Abstract
Diabetes is one of the most impactful diseases worldwide. The most commonly prescribed anti-diabetic drug is metformin. In this study, we identified an endosomal Na+/H+ exchanger (NHE) as a new potential target of metformin from an unbiased screen in Caenorhabditis elegans. The same NHE homolog also exists in flies, where it too mediates the effects of metformin. Our results suggest that endosomal NHEs could be a metformin target and provide an insight into a novel mechanism of action of metformin on regulating the endocytic cycle.
Keywords: autophagy, Caenorhabditis elegans (C. elegans), drug action, endosome, metformin, sodium-proton exchange, Type 2 diabetes, sodium-proton exchange
Introduction
Diabetes, a common consequence of the growing obesity epidemic, has emerged as a global health threat with over 422 million people worldwide being diagnosed in 2014 (1). Type 2 diabetes mellitus, which is characterized by insulin resistance, is the most common type of diabetes (2).
The most prescribed anti-diabetic drug is metformin, 1,1-dimethylbiguanide (3). Metformin reduces glucose production in the liver and increases glucose uptake in muscle and adipose tissue, ameliorating diabetes (4). In addition, metformin exerts beneficial effects on other diseases such as polycystic ovary syndrome, certain cancers, and neurological disorders (2, 5). Several studies identified molecular targets of metformin: it inhibits complex I in mitochondria and decreases NADH oxidation, while reducing proton pumping across the inner mitochondrial membrane and decreasing oxygen consumption rate (6, 7). These effects in turn reduce the proton gradient and increase the AMP/ATP ratio, resulting in activation of AMP-activated protein kinase (AMPK)2 and triggering a cascade that inhibits gluconeogenic gene expression and energy-consuming processes such as lipogenesis (5, 8). Metformin increases AMP and inhibits adenylate cyclase activity, leading to a reduction in cAMP levels, thereby inhibiting PKA activity and glucagon-dependent glucose production in mice (9). Metformin in physiological doses also suppresses hepatic gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase (10). Given its versatile effects, it is likely that there are multiple targets of metformin with a wide range of affinities. A better understanding of the targets of metformin will not only allow for a more fine-tuned control of treatment with the drug, but also potentially provide insight into new therapeutic targets for diabetes and other metabolic disorders.
Caenorhabditis elegans has been used for studying metabolism, reproduction, aging and other physiological processes, providing useful knowledge of the biology and molecular pathways underpinning human diseases. In this study, we report that metformin reduces C. elegans L1 longevity, a survival span of starvation. Using this phenotype, we performed an unbiased genetic screen and found an endosomal Na+/H+ exchanger (NHE) as a potential molecular target of metformin. Moreover, the same NHE homolog in flies also mediates the effects of metformin, suggesting that an NHE could be a conserved target for metformin.
Results
Metformin Reduces L1 Longevity in C. elegans via NHX-5
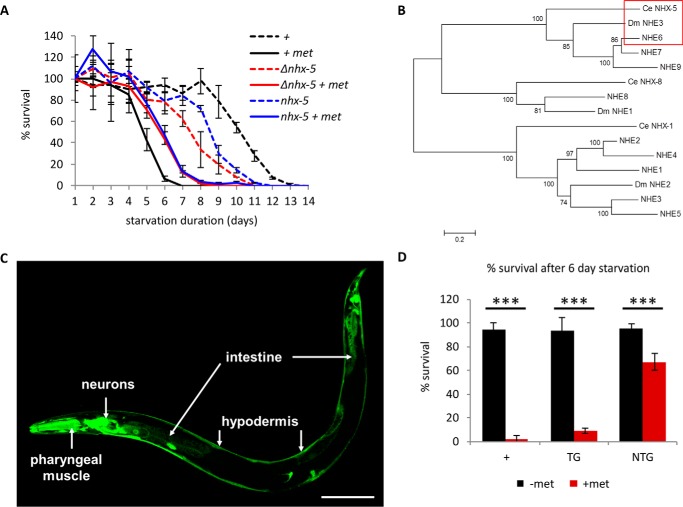
It has been shown that in Drosophila, metformin treatment (100 mm) reduces lifespan (11). We discovered that metformin (100 mm) also reduces a certain type of lifespan, L1 survival span or L1 longevity, in C. elegans. Hatching without food, the first stage larva (L1) survives ∼14 days at 25 °C and 21 days at 20 °C (12). Metformin (100 mm) reduces L1 longevity to 7 days at 25 °C (Fig. 1A) and to 10 days at 20 °C (Fig. 2A). To identify a molecular target of metformin, we performed an unbiased genetic screen in C. elegans, using this metformin sensitivity phenotype. We screened ∼105 ethyl methanesulfonate (EMS)-mutagenized haploid genomes and isolated a mutant with better survival in the presence of metformin. Subsequent mapping and whole genome sequencing revealed a single nucleotide substitution in the nhx-5 gene that led to an amino acid substitution of the conserved glycine 207 to glutamate (G207E). The C. elegans genome contains nine NHEs, NHX-1 through NHX-9, all of which exchange a vesicular or extracellular sodium for a luminal or intracellular proton (13). Some are expressed on the plasma membrane of cells, whereas others are expressed on the membranes of intracellular organelles. nhx-5 encodes the sole homolog of endosomal Na+/H+ exchangers 6, 7, and 9, which reside in the endosomal membranes to modulate endosome pH and regulate the rate of the endocytic cycle in mammals (Fig. 1B) (14). NHX-5 is expressed in many cells including neurons, hypodermis, and intestine in C. elegans (Fig. 1C). NHX-5 proteins in C. elegans are also localized in the membranes of intracellular vesicles of the endocytic cycle (13), suggesting its conserved cellular function. An existing deletion mutant (ok661, obtained from the Caenorhabditis Genetic Center) also survived metformin treatment better (Fig. 1A, shown as Δnhx-5), validating our results from the screen. Both mutants are slightly short-lived when compared with wild type in the absence of metformin (Fig. 1A). The changes in 50% survival due to metformin treatment are 52% for wild type, 29% for nhx-5, and 20% for Δnhx-5. Thus they survive the metformin treatment better than wild type, showing that the better survival by nhx-5 mutations is not due to the intrinsic longevity of nhx-5 mutants.
FIGURE 1.
NHX-5 mediates metformin-induced L1 longevity phenotype in C. elegans. A, metformin (100 mm) reduces L1 longevity in wild type, whereas two nhx-5 mutants survive better in the presence of metformin (+ met) (Δnhx-5 and nhx-5). p < 0.001 between metformin-treated wild type and metformin-treated nhx-5 mutant by log-rank test (see “Experimental Procedures”). Each data point is plotted as the mean of triplicated samples and standard deviation. B, phylogenetic tree to compare similarity among endosomal NHEs with C. elegans (Ce) nhx-5 and D. melanogaster (Dm) nhe3. nhe6 and nhe7 are the closest homologs of CeNHX-5 and DmNHE3 (red box). However, NHE6 and CeNHX-5 do not contain the Golgi retention motif, whereas NHE7 does (46). This suggests that NHE6 is the closest functional homolog to ceNHX-5. Five CeNHXs (CeNHX1–5) localize in the plasma membrane and belong to the same phylogenetic group as the human NHE1 family, The phylogenetic tree was created using the MEGA program (47). The p-distance method was used to calculate distance, and the neighbor-joining method was used to create a tree. The tree was evaluated by using the bootstrap method (1000 times). Neighbor-joining, unweighted pair group method with arithmetic mean (UPGMA), and minimum evolution methods created similar trees. C, NHX-5 is expressed in many cells including neurons, pharyngeal muscle, intestine and hypodermis in C. elegans (scale bar = 100 μm). D, transgenic nhx-5 mutant worms carrying a wild-type transgenic copy of nhx-5 (TG for transgenic, black square for without metformin (−met), red square for with metformin (+met) (100 mm)) restore sensitivity to metformin, whereas non-transgenic siblings (NTG, black square for without metformin (−met), red square for with metformin (+met) (100 mm)) are still resistant. Because the difference between wild type and nhx-5 mutants is most obvious after 6 days of starvation (panel A), we compared the percentage of survival between transgenic siblings and non-transgenic siblings after 6 days of starvation. Approximately 120 transgenic worms and 1000 non-transgenic worms were used. The numbers are the mean ± S.D. ***, p < 0.001, by unpaired two-sample t test.
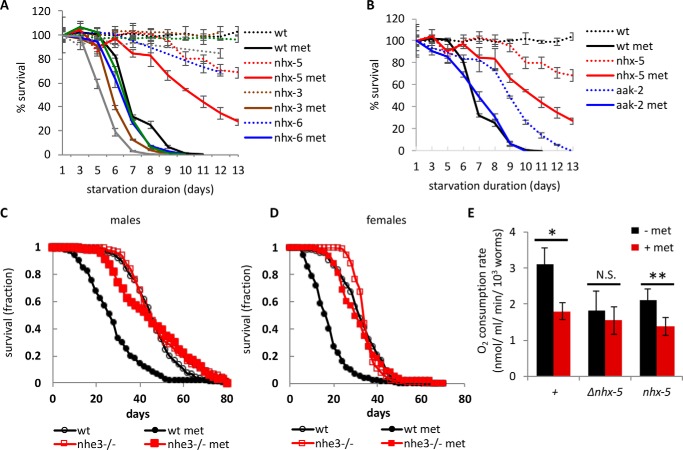
FIGURE 2.
The homologs of NHX-5 mediate metformin actions. A, the L1 longevities of all available and viable nhx mutants (nhx-3, nhx-6, nhx-8, and nhx-9) were measured and compared with those of a wild-type control and nhx-5 mutants. L1 longevity in the absence (dotted lines) and in the presence of 100 mm metformin (met) (solid lines) was measured at 20 °C to better distinguish the difference. Each data point is plotted as the average of triplicated samples and standard error. B, the L1 longevities of aak-2(ok524) mutants were compared with those of a wild-type control and nhx-5 mutants. The data of wild-type control and nhx-5 are re-used from panel A. L1 longevity in the absence (dotted lines) and in the presence of 100 mm metformin (solid lines) was measured at 20 °C. C, metformin reduces the lifespan of male flies (black circle). This metformin sensitivity is completely abolished in nhe3KG08307 null mutants (red square). Median survival times were 44 days for wild type (n = 257) without metformin, 26 days with metformin (100 mm, n = 70), and 44 days for nhe3 mutants without (n = 290) or with metformin (n = 276). D, metformin reduces the lifespan of female flies. Median survival times were 32 days for wild type (n = 267) without metformin, 16 days with metformin (n = 275), 34 days for nhe3 mutants without metformin (n = 294), and 30 days (n = 291) with metformin. There was no significant difference in survival between metformin-treated and non-treated nhe3 mutant flies by log-rank test from two independent experiments (p = 0.2 for both sexes, log-rank test), whereas there was significant difference between metformin-treated and non-treated wild-type flies (p < 0.0001, for both sexes). E, metformin affects mitochondrial function in part through NHX-5. The isolated mutants of nhx-5 and an existing deletion mutant of nhx-5 (Δnhx-5) were tested. Approximately 3000 adult worms were treated with 100 mm metformin for 30 min, and the rates of oxygen consumption were measured (see “Experimental Procedures”). Each experiment was done in triplicate, and the experiment was repeated at least three times. N.S., not significant, *, p < 0.05, **, p < 0.01, by paired Student's t test.
The resistance of nhx-5 mutants to metformin was rescued by restoring the expression of nhx-5 using its endogenous promoter, confirming that nhx-5 mediated metformin sensitivity (Fig. 1D, TG: transgenic siblings that carry nhx-5 gene; NTG: non-transgenic siblings that do not carry nhx-5 gene; TG looked similar to wild type, so therefore it rescued the phenotype.). These results are consistent with the idea that metformin could act on multiple targets.
Among nine Na+/H+ exchangers in C. elegans, deletion mutants of five other nhxs were available to test (nhx-3, nhx-4, nhx-6, nhx-8, and nhx-9). On the other hand, nhx-2 deletion mutants are embryonic lethal or arrested at L1. When we tested the available nhx mutants for metformin sensitivity, all showed sensitivity to metformin similar to wild type (Fig. 2A). We tested the sensitivity at 20 °C so that even a small difference can be detected because of longer L1 lifespan at 20 °C. This result supports the inference that among these NHXs, NHX-5 is specific for conferring metformin sensitivity.
Because AMPK is one of the known molecular targets of metformin, we tested the L1 longevity of aak-2 mutants. aak-2 encodes an α subunit of C. elegans AMPK. Although aak-2 mutants already have reduced L1 longevity when compared with wild type (12), we could still observe a consistent effect of metformin in L1 longevity of aak-2 mutants (Fig. 2B).
An Endosomal NHE Mediates Metformin Sensitivity in Drosophila melanogaster
Consistent with a previous study, metformin (100 mm) greatly reduced the lifespan of D. melanogaster (11) (Fig. 2, C and D). We found that this metformin sensitivity was entirely dependent on NHE3, the fly homolog of worm NHX-5 (Fig. 2C for males, Fig. 2D for females). Remarkably, the nhe3KG08307 mutant flies, which have a P element insertion in the first exon and have no detectable mRNA, are almost completely resistant to metformin, suggesting that NHE3 may be a major molecular target of metformin for metformin-induced lifespan reduction in flies. These results from worms and flies suggest that endosomal NHEs can be a conserved target of metformin action.
Endosomal NHEs Mediate Metformin Sensitivity Potentially by Regulating Mitochondrial Function
Although the major site of action for metformin is somewhat controversial (15, 16), one of the sites is mitochondria, where metformin inhibits complex I, thus increasing AMP levels and reducing oxygen consumption (6). To examine whether an endosomal NHE is involved in the inhibition of mitochondrial function by metformin, we measured oxygen consumption rate in C. elegans in the presence of metformin. It has been shown that metformin indirectly affects C. elegans lifespan via changing the metabolism of bacteria (C. elegans food) (17). To avoid this complication, we treated worms with metformin in the solution in which they were suspended without food for 30 min. We did not treat the animals longer than 30 min to avoid starvation effect. We found that metformin reduced oxygen consumption significantly in wild-type worms, suggesting that metformin inhibits mitochondrial function in C. elegans as it does in mammals (Fig. 2E, shown as +). Next we asked whether this reduction of oxygen consumption rate by metformin is dependent on NHX-5. First we noticed that both nhx-5 mutants showed reduced baseline oxygen consumption, suggesting that endosomal NHEs are important in mitochondrial function (black bars in Fig. 2E). Metformin failed to reduce oxygen consumption rate in the deletion mutant of nhx-5 (shown as Δnhx-5 in Fig. 2E) but still reduced it in the G270E mutant (shown as nhx-5). Because the deletion mutation likely produces no functional NHX-5, whereas the G270E mutation could produce proteins with residual function, this result could suggest that at least a part of metformin action in mitochondria could be mediated by endosomal NHEs. We do not know the reason behind the difference between the two mutations. However, this kind of inconsistency in phenotypes depending on the nature of mutations is not uncommon. One possible interpretation in this case could be that because the deletion mutant of nhx-5 already has low basal O2 consumption without metformin, any further reduction would be harder to detect. The point mutation, however, may have residual function, so it has room for metformin to reduce it further.
Metformin Inhibits Autophagosome Formation during Starvation in C. elegans
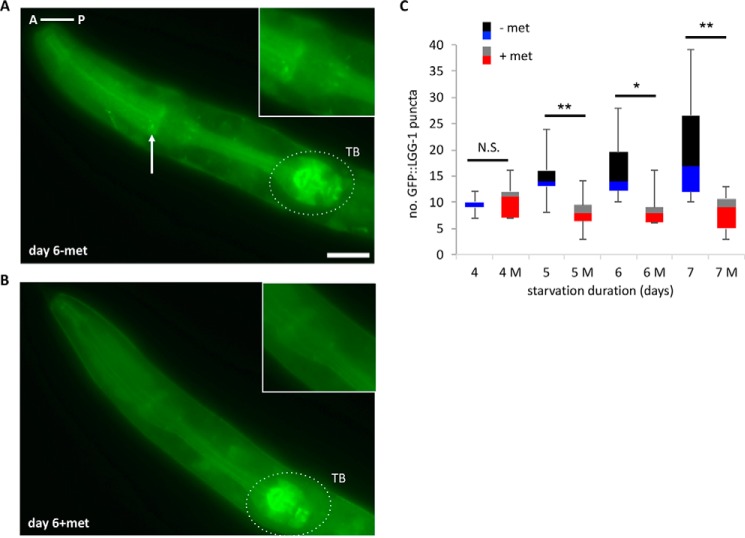
One of the major catabolic processes necessary for C. elegans L1s to survive long-term starvation is autophagy (18), whose final step is to fuse autophagosomes with lysosomes to form autolysosomes, which degrade organelles as well as macromolecules to acquire energy (19). In the absence of lysosomal degradation, autophagy does not progress normally and results in pathologic accumulation of autophagic vesicles, misfolded proteins, and abnormal organelles (2). Interestingly, knock-out mice that lack NHE6, the mammalian homolog of C. elegans NHX-5, show Angelman-like syndrome (20) in which the lysosome becomes dysfunctional, mimicking lysosomal storage disorders (21, 22). This indicates that NHE6 functions at the interface of late endosomes and lysosomes. We previously showed that too little autophagy in C. elegans during long-term starvation greatly reduces L1 longevity (18). Based on that and also because the lysosomal dysfunction will affect autophagy, we hypothesized that metformin dysregulated endosome trafficking and autophagy during L1 starvation by regulating NHX-5, which in turn reduced L1 longevity. When we measured autophagosomal puncta using an LC3 homolog fused with GFP (GFP-LGG-1), a marker widely used to label autophagosomes (23, 24), we found that the number of puncta increased as starvation continued. However, we found that in the presence of metformin, the number of puncta did not increase but stayed at the basal level as starvation continued (Fig. 3, 4, A and B). This result suggests that in the presence of metformin, the level of autophagy does not increase even if starvation persists, suggesting that metformin inhibits autophagy during starvation. Because autophagy is a continuous process of influx and efflux, measuring GFP puncta at a given time shows only a static state of autophagy level, and therefore it does not rule out the other possibility that metformin might enhance the efflux, i.e. faster degradation of GFP-LGG-1 in the autolysosomes, which in turn causes a lower number of GFP puncta. However, the number of puncta in the metformin-treated animals remains at the basal level (about 10 throughout the starvation period) both in the intestine and in the pharyngeal muscle even if the intestine has a higher autophagy level than the pharyngeal muscle in the absence of metformin (an average of 15 puncta for pharyngeal muscle in Fig. 3C, when compared with an average of 30 puncta for intestine in Fig. 4F on day 5). Also, although the number of puncta continues to increase in non-treated animals as starvation continues, the number of puncta in metformin-treated animals does not change throughout starvation. These two observations suggest that the level of autophagy in the presence of metformin does not increase however long starvation continues, suggesting that the influx of autophagy is inhibited. It would be harder to maintain the consistent level of autophagy by controlling efflux rate throughout the starvation period in the two different organs. Supporting this notion, we were not able to capture an increase in GFP-LGG-1 puncta or any fluctuation in numbers of puncta in metformin-treated animals regardless of how early we looked in the treatment time course. These results suggest that it is unlikely that metformin enhances efflux but more likely that it inhibits influx.
FIGURE 3.
Metformin reduces L1 longevity and dysregulates autophagy. A and B, 6 days of L1 starvation increases GFP-LGG-1 puncta in the head (A), but metformin (met) treatment reduces puncta formation after 6 days of starvation (B). An arrow indicates a punctum. Scale bar = 10 μm. A is anterior (left), and P is posterior (right) of the animal. TB, terminal bulb. C, quantification of the puncta after each day of starvation (for the x axis, 4, 5, 6, 7: starvation days without metformin, 4 M, 5 M, 6 M, 7 M: starvation days with metformin (100 mm)). The box plot represents the median (the line between two boxes) and interquartile ranges (the bottom box is for the 25th to 50th percentile, and the upper box is for the 50th to 75th percentile). Whiskers (T-shaped projection) represent the minimum and the maximum values. N.S., not significant, *, p < 0.05, **, p < 0.01, by Student's t test.
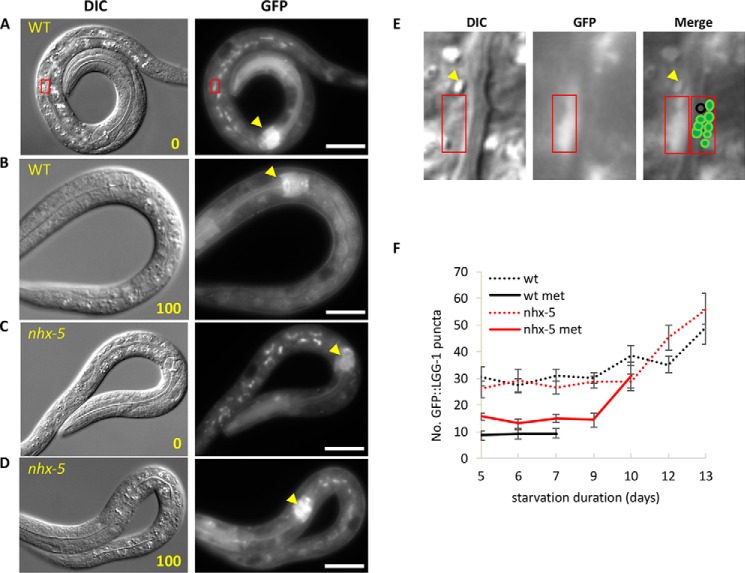
FIGURE 4.
Metformin interferes with autophagy in the intestine during starvation. A–D, after 9 days of starvation, both wild-type worms and nhx-5 mutants showed GFP-LGG-1 puncta in the intestine (A and C, 0 mm metformin). The number of puncta was greatly reduced in wild type by metformin treatment but not as much in nhx-5 mutants (B and D, 100 mm metformin). Yellow arrowheads show that the overall intensity of GFP-LGG-1 in the terminal bulb is not significantly reduced in all the animals. Scale bar = 20 μm. The left panel shows DIC images, and the right panel shows the corresponding GFP images. E, enlarged images of DIC, GFP, and both (from left to right) of the red box in panel A. These images show an example of how individual GFP-LGG-1 puncta looked and how they were counted. The graphic next to the red box in the merged image was drawn to help visualization of individual puncta. A vesicle not labeled by GFP-LGG-1 is marked as a black circle. The yellow arrowhead indicates a birefringent gut granule (no GFP and not an autophagosome), known to be stained with lysosomal dyes (48). GFP-LGG-1 puncta were counted under a GFP filter (GFP, right panel, 63×). DIC (left panel, 63×) is used to show the animals as well as to validate the location of the individual puncta. F, GFP-LGG-1-labeled puncta from starved worms were determined at the given time. Metformin-treated wild type (wt met) showed a significantly reduced number of puncta when compared with the nhx-5 mutants in the presence or absence of metformin (p < 0.05, unpaired two-sample t test). In addition, metformin-treated nhx-5 showed a significantly reduced number of puncta when compared with cohorts in the absence of metformin (p < 0.01). However, wild type and nhx-5 controls are not different in starvation-induced GFP-LGG-1-labeled puncta. To avoid a sampling error, the days were cut off when fewer than 30% of animals were alive because the dead animals did not show autophagy puncta. The numbers are the mean ± S.E.
Metformin treatment did not inhibit autophagy in nhx-5 (ok661) mutants as much as in the wild type, suggesting that metformin affects autophagy mainly through NHX-5 (Fig. 4, C–F). Although metformin-treated nhx-5 mutants showed higher numbers of GFP-LGG-1 puncta than metformin-treated wild type, they showed a significantly lower number of puncta than non-treated wild type, suggesting additional targets for the metformin effect on autophagy. Because the overall intensity of GFP-LGG-1 is not significantly reduced in all the animals (for example, compare the intensity of GFP in the terminal bulb, indicated as dotted circles in Fig. 3, A and B, and yellow arrowheads in Fig. 4, A–D), the reduced number of puncta in intestinal cells in the metformin-treated wild type is unlikely to be due to a reduction in GFP-LGG-1 protein level.
Interestingly, most of the GFP-LGG-1-labeled puncta were located specifically on the intestinal lumen side rather than on the hypodermis side or randomly distributed (Fig. 4, A–E). Also they were clustered in one or two regions in an intestinal cell (Fig. 4E). These regions are mostly perinuclear regions where macroautophagy occurs during starvation.
nhx-5 mutants do not show any gross abnormality. However, their L1 longevity is slightly shorter than wild type (Fig. 1A), and after long-term starvation as L1s, they recovered more slowly than wild type when they were given food. In addition, the GFP-LGG-1-labeled puncta in wild-type animals disappeared within 6 h after the animals were fed, whereas those in nhx-5 mutants did not. These observations suggest an autolysosomal defect in nhx-5 mutants that could lead to the reduced L1 longevity of nhx-5 mutants in the absence of metformin.
Although metformin generally increases autophagy in cancer cell lines or pathological conditions (25, 26), our results suggested a potentially opposite effect of metformin in autophagy, probably due to the long-term starvation effect. Indeed, metformin treatment combined with 2-deoxyglucose, which could mimic certain aspects of extreme starvation by depleting ATP, inhibits autophagy (27). Similarly, metformin suppresses autophagy in H4IIE cells upon glucose starvation (28). Taken together, our results suggest that inhibition of autophagy and consequential insufficient energy could contribute to the reduced L1 longevity by metformin and that NHX-5 mediates a part of this action.
Discussion
From an unbiased genetic screen in C. elegans, we identified an endosomal NHE as a potential target of metformin. Metformin reduces oxygen consumption, and reduces autophagosome formation during long-term starvation, suggesting a connection between mitochondria and the endocytic cycle. NHE6, the homolog of C. elegans NHX-5, was identified as a novel insulin-responsive gene, suggesting that the trafficking of NHE6-loaded endosomes by insulin could play an important role in glucose homeostasis (29). It is possible that metformin could regulate NHE6 function so as to restore insulin sensitivity. It has been shown that insulin-induced GLUT4 translocation to plasma membrane occurs by stimulating GLUT4 storage vesicles that contain NHE6. Based on our results, we suggest that endosomal NHEs, such as C. elegans NHX-5 and D. melanogaster NHE3, are targets of metformin action to regulate the endocytic cycle and also potentially lysosomal function.
Several interesting studies may indirectly support the idea that metformin regulates the endocytic cycle. Two unrelated studies showed that metformin treatment cured flu patients (30) and blocked Src-induced cellular transformation (31). Both processes (viral infection and Src activation) require an intact endocytic cycle. Therefore, it is possible that metformin prevents viral infection and Src activation, two seemingly unrelated phenomena, simply by interfering with the endocytic cycle. Indeed, it is well known that dysregulation of endosome-lysosome fusion affects autophagy. For example, lack of endosomal sorting complexes required for transport (ESCRT) protein functions causes the accumulation of autophagosomes in mammals and flies (32) or elevated autophagy in C. elegans (33). Moreover, endosomes are known to be a place for starvation-induced AMPK phosphorylation by LKB1 (34) and nutrient-induced mTORC1 activation, which is inhibited by metformin (34). Because genetic mutations in NHE6 cause abnormal lysosome function and lead to Angelman-like syndrome in humans (20), which is similar to lysosomal storage disorders (21, 22), dysregulation of endosomal NHE activity by metformin may inhibit mammalian target of rapamycin (mTOR) translocation to the lysosome.
It can be speculated that metformin might enhance insulin sensitivity by acting directly in the endocytic cycle, a part of the insulin signaling pathway. Finally, we also found that phenformin reduced the L1 longevity of C. elegans and that the two nhx-5 mutants were partially resistant to phenformin, suggesting that the NHE is a possible target of both metformin and phenformin (data not shown). Indeed, various drugs, such as galegine (35) and phenformin, previously prescribed to treat diabetes share the guanidine backbone with metformin. These studies suggest that the backbone structure of these drugs may play a significant role in drug action. Notably, amiloride, a guanidine that shares the basic backbone structure with metformin, inhibits several paralogs of NHE6, namely NHE1, NHE2, NHE3, and/or NHE4 (36, 37).
Taken together, our study suggests a new way to understand metformin action through a new molecular target, and thus provides new insight into treating diabetes.
Experimental Procedures
C. elegans Strains and Culture Conditions
Worms were maintained as described (38) with the following modifications. Worms were routinely grown on NGMSR plates. (NGMSR plates differ from NGM (nematode growth medium) plates as described in Ref. 39.) Worms were maintained at 20 °C on Escherichia coli strain HB101. The wild-type strain was C. elegans variant Bristol, N2. Strains used were YJ49 nhx-5(uy48) X, YJ53 nhx-5(ok661) X, YJ70 aak-2(ok524) X, YJ97 nhx-8(ok549) I, YJ51 nhx-5(uy48) X; uyEx51[F57C7.2 rol-6::GFP]. YJ49 was three times outcrossed against N2 from the original isolated mutant from the screen, and YJ53 was three times outcrossed against N2 from RB836 (ok661) acquired from the Caenorhabditis Genetics Center (CGC). YJ70 aak-2(ok524) was six times outcrossed against N2 from RB754. YJ66 nhx-3(ok1049), YJ79 nhx-4 (ok668), YJ97 nhx-8(ok549), YJ205 nhx-6(ok609), and YJ206 nhx-9(ok847) was twice outcrossed against N2 from VC717 nhx-3(ok1049), RB841 nhx-4 (ok668), RB770 nhx-8(ok549), VC383 nhx-6(ok609), and RB952 nhx-9(ok847), respectively. YJ220 adIs2123 [GFP::lgg-1 rol-6 (df)]; nhx-5(ok661) X was constructed by using DA2123 adIs2123[GFP::lgg-1 rol-6 (df)].
D. melanogaster Strains
An nhe3 mutant fly (#14715; y1 w67c23; P{SUPor-P} Nhe3KG08307) was obtained from the Bloomington Drosophila Stock Center. This mutant carries a variant version of the P element at the first exon of the nhe3 gene that causes abnormal transcription of the nhe3 gene. Quantitative RT-PCR using two primers, 5′-AGCAAAGGAAAGTCTCGAACCC-3′ and 5′-CTACAGCTTTTCTACGCTCGTTG-3′, confirmed that unlike wild-type w1118, the nhe3KG08307 mutant had no detectable mRNA (data not shown). Five males were used for preparing total RNA samples.
Fly Husbandry and Preparation of Diet
Larvae of the w1118 strain or an nhe3KG08307 mutant were grown separately until emergence on standard cornmeal-sugar-yeast medium (5.2 g of cornmeal, 11 g of sucrose, 2.5 g of yeast (MP Biomedicals, Solon, OH), 1.1 ml of 20% tegosept, and 0.79 g of agar per 100 ml of water). The rearing room was maintained at 25 °C with 40% humidity on a 12-h light and 12-h dark cycle. Once eclosed as adults, flies were fed with sugar-yeast diet supplemented with or without metformin. Adult sugar-yeast diets were prepared using 10 g of sucrose, 10 g of yeast, 1.1 ml of 20% tegosept, and 0.79 g of agar per 100 ml of water. Metformin (Sigma-Aldrich) was dissolved in water to make 1 m stock solutions.
Measuring Lifespan of D. melanogaster
Newly eclosed adults were collected over 48 h and were transferred to demography cages to the final density of 100 males and 100 females per cage. Food vials (25 × 95 mm) containing 0 or 100 mm metformin were attached to each cage via a 25-mm plastic tube and were changed every 2 days, at which time dead flies were removed and recorded (40). Three replicates were done for each dose concentration, and the experiments were done twice independently.
Measuring C. elegans L1 Longevity, Metformin Treatment, and Fluorescence Microscopy
L1 longevity was measured as described (12, 41). Briefly, eggs were isolated by basic hypochlorite treatment and then harvested in M9 buffer to get synchronous populations. The collected eggs were immediately treated with 100 mm metformin in M9 (drug-treated) or with M9 only (control). Tubes containing the eggs were kept rocking at indicated temperature. 20-μl aliquots of control and drug-treated samples were taken and placed on three E. coli HB101 seeded plates for each group. Three days after plating, the numbers of adult worms were counted to measure survival. The percentage of survival was calculated based on the number of survivals on day 1 starvation. For transgenic rescue lines, the samples were prepared in the same way. Then surviving transgenic and non-transgenic siblings were counted using a fluorescence microscope, and the percentage of survival was calculated by separately normalizing by the numbers of each genotype on day 1. The number of worms used for each day is ∼500–600. All the experiments were repeated at least three times, and the graphs represent one of the replicates. GFP-LGG-1 fluorescence and localization were measured by using Zeiss Axio Imager A.2 microscope (GFP filter with 63× magnification), while autofluorescence was measured by using DAPI and RFP filters. Images were taken by using a Zeiss AxioCam MRm camera.
EMS Mutagenesis and Whole Genome Sequencing
Approximately 104 P0 L4s were mutagenized with EMS (Sigma-Aldrich) for 4 h, and ∼105 haploid genomes of F2s were screened for resistance to metformin (100 mm) (38). Eleven candidates were isolated from the primary screen. Only one of them showed consistent resistance to metformin in later tests. To map this mutation, we used both whole genome sequencing and SNP mapping methods (42). Whole genome sequencing was performed by the University of Virginia Core Facility. The Whole Genome Sequencing dataset was analyzed using MAQGene (43).
DNA Constructs to Generate Transgenic Animals
The F57C7 cosmid (obtained from the Wellcome Trust Sanger Institute) was used to rescue metformin resistance of nhx-5 mutants. The construct was injected as described previously (41) using rol-6::GFP as a co-injection marker. Transgenic worms carrying pJP113-nhx-5 (a gift from Dr. Nehrke) also restored metformin sensitivity similarly. NHX-5 expression was examined by using transgenic worms carrying a DNA construct that has an nhx-5 promoter (4.0 kb upstream from ATG start codon) fused with GFP.
These two primers, 5′-TGAGGACGAGCGTATCAACAG-3′ and 5′-TCCTTTACTCATTATATCGACTGGGAAGAGTGG-3′, were used to amplify nhx-5 promoter region, and these two primers, 5′-CCAGTCGATATAATGAGTAAAGGAGAAGAACTTTTC-3′ and 5′-GTACGGCCGACTAGTAGGA-3′, were used to amplify the GFP coding region as well as the 3′-UTR region. Finally, the two PCR products were fused using primers 5′-CAACTTCGTAGCATGAGTGAAGG-3′ and 5′-GGAAACAGTTATGTTTGGTATATTGGG-3′. Confocal images were obtained using a Zeiss LSM510.
Oxygen Consumption Measurement
Wild type and two nhx-5 mutants, YJ49 and YJ53, were prepared and synchronized by egg preparation (44). Synchronized L1s were grown to young adulthood for 48–50 h at 20 °C. Young adults were collected with M9 and were resuspended in either 2 ml of 100 mm metformin in buffer or 2 ml of buffer only (control) for 30 min in liquid. Oxygen consumption rates were measured for 7 min and normalized to the number of worms counted after the recording. Approximately 2000–3000 worms were used. The measurements were performed in triplicate for each treatment, and each experiment was repeated at least three times. Oxygen consumption rates were measured using the Hansatech Instruments Oxytherm system with S1/MINI electrode and Peltier Electrode Chamber. The data were acquired using the O2 view data acquisition and system configuration software (Hansatech Instruments).
Statistical Analyses
Statistical log-rank tests for the fly demographic data were carried out using survival models in the JMP statistical package (SAS, Cary, NC). Statistical log-rank tests for the C. elegans wild type and mutants were carried out by the same method as the flies with a small modification of smoothing (12, 45). For the unpaired two-sample t test, Excel in Microsoft Office 2010 was used.
Author Contributions
J. K. and Y. J. Y. designed the research. J. K., I. L., J. A., M. H., H. Y. L,. and K. J. M. performed research. J. K. and Y. J. Y. wrote the manuscript.
Acknowledgments
We thank Dr. L. Avery for reviewing the manuscript and Dr. K.-C. Kim for helping with WGS analysis. Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by National Institutes of Health Office of Research Infrastructure Programs (Grant P40 OD010440) and the Bloomington Drosophila Stock Center.
This work was supported by Inha University (to J. K.), Korean MEST Grant 2012R1A1A2041099 (to K. J. M.), Virginia Commonwealth University School of Medicine (to Y. J. Y.), and Nagoya Research Center for Brain & Neural Circuits (to Y. J. Y.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- AMPK
- AMP-activated protein kinase
- NHE
- NHX, Na+/H+ exchanger
- EMS
- ethyl methanesulfonate
- DIC
- differential interference contrast.
References
- 1. NCD Risk Factor Collaboration (NCD-RisC) (2016) Worldwide trends in diabetes since 1980: a pooled analysis of 751 population-based studies with 4.4 million participants. Lancet 387, 1513–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wilson C. M., Magnaudeix A., Yardin C., and Terro F. (2014) Autophagy dysfunction and its link to Alzheimer's disease and type II diabetes mellitus. CNS Neurol. Disord. Drug Targets 13, 226–246 [DOI] [PubMed] [Google Scholar]
- 3. Danaei G., Finucane M. M., Lu Y., Singh G. M., Cowan M. J., Paciorek C. J., Lin J. K., Farzadfar F., Khang Y. H., Stevens G. A., Rao M., Ali M. K., Riley L. M., Robinson C. A., Ezzati M., and Global Burden of Metabolic Risk Factors of Chronic Diseases Collaborating Group (Blood Glucose) (2011) National, regional, and global trends in fasting plasma glucose and diabetes prevalence since 1980: systematic analysis of health examination surveys and epidemiological studies with 370 country-years and 2.7 million participants. Lancet 378, 31–40 [DOI] [PubMed] [Google Scholar]
- 4. Bailey C. J., and Turner R. C. (1996) Metformin. N. Engl. J. Med. 334, 574–579 [DOI] [PubMed] [Google Scholar]
- 5. Quinn B. J., Kitagawa H., Memmott R. M., Gills J. J., and Dennis P. A. (2013) Repositioning metformin for cancer prevention and treatment. Trends Endocrinol. Metab. 24, 469–480 [DOI] [PubMed] [Google Scholar]
- 6. El-Mir M. Y., Nogueira V., Fontaine E., Avéret N., Rigoulet M., and Leverve X. (2000) Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J. Biol. Chem. 275, 223–228 [DOI] [PubMed] [Google Scholar]
- 7. Owen M. R., Doran E., and Halestrap A. P. (2000) Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem. J. 348, 607–614 [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou G., Myers R., Li Y., Chen Y., Shen X., Fenyk-Melody J., Wu M., Ventre J., Doebber T., Fujii N., Musi N., Hirshman M. F., Goodyear L. J., and Moller D. E. (2001) Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Invest. 108, 1167–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miller R. A., Chu Q., Xie J., Foretz M., Viollet B., and Birnbaum M. J. (2013) Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature 494, 256–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Madiraju A. K., Erion D. M., Rahimi Y., Zhang X. M., Braddock D. T., Albright R. A., Prigaro B. J., Wood J. L., Bhanot S., MacDonald M. J., Jurczak M. J., Camporez J. P., Lee H. Y., Cline G. W., Samuel V. T., et al. (2014) Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature 510, 542–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Slack C., Foley A., and Partridge L. (2012) Activation of AMPK by the putative dietary restriction mimetic metformin is insufficient to extend lifespan in Drosophila. PLoS ONE 7, e47699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee I., Hendrix A., Kim J., Yoshimoto J., and You Y. J. (2012) Metabolic rate regulates L1 longevity in C. elegans. PLoS ONE 7, e44720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nehrke K., and Melvin J. E. (2002) The NHX family of Na+-H+ exchangers in Caenorhabditis elegans. J. Biol. Chem. 277, 29036–29044 [DOI] [PubMed] [Google Scholar]
- 14. Ohgaki R., van IJzendoorn S. C., Matsushita M., Hoekstra D., and Kanazawa H. (2011) Organellar Na+/H+ exchangers: novel players in organelle pH regulation and their emerging functions. Biochemistry 50, 443–450 [DOI] [PubMed] [Google Scholar]
- 15. Foretz M., Guigas B., Bertrand L., Pollak M., and Viollet B. (2014) Metformin: from mechanisms of action to therapies. Cell Metab. 20, 953–966 [DOI] [PubMed] [Google Scholar]
- 16. Rena G., Pearson E. R., and Sakamoto K. (2013) Molecular mechanism of action of metformin: old or new insights? Diabetologia 56, 1898–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cabreiro F., Au C., Leung K. Y., Vergara-Irigaray N., Cochemé H. M., Noori T., Weinkove D., Schuster E., Greene N. D., and Gems D. (2013) Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell 153, 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kang C., You Y. J., and Avery L. (2007) Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Genes Dev. 21, 2161–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rubinsztein D. C., Gestwicki J. E., Murphy L. O., and Klionsky D. J. (2007) Potential therapeutic applications of autophagy. Nat. Rev. Drug Discov. 6, 304–312 [DOI] [PubMed] [Google Scholar]
- 20. Gilfillan G. D., Selmer K. K., Roxrud I., Smith R., Kyllerman M., Eiklid K., Kroken M., Mattingsdal M., Egeland T., Stenmark H., Sjøholm H., Server A., Samuelsson L., Christianson A., Tarpey P., et al. (2008) SLC9A6 mutations cause X-linked mental retardation, microcephaly, epilepsy, and ataxia, a phenotype mimicking Angelman syndrome. Am. J. Hum. Genet. 82, 1003–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Strømme P., Dobrenis K., Sillitoe R. V., Gulinello M., Ali N. F., Davidson C., Micsenyi M. C., Stephney G., Ellevog L., Klungland A., and Walkley S. U. (2011) X-linked Angelman-like syndrome caused by Slc9a6 knockout in mice exhibits evidence of endosomal-lysosomal dysfunction. Brain 134, 3369–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ouyang Q., Lizarraga S. B., Schmidt M., Yang U., Gong J., Ellisor D., Kauer J. A., and Morrow E. M. (2013) Christianson syndrome protein NHE6 modulates TrkB endosomal signaling required for neuronal circuit development. Neuron 80, 97–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matsushita M., Suzuki N. N., Obara K., Fujioka Y., Ohsumi Y., and Inagaki F. (2007) Structure of Atg5·Atg16, a complex essential for autophagy. J. Biol. Chem. 282, 6763–6772 [DOI] [PubMed] [Google Scholar]
- 24. Meléndez A., Tallóczy Z., Seaman M., Eskelinen E. L., Hall D. H., and Levine B. (2003) Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301, 1387–1391 [DOI] [PubMed] [Google Scholar]
- 25. Xie Z., Lau K., Eby B., Lozano P., He C., Pennington B., Li H., Rathi S., Dong Y., Tian R., Kem D., and Zou M. H. (2011) Improvement of cardiac functions by chronic metformin treatment is associated with enhanced cardiac autophagy in diabetic OVE26 mice. Diabetes 60, 1770–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tomic T., Botton T., Cerezo M., Robert G., Luciano F., Puissant A., Gounon P., Allegra M., Bertolotto C., Bereder J. M., Tartare-Deckert S., Bahadoran P., Auberger P., Ballotti R., and Rocchi S. (2011) Metformin inhibits melanoma development through autophagy and apoptosis mechanisms. Cell Death Dis. 2, e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ben Sahra I., Tanti J. F., and Bost F. (2010) The combination of metformin and 2-deoxyglucose inhibits autophagy and induces AMPK-dependent apoptosis in prostate cancer cells. Autophagy 6, 670–671 [DOI] [PubMed] [Google Scholar]
- 28. Park D. B. (2015) Metformin promotes apoptosis but suppresses autophagy in glucose-deprived H4IIE hepatocellular carcinoma cells. Diabetes Metab. J. 39, 518–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Prior M. J., Larance M., Lawrence R. T., Soul J., Humphrey S., Burchfield J., Kistler C., Davey J. R., La-Borde P. J., Buckley M., Kanazawa H., Parton R. G., Guilhaus M., and James D. E. (2011) Quantitative proteomic analysis of the adipocyte plasma membrane. J. Proteome Res. 10, 4970–4982 [DOI] [PubMed] [Google Scholar]
- 30. Doms R. W., Helenius A., and White J. (1985) Membrane fusion activity of the influenza virus hemagglutinin: the low pH-induced conformational change. J. Biol. Chem. 260, 2973–2981 [PubMed] [Google Scholar]
- 31. Hirsch H. A., Iliopoulos D., and Struhl K. (2013) Metformin inhibits the inflammatory response associated with cellular transformation and cancer stem cell growth. Proc. Natl. Acad. Sci. U.S.A. 110, 972–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee J. A., Beigneux A., Ahmad S. T., Young S. G., and Gao F. B. (2007) ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol. 17, 1561–1567 [DOI] [PubMed] [Google Scholar]
- 33. Djeddi A., Michelet X., Culetto E., Alberti A., Barois N., and Legouis R. (2012) Induction of autophagy in ESCRT mutants is an adaptive response for cell survival in C. elegans. J. Cell Sci. 125, 685–694 [DOI] [PubMed] [Google Scholar]
- 34. Zhang C. S., Jiang B., Li M., Zhu M., Peng Y., Zhang Y. L., Wu Y. Q., Li T. Y., Liang Y., Lu Z., Lian G., Liu Q., Guo H., Yin Z., Ye Z., et al. (2014) The lysosomal v-ATPase-Ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metab. 20, 526–540 [DOI] [PubMed] [Google Scholar]
- 35. Hawley S. A., Ross F. A., Chevtzoff C., Green K. A., Evans A., Fogarty S., Towler M. C., Brown L. J., Ogunbayo O. A., Evans A. M., and Hardie D. G. (2010) Use of cells expressing γ subunit variants to identify diverse mechanisms of AMPK activation. Cell Metab. 11, 554–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sardet C., Franchi A., and Pouysségur J. (1989) Molecular cloning, primary structure, and expression of the human growth factor-activatable Na+/H+ antiporter. Cell 56, 271–280 [DOI] [PubMed] [Google Scholar]
- 37. Counillon L., Franchi A., and Pouysségur J. (1993) A point mutation of the Na+/H+ exchanger gene (NHE1) and amplification of the mutated allele confer amiloride resistance upon chronic acidosis. Proc. Natl. Acad. Sci. U.S.A. 90, 4508–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sulston J., and Hodgkin J. (1988) Methods. in The Nematode Caenorhabditis elegans. pp. 594–595, Cold Spring Harbor Monograph Series, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 39. Avery L. (1993) The genetics of feeding in Caenorhabditis elegans. Genetics 133, 897–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee S. H., Do H. S., and Min K. J. (2015) Effects of essential oil from Hinoki cypress, Chamaecyparis obtusa, on physiology and behavior of flies. PLoS ONE 10, e0143450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. You Y. J., Kim J., Cobb M., and Avery L. (2006) Starvation activates MAP kinase through the muscarinic acetylcholine pathway in Caenorhabditis elegans pharynx. Cell Metab. 3, 237–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wicks S. R., Yeh R. T., Gish W. R., Waterston R. H., and Plasterk R. H. (2001) Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28, 160–164 [DOI] [PubMed] [Google Scholar]
- 43. Bigelow H., Doitsidou M., Sarin S., and Hobert O. (2009) MAQGene: software to facilitate C. elegans mutant genome sequence analysis. Nat. Methods 6, 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lewis J. A., and Fleming J. T. (1995) Basic culture methods. Methods Cell Biol. 48, 3–29 [PubMed] [Google Scholar]
- 45. Lee B. H., and Ashrafi K. (2008) A TRPV channel modulates C. elegans neurosecretion, larval starvation survival, and adult lifespan. PLoS Genet. 4, e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Numata M., and Orlowski J. (2001) Molecular cloning and characterization of a novel (Na+,K+)/H+ exchanger localized to the trans-Golgi network. J. Biol. Chem. 276, 17387–17394 [DOI] [PubMed] [Google Scholar]
- 47. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., and Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hermann G. J., Schroeder L. K., Hieb C. A., Kershner A. M., Rabbitts B. M., Fonarev P., Grant B. D., and Priess J. R. (2005) Genetic analysis of lysosomal trafficking in Caenorhabditis elegans. Mol. Biol. Cell 16, 3273–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]