Abstract
cis-Prenyltransferases (cis-PTs) constitute a large family of enzymes conserved during evolution and present in all domains of life. cis-PTs catalyze consecutive condensation reactions of allylic diphosphate acceptor with isopentenyl diphosphate (IPP) in the cis (Z) configuration to generate linear polyprenyl diphosphate. The chain lengths of isoprenoid carbon skeletons vary widely from neryl pyrophosphate (C10) to natural rubber (C>10,000). The homo-dimeric bacterial enzyme, undecaprenyl diphosphate synthase (UPPS), has been structurally and mechanistically characterized in great detail and serves as a model for understanding the mode of action of eukaryotic cis-PTs. However, recent experiments have revealed that mammals, fungal, and long-chain plant cis-PTs are heteromeric enzymes composed of two distantly related subunits. In this review, the classification, function, and evolution of cis-PTs will be discussed with a special emphasis on the role of the newly described NgBR/Nus1 subunit and its plants' orthologs as essential, structural components of the cis-PTs activity.
Keywords: enzyme, genetic disease, glycosylation, glycosylphosphatidylinositol (GPI anchor), lipid, dolichol, polyprenol, cis-prenyltransferase, polyprenol synthesis, NgBR, hCIT, NUS1, RER2, DHDDS
Introduction
The synthesis of polyprenols in all living organisms is essential for many cellular functions. The five-carbon building block, isopentenyl diphosphate (IPP),2 and its isomer, dimethylallyl diphosphate (DMAPP), are precursors for the synthesis of over 55,000 structurally and chemically diverse isoprenoids including dolichols, sterols, the side chain of ubiquinone and chlorophyll, prenyl groups of certain proteins and tRNA molecules, carotenoids, plant and animal steroid hormones, and aromatic compounds (1, 2). In a majority of eukaryotic cells, Archaea, and some Eubacteria, IPP and DMAPP are synthesized via the mevalonate pathway (MVA). In most bacteria, in some Protozoa (Plasmodium sp.), and in chloroplasts of all phototrophic organisms, IPP and DMAPP are made via methylerythritol phosphate (MEP) pathway (3).
Linear isoprenoids are synthesized by a group of enzymes called prenyltransferases (PTs). PTs catalyze the head-to-tail condensation reactions of allylic primer (for example, DMAPP, farnesyl diphosphate (FPP), geranyl diphosphate, or geranylgeranyl diphosphate) with specific numbers of IPP units. Depending on the stereochemistry of the double bonds formed during IPP condensation, PTs belong to two structurally different families: trans- or (E)-prenyltransferase and cis- or (Z)-prenyltransferase. These enzymes utilize similar allylic and homo-allylic substrates but differ completely in their primary amino acid sequences, tertiary structures, and mechanisms for substrate binding and catalysis (2, 4, 5).
The carbon skeletons of the majority of isoprenoids are derived from products of trans-PTs. trans-PTs share a common protein fold and two conserved, functionally important asparagine-rich (DDXX(XX)D) motifs. trans-PTs synthesize linear allylic diphosphates ranging in size from 2 (C10) to 10 (C50) isoprene units (1). The precise mechanisms of prenyl chain elongation are well established based on structural and site-directed mutagenesis studies of a number of enzymes in this class (1, 2, 5).
On the other hand, cis-PTs synthesize allylic diphosphates ranging in size from C10 polyprenyl diphosphate to natural rubber (C>10,000). While searching for the enzymatic machinery involved in the formation of crucial intermediates in bacterial cell wall synthesis, researchers discovered UPPS over 40 years ago (6–12). Following the studies in bacteria, a description of an ER-localized enzymatic activity was discovered in mammals and in yeast (13–15). UPPS utilizes FPP as a substrate to sequentially add eight IPP units to form undecaprenyl diphosphate (C11). In eukaryotic cells, cis-PT (known also as polyprenyl diphosphate synthase or dehydrodolichyl diphosphate synthase (DHDDS)) provides cells with the isoprenoid backbone essential for the formation of dolichol phosphate (C55–100; DolP), an obligate lipid carrier necessary for protein glycosylation reactions in the ER (Fig. 1).
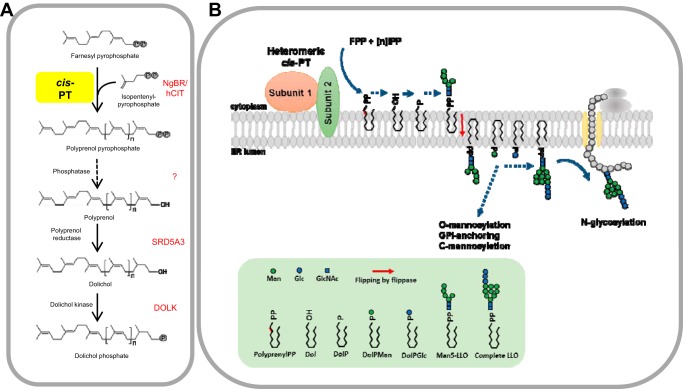
FIGURE 1.
The role cis-PTs in dolichol phosphate synthesis and protein glycosylation. A, DolP synthesis de novo in eukaryotic cell. Genes encoding enzymes involved in DolP synthesis in human cell are marked in red. DOLK, dolichol kinase. B, the role of heteromeric cis-PT in protein glycosylation. cis-PT generates polyprenol diphosphate on the cytoplasmic leaflet of the ER membrane. Polyprenol diphosphate is used as an intermediate in the synthesis of dolichol-linked saccharides (LLO). LLO is intermediate in protein N-glycosylation reactions. Dolichol-phosphate mannose (DolPMan) is also involved in O-mannosylation, glycosylphosphatidylinositol (GPI) anchor synthesis, and C-mannosylation. DolPGlc, dolichol-phosphate glucose; circled P, phosphorylation.
Classification of cis-PTs
cis-PTs are classified according to the product chain length into three subfamilies: short-chain (C15), medium-chain (C50–55), and long-chain (C70–120) cis-PTs (2, 16). In addition to chain length, we propose that cis-PTs should be classified based on enzyme composition into two classes: 1) homo-dimeric enzymes (short- and medium-chain enzymes) and 2) heteromeric enzymes (long-chain enzymes and rubber synthases) as seen in Fig. 2A.
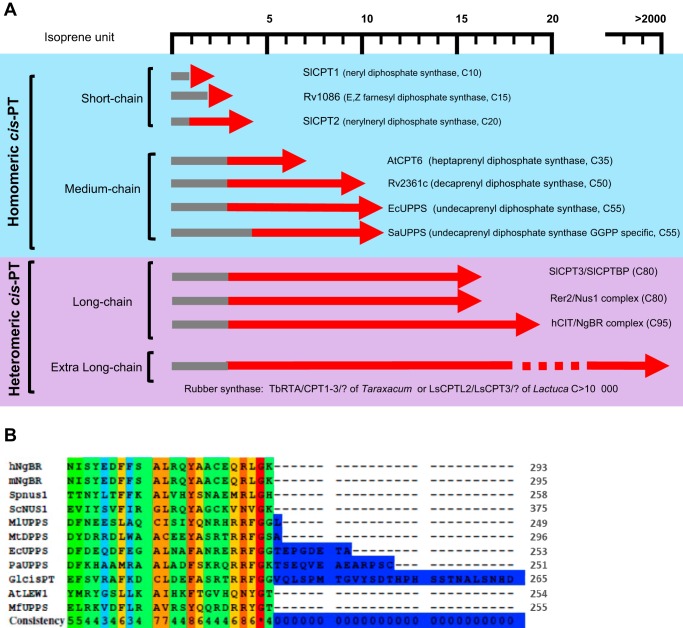
FIGURE 2.
Classification of cis-prenyltransferases. A, classification of cis-PTs according to product chain length and subunit structure. Gray lines indicate numbers of isoprene units of representative allylic substrates for each cis-PT. Red arrows indicate numbers of isoprene units of representative final products. Enzymes represented in the figure are: SlCPT1 (Solanum lycopersicum, GenBankTM NM_001247704), Rv1086 (M. tuberculosis, GenBank: CFB23420.1), SlCPT2 (S. lycopersicum GenBank JX943884), AtCPT6 (A. thaliana, GenBank NP_568882), Rv2361c (M. tuberculosis, GenBank: WP_031739650), EcUPPS (Escherichia coli, GenBank: P60472), SaUPPS (Sulfolobus acidocaldarius, GenBank WP_011277635.1), SlCPT3/SlCPTBP (S. lycopersicum GenBank: JX943885/XP_004241992), hCIT/NgBR (Homo sapiens GenBank accession NP_612468/BAB14439), RER2/NUS1 (S. cerevisiae, GenBank P35196/NP_010088), TbRTA/CPT1–3 (T. brevicorniculatum, GenBank ALX37963/AGE89403/AGE89404/AGE89405), and LsCPTL2/LsCPT3 (L. sativa, GenBank AIQ81190/AIQ81186). B, alignment of the C terminus of NgBR orthologs (hNgBR, human; mNgBR, mouse; SpNus1, S. pombe; ScNus1, S. cerevisiae; AtLEW1, A. thaliana) and single subunit cis-PTs (MlUPPS, Micrococcus luteus; MtDPPS, M. tuberculosis; EcUPPS, E. coli; PaUPPS, P. aeruginosa; GlcisPT, G. lamblia; and MfUPPS, Methanobacterium formicicum). The conservation scoring was performed by PRALINE. The scoring scheme works from 0 for the least conserved alignment position, up to 10 (*) for the most conserved alignment position.
The homo-dimeric enzymes synthesize short- and medium-chain prenols and are encoded by a single gene product that generates a dimeric protein. The first described short-chain cis-PT from Mycobacterium tuberculosis (Rv1086) catalyzes cis-condensation of geranyl diphosphate with one IPP forming Z,E-FPP (C15) (17). Z,E-FPP then serves as an allylic primer in the synthesis of decaprenyl diphosphate. Solanum sp. have numbers of short-chain enzymes localized to plastids, and these enzymes generate products ranging in size form neryl diphosphate (C10) to nerylneryl diphosphate (C20) (18–20). Lavandula x intermedia lavandulyl diphosphate synthase is structurally similar to other short cis-PTs. Products of plant shortcis-PTs are intermediates in the synthesis of monoterpenes and sesquiterpenes (21, 22).
Medium-chain cis-PTs are represented by UPPS purified from a number of bacterial species (5, 16, 23–30), Z,E-mixed decaprenyl diphosphate synthase (Z,E-DecPP, C50) from M. tuberculosis (Rv2361), as well as plant, protozoan, and archaeal enzymes. UPPS is responsible for the biogenesis of undecaprenyl phosphate, an indispensable glycosyl carrier lipid in bacterial cell wall biosynthesis. Z,E-DecPP from M. tuberculosis was the first described cis-PT utilizing a different allylic substrate other than FPP (17, 31). In Arabidopsis thaliana, AtCPT6 functions in response to abiotic stress (32, 33); however, the AtCPT6 product most likely does not serve as lipid carrier in glycosylation reactions.
The medium-chain cis-PT of Giardia lamblia,which is phylogenetically related to bacterial UPPS, is the only molecularly characterized protozoan enzyme (34). Archaeal enzymes make polyprenol diphosphates ranging in size from C30 to C60, which serve as precursors for DolP utilization in N-linked glycosylation reactions (35–38).
Long-chain cis-PTs involved in dolichol synthesis and dolichol-dependent glycosylation of proteins in yeast, mammals, and plants were predicted to be similar to homo-dimeric UPPS. This assumption was supported by the fact that overexpression of predicted eukaryotic cis-PTs (RER2 and SRT1 of bakers' yeast, human hCIT, and A. thaliana LEW1) in homologous and/or heterologous expression systems was able to increase cis-PT enzymatic activity and dolichol content (39–46). However, recent data have challenged this paradigm because mammalian, fungal, and long-chain plant cis-PTs are composed of two subunits critical for enzymatic activity. The two-component system is composed of NgBR (Nogo-B receptor) and hCIT in mammals (47); Nus1 and Rer2 or Nus1 and Srt1 in Saccharomyces cerevisiae; SpNus1 and SpRer2 in Schizosaccharomyces pombe (47); SlCPT3 and SlCPTBP in tomato (48); Lew1 and At2g17570 in Arabidopsis (49); and LsCPTL1 and LsCPT1 in lettuce (50), respectively. Rubber synthase of Taraxacum brevicorniculatum and Lactuca sativa are also heteromeric enzymes composed of at least two subunits (50, 51). Interestingly, when comparing the primary amino acid sequences of known experimentally analyzed single- and two-component enzymes, the NgBR/Nus1 class does not have a catalytic motif present in UPPS. However, it shares with UPPS, but not with hCIT, an -RXG- C-terminal conserved motif (Fig. 2B), implicating an important role of the C terminus in activity. In the context of the crystal structure of homo-dimeric UPPS, Arg242 in the -RXG- motif is involved IPP binding (26), and the very C terminus of one monomer of decaprenyl diphosphate synthase in M. tuberculosis interacts with the active site of the other subunit (52). Interestingly, patients harboring an R290H mutation in NgBR, and the corresponding mutations in ScNus1 and SpNus1, show reduced cis-PT activity and chain length of the polyprenol diphosphate (47). Finally C-terminal tagging of L. sativa CPT-like (CPTL-NgBR ortholog) (50) severely reduces cis-PTs activity, thereby supporting the idea that the C terminus is highly conserved and important for cis-PT activity.
The molecular elements determining how heteromeric enzymes regulate prenol chain length are unknown. However, a number of mutagenesis, structural, and modeling studies of monomeric enzymes show that the specificities of product chain lengths primarily depend on different enzymatic properties (16, 20, 28, 52–55). In addition, chain length distribution of products from cis-PT could be affected by reaction conditions in vitro such as detergents and phospholipids, which may influence co-substrate binding or accessibility of intermediates (56, 57). In the context of heteromeric enzymes, the different gene products(NgBR/Nus1 and hCIT/Rer2/Srt1) both determine polyprenol chain length (47), as recently found during natural rubber synthesis (50, 51). Interestingly, components of rubber synthases are able to form only long-chain polyprenols in vitro or expressed in yeast, and additional structural components or chaperones may be necessary to fully synthesize rubber. Other factors that may influence the chain length of cis-PT could be the degree of subunit oligomerization or the specific lipidic environment of the heteromeric subunits.
Phylogenetic Distribution of cis-PTs
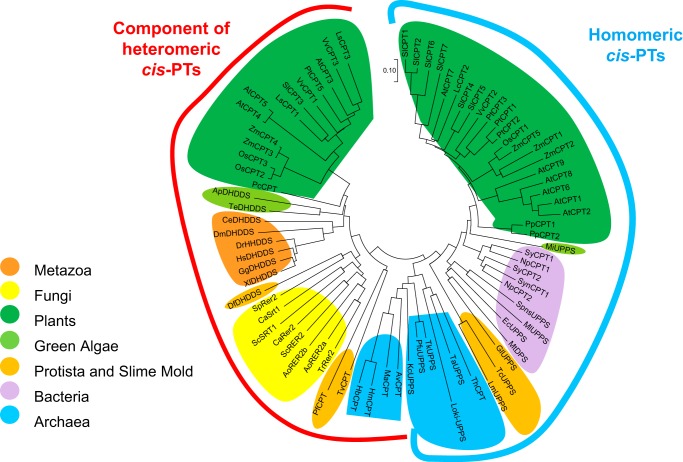
Phylogenetic analysis of representative eukaryotic and prokaryotic UPPS enzymes reveals two main clusters: one composed of hCIT/Rer2 orthologs that require NgBR/Nus1 to form active enzyme and the other composed of mainly single subunit enzymes (Fig. 3) (34).
FIGURE 3.
Phylogenetic distribution of known and predicted single-component cis-PTs and hCIT/Rer2 orthologs from animals, plants and microbes. The evolutionary history was inferred using the neighbor-joining method and conducted in MEGA7(90). Species abbreviations are: Ao, Aspergillus oryzae; Ap, Auxenochlorella protothecoides; At, A. thaliana; Av, Archaeoglobus veneficus; Ca, Candida albicans; Ce, Caenorhabditis elegans; Df, Dictyostelium fasciculatum; Dm, Drosophila melanogaster; Dr, Danio rerio; Ec, E. coli; Gg, Gallus gallus; Gl, G. lamblia; Hb, Halogeometricum borinquense; Hm, Haloferax mediterranei; Hs, H. sapiens; Kc, Korarchaeum cryptofilum; Lc, L. sativa; Lm, Leishmania major; Loki, Lokiarchaeum sp.; Ma, Methanosarcina acetivorans; Mi, Micromonas sp.; Mt, M. tuberculosis; Np, Nostoc punctiforme; Os, Oryza sativa; Pc, Picea sitchensis; Pf, Plasmodium falciparum; Pfu, Pyrococcus furiosus; Pp, Physcomitrella patens; Pt, Populus trichocarpa; Sc, S. cerevisiae; Si, Setaria italica; Sl, S. lycopersicum; Sp, S. pombe; Spns, Streptococcus pneumonia; Sy, Synechococcus; Syc, Synechocystis; Ta, Thermosphaera aggregans; Tc, Trypanosoma cruzi; Te, Tetraselmis sp.; Tg, Toxoplasma gondii; Th, Thaumarchaeote; Tk; Thermococcus kodakarensis; Tr, T. reesei; Tv, Trichomonas vaginalis; Vv, Vitis vinifera; Xl, Xenopus laevis; Zm, Zea mays. DHDDS, subunit of dehydrodolichyl diphosphate synthase.
Fungi, animals, and slime mold have only the heteromeric, NgBR/hCIT class of cis-PT. In plants, it is possible to distinguish homo-dimeric from heteromeric cis-PTs (19, 48–51, 58). Functionally, heteromeric enzymes are critical for dolichol synthesis and N-glycosylation reactions in the ER, as well as in natural rubber biosynthesis in rubber-producing plants. Single-subunit cis-PTs of endosymbiotic origin are phylogenetically related to bacteria cis-PTs and are very often localized to the chloroplast.
Trichomonas vaginalis and Apicomplexa, including parasitic Plasmodium sp., have only one gene per species resembling hCIT/Rer2; however, they lack an NgBR ortholog. However, functional studies are lacking examining whether these hCIT-like proteins are unusual homo-dimeric cis-PTs or, as we predict, require an additional subunit.
Only single homo-dimeric cis-PTs are present in several distantly related protists including Giardia and Trypanosomatida (34). UPPS such as cis-PTs in protists are probably the result of horizontal gene transfer, which is a major force in the evolution of Giardia (34, 59). This assumption is supported by the fact that unlike Giardia, other Metamonada, T. vaginalis, and recently reported Monocercomonoides sp. (60) have the hCIT type of cis-PT.
Finally, the genomes of Entamoeba sp. lack orthologs of cis-PTs. Because Entamoeba use dolichol in glycosylation reactions, the lack of a homolog of cis-PT could be explained by an alternative enzyme or co-opting dolichol from the host (34, 61).
Eubacteria and the majority of Archaea, including recently discovered Lokiarchaeota, the nearest relative of eukaryotes (62), possess a typical single-subunit enzyme. Surprisingly, Halomebacteria and Archaeoglobaceae, belonging to Euryarchaeota, the species less closely related to the Eukaryote phylum of Archaea, have clear NgBR/Nus1 orthologs (63). Their UPPS orthologs lack a C-terminal conserved RXG sequence, implying that ancestral genes of NgBR/Nus1 and hCIT orthologs may have emerged in Euryarchaeota after being acquired via horizontal gene transfer by a common ancestor of eukaryotes.
Discovery of Heteromeric Eukaryotic cis-PTs
Mammalian NgBR as a Component of cis-PT
NgBR was identified as a receptor for the N terminus of Nogo-B (amino acids 1–200; AmNogo-B) (64). Nogo-B, also known as reticulon 4B, is highly abundant in endothelial cells and vascular smooth muscle cells, and mice lacking Nogo-A and -B have accelerated neointima after vascular injury, defective wound healing, and impaired blood flow recovery after ischemia (65). Soluble AmNogo-B promotes the adhesion and chemotaxis of endothelial cells (65), but the mechanism of this effect was unclear. To understand the mechanisms of soluble AmNogo-B function, an expression cloning strategy identified potential binding proteins, and a cDNA encoding NgBR was isolated (64). Analysis of the primary sequence of NgBR revealed a single C-terminal domain with homology to cis-PTs; however, semi-purified NgBR was not active in cis-PT assays.
Insights into the biology of NgBR as a component of cis-PT activity stemmed from experiments showing the C terminus of NgBR interacts with Niemann Pick C2 (NPC2) protein, a lysosomal protein critical for cholesterol transfer from the lysosome to the ER or plasma membrane (66). The loss of NgBR increased cellular free cholesterol content due to the destabilization of NPC2, a phenotype similar to loss-of-function mutants of NPC2. When studying how NgBR interacts with NPC2, researchers used pulse-chase studies examining protein glycosylation to demonstrate that the loss of NgBR markedly reduced protein N-glycosylation by reducing cis-PT activity and dolichol-linked sugars. Co-immunoprecipitation experiments showed that the C terminus of NgBR interacts with hCIT and that NgBR is necessary for protein N-glycosylation and cis-PT activity (67).
Overexpression of NgBR/Nus1 shows a minor increase in cis-PT enzymatic activity and dolichol levels; however, hCIT overexpression stimulates enhanced accumulation of dolichol, implying that hCIT may be a limiting factor in dolichol synthesis and may recruit NgBR and its orthologs to active complexes (41, 43, 67, 68). The first evidence supporting eukaryotic cis-PT activity is due to a heteromeric complex of NgBR with hCIT was from studies in yeast (S. cerevisiae). A triple deletion strain lacking NUS1 (ortholog of NgBR), RER2, and SRT1 (orthologs of hCIT) in S. cerevisiae (nus1Δ, rer2Δ, srt1Δ) is lethal; however, the survival of the strain was accomplished by expression of a single subunit enzyme from G. lamblia. Moreover, co-expression of hCIT and NgBR, but not individual genes, reconstituted cis-PT activity in the triple mutant strain. These data suggest that both genes are required for cis-PT activity; however, these experiments did not address the contribution of each partner to cis-PT activity. Utilizing an in vitro translation (IVT) system to express either hCIT or NgBR (and yeast orthologs) did not result in cis-PT activity, nor did mixing of the IVT reaction products. Surprising, co-translation of NgBR/hCIT is required for cis-PT activity (47), rationalizing prior biochemical data showing that NgBR/hCIT stabilize each other (66). Thus, hCIT and NgBR form an active cis-PT complex that is assembled during translation.
Yeast Nus1and NgBR Are Orthologs
NUS1 was described as an essential gene needed for cell division in S. cerevisiae (69) contemporaneously with the identification of NgBR in human cells (64). Yeast strains were constructed by replacement of native promoters with a TetO7 cassette (Tet-off system), which was examined for defects after doxycycline addition. NUS1 was found in a cluster of genes affecting 3C/4C DNA content, an unexpected cell cycle profile likely related to mitotic defects. Moreover, suppression of NUS1 results in defective glycosylation of carboxypeptidase Y (CPY), disrupted secretion of alkaline phosphatase, and impaired production of the glycosylphosphatidylinositol-linked protein Gas1. To examine whether Nus and NgBR are functional orthologs (47), co-expression of either NgBR/hCIT, Nus1/Rer2, or Spnus1/Sp-rer2 in the triple deletion strain lacking Nus1/Rer2 and Srt1 yielded viable cells (47). Similar to the requirements of NgBR/hCIT for mammalian cis-PT activity, Nus1, Rer2, or mixtures of Nus1 with Rer2 IVT products were unable to form polyprenols, whereas co-translation of Nus1 and Rer2 formed an active cis-PT complex producing polyprenols of expected lengths, supporting the heteromeric structure of yeast cis-PT.
Plant Orthologs of NgBR and Rubber Synthesis
Functional conservation of NgBR/Nus1 orthologs from distantly related organisms is supported by the studies of Lew1, an ortholog of NgBR in A. thaliana. Depletion of Lew1 led to defects in dolichol production and protein glycosylation (39) Recently, it was proven that AtLew1 forms active cis-PT with hCIT/Rer2 ortholog AtCPT3 (At2g17570) (49). The first evidence for the existence of heteromeric plant cis-PT complex was supported by results from CPT-like (CPTL-NgBR ortholog) and CPT (hCIT ortholog) protein in L. sativa. CPTL2 is predominantly expressed in latex, and silencing CPTL2 coincides with a reduction of natural rubber synthesis. Yeast microsomes containing CPTL2/CPT3 show enhanced synthesis of short cis-polyisoprenes, and microsomes containing recombinant CPT1/CPTL1 or CPT1/CPTL2 show a strong increase of polyprenol biosynthetic activity (50). In T. brevicorniculatum, the NgBR-like protein (TbRTA) was also found in the rubber particle in a proteomics study. TbRTA interaction with the rubber synthase subunits TbCPT1–3 was confirmed by co-immunoprecipitation experiments (51). TbCPT1–3 or TbRTA expression in a S. cerevisiae triple deletion strain (51) fails to support cell survival. However, co-expression of TbRTA with TbCPT1–3 complements the survival of the mutants, suggesting a requirement of both TbRTA and TbCPT for cis-PT activity similar to studies with human and yeast components (51). Additional evidence for a heteromeric cis-PT complex was found in tomato plants that express SICPTBP, a close homolog of NgBR and Lew1, and SICPT3 is a homolog of hCIT/RER2. Similar to other studies, co-expression of SICPTBP and SICPT3 was required to complement the survival of rer2Δ yeast strain and dolichol biosynthesis (48).
Regulation of cis-PT Activity and Expression
The knowledge about regulation of cis-PT activity is still limited despite its critical importance in protein N-glycosylation. Cyclic nucleotides such as cAMP increase microsomal cis-PT activity (70); however, the mechanism is unknown. Stimulation of the unfolded protein response induces the expression of several genes involved in N-glycosylation reactions including NgBR and hCIT genes in mammalian cells as well as NUS1 and RER2 in yeast (71). Blockade of sterol biosynthesis up-regulates dolichol synthesis in yeast and mammalian cells. Yeast lacking functional squalene synthase have elevated cis-PT activity and dolichol levels (72). Inhibition of squalene synthase in mammalian cells deficient in dolichol phosphate mannose activity was proposed as therapeutic intervention, because it increases both dolichol phosphate and dolichol phosphate mannose levels and corrects glycosylation defects (73). Both Rer2-dependent and Srt1-dependent cis-PT activity in yeast S. cerevisiae are stimulated by overexpression of farnesyl diphosphate synthase (FPPS) (74). Overexpression of FPPS not only increases the level of allylic substrate available for the cis-PT but also stimulates the expression of both RER2 and SRT1 (42), and in Trichoderma reesei, expression of FPPS synthase enhances cis-PT activity (75).
It is clear that NgBR binding to hCIT is critical for cis-PT activity; however, the functional role of NgBR in complex with Nogo-B or NPC2 is less obvious. Although NgBR was discovered as receptor or binding protein for the N terminus of Nogo-B, the release of soluble Nogo-B or exposure of the N terminus of the reticulon-4 family of proteins on the cell surface is controversial. Also, a majority of NgBR is found on the ER membrane, and topology studies have not convincingly shown NgBR on the cell surface. In genetic experiments, the loss of Nogo-A/B clearly affects ER morphology (76) but does not affect cis-PT activity in vitro or protein glycosylation (68). Additional fine mapping studies of Nogo-B/NgBR interacting domains may permit additional understanding of this protein-protein interaction. In addition to Nogo-B, NgBR can interact with NPC2 in vivo and in vitro, and studies in cells using siRNA depletion of NgBR or genetic loss of NgBR document impaired sterol sensing and elevations in free cellular cholesterol. This may occur via NgBR binding to NPC2 (66) or NgBR modifying the N-glycosylation of NPC2. Also, a direct interaction of NgBR and NPC2 is supported by the dual topology of NgBR in the ER (67) and the co-existence of NPC2 and NgBR orthologs in the same organisms during evolution.
Genetic Validation of Two-component NgBR/hCIT Complex in Humans
The essential nature of cis-PT activity for dolichol biosynthesis is clear based on lethality in yeast and mice when Nus1, Rer2/Srt1, or NgBR is deleted (34, 46, 47, 69, 77). Ablation of NgBR in the mouse results in early embryonic lethality around embryonic day 6.5. In addition, conditional NgBR deletion in vascular endothelial cells also demonstrates embryonic vascular development due to defects in glycosylation of important endothelial proteins (68). The importance NgBR/hCIT in humans is shown by recent studies documenting loss-of-function mutations via exome sequencing. A NgBR-R290H loss-of-function mutation was identified in a family of Roma origin with a constellation of symptoms consistent with a congenital disorder of glycosylation (47), and hCIT K42E and T206A missense mutations were identified in a family of Ashkenazi Jewish origin with retinitis pigmentosa (78–80), Homozygous mutation in NgBR causes a broad and severe congenital disorder of glycosylation phenotype including retinitis pigmentosa, severe neurological impairment, and refractory epilepsy, whereas mutations in hCIT cause retinitis pigmentosa. Indeed, experiments in a reconstituted system shows that loss-of-function mutations in NgBR or hCIT reduce cis-PT activity and are additive when combined. In both instances, mutations in NgBR or hCIT show altered ratios of dolichol chain lengths when measured in urine and plasma from affected carriers (47, 80). Finally, chromosomal deletion within NgBR locus may play a role in driving susceptibility to pediatric epilepsy and congenital anomalies (81, 82). Because epilepsy is commonly observed in congenital disorders of glycosylation (83), observed symptoms may be consistent with defects in dolichol synthesis and hypoglycosylation.
In addition to rare variants, recent data suggest that cis-PT activity and dolichol biosynthesis may influence cancer progression based on altered NgBR expression in cancerous tissues. NgBR mRNA levels are increased in ductal adenocarcinoma and non-small cell lung carcinoma specimens (84, 85). Clearly, regulation of glycosylation influences cell growth because the loss of ALG genes required for protein N-glycosylation triggers cell cycle arrest in yeast (86). In addition, blocking protein glycosylation using tunicamycin or NgBR deletion inhibits endothelial cell proliferation (68, 87) and triggers apoptosis.
Recently, several studies have shown that bacterial UPPS is an attractive antibiotic target (4, 88) and that inhibitors of UPPS are protective in a mouse model of infection (89). Based on the experimental data concerning G. lamblia cis-PT and phylogenetic analysis of cis-PT from Trypanosoma sp., Leishmania sp., and Plasmodium sp., we predict that targeting protozoan cis-PT may also be a reasonable strategy to treat and control infections such as malaria, sleeping sickness, Chagas disease, leishmaniasis, and intestinal infections. Solving the structure and understanding the mechanism of action of the mammalian NgBR/hCIT complex would complement studies on the precise characterization of novel antimicrobials.
In summary, the synthesis of polyprenols is essential for all life forms. Recent advances discussed herein were not predicted based on genomic similarities of prokaryotic and eukaryotic cis-PT. The discovery of a two-component system for the synthesis of the long-chain polyprenols in yeast, plants, and humans provides new opportunities to define the similarities and differences across phyla and examine the mechanisms of polyprenol synthesis.
Acknowledgment
We thank Dr. Anna Karnkowska for help searching the genome of Monocercomonoides sp. for cis-PTs.
This work was supported by National Institutes of Health Grants R01 HL64793, R01 HL61371, R01 HL081190, and RO1 HL133018 (to W. C. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- IPP
- isopentenyl diphosphate
- DMAPP
- dimethylallyl diphosphate
- FPP
- farnesyl diphosphate
- FPPS
- farnesyl diphosphate synthase
- cis-PT
- cis-prenyltransferase
- UPPS
- undecaprenyl diphosphate synthase
- NgBR
- Nogo-B receptor
- CPTL
- CPT-like
- CPT
- cis-prenyltransferase
- NPC2
- Niemann Pick C2
- DolP
- dolichol phosphate
- ER
- endoplasmic reticulum
- IVT
- in vitro translation.
References
- 1. Wallrapp F. H., Pan J.-J., Ramamoorthy G., Almonacid D. E., Hillerich B. S., Seidel R., Patskovsky Y., Babbitt P. C., Almo S. C., Jacobson M. P., and Poulter C. D. (2013) Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily. Proc. Natl. Acad. Sci. U.S.A. 110, E1196–E1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Takahashi S., and Koyama T. (2006) Structure and function of cis-prenyl chain elongating enzymes. Chem. Rec. 6, 194–205 [DOI] [PubMed] [Google Scholar]
- 3. Rohmer M., Grosdemange-Billiard C., Seemann M., and Tritsch D. (2004) Isoprenoid biosynthesis as a novel target for antibacterial and antiparasitic drugs. Curr. Opin. Investig. Drugs 5, 154–162 [PubMed] [Google Scholar]
- 4. Teng K.-H., and Liang P.-H. (2012) Structures, mechanisms and inhibitors of undecaprenyl diphosphate synthase: a cis-prenyltransferase for bacterial peptidoglycan biosynthesis. Bioorg. Chem. 43, 51–57 [DOI] [PubMed] [Google Scholar]
- 5. Shimizu N., Koyama T., and Ogura K. (1998) Molecular cloning, expression, and purification of undecaprenyl diphosphate synthase: no sequence similarity between E- and Z-prenyl diphosphate synthases. J. Biol. Chem. 273, 19476–19481 [DOI] [PubMed] [Google Scholar]
- 6. Baba T., and Allen C. M. (1980) Prenyl transferases from Micrococcus luteus: characterization of undecaprenyl pyrophosphate synthetase. Arch. Biochem. Biophys. 200, 474–484 [DOI] [PubMed] [Google Scholar]
- 7. Koyama T., Yoshida I., and Ogura K. (1988) Undecaprenyl diphosphate synthase from Micrococcus luteus BP 26: essential factors for the enzymatic activity. J. Biochem. 103, 867–871 [DOI] [PubMed] [Google Scholar]
- 8. Allen C. M., Keenan M. V., and Sack J. (1976) Lactobacillus plantarum undecaprenyl pyrophosphate synthetase: purification and reaction requirements. Arch. Biochem. Biophys. 175, 236–248 [DOI] [PubMed] [Google Scholar]
- 9. Takahashi I., and Ogura K. (1982) Prenyltransferases of Bacillus subtilis: undecaprenyl pyrophosphate synthetase and geranylgeranyl pyrophosphate synthetase. J. Biochem. 92, 1527–1537 [DOI] [PubMed] [Google Scholar]
- 10. Higashi Y., Strominger J. L., and Sweeley C. C. (1967) Structure of a lipid intermediate in cell wall peptidoglycan synthesis: a derivative of a C55 isoprenoid alcohol. Proc. Natl. Acad. Sci. U.S.A. 57, 1878–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wright A., Dankert M., Fennessey P., and Robbins P. (1967) Characterization of a polyisoprenoid compound functional in O-antigen biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 57, 1798–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Christenson J. G., Gross S. K., and Robbins P. W. (1969) Enzymatic synthesis of the antigen carrier lipid. J. Biol. Chem. 244, 5436–5439 [PubMed] [Google Scholar]
- 13. Adair W. L Jr., Cafmeyer N., and Keller R. K. (1984) Solubilization and characterization of the long chain prenyltransferase involved in dolichyl phosphate biosynthesis. J. Biol. Chem. 259, 4441–4446 [PubMed] [Google Scholar]
- 14. Adair W. L Jr., and Cafmeyer N. (1987) Characterization of the Saccharomyces cerevisiae cis-prenyltransferase required for dolichyl phosphate biosynthesis. Arch. Biochem. Biophys. 259, 589–596 [DOI] [PubMed] [Google Scholar]
- 15. Ericsson J., Thelin A., Chojnacki T., and Dallner G. (1991) Characterization and distribution of cis-prenyl transferase participating in liver microsomal polyisoprenoid biosynthesis. Eur. J. Biochem. 202, 789–796 [DOI] [PubMed] [Google Scholar]
- 16. Kharel Y., Takahashi S., Yamashita S., and Koyama T. (2006) Manipulation of prenyl chain length determination mechanism of cis-prenyltransferases. FEBS J. 273, 647–657 [DOI] [PubMed] [Google Scholar]
- 17. Schulbach M. C., Brennan P. J., and Crick D. C. (2000) Identification of a short (C15) chain Z-isoprenyl diphosphate synthase and a homologous long (C50) chain isoprenyl diphosphate synthase in Mycobacterium tuberculosis. J. Biol. Chem. 275, 22876–22881 [DOI] [PubMed] [Google Scholar]
- 18. Schilmiller A. L., Schauvinhold I., Larson M., Xu R., Charbonneau A. L., Schmidt A., Wilkerson C., Last R. L., and Pichersky E. (2009) Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc. Natl. Acad. Sci. U.S.A. 106, 10865–10870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Akhtar T. A., Matsuba Y., Schauvinhold I., Yu G., Lees H. A., Klein S. E., and Pichersky E. (2013) The tomato cis-prenyltransferase gene family. Plant J. 73, 640–652 [DOI] [PubMed] [Google Scholar]
- 20. Kang J.-H., Gonzales-Vigil E., Matsuba Y., Pichersky E., and Barry C. S. (2014) Determination of residues responsible for substrate and product specificity of Solanum habrochaites short-chain cis-prenyltransferases. Plant Physiol. 164, 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Demissie Z. A., Erland L. A., Rheault M. R., and Mahmoud S. S. (2013) The biosynthetic origin of irregular monoterpenes in Lavandula isolation and biochemical characterization of a novel cis-prenyl diphosphate synthase gene, lavandulyl diphosphate synthase. J. Biol. Chem. 288, 6333–6341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu M., Chen C. C., Chen L., Xiao X., Zheng Y., Huang J. W., Liu W., Ko T. P., Cheng Y. S., Feng X., Oldfield E., Guo R. T., and Ma Y. (2016) Structure and function of a “head-to-middle” prenyltransferase: lavandulyl diphosphate synthase. Angew. Chem. Int. Ed. Engl. 55, 4721–4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kato J.-i., Fujisaki S., Nakajima K.-i., Nishimura Y., Sato M., and Nakano A. (1999) The Escherichia coli homologue of yeast RER2, a key enzyme of dolichol synthesis, is essential for carrier lipid formation in bacterial cell wall synthesis. J. Bacteriol. 181, 2733–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Apfel C. M., Takács B., Fountoulakis M., Stieger M., and Keck W. (1999) Use of genomics to identify bacterial undecaprenyl pyrophosphate synthetase: cloning, expression, and characterization of the essential uppS gene. J. Bacteriol. 181, 483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chang S. Y., Ko T. P., Chen A. P. C., Wang A. H. J., and Liang P. H. (2004) Substrate binding mode and reaction mechanism of undecaprenyl pyrophosphate synthase deduced from crystallographic studies. Protein Sci. 13, 971–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chang S.-Y., Ko T.-P., Liang P.-H., and Wang A. H.-J. (2003) Catalytic mechanism revealed by the crystal structure of undecaprenyl pyrophosphate synthase in complex with sulfate, magnesium, and Triton. J. Biol. Chem. 278, 29298–29307 [DOI] [PubMed] [Google Scholar]
- 27. Chen Y.-H., Chen A. P.-C., Chen C.-T., Wang A. H.-J., and Liang P.-H. (2002) Probing the conformational change of Escherichia coli undecaprenyl pyrophosphate synthase during catalysis using an inhibitor and tryptophan mutants. J. Biol. Chem. 277, 7369–7376 [DOI] [PubMed] [Google Scholar]
- 28. Ko T.-P., Chen Y.-K., Robinson H., Tsai P.-C., Gao Y.-G., Chen A. P.-C., Wang A. H.-J., and Liang P.-H. (2001) Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli undecaprenyl-pyrophosphate synthase catalysis. J. Biol. Chem. 276, 47474–47482 [DOI] [PubMed] [Google Scholar]
- 29. Kharel Y., Zhang Y.-W., Fujihashi M., Miki K., and Koyama T. (2001) Identification of significant residues for homoallylic substrate binding of Micrococcus luteus BP 26 undecaprenyl diphosphate synthase. J. Biol. Chem. 276, 28459–28464 [DOI] [PubMed] [Google Scholar]
- 30. Fujihashi M., Zhang Y.-W., Higuchi Y., Li X.-Y., Koyama T., and Miki K. (2001) Crystal structure of cis-prenyl chain elongating enzyme, undecaprenyl diphosphate synthase. Proc. Natl. Acad. Sci. U.S.A. 98, 4337–4342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kaur D., Brennan P. J., and Crick D. C. (2004) Decaprenyl diphosphate synthesis in Mycobacterium tuberculosis. J. Bacteriol. 186, 7564–7570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kera K., Takahashi S., Sutoh T., Koyama T., and Nakayama T. (2012) Identification and characterization of a cis,trans-mixed heptaprenyl diphosphate synthase from Arabidopsis thaliana. FEBS J. 279, 3813–3827 [DOI] [PubMed] [Google Scholar]
- 33. Surmacz L., Plochocka D., Kania M., Danikiewicz W., and Swiezewska E. (2014) cis-Prenyltransferase AtCPT6 produces a family of very short-chain polyisoprenoids in planta. Biochim. Biophys. Acta 1841, 240–250 [DOI] [PubMed] [Google Scholar]
- 34. Grabińska K. A., Cui J., Chatterjee A., Guan Z., Raetz C. R., Robbins P. W., and Samuelson J. (2010) Molecular characterization of the cis-prenyltransferase of Giardia lamblia. Glycobiology 20, 824–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hemmi H., Yamashita S., Shimoyama T., Nakayama T., and Nishino T. (2001) Cloning, expression, and characterization of cis-polyprenyl diphosphate synthase from the thermoacidophilic archaeon Sulfolobus acidocaldarius. J. Bacteriol. 183, 401–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mori T., Ogawa T., Yoshimura T., and Hemmi H. (2013) Substrate specificity of undecaprenyl diphosphate synthase from the hyperthermophilic archaeon Aeropyrum pernix. Biochem. Biophys. Res. Commun. 436, 230–234 [DOI] [PubMed] [Google Scholar]
- 37. Yamada Y., Fukuda W., Hirooka K., Hiromoto T., Nakayama J., Imanaka T., Fukusaki E., and Fujiwara S. (2009) Efficient in vitro synthesis of cis-polyisoprenes using a thermostable cis-prenyltransferase from a hyperthermophilic archaeon Thermococcus kodakaraensis. J. Biotechnol. 143, 151–156 [DOI] [PubMed] [Google Scholar]
- 38. Taguchi Y., Fujinami D., and Kohda D. (2016) Comparative analysis of archaeal lipid-linked oligosaccharides that serve as oligosaccharide donors for Asn glycosylation. J. Biol. Chem. 291, 11042–11054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang H., Ohyama K., Boudet J., Chen Z., Yang J., Zhang M., Muranaka T., Maurel C., Zhu J.-K., and Gong Z. (2008) Dolichol biosynthesis and its effects on the unfolded protein response and abiotic stress resistance in Arabidopsis. Plant Cell 20, 1879–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Perlińska-Lenart U., Bańkowska R., Palamarczyk G., and Kruszewska J. S. (2006) Overexpression of the Saccharomyces cerevisiae RER2 gene in Trichoderma reesei affects dolichol dependent enzymes and protein glycosylation. Fungal Genet. Biol. 43, 422–429 [DOI] [PubMed] [Google Scholar]
- 41. Jones J., Viswanathan K., Krag S. S., and Betenbaugh M. J. (2005) Polyprenyl lipid synthesis in mammalian cells expressing human cis-prenyl transferase. Biochem. Biophys. Res. Commun. 331, 379–383 [DOI] [PubMed] [Google Scholar]
- 42. Grabińska K., Sosińska G., Orłowski J., Swiezewska E., Berges T., Karst F., and Palamarczyk G. (2005) Functional relationships between the Saccharomyces cerevisiae cis-prenyltransferases required for dolichol biosynthesis. Acta Biochim. Pol. 52, 221–232 [PubMed] [Google Scholar]
- 43. Shridas P., Rush J. S., and Waechter C. J. (2003) Identification and characterization of a cDNA encoding a long-chain cis-isoprenyltranferase involved in dolichyl monophosphate biosynthesis in the ER of brain cells. Biochem. Biophys. Res. Commun. 312, 1349–1356 [DOI] [PubMed] [Google Scholar]
- 44. Schenk B., Rush J. S., Waechter C. J., and Aebi M. (2001) An alternative cis-isoprenyltransferase activity in yeast that produces polyisoprenols with chain lengths similar to mammalian dolichols. Glycobiology 11, 89–98 [DOI] [PubMed] [Google Scholar]
- 45. Sato M., Fujisaki S., Sato K., Nishimura Y., and Nakano A. (2001) Yeast Saccharomyces cerevisiae has two cis-prenyltransferases with different properties and localizations: implication for their distinct physiological roles in dolichol synthesis. Genes Cells 6, 495–506 [DOI] [PubMed] [Google Scholar]
- 46. Sato M., Sato K., Nishikawa S.-i., Hirata A., Kato J.-i., and Nakano A. (1999) The yeast RER2 gene, identified by endoplasmic reticulum protein localization mutations, encodes cis-prenyltransferase, a key enzyme in dolichol synthesis. Mol. Cell. Biol. 19, 471–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Park E. J., Grabińska K. A., Guan Z., Stránecký V., Hartmannová H., Hodaňová K., Barešová V., Sovová J., Jozsef L., Ondrušková N., Hansíková H., Honzík T., Zeman J., Hůlková H., Wen R., et al. (2014) Mutation of Nogo-B receptor, a subunit of cis-prenyltransferase, causes a congenital disorder of glycosylation. Cell Metab. 20, 448–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brasher M. I., Surmacz L., Leong B., Pitcher J., Swiezewska E., Pichersky E., and Akhtar T. A. (2015) A two-component enzyme complex is required for dolichol biosynthesis in tomato. Plant J. 82, 903–914 [DOI] [PubMed] [Google Scholar]
- 49. Kwon M., Kwon E.-J., and Ro D. (2016) cis-Prenyltransferase and polymer analysis from a natural rubber perspective. Methods Enzymol. 10.1016/bs.mie.2016.02.026 [DOI] [PubMed] [Google Scholar]
- 50. Qu Y., Chakrabarty R., Tran H. T., Kwon E.-J. G., Kwon M., Nguyen T.-D., and Ro D.-K. (2015) A lettuce (Lactuca sativa) homolog of human Nogo-B receptor interacts with cis-prenyltransferase and is necessary for natural rubber biosynthesis. J. Biol. Chem. 290, 1898–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Epping J., van Deenen N., Niephaus E., Stolze A., Fricke J., Huber C., Eisenreich W., Twyman R. M., Prüfer D., and Gronover C. S. (2015) A rubber transferase activator is necessary for natural rubber biosynthesis in dandelion. Nat. Plants 1, 1–9 [Google Scholar]
- 52. Wang W., Dong C., McNeil M., Kaur D., Mahapatra S., Crick D. C., and Naismith J. H. (2008) The structural basis of chain length control in Rv1086. J. Mol. Biol. 381, 129–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Poznański J., and Szkopinska A. (2007) Precise bacterial polyprenol length control fails in Saccharomyces cerevisiae. Biopolymers 86, 155–164 [DOI] [PubMed] [Google Scholar]
- 54. Noike M., Ambo T., Kikuchi S., Suzuki T., Yamashita S., Takahashi S., Kurokawa H., Mahapatra S., Crick D. C., and Koyama T. (2008) Product chain-length determination mechanism of Z,E-farnesyl diphosphate synthase. Biochem. Biophys. Res. Commun. 377, 17–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ambo T., Noike M., Kurokawa H., and Koyama T. (2008) Cloning and functional analysis of novel short-chain cis-prenyltransferases. Biochem. Biophys. Res. Commun. 375, 536–540 [DOI] [PubMed] [Google Scholar]
- 56. Matsuoka S., Sagami H., Kurisaki A., and Ogura K. (1991) Variable product specificity of microsomal dehydrodolichyl diphosphate synthase from rat liver. J. Biol. Chem. 266, 3464–3468 [PubMed] [Google Scholar]
- 57. Troutman J. M., Erickson K. M., Scott P. M., Hazel J. M., Martinez C. D., and Dodbele S. (2015) Tuning the production of variable length, fluorescent polyisoprenoids using surfactant-controlled enzymatic synthesis. Biochemistry 54, 2817–2827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Surmacz L., Wojcik J., Kania M., Bentinger M., Danikiewicz W., Dallner G., Surowiecki P., Cmoch P., and Swiezewska E. (2015) Short-chain polyisoprenoids in the yeast Saccharomyces cerevisiae: new companions of the old guys. Biochim. Biophys. Acta 1851, 1296–1303 [DOI] [PubMed] [Google Scholar]
- 59. Andersson J. O., Sjögren A. M., Davis L. A., Embley T. M., and Roger A. J. (2003) Phylogenetic analyses of diplomonad genes reveal frequent lateral gene transfers affecting eukaryotes. Curr. Biol. 13, 94–104 [DOI] [PubMed] [Google Scholar]
- 60. Karnkowska A., Vacek V., Zubáčová Z., Treitli Sebastian C., Petrželková R., Eme L., Novák L., Žárský V., Barlow Lael D., Herman Emily K., Soukal P., Hroudová M., Doležal P., Stairs Courtney W., Roger Andrew J., et al. (2016) A eukaryote without a mitochondrial organelle. Curr. Biol. 26, 1274–1284 [DOI] [PubMed] [Google Scholar]
- 61. Magnelli P., Cipollo J. F., Ratner D. M., Cui J., Kelleher D., Gilmore R., Costello C. E., Robbins P. W., and Samuelson J. (2008) Unique Asn-linked oligosaccharides of the human pathogen Entamoeba histolytica. J. Biol. Chem. 283, 18355–18364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Spang A., Saw J. H., Jørgensen S. L., Zaremba-Niedzwiedzka K., Martijn J., Lind A. E., van Eijk R., Schleper C., Guy L., and Ettema T. J. (2015) Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ogawa T., Emi K.-i., Koga K., Yoshimura T., and Hemmi H. (2016) A cis-prenyltransferase from Methanosarcina acetivorans catalyzes both head-to-tail and nonhead-to-tail prenyl condensation. FEBS J. 283, 2369–2383 [DOI] [PubMed] [Google Scholar]
- 64. Miao R. Q., Gao Y., Harrison K. D., Prendergast J., Acevedo L. M., Yu J., Hu F., Strittmatter S. M., and Sessa W. C. (2006) Identification of a receptor necessary for Nogo-B stimulated chemotaxis and morphogenesis of endothelial cells. Proc. Natl. Acad. Sci. U.S.A. 103, 10997–11002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Acevedo L., Yu J., Erdjument-Bromage H., Miao R. Q., Kim J.-E., Fulton D., Tempst P., Strittmatter S. M., and Sessa W. C. (2004) A new role for Nogo as a regulator of vascular remodeling. Nat. Med. 10, 382–388 [DOI] [PubMed] [Google Scholar]
- 66. Harrison K. D., Miao R. Q., Fernandez-Hernándo C., Suárez Y., Dávalos A., and Sessa W. C. (2009) Nogo-B receptor stabilizes Niemann-Pick type C2 protein and regulates intracellular cholesterol trafficking. Cell Metab. 10, 208–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Harrison K. D., Park E. J., Gao N., Kuo A., Rush J. S., Waechter C. J., Lehrman M. A., and Sessa W. C. (2011) Nogo-B receptor is necessary for cellular dolichol biosynthesis and protein N-glycosylation. EMBO J. 30, 2490–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Park E. J., Grabińska K. A., Guan Z., and Sessa W. C. (2016) NgBR is essential for endothelial cell glycosylation and vascular development. EMBO Rep. 17, 167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yu L., Peña Castillo L., Mnaimneh S., Hughes T. R., and Brown G. W. (2006) A survey of essential gene function in the yeast cell division cycle. Mol. Biol. Cell 17, 4736–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Konrad M., and Merz W. E. (1996) Long-term effect of cyclic AMP on N-glycosylation is caused by an increase in the activity of the cis-prenyltransferase. Biochem. J. 316, 575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., Storz G., Botstein D., and Brown P. O. (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Grabowska D., Karst F., and Szkopińska A. (1998) Effect of squalene synthase gene disruption on synthesis of polyprenols in Saccharomyces cerevisiae. FEBS Lett. 434, 406–408 [DOI] [PubMed] [Google Scholar]
- 73. Haeuptle M. A., Welti M., Troxler H., Hülsmeier A. J., Imbach T., and Hennet T. (2011) Improvement of dolichol-linked oligosaccharide biosynthesis by the squalene synthase inhibitor zaragozic acid. J. Biol. Chem. 286, 6085–6091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Szkopińska A., Grabińska K., Delourme D., Karst F., Rytka J., and Palamarczyk G. (1997) Polyprenol formation in the yeast Saccharomyces cerevisiae: effect of farnesyl diphosphate synthase overexpression. J. Lipid Res. 38, 962–968 [PubMed] [Google Scholar]
- 75. Piłsyk S., Perlińska-Lenart U., Górka-Nieć W., Graczyk S., Antosiewicz B., Zembek P., Palamarczyk G., and Kruszewska J. S. (2014) Overexpression of erg20 gene encoding farnesyl pyrophosphate synthase has contrasting effects on activity of enzymes of the dolichyl and sterol branches of mevalonate pathway in Trichoderma reesei. Gene 544, 114–122 [DOI] [PubMed] [Google Scholar]
- 76. Jozsef L., Tashiro K., Kuo A., Park E. J., Skoura A., Albinsson S., Rivera-Molina F., Harrison K. D., Iwakiri Y., Toomre D., and Sessa W. C. (2014) Reticulon 4 is necessary for endoplasmic reticulum tubulation, STIM1-Orai1 coupling, and store-operated calcium entry. J. Biol. Chem. 289, 9380–9395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Blomen V. A., Májek P., Jae L. T., Bigenzahn J. W., Nieuwenhuis J., Staring J., Sacco R., van Diemen F. R., Olk N., Stukalov A., Marceau C., Janssen H., Carette J. E., Bennett K. L., Colinge J., et al. (2015) Gene essentiality and synthetic lethality in haploid human cells. Science 350, 1092–1096 [DOI] [PubMed] [Google Scholar]
- 78. Zelinger L., Banin E., Obolensky A., Mizrahi-Meissonnier L., Beryozkin A., Bandah-Rozenfeld D., Frenkel S., Ben-Yosef T., Merin S., Schwartz S. B., Cideciyan A. V., Jacobson S. G., and Sharon D. (2011) A missense mutation in DHDDS, encoding dehydrodolichyl diphosphate synthase, is associated with autosomal-recessive retinitis pigmentosa in Ashkenazi Jews. The Am. J. Hum. Genet. 88, 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Züchner S., Dallman J., Wen R., Beecham G., Naj A., Farooq A., Kohli M. A., Whitehead P. L., Hulme W., Konidari I., Edwards Y. J., Cai G., Peter I., Seo D., Buxbaum J. D., et al. (2011) Whole-exome sequencing links a variant in DHDDS to retinitis pigmentosa. Am. J. Hum. Genet. 88, 201–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wen R., Lam B. L., and Guan Z. (2013) Aberrant dolichol chain lengths as biomarkers for retinitis pigmentosa caused by impaired dolichol biosynthesis. J. Lipid Res. 54, 3516–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Milani D., Cagnoli G. A., Baccarin M., Alfei E., Guerneri S., and Esposito S. (2016) Insights into 6Q21-Q22: refinement of the critical region for acro-cardio-facial syndrome. Congenit Anom (Kyoto) 56, 187–189 [DOI] [PubMed] [Google Scholar]
- 82. Szafranski P., Von Allmen G. K., Graham B. H., Wilfong A. A., Kang S.-H. L., Ferreira J. A., Upton S. J., Moeschler J. B., Bi W., Rosenfeld J. A., Shaffer L. G., Wai Cheung S., Stankiewicz P., and Lalani S. R. (2015) 6q22.1 microdeletion and susceptibility to pediatric epilepsy. Eur. J. Hum. Genet. 23, 173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Barba C., Darra F., Cusmai R., Procopio E., Dionisi Vici C., Keldermans L., Vuillaumier-Barrot S., Lefeber D. J., Guerrini R., and CDG Group (2016) Congenital disorders of glycosylation presenting as epileptic encephalopathy with migrating partial seizures in infancy. Dev. Med. Child Neurol. 10.1111/dmcn.13141 [DOI] [PubMed] [Google Scholar]
- 84. Pula B., Werynska B., Olbromski M., Muszczynska-Bernhard B., Chabowski M., Janczak D., Zabel M., Podhorska-Okolow M., and Dziegiel P. (2014) Expression of Nogo isoforms and Nogo-B receptor (NgBR) in non-small cell lung carcinomas. Anticancer Res. 34, 4059–4068 [PubMed] [Google Scholar]
- 85. Wang B., Zhao B., North P., Kong A., Huang J., and Miao Q. R. (2013) Expression of NgBR is highly associated with estrogen receptor α and survivin in breast cancer. PLoS One 8, e78083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lennon K., Pretel R., Kesselheim J., te Heesen S., and Kukuruzinska M. A. (1995) Proliferation-dependent differential regulation of the dolichol pathway genes in Saccharomyces cerevisiae. Glycobiology 5, 633–642 [DOI] [PubMed] [Google Scholar]
- 87. Martínez J. A., Torres-Negrón I., Amigó L. A., Roldán R. A., Mendéz A., and Banerjee D. K. (2000) Tunicamycin inhibits capillary endothelial cell proliferation by inducing apoptosis. in Angiogenesis, pp. 197–208, Springer, New York: [PubMed] [Google Scholar]
- 88. Chan H.-C., Feng X., Ko T.-P., Huang C.-H., Hu Y., Zheng Y., Bogue S., Nakano C., Hoshino T., Zhang L., Lv P., Liu W., Crick D. C., Liang P. H., Wang A. H., et al. (2014) Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis. J. Am. Chem. Soc. 136, 2892–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zhu W., Zhang Y., Sinko W., Hensler M. E., Olson J., Molohon K. J., Lindert S., Cao R., Li K., Wang K., Wang Y., Liu Y. L., Sankovsky A., de Oliveira C. A., Mitchell D. A., et al. (2013) Antibacterial drug leads targeting isoprenoid biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 110, 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kumar S., Stecher G., and Tamura K. (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]