chloride following the procedure of Bank et al. (49). Derivatized amino acid samples were analyzed by reverse phase HPLC (13).
Circular Dichroism
Purified collagens were diluted to 0.1 mg/ml in 0.1 m acetic acid. Ellipticity was measured between 210 and 250 nm in a spectropolarimeter (J-810, Jasco) with a thermostatted quartz cuvette with 1-mm length. Thermal stability was analyzed at 222 nm under heating at a rate of 0.5 °C/min from 30 to 50 °C. For refolding, collagen (1 mg/ml) in PBS was denatured for 5 min at 50 °C, and ellipticity was monitored at 222 nm for 20 h.
Immunofluorescence
The cells were grown on sterile 11-mm glass coverslips for 48 h, then fixed in 4% paraformaldehyde in PBS for 15 min at room temperature and then permeabilized with 0.5% saponin for 10 min at room temperature. The cells were incubated with 5% bovine serum albumin (Sigma-Aldrich) for 1 h and then with primary antibodies diluted in PBS, 0.05% Tween, 1% bovine serum albumin for 1 h. After three wash steps in PBS, 0.05% Tween, the cells were incubated with secondary antibodies for 1 h at room temperature. The cells were washed three times in PBS, and the nuclei were stained with DAPI (Biotium, Hayward, CA). Coverslips were mounted on ProLong Gold antifade medium (Life Technologies, Inc.). Antibodies used were mouse anti-collagen type I (ab6308; Abcam, Cambridge, UK), rabbit anti-collagen type I (ab34710; Abcam), goat anti-collagen type III (ab24129; Abcam), rabbit anti-collagen type V (ab7046; Abcam), rabbit anti-GLT25D1 (ab151011; Abcam), mouse anti-protein disulfide-isomerase (RL90) (Alexis 804-012; Enzo Life Sciences, Lausen, Switzerland), and rabbit anti-giantin (ab24586; Abcam). The secondary antibodies used were goat anti-mouse 647 (ab150119; Abcam), goat anti-rabbit 488 (ab150077; Abcam,) and donkey anti-goat 488 (ab150129; Abcam).
Image Acquisition and Channel Intensity Quantification
Coverslips were imaged using a confocal laser scanning microscope type Leica TCS SP8 using a HCX PL APO CS2 oil objective lens at 63× magnification, f1.4 numerical aperture at room temperature. The fluorophores DAPI, Alexa 488, and Alexa 647 were excited sequentially at 405, 488, and 638 nm, respectively. The spectral emission detection unit was set between 415 and 525 nm for DAPI, between 495 and 590 nm for Alexa 488, and between 645 and 750 nm for Alexa 647. Images were recorded at a resolution of 1400 × 1400 pixels at 1000 Hz, with a line average of 3, and z-stack (∼25) height was set to system optimized by the software. Images were recorded using two-hybrid detectors using the Leica LAS X software (Leica Microsystems AG, Heerbrugg, Switzerland). Three-dimensional images were projected to two dimensions using Imaris 7 (Bitplane AG, Switzerland) with the according MATLAB plugin (MathWorks, Bern, Switzerland) using MIP projection on the x-y plane. Channel intensities were quantified using ImageJ 1.50B (50) as described (51).
Western Blotting Analysis
5 × 106 cells were lysed in radioimmune precipitation assay lysis buffer for 30 min on ice and then spun for 15 min at 13,000 × g at 4 °C. Amounts of 20 μg of whole protein lysates were resolved on 10% SDS-PAGE. Proteins were blotted on a nitrocellulose membrane (GE Healthcare) and blocked in 5% skim milk powder in PBS for 1 h. The membranes were incubated overnight at 4 °C with anti-GLT25D1 antibody at 1:250 (ab151011; Abcam), anti-collagen type I at 1:10,000 (ab138492; Abcam), or anti-β-tubulin I at 1:10,000 (Sigma-Aldrich) in Tris-buffered saline, 0.05% Tween, 1% bovine serum albumin. Secondary goat anti-rabbit or goat anti-mouse antibodies coupled to horseradish peroxidase at 1:10,000 (Sigma-Aldrich) were diluted in Tris-buffered saline, 0.05% Tween and incubated for 1 h at room temperature.
Pulse-Chase Labeling of Collagens
The cells were plated in 6-well plates at 2.5 × 105 cells/well and incubated for 24 h at 37 °C. Medium was exchanged with DMEM (Sigma-Aldrich) with 10% fetal bovine serum (Biochrom AG) containing 50 μg/ml ascorbate and 50 μg/ml catalase, and the cells were incubated overnight at 37 °C. The cells were washed once in PBS and then pulsed in 1 ml of DMEM, 50 μg/ml ascorbate, 20 μCi/ml l-[14C(U)]-proline (PerkinElmer Life Sciences) for 4 h. Chase was initiated by changing medium to DMEM, 50 μg/ml ascorbate, 30 μg/ml l-proline. Secreted and cellular collagens were digested in the cell medium using 25 μg/ml pepsin in 1 m HCl for 2 h at 4 °C. Neutral pH was restored by the addition of 1 m NaOH, and collagens were precipitated in 50% ethanol at −20 °C overnight and then resuspended in Laemmli buffer. The collagens were separated in 9% SDS-PAGE and blotted on nitrocellulose membrane (GE Healthcare) prior to autoradiography. Band intensity was quantified using the open source software ImageJ (50) with the gel analyzer plugin.
Author Contributions
S. B. and T. H. designed and coordinated the study, S. B. performed the experiments, and S. B. and T. H. wrote the manuscript.
Acknowledgment
We thank Prof. Dr. Roman Muff for providing us with SaOS-2, MG63, and U2OS cells.
This work was supported by the Research Credit of the University of Zurich (to S. B.) and by Swiss National Foundation Grant 310030_149949 (to T. H.). The authors declare that they have no conflicts of interest with the contents of this article.
- Hyl
- hydroxylysine
- uPARAP
- urokinase-type plasminogen activator receptor associated protein
- ER
- endoplasmic reticulum
- gRNA
- guide RNA.
References
- 1. Myllyharju J., and Kivirikko K. I. (2004) Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 20, 33–43 [DOI] [PubMed] [Google Scholar]
- 2. Kukkola L., Hieta R., Kivirikko K. I., and Myllyharju J. (2003) Identification and characterization of a third human, rat, and mouse collagen prolyl 4-hydroxylase isoenzyme. J. Biol. Chem. 278, 47685–47693 [DOI] [PubMed] [Google Scholar]
- 3. Vranka J. A., Sakai L. Y., and Bächinger H. P. (2004) Prolyl 3-hydroxylase 1, enzyme characterization and identification of a novel family of enzymes. J. Biol. Chem. 279, 23615–23621 [DOI] [PubMed] [Google Scholar]
- 4. Myllyharju J. (2008) Prolyl 4-hydroxylases, key enzymes in the synthesis of collagens and regulation of the response to hypoxia, and their roles as treatment targets. Ann. Med. 40, 402–417 [DOI] [PubMed] [Google Scholar]
- 5. Spiro R. G. (1967) The structure of the disaccharide unit of the renal glomerular basement membrane. J. Biol. Chem. 242, 4813–4823 [PubMed] [Google Scholar]
- 6. Harwood R., Grant M. E., and Jackson D. S. (1975) Studies on the glycosylation of hydroxylysine residues during collagen biosynthesis and the subcellular localization of collagen galactosyltransferase and collagen glucosyltransferase in tendon and cartilage cells. Biochem. J. 152, 291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cabral W. A., Chang W., Barnes A. M., Weis M., Scott M. A., Leikin S., Makareeva E., Kuznetsova N. V., Rosenbaum K. N., Tifft C. J., Bulas D. I., Kozma C., Smith P. A., Eyre D. R., and Marini J. C. (2007) Prolyl 3-hydroxylase 1 deficiency causes a recessive metabolic bone disorder resembling lethal/severe osteogenesis imperfecta. Nat. Genet. 39, 359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brinckmann J., Açil Y., Feshchenko S., Katzer E., Brenner R., Kulozik A., and Kügler S. (1998) Ehlers-Danlos syndrome type VI: lysyl hydroxylase deficiency due to a novel point mutation (W612C). Arch Derm. Res. 290, 181–186 [DOI] [PubMed] [Google Scholar]
- 9. van der Slot A. J., Zuurmond A. M., Bardoel A. F., Wijmenga C., Pruijs H. E., Sillence D. O., Brinckmann J., Abraham D. J., Black C. M., Verzijl N., DeGroot J., Hanemaaijer R., TeKoppele J. M., Huizinga T. W., and Bank R. A. (2003) Identification of PLOD2 as telopeptide lysyl hydroxylase, an important enzyme in fibrosis. J. Biol. Chem. 278, 40967–40972 [DOI] [PubMed] [Google Scholar]
- 10. Salo A. M., Cox H., Farndon P., Moss C., Grindulis H., Risteli M., Robins S. P., and Myllylä R. (2008) A connective tissue disorder caused by mutations of the lysyl hydroxylase 3 gene. Am. J. Hum. Genet. 83, 495–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rauch F., Fahiminiya S., Majewski J., Carrot-Zhang J., Boudko S., Glorieux F., Mort J. S., Bächinger H. P., and Moffatt P. (2015) Cole-Carpenter syndrome is caused by a heterozygous missense mutation in P4HB. Am. J. Hum. Genet. 96, 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liefhebber J. M., Punt S., Spaan W. J., and van Leeuwen H. C. (2010) The human collagen β(1-O)galactosyltransferase, GLT25D1, is a soluble endoplasmic reticulum localized protein. BMC Cell Biol. 11, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schegg B., Hülsmeier A. J., Rutschmann C., Maag C., and Hennet T. (2009) Core glycosylation of collagen is initiated by two β(1-O)galactosyltransferases. Mol. Cell Biol. 29, 943–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heikkinen J., Risteli M., Wang C., Latvala J., Rossi M., Valtavaara M., and Myllylä R. (2000) Lysyl hydroxylase 3 is a multifunctional protein possessing collagen glucosyltransferase activity. J. Biol. Chem. 275, 36158–36163 [DOI] [PubMed] [Google Scholar]
- 15. Wang C., Kovanen V., Raudasoja P., Eskelinen S., Pospiech H., and Myllylä R. (2009) The glycosyltransferase activities of lysyl hydroxylase 3 (LH3) in the extracellular space are important for cell growth and viability. J. Cell Mol. Med. 13, 508–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sricholpech M., Perdivara I., Nagaoka H., Yokoyama M., Tomer K. B., and Yamauchi M. (2011) Lysyl hydroxylase 3 glucosylates galactosylhydroxylysine residues in type I collagen in osteoblast culture. J. Biol. Chem. 286, 8846–8856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Junqua S., Robert L., Garrone R., Pavans de Ceccatty M., and Vacelet J. (1974) Biochemical and morphological studies on collagens of horny sponges: ircinia filaments compared to spongines. Connect. Tissue Res. 2, 193–203 [DOI] [PubMed] [Google Scholar]
- 18. Jürgensen H. J., Madsen D. H., Ingvarsen S., Melander M. C., Gårdsvoll H., Patthy L., Engelholm L. H., and Behrendt N. (2011) A novel functional role of collagen glycosylation: interaction with the endocytic collagen receptor uparap/ENDO180. J. Biol. Chem. 286, 32736–32748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lauer-Fields J. L., Malkar N. B., Richet G., Drauz K., and Fields G. B. (2003) Melanoma cell CD44 interaction with the α1(IV)1263–1277 region from basement membrane collagen is modulated by ligand glycosylation. J. Biol. Chem. 278, 14321–14330 [DOI] [PubMed] [Google Scholar]
- 20. Stawikowski M. J., Aukszi B., Stawikowska R., Cudic M., and Fields G. B. (2014) Glycosylation modulates melanoma cell α2β1 and α3β1 integrin interactions with type IV collagen. J. Biol. Chem. 289, 21591–21604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peake P. W., Hughes J. T., Shen Y., and Charlesworth J. A. (2007) Glycosylation of human adiponectin affects its conformation and stability. J. Mol. Endocrinol. 39, 45–52 [DOI] [PubMed] [Google Scholar]
- 22. Colley K. J., and Baenziger J. U. (1987) Identification of the post-translational modifications of the core-specific lectin. The core-specific lectin contains hydroxyproline, hydroxylysine, and glucosylgalactosylhydroxylysine residues. J. Biol. Chem. 262, 10290–10295 [PubMed] [Google Scholar]
- 23. Heise C. T., Nicholls J. R., Leamy C. E., and Wallis R. (2000) Impaired secretion of rat mannose-binding protein resulting from mutations in the collagen-like domain. J. Immunol. 165, 1403–1409 [DOI] [PubMed] [Google Scholar]
- 24. Pautke C., Schieker M., Tischer T., Kolk A., Neth P., Mutschler W., and Milz S. (2004) Characterization of osteosarcoma cell lines MG-63, Saos-2 and U-2 OS in comparison to human osteoblasts. Anticancer Res. 24, 3743–3748 [PubMed] [Google Scholar]
- 25. McQuillan D. J., Richardson M. D., and Bateman J. F. (1995) Matrix deposition by a calcifying human osteogenic sarcoma cell line (SAOS-2). Bone 16, 415–426 [DOI] [PubMed] [Google Scholar]
- 26. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chang Y. F., Imam J. S., and Wilkinson M. F. (2007) The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 76, 51–74 [DOI] [PubMed] [Google Scholar]
- 28. Rossi A., Kontarakis Z., Gerri C., Nolte H., Hölper S., Krüger M., and Stainier D. Y. (2015) Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230–233 [DOI] [PubMed] [Google Scholar]
- 29. Pace J. M., Kuslich C. D., Willing M. C., and Byers P. H. (2001) Disruption of one intra-chain disulphide bond in the carboxyl-terminal propeptide of the pro-α1(I) chain of type I procollagen permits slow assembly and secretion of overmodified, but stable procollagen trimers and results in mild osteogenesis imperfecta. J. Med. Genet. 38, 443–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ruotsalainen H., Sipilä L., Vapola M., Sormunen R., Salo A. M., Uitto L., Mercer D. K., Robins S. P., Risteli M., Aszodi A., Fässler R., and Myllylä R. (2006) Glycosylation catalyzed by lysyl hydroxylase 3 is essential for basement membranes. J. Cell Sci. 119, 625–635 [DOI] [PubMed] [Google Scholar]
- 31. Yoshikawa K., Takahashi S., Imamura Y., Sado Y., and Hayashi T. (2001) Secretion of non-helical collagenous polypeptides of α1(IV) and α2(IV) chains upon depletion of ascorbate by cultured human cells. J. Biochem. 129, 929–936 [DOI] [PubMed] [Google Scholar]
- 32. Mizuno K., Adachi E., Imamura Y., Katsumata O., and Hayashi T. (2001) The fibril structure of type V collagen triple-helical domain. Micron 32, 317–323 [DOI] [PubMed] [Google Scholar]
- 33. Hetz C. (2012) The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 13, 89–102 [DOI] [PubMed] [Google Scholar]
- 34. Oslowski C. M., and Urano F. (2011) Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol 490, 71–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gu Z., Steinmetz L. M., Gu X., Scharfe C., Davis R. W., and Li W. H. (2003) Role of duplicate genes in genetic robustness against null mutations. Nature 421, 63–66 [DOI] [PubMed] [Google Scholar]
- 36. Rowe D. W., Shapiro J. R., Poirier M., and Schlesinger S. (1985) Diminished type-1 collagen-synthesis and reduced α-1(1) collagen messenger-RNA in cultured fibroblasts from patients with dominantly inherited (type-1) osteogenesis imperfecta. J. Clin. Invest. 76, 604–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van Dijk F. S., Cobben J. M., Kariminejad A., Maugeri A., Nikkels P. G., van Rijn R. R., and Pals G. (2011) Osteogenesis imperfecta: a review with clinical examples. Mol. Syndromol 2, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Malhotra V., and Erlmann P. (2011) Protein export at the ER: loading big collagens into COPII carriers. EMBO J. 30, 3475–3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Helenius A., and Aebi M. (2001) Intracellular functions of N-linked glycans. Science 291, 2364–2369 [DOI] [PubMed] [Google Scholar]
- 40. Nichols W. C., Seligsohn U., Zivelin A., Terry V. H., Hertel C. E., Wheatley M. A., Moussalli M. J., Hauri H. P., Ciavarella N., Kaufman R. J., and Ginsburg D. (1998) Mutations in the ER-Golgi intermediate compartment protein ERGIC-53 cause combined deficiency of coagulation factors V and VIII. Cell 93, 61–70 [DOI] [PubMed] [Google Scholar]
- 41. Rossert J., Terraz C., and Dupont S. (2000) Regulation of type I collagen genes expression. Nephrol. Dial Transplant. 15, 66–68 [DOI] [PubMed] [Google Scholar]
- 42. Gardner H., Broberg A., Pozzi A., Laato M., and Heino J. (1999) Absence of integrin α1β1 in the mouse causes loss of feedback regulation of collagen synthesis in normal and wounded dermis. J. Cell Sci. 112, 263–272 [DOI] [PubMed] [Google Scholar]
- 43. Engelholm L. H., List K., Netzel-Arnett S., Cukierman E., Mitola D. J., Aaronson H., Kjøller L., Larsen J. K., Yamada K. M., Strickland D. K., Holmbeck K., Danø K., Birkedal-Hansen H., Behrendt N., and Bugge T. H. (2003) uPARAP/Endo180 is essential for cellular uptake of collagen and promotes fibroblast collagen adhesion. J. Cell Biol. 160, 1009–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Terajima M., Perdivara I., Sricholpech M., Deguchi Y., Pleshko N., Tomer K. B., and Yamauchi M. (2014) Glycosylation and cross-linking in bone type I collagen. J. Biol. Chem. 289, 22636–22647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shinkai H., and Yonemasu K. (1979) Hydroxylysine-linked glycosides of human complement subcomponent C1q and of various collagens. Biochem. J. 177, 847–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Håkansson K., and Reid K. B. (2000) Collectin structure: a review. Protein Sci. 9, 1607–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Qiu P., Shandilya H., D'Alessio J. M., O'Connor K., Durocher J., and Gerard G. F. (2004) Mutation detection using Surveyor (TM) nuclease. BioTechniques 36, 702–707 [DOI] [PubMed] [Google Scholar]
- 48. Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., and Madden T. L. (2012) Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bank R. A., Jansen E. J., Beekman B., and te Koppele J. M. (1996) Amino acid analysis by reverse-phase high-performance liquid chromatography: improved derivatization and detection conditions with 9-fluorenylmethyl chloroformate. Anal. Biochem. 240, 167–176 [DOI] [PubMed] [Google Scholar]
- 50. Burger W., and Burge M. J. (2008) In Digital Image Processing-An Algorithmic Approach using Java, 1st Ed., pp 469–523, Springer-Verlag, New York [Google Scholar]
- 51. McCloy R. A., Rogers S., Caldon C. E., Lorca T., Castro A., and Burgess A. (2014) Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle 13, 1400–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]