Abstract
A subset of bovine antibodies have an exceptionally long third heavy-chain complementarity determining region (CDR H3) that is highly variable in sequence and includes multiple cysteines. These long CDR H3s (up to 69 residues) fold into a long stalk atop which sits a knob domain that is located far from the antibody surface. Three new bovine Fab crystal structures have been determined to decipher the conserved and variable features of ultralong CDR H3s that lead to diversity in antigen recognition. Despite high sequence variability, the stalks adopt a conserved β-ribbon structure, while the knob regions share a conserved β-sheet that serves as a scaffold for two connecting loops of variable length and conformation, as well as one conserved disulfide. Variation in patterns and connectivity of the remaining disulfides contribute to the knob structural diversity. The unusual architecture of these ultralong bovine CDR H3s for generating diversity is unique in adaptive immune systems.
Introduction
The human adaptive immune system uses V(D)J recombination to construct germline antibodies that are then somatically mutated in response to antigen challenge. The classic scaffold for generating such diversity is the immunoglobulin (Ig) fold. Antibodies are assembled from heavy and light chains, where diversity is mainly found in the complementarity determining region (CDR) loops, which connect the core β-strands of the Ig-fold of the heavy and light variable regions and form the antigen binding surface. CDR H3 is normally the most variable CDR in both sequence and length, as it is encoded by V, D, and J gene recombination where combinatorial diversity can produce more than 106–107 germline antibodies. About 15 years ago, it was discovered that 10% of cow antibodies have ‘exceptionally long’ CDR H3 regions, with lengths of up to 69 residues (1), while more conventional cow Abs have CDR H3s of around 23 residues, which also are quite long (n.b. we also use the term ‘ultralong’ CDR to correspond to the original genetic description of ‘exceptionally long’ CDRs). The ultralong CDR H3s contain a large, usually even, number of cysteine residues (from 2–12) (1–2). The longest CDR H3 regions found to date in the human germline repertoire contain up to 35 residues (3), while camelid single-chain antibodies have up to 24 residues (4) and shark IgNAR antibodies up to 27 residues (5). While CDR H3 cysteines are rare in humans and mice, they are found more extensively in sharks and camelids, and tether the long CDR H3s to the framework region or to a neighboring CDR. Cysteines are also commonly found in CDR H3 of chickens (6), pigs (7), and the duck-billed platypus (8). The ultralong CDR H3s are found in bovine immunoglobulins of all classes (9), arise early in fetal development (10), and are encoded by the VHBUL, DH2 and JH1 gene segments (2, 11–12). Their length is due to a remarkably long, germline DH2 segment that encodes four cysteines, and also repeated glycine, serine, and tyrosine residues, whose codons are biased for mutation to cysteine with just one base change (Fig. 1). Furthermore, CDR H3 size can be increased via N and P additions, and ‘conserved short nucleotide sequence’ insertions at the V-D junction (13). The gene segments also contain a large number of RGYW hotspots, which are recognition sites for activation-induced cytidine deaminase, which produces somatic hypermutation (SH).
Fig. 1.
Bovine ultralong CDR H3 sequence features and numbering scheme. Sequence alignment of bovine CDR H3 regions in this study with the germline-encoded VHBUL, DH2, and JH1 gene segments. The highly abundant Tyr, Gly, Ser residues encoded in the DH2 gene are colored in purple. The conserved canonical CysH92 (yellow) and TrpH103 (cyan) that generally define the start and end of CDR H3 are indicated in all sequences. The length of each CDR and number of non-canonical Cys residues (also yellow) are shown on the far right. The numbering scheme (below the sequences in red) used here for bovine ultralong CDR H3s will follow Kabat numbering until the region encoded by DH2 (shown in blue), which is easily identifiable by a highly conserved CPD motif with cysteine typically at position D2. This D region is numbered sequentially with a D identifier, from D1 to D50, then the region encoded by the JH1 gene is numbered J1, J2, before returning to Kabat numbering at position H101 (two residues preceding the conserved TrpH103 highlighted in cyan).
Thus, the repertoire of most ultralong CDR H3 antibodies appear to derive from a single germline rearrangement, with diversity arising mainly at the V-D and D-J junctions (through N and P additions and deletions) and through extensive SH. As only a single framework is used for both the heavy and light chain variable regions, only CDR H3 appears to be significantly diversified in the repertoire. Thus, this bovine system differs from that of classical (human or mouse) antibodies in two respects: (i) the diversity mechanism used to create the repertoire, and (ii) the CDR H3 structural scaffold for binding antigen. The first two crystal structures for bovine antibodies with ultralong CDR H3 regions (2) revealed that the CDR H3s fold into an extended, β-ribbon ‘stalk’ topped by a ‘knob’ domain containing different combinations of cysteine residues that pair to form disulfides. These structures, along with deep sequencing data, suggested that the cow repertoire uses different disulfide patterns, generated through SH, in the knob region of CDR H3 as the basis for creating structural diversity for antigen binding. Here we determined three further crystal structures of bovine ‘ultralong’ CDR H3 antibodies and, with this larger dataset, have elucidated the key conserved features that shed insight into how a remarkably structurally diverse repertoire can be created from limited genetic information.
Results
Overall structure/sequence
Bovine heavy chain IgG variable region cDNAs were obtained from a cow immunized with bovine viral diarrhea virus (2). From 133 available heavy chain sequences, several were chosen for expression as Fab fragments, and three of these Fabs crystallized in forms suitable for x-ray diffraction studies (Table 1). Crystal structures were determined for bovine Fabs B11 (2.06 Å), E03 (2.20 Å), and A01 (2.00 Å). Fab B11 has eight cysteines within its 63-residue CDR H3, which is the longest CDR H3 structure determined to date, Fab E03 has one of the shortest ‘ultralong’ CDR H3s of 44 residues with only two cysteines, and Fab A01 has a 61-residue H3 with 6 cysteines, but not the first, highly conserved cysteine residue in the knob domain (Figs. 1, 2). The CDR H3s of these five Fabs are highly dissimilar from one another in terms of aminoacid sequence (Fig. 1) with less than 35% identity between the sequences. Thus, these Fabs, along with the previously crystallized BLV1H12 and BLV5B8, display a range of features (e.g. CDR H3 length, sequence diversity, disulfide content and position) that provide insights into the structural and functional diversity of the ultralong CDR H3 repertoire as well as conserved motifs and scaffolds.
Table 1.
Data collection and refinement statistics for bovine Fabs.
| Fab B11 | Fab E03 | Fab A01 | |
|---|---|---|---|
| Beamline | SSRL 12-2 | APS 23-ID-B | SSRL 12-2 |
| Wavelength (Å) | 0.9795 | 1.0331 | 1.0332 |
| Resolution (Å)a | 38.9 - 2.06 (2.13 - 2.06) | 49.1 - 2.20 (2.24 - 2.20) | 38.4 - 2.00 (2.07 - 2.00) |
| Space group | C2 | P21 | C2 |
| Unit cell (Å) | 78.41, 71.46, 88.32 | 62.24, 221.74, 67.95 | 80.49, 72.49, 87.42 |
| Unit cell (°) | 90, 97.27, 90 | 90, 104.81, 90 | 90, 107.31, 90 |
| Total reflections | 126,115 (12,027) | 357,535 (17,161) | 206,132 (21,059) |
| Unique reflections | 28,518 (2791) | 90,089 (4474) | 31,945 (3179) |
| Multiplicity | 4.4 (4.3) | 4.0 (3.9) | 6.5 (6.6) |
| Completeness (%) | 95.0 (94.1) | 100.0 (100.0) | 98.4 (98.8) |
| Mean (I/σI) | 13.8 (1.6) | 16.5 (1.6) | 11.2 (1.8) |
| Rmergeb | 0.054 (0.968) | 0.088 (0.886) | 0.106 (1.34) |
| Rmeasc | 0.061 (1.12) | 0.097 (1.00) | 0.115(1.45) |
| Rpimd | 0.03 (0.53) | 0.051 (0.518) | 0.045 (0.612) |
| CC1/2e | 0.999 (0.733) | 0.910 (0.685) | 0.997 (0.675) |
| Rwork | 0.194 (0.381) | 0.217 (0.310) | 0.211 (0.331) |
| Rfree | 0.235 (0.376) | 0.251 (0.380) | 0.254 (0.353) |
| # reflections used in refinement (work/free) | 26,133/2372 | 85,377/4416 | 30,047/1883 |
| # Protein atoms | 3562 | 13583 | 3592 |
| # Waters | 55 | 339 | 37 |
| # Protein residues | 486 | 1842 | 484 |
| RMS (bonds) | 0.003 | 0.003 | 0.003 |
| RMS (angles) | 0.85 | 0.61 | 0.66 |
| Ramachandran favored, allowed, outliers (%) | 94.2, 5.2, 0.6 | 95.9, 3.8, 0.3 | 96.0, 4.1, 0.0 |
| Clashscoref | 1.86 | 1.87 | 1.42 |
| Wilson B (Å2) | 45.1 | 36.6 | 39.7 |
| Average B (Å2) | 73.3 | 65.9 | 60.1 |
| Protein | 73.5 | 66.8 | 60.2 |
| Solvent | 55.8 | 42.5 | 45.9 |
Numbers in parentheses are for highest resolution shell
Rmerge = Σhkl Σi=1,n|Ii(hkl)−<I(hkl)>|/Σhkl Σi=1,n Ii(hkl)
Rmeas = Σhkl √(n/n−1) Σi=1,n|Ii(hkl)−<I(hkl)>|/Σhkl Σi=1,n Ii(hkl)
Rpim = Σhkl √(1/n−1) Σi=1,n|Ii(hkl)−<I(hkl)>|/Σhkl Σi=1,n Ii(hkl)
CC1/2 = Pearson Correlation Coefficient between two random half datasets
Number of unfavorable all-atom steric overlaps ≥ 0.4Å per 1000 atoms
Fig. 2.
Bovine Fab structures and sequence alignments. (A) Crystal structures of bovine Fabs E03, B11, BLV1H12 (PDB 4K3D), BLV5B8 (PDB 4K3E), and A01 in a schematic representation. The light chains are in pink and heavy chains in light blue. The core β-strands (as defined by DSSP) in the distal CDR H3 knob domains are colored yellow, green, and blue. (B) The bovine Vλ sequence (top) is aligned with the Vλ sequence from human antibody KOL (PDB 2FB4) (61). The five bovine VH sequences (bottom) are aligned with the VH sequence from human Fab PGT121 (PDB 4JY4) (24). The bovine H3 regions are aligned based on structural conservation, with the type I β-turn residues in red and β-strand residues colored as in (A).
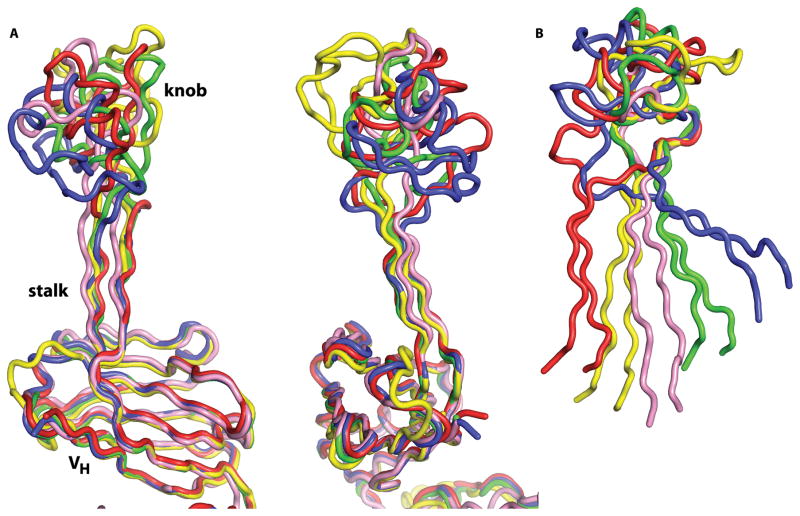
As with Fabs BLV1H12 and BLV5B8 (PDB codes 4k3d and 4k3e (2)), all three of the newly characterized bovine Fabs have a long β-ribbon stalk region supporting a compact knob domain, with the distal tip of the knob rising far above the tips of the other CDR loops. Alignment of these three Fabs with the two previously determined bovine Fab structures reveals little overall variation or movement of the stalk region (Fig. 2) and surrounding CDRs, suggesting the long β-hairpin is quite stable. When variable heavy chain framework residues are superimposed, and the rotation necessary to superimpose the CDR H3 stalks (which were not included in the first superposition) is measured, we see only small differences of 3.5°, 5.0°, 5.5°, and 10.7° for BLV5B8, BLV1H12, C07, and E03 with respect to B11 (Fig. 3A). However, the knob can adopt different relative positions and orientations with respect to its stalk. When only the knob domains are superimposed, the rotations required to superimpose the stalk domains are quite large being 23.0°, 31.5°, 57.1° and 57.3° for BLV5B8, BLV1H12, C07, and E03 when compared to B11 as a reference (Fig. 3B). The overall lengths of the CDR H3 also differ slightly and contribute to variation in the distance of the knob domain to the Fab surface. We approximated the CDR H3 vertical height by measuring from the Cα atom of residue H98, which is roughly coplanar with the tips of the neighboring CDR loops, to the most distal Cα atom of the knob domain. BLV1H12 CDR H3 extends the farthest around 41 Å, while A01 extends only 32 Å. BLV5B8, B11, and E03 all have similar, intermediate CDR H3 lengths of 36 Å, 36 Å, and 37 Å. The distance above the tips of the other CDR loops is not simply a function of the number of residues in CDR H3, but also of how the knob domain is positioned with respect to its stalk. The rest of the heavy-chain variable region is very similar in sequence, with differences at only 13 positions among the five Fabs for residues H1-H92 (Fig. 2B). The variable regions also superimpose with low root mean square deviations (RMSDs) on Cα residues (compared to B11) of between 0.43–0.59 Å for A01, E03, and BLV5B8. BLV1H12 has a larger RMSD of 1.43 Å due to a change in CDR H1 conformation, and a slight shift at the tip of a loop in the heavy Framework Region 3 (residues H72-H76) that corresponds to HV4 in TCRs and shark IgNAR. The ultralong CDR H3 antibodies have also been reported to use a largely invariant λ light chain (14), and all heavy chains are paired with an identical λ chain in the five structures (Fig. 2B). The five λ chains are also highly structurally similar with RMSDs ranging from 0.28 to 0.50 Å for Vλ residues (when compared pairwise to B11).
Fig. 3.
Architecture and relative disposition of bovine CDR H3 stalk and knob domains. (A) When the VH domains of five bovine antibodies are superimposed (left), very little residual movement is seen in the stalk regions. CDR H3’s from bovine antibodies B11, BLV1H12, BLV5B8, A01, and E03 are shown in red, yellow, green, blue, and pink, respectively, with orthogonal views on left and right. (B) In contrast, when the knob domains are superimposed by their conserved type I β-turn and β-strand core, the difference in the position of the supporting stalks is much larger, showing that different knob domains have different relative orientations with respect to the conserved stalk and VH core.
There are many different schemes for numbering of antibody variable regions, such as Kabat (15), Chothia (16), and more recent versions from Martin (17) and Honegger (18). However, the ultralong bovine CDR H3 regions are far too long to be accommodated by any of these numbering schemes. For clarity, we will use Kabat numbering here for the light chain (L), and for heavy chain (H) residues 1–100, and 101–228, but residues in CDR H3 encoded by the DH2 and JH1 genes will be numbered sequentially. The DH2 encoded region begins with a highly conserved CPD motif at residues 2–4 compared to the germline-encoded DH2 sequence; therefore in our numbering scheme, the conserved Cys is residue ‘D2’ (Fig. 1), followed by D3, D4, etc., and then the JH1-encoded residues J1 and J2 followed by H101.
As noted previously for human and mouse Fabs with λ light chains (19), bovine Fabs can also adopt a wide range of elbow angles, even within the same crystallographic asymmetric unit. In Fab E03 with four Fabs in the asymmetric unit, two Fabs have similar elbow angles (109°, 118°), while the other two Fabs are also similar (205° and 208°) but differ from the first pair. Fabs B11 and A01 with one Fab in the asymmetric unit have elbow angles of 139° and 136°, BLV5B8 and BLV1H12, with two Fabs in the asymmetric unit, have elbow angles of 131° and 134°, and 150° and 139°, respectively. Thus, the elbow angles are extremely flexible despite the high sequence conservation in the variable (except CDR H3) and constant regions of these bovine antibodies.
Knob structure
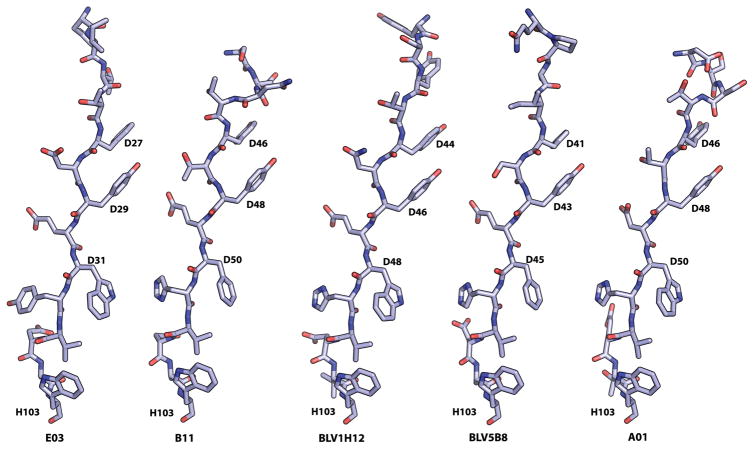
The knob domains of the five Fabs appear dissimilar upon initial inspection, but some conserved structural features can be identified after their superposition. Each knob domain is initiated by a type I β-turn, followed by three anti-parallel β-strands, with the strands connected by loops of differing lengths and conformations (Fig. 4) varying from short two-residue connecting loops in E03, to longer helical or extended loops in the other Fabs. The strands (as defined by DSSP (20)) also differ slightly in length (Fig. 4). The sequence around the type I β-turn is fairly well conserved, with the highly conserved CysD2 followed by Pro/SerD3, AspD4, Gly/AspD5, Tyr/SerD6, where D4 and D5 are the i+1 and i+2 residues of the β-turn. CysD2 always forms a spatially conserved disulfide with a residue in the second β-strand (CysD13, CysD19, CysD23, CysD20, in E03, B11, BLV1H12, and BLV5B8, respectively); however, we can discern no other conserved sequence patterns within the knob domains (Fig. 4). The second cysteine in the conserved disulfide bond (CysD13, CysD19, CysD23 and CysD20) is also separated in the linear sequence from its conserved partner CysD2 by anywhere from 10 to 20 residues depending on the length of the loop connecting the first and second strands. All cysteine residues form disulfide bonds within the knob domains and, with the exception of the spatially conserved disulfide involving CysD2, little conservation is observed in the disulfide positions. Notwithstanding, Fabs BLV1H12 and BLV5B8 share spatial conservation of one other disulfide, but the actual residues on the C-terminal side that form the disulfide differ; the BLV1H12 disulfide forms between CysD12 and CysD32, while that of BLV5B8 is between CysD12 and CysD24 (Fig. 4). While high sequence conservation makes it easy to identify residues D2-D5 from aligned sequence data, assigning the actual residues that form the second and third β-strands and the disulfide connectivity is very difficult without a crystal structure. Fab A01 was also included in this study because it has a very rare sequence that lacks the highly conserved CysD2. However, its crystal structure reveals the same type I β-turn and a similar, three-stranded knob structure, despite lacking the conserved CysD2 disulfide. The disulfide connectivities in each knob domain differ, so that E03, with just one disulfide has 1–2 connectivity, A01, BLV1H12 and BLV5B8, all with three disulfides, have 1–4, 2–5, 3–6 (A01), 1–4, 2–6, 3–5 (BLV1H12) and 1–3, 2–4, 5–6 (BLV5B8) connectivity, and B11 with four disulfides has 1–4, 2–7, 3–8, and 5–6 connectivity. Interestingly, Fab A01, which lacks the conserved D2 cysteine, has weak electron density for an alternate disulfide connection between CysD24 and CysD38 (present at about 40% occupancy), in addition to the CysD10/CysD24 and CysD19/CysD38 disulfides with stronger electron density. In the alternate configuration, two cysteines would be left unpaired (D10 and D19). Thus, the cysteines in CDR H3 of Fab A01 may form 1–4, 2–5, 3–6 and, less often, 2–5, 1–3 disulfide patterns. Although we do not see electron density supporting alternate disulfides in the other structures, each knob domain has some cysteine residues that are within disulfide bonding distance to more than one cysteine (table S1) with many Cα-Cα distances between 3.4–7.6 Å, which corresponds to the range of Cα-Cα distances found in a survey of 351 disulfide bridges (21). The close spacing of these cysteine residues may be beneficial during SH, where alternate disulfide bonds could be readily formed as cysteines are lost or gained.
Fig. 4.
Secondary and tertiary structure and topology of the bovine CDR H3 knob domain. The knob domain secondary and tertiary structure (A) and primary structure topology diagrams (B) are shown from left to right for bovine Fabs E03, B11, BLV1H12, BLV5B8, and A01. The spatially conserved disulfide from CysD2 to a Cys in the second β-strand in the knob domain is colored in red, and the other disulfides are color coded to match the topology diagrams. The β-strands 1, 2, and 3 are colored yellow, green and blue in all panels. GlyD2 in A01 (that is usually a Cys in other bovine Fabs) is highlighted in red on a white background.
Stalk structure
All of the bovine Fab CDR H3 stalk regions fold into long, extended β-ribbons with a number of aromatic residues, especially in the descending strands, that may aid in stabilization of the long stalks (Fig. 5, fig. S1). Interestingly, some human antibodies also have multiple aromatic residues in this region (for example 4JY4 in Fig. 2), especially those using the human IgH J6*03 germline gene coding for the sequence ‘YYYYYYMDVWGKG’. Several of these human antibodies have extended, β-hairpin CDR H3s whose bases (stalks) are similar in conformation to the corresponding region of the bovine CDR H3 stalk regions. These antibodies include anti-HIV-1 antibodies 10–1074 (PDB 4FQ2) (22), PGT121 (PDB 4NPY) (23), PGT122 (PDB 4JY5) (24), Z13e1 (PDB 3FN0) (25) and anti-Marburg virus antibody MR78 (PDB 3X2D) (26) (fig. S2). The bovine CDR β-ribbons start with an ascending strand, which emanates from the base of CDR H3 and leads into the stalk domain, and then returns via the descending strand, in which all five structures share a kink at its base around residues Kabat residues H100-H103 (Val directly preceding H101, which is called here J2, Asp/GluH101, Ala/Thr/ValH102, TrpH103). The kink differs slightly from those seen in human and mouse Fabs, where a hydrogen bond is usually formed between TrpH103 NE1 and the carbonyl oxygen from the residue preceding H101. In the five bovine structures, TrpH103 NE1 instead hydrogen bonds with the conserved ThrL46 side-chain hydroxyl (Fig. 6A). L46 is Thr in 15 non-bovine Fab PDB depositions, and those structures show a mixture of kinked and extended bases; thus ThrL46 is not sufficient to always enforce a kinked H3 base in non-bovine heavy chains. The residue at position 101 in the five bovine heavy chains is Asp or Glu. In mouse and human kinked H3 bases, Asp at this position often hydrogen bonds with ArgH94 on the opposing H3 strand; however, in the bovine structures, H94 is Thr or Ser. Thus, H101 either: (i) does not interact with any other residues (E03 and BLV1H12), (ii) forms a salt bridge with conserved LysH32 (A01 and B11), or (iii) hydrogen bonds with HisD46 (BLV5B8). ThrH94 is germline encoded in the VHBUL gene (H92CTTVHQH97) (Fig. 1), while most mouse or human antibodies have H92CARH94 or H92CAKH94 rather than H92CTTH94 (2). The length of the extended, hydrogen-bonded ribbon connecting the kinked H3 base and knob domains can differ, with long (~12 residues) ribbons in E03, BLV1H12 and BLV5B8 and shorter ribbons (~ 8 residues) in B11 and A01. The shorter ribbons are separated from the knob domain by a 5-residue loop region on the descending strand involving residues D41 to D45 (SSDNT or LDSST) that disrupts the hydrogen bond ladder with the opposing strand and that may allow the knob domain to have more flexibility around its stalk (Figs. 3 and 5). In B11, residues SSDNT form an undefined 5-residue turn, while DSST in A01 forms a type I β-turn. Some side-chain hydrogen bonds within the stalk may also aid its stabilization (table S2), with one fairly well conserved hydrogen bond between His/TyrH96 and a Glu six residues before the conserved TrpH103 (Glu D49, D30, D49, and D44 in B11, E03, BLV1H12, and BLV5B8, respectively). This conserved hydrogen bond is not present in A01, instead, AspD49 hydrogen bonds to ArgH100b, while HisH96 hydrogen bonds to GluH98. Thus, the conserved rigid stalk structures share potentially stabilizing hydrogen bonds and aromatic residues, a kinked base, and potentially flexible junction with the knob domain.
Fig. 5.
Descending strand of the bovine CDR H3 stalk regions. Each Fab has conserved aromatic residues in the descending strand of the stalk that likely help stabilize the stalk β-ribbon. A kinked base region around H103 is found at the H3 base in all five bovine antibodies.
Fig. 6.
Comparison of bovine CDR loop conformations with CDR canonical classes. (A) The H3 ‘kink’ or ‘bulge’ in 2FB4 (left) and B11 (right) are shown. In a ‘normal’ kinked base, Trp103 NE1 hydrogen bonds to a neighboring carbonyl as in 2FB4, but in the bovine CDR H3, Trp103 NE1 hydrogen bonds to the side chain of ThrL46. (B) The CDR regions L1, L2, L3, H1 and H2 are shown superimposed with canonical reference CDRs from published structures. Bovine CDRs are shown in gray, and reference CDRs in red. Sequences for the CDRs are shown below the superpositions. The sequences for CDR H1 are identical in all five bovine structures, but differ for CDR H2, while the λ chain CDR sequences for all five bovine Fabs are identical. As the bovine λ chains are therefore also highly similar in structure, only one bovine CDR loop is shown in each of the superpositions. The L1 loops are very similar to canonical class 5λ/13a, except at GlyL31 where a difference is observed in the position and orientation of its carbonyl oxygen. The L2 loops belong to class 1λ/7a. The L3 loops would be predicted to belong to class 5λ/11a, but the bovine loop tip bends away from the canonical structure to support the H3 stalk. The H1 loops for B11, A01, E03, and BLV5B8 (left) align fairly well with canonical class 1/10a, but both Fabs in BLV1H12 (center) have distorted H1 loops that interact with each other via crystal packing interactions. The H2 loops belong to class 1/9a.
Canonical CDRs
Human and mouse CDR regions L1, L2, L3, H1, and H2 are encoded by the diverse germline variable region genes but adopt canonical structures that can often be predicted on the basis of key residues within the loops (16, 27–29). In typical antibodies, these CDRs contribute to antibody diversity and to antigen contacts. However bovine ultralong CDR H3 antibodies have most of their sequence diversity in CDR H3, and appear to bind antigen mainly or only through CDR H3 (2), suggesting that the other CDRs may play a purely structural role. Thus, we compared the bovine CDR sequences and conformations to determine whether they adopt canonical structures found in other conventional antibodies (Fig. 6B). Indeed, most of the bovine CDRs adopt structures consistent with the previously established canonical classes. L1 resembles class 5λ/13a, with a small deviation and carbonyl shift around GlyL31, L2 is in the 1λ/7A class, and L3 is predicted to belong to the 5λ/11a class, but is bent over at its tip to accommodate the descending strand of the stalk (2). CDR H1 from B11, A01, and E03 belongs to canonical class 1/10A; however, in BLV1H12, the CDR H1s of the two Fabs in the asymmetric unit do not adopt the expected canonical structure as they interact with one another in the crystal lattice. CDR H2 belongs to class 1/9a. However, the main role of the bovine light chain CDRs appears to be to lend structural support to CDR H3: stabilizing contacts include stacking of CDR L1 TyrL32 against CDR H3 Glu/Asp (three residues before H100), stacking of CDR L2 TyrL49 with His/Tyr (one residue before H100), stacking of the CDR L3 tip (L92AEDSSL95) with Tyr (four residues before H100) (fig. S3), and CDR L1 AsnL30 hydrogen bonds to the main-chain of Tyr (four residues before H100). As originally proposed based on a homology model (14), the small Ser side chain at position L90 is necessary, as this side chain is tightly packed between CDR L1 AsnL27b and ValL28 and a Trp/Phe in the descending strand of CDR H3 (TrpD31, PheD50, TrpD48, PheD45, TrpD50, in E03, B11, BLV1H12, BLV5B8, or A01, respectively). Contacts from CDR H3 to the heavy-chain CDR H1 and H2 are less extensive with one potentially stabilizing hydrogen bond between CDR H2 Ser/AsnH50 and CDR H3 GlnH97.
Based on the two original structures (2), it was proposed that CDR H1 AspH31 and LysH32 stabilize H3 by interacting with HisH96 via a water molecule (2). However, this interaction is only found in BLV1H12, due to the unusual conformation of CDR H1 (likely induced by crystal packing). With the exception of the bent over CDR L3, the canonical structures and residue composition of the bovine CDR’s are not notably different from those in classical human and mouse antibodies. Thus, these bovine CDR’s have not evolved unique structures or unusual sequences, but have co-evolved nevertheless with CDR H3 to optimize the rigidity and stability of the extended bovine H3 stalk. Non-bovine antibodies with long, protruding CDR H3 regions may also use their CDR loops, especially from the light chain, for H3 stabilization (30), while human anti-hemagglutinin antibody C05 that binds antigen primarily with its long 24-residue CDR H3 (31), uses its CDRs H1 and H2 to make potentially stabilizing contacts to rigidify and extend its protruding H3.
Comparison of knob with other protein folds
We searched for proteins with structural homology to the knob domains using the PDBeFOLD server (32) and the closest matches were the small cyclic peptide (cyclotide) Kalata B1 (PDB 1N1U) (33), WW domains such as FE65 (PDB 2HO2) (34), and a segment from the tumor necrosis factor receptor superfamily member 13C (PDB 1OQE) (35). The best match was between the E03 knob and Kalata B1, a small (29-residue) cyclic protein with antimicrobial activities thought to result from disruption of membranes. Kalata B1 has three disulfides and 16 of its residues overlap with the E03 knob with an RMSD on Cα of 1.6Å. The conserved CysD2 disulfide in E03 is spatially conserved with the 3–6 disulfide in Kalata B1 (Fig. 7). Strands 1 and 2 in E03 (SRLI and DCRN) are the same length as the corresponding strands in Kalata B1 (CTCS and VCTR), whereas strand 3 in E03 has two residues (DV) that overlap fairly well with the corresponding region in Kalata B1; however, that region in Kalata B1 is not defined as a strand by DSSP.
Fig. 7.
Structural homology of the bovine CDR H3 knob domain with the cyclotide Kalata B1. (A) The closest structural homologue to the knob domain, as identified with the PDBeFold server, is the small, cyclic peptide (cyclotide) Kalata B1. Superposition of the bovine Fab E03 knob domain (blue) with Kalata B1 (gray, PDB 1N1U) is shown. The E03 CysD2-D13 disulfide is shown in red at the same spatial location as the Kalata B1 disulfide. In the sequence alignment (below), the 16 underlined residues superimpose with an RMSD on Cα atoms of 1.6 Å. The red Cys residues in the sequence alignment represent the spatially conserved disulfide bond. (B) The E03 knob with core β-strands (yellow, green, and blue) and conserved disulfide (red). (C) Kalata B1 with homologous regions colored as for E03.
Discussion
The paradigm for antigen recognition by most vertebrate antibodies is to make specific contacts with antigen through many or all of the hypervariable CDR loops that emanate from and are supported by the framework β-strands within the immunoglobulin domains of the heavy and light chain variable regions. While unusual antibodies, such as the human anti-hemagglutinin (HA) antibody C05, can bind antigen primarily using CDR H3 (31), or only the heavy-chain CDRs as for HA stem binding VH1–69 antibodies (36–37), most anti-protein antibodies use 5–6 CDR loops for antigen contact (38). However, the component of the cow antibody repertoire that is characterized by ultralong CDR H3s appears to utilize only CDR H3 for antigen recognition, as shown previously in mutation studies of the unusual H3 knob domain (2).
The five bovine Fab structures that are now available enable us to compare and analyze these structures to identify any conserved features and motifs in these ultralong CDR H3s. Such a study has now revealed new features and mechanisms to attain structural diversity for antigen recognition. First, three conserved short β-strands within the knob domain provide a scaffold for two hypervariable loops that can differ in length, structure and disulfide connectivity. Second, the stalk can protrude at slightly variable distances from the surface of the antibody. Third, the orientation of the knob in relation to the stalk can change between antibodies, with rotational differences by nearly 60°. The variability in length and structure of the loops connecting β-strands 1 and 2, and 2 and 3 in the knob domains mirrors the diversity in antibody CDR regions, where varying the length and composition of the CDRs displayed on a largely conserved Fab immunoglobulin framework result in innumerable different binding site architectures. Thus, the bovine antibody has introduced a new level of structural diversity to its binding site by incorporating two pseudo-CDR-like loops within its novel CDR H3 knob domain, with the ability to further diversify through incorporation of diverse disulfide patterns and extensive somatic mutation. Additionally, the knob domain itself can be oriented closer or further from the antibody surface by varying the stalk length and its twist in relation to the stalk. Thus, fine-tuning of the flexible knob may allow it to optimally reach into and bind epitopes that are challenging for conventional antibodies.
Most ultralong CDR H3 antibodies maintain a conserved cysteine at position D2 that forms a disulfide bond with a Cys in the second, core β-strand. However, while conserved in the 3-dimensional structure, this second cysteine is not conserved in the linear sequence of the five Fab fragments. While the germline DH2 region encodes four cysteines, only the Cys encoded at D2 is highly conserved in deep sequence analysis of CDR H3 (2). Thus, diversity generation mechanisms may place the second cysteine in the appropriate location to form a disulfide bond with D2.
Tyrosine, glycine, and serine are highly abundant in the bovine DH2-encoded germline sequence ‘SCPDGYSYGYGCGYGYGCSGYDCYGYGGYGGYGGYGYSSYSYSYTYEY’ and account for 6.7% (Tyr), 10.0% (Gly) and 9.5% (Ser) of the residues from a set of 52 mature bovine CDR H3 sequences. These amino acids are also found frequently in human and murine CDR H3 regions (3, 39), but in a survey of the complete UniProtKB/SwissProt sequence database as of February 2016 (http://web.expasy.org/docs/relnotes/relstat.html), frequencies for these particular amino acids in all available protein sequences are 2.9% (Tyr), 7.1% (Gly), and 6.6% (Ser), so that they are indeed over-represented in these ultralong CDR H3s. Synthetic libraries utilizing antibody scaffolds as well as other protein folds have been developed using very small subsets of amino acids as recognition elements, with tyrosine, serine, and glycine proving the best minimal combination (40) for effective antigen recognition. Tyrosines are typically responsible for forming most of the molecular contacts, while the smaller serine and glycine residues can add flexibility and act as spacers between the larger tyrosine residues (41). Interestingly, some proteins including ‘keratin associated proteins’ (KAP), and the shell matrix protein prismalin-14, also have high glycine/tyrosine content, and share sequence motifs such as ‘YGGYGGYGGYGGY’ that are also encoded by the germline DH2 gene. The KAP proteins crosslink with keratin filaments, and also can contain a number of cysteines, while prismalin-14 is involved in mollusk shell formation. Whether there are similar binding functions between these motifs and the cow ultralong CDR H3 germline antibody requires further study.
The burning question remains as to how the bovine ultralong CDR H3 antibodies recognize antigen, and what types of epitopes they recognize. Earlier work has indicated that the IgM antibodies BLV1H12 and BLV5B8 are multispecific, recognizing antigens such DNA, synthetic haptens, and cytoskeleton proteins and may be natural autoantibodies (1, 42). Previous experiments with one ultralong antibody to the immunogen bovine viral diarrhea virus indicate that only CDR H3 is necessary for binding antigen (2). Bovine heavy chains from deep sequencing show high sequence homology in all but the CDR H3 region suggesting that CDR H3 is the sole, or at least the most critical element for antigen recognition, with decreased variability in other locations (Smider, personal communication). The longest human CDR H3 regions have mostly been found in human anti-HIV-1 antibodies where they can insert between the densely packed carbohydrates of the glycan shield to attain contact with the protein surface below; thus, the bovine CDRs might be used in a similar fashion to reach into deep clefts or pores. The structural similarity to Kalata B1 also suggests that the antibodies might use their CDR knob regions to disrupt membranes; however, extremely limited tests in our laboratory have not so far been able to support this notion. Indeed, the plethora of different CDR H3 sequences in cows argues against such an innate immune function for these antibodies, suggesting instead that they recognize many different antigens. It is remarkable that cows have evolved such a unique scaffold and architecture for antigen recognition, but which is still comprised within CDR H3 of a typical immunoglobulin domain framework.
Cyclotides such as Kalata B1 are being used as novel scaffolds for the design of small binding proteins (43–46), and their structural homology with the knob domains on the bovine antibodies suggests that libraries of bovine antibodies with extensive natural variation in the loop regions and disulfide pairings could be used to search for small knob domains with specific affinities or functions. Antibody engineering inspired by the bovine antibody template has already taken off at a rapid pace, with knob domains replaced with other proteins, stalk regions replaced by stable structures such as a coiled-coiled domain, or the stalk/knob structures transplanted into the CDR regions of human antibodies (47–54). Indeed, antibodies with novel activities have also been created through insertion of large protein domains into the CDR H3 of conventional antibodies by functional selection using libraries of permissive junctions (55). The structural insights presented here may aid in future immunization or engineering efforts based on these interesting antibodies, and could further our understanding of their unique place in the bovine immune system, as well as the unusual paradigm they represent in creating genetic and structural diversity.
Materials and Methods
Protein expression and purification
The light and heavy chain DNA for each bovine IgG was cloned separately into vector pcDNA3, and a stop codon was introduced into the heavy chain DNA after residue H228 for Fab production. Mammalian 293Freestyle cells (1 liter) were transformed with light (0.25 mg) and heavy chain (0.50 mg) DNA, and incubated in a shaking CO2 incubator for 6 days. The cells were centrifuged at 4000 rpm for 20 minutes and the media supernatant was then filtered with a 0.22 micron filter. The filtered media was loaded onto a CaptureSelect LC-lambda (ungulate) resin (Life Technologies) at 1ml/min, washed with phosphate-buffered saline, pH 7.4, and Fab was eluted with 0.1M glycine, pH 3, into fractions containing 1M Tris, pH 8.0. Each Fab was further purified by size exclusion chromatography on a Superdex 75, 16/600 column (GE Healthcare Life Sciences) with running buffer 20mM HEPES, pH 8, 150mM NaCl. Fabs were concentrated in this buffer for crystallization trials: Fab A01, 16.9 mg/ml, Fab B11, 16.1 mg/ml, and Fab E03, 12.3 mg/ml.
Crystallization
All Fabs were screened for crystallization using 384 conditions (JCSG Core Suites I, II, III, and IV) at 4°C and 20°C using the IAVI/JCSG/TSRI CrystalMation robot. The Fab B11 crystal for data collection was obtained using a precipitant of 5% PEG3000, 30% PEG400, 0.1M HEPES, pH 7.5, 10% glycerol (20° C, JCSG Core Suite III, condition C9). The crystal was flash-cooled in mother liquor with no additional cryoprotectant. Fab E03 crystals were obtained in JCSG Core suite II, condition H5, and that condition was refined by fine screening in CrysChem sitting drop plates. The Fab E03 crystal for data collection was grown at 22°C with a precipitant of 0.2M NaCl, 16.5% PEG8000, 0.1M phosphate-citrate buffer, pH 4.2, and flash-cooled after brief immersion in the well solution augmented with 30% ethylene glycol. The Fab A01 crystal was taken directly from the screening tray, with precipitant 20% PEG6000, 0.1M bicine, pH 9 (4°C, JCSG Core Suite I, condition A2), and flash-cooled after immersion in well solution augmented with 30% glycerol.
Data collection
Diffraction data for Fab B11 were collected at SSRL beamline 12-2 and processed with XDS (56). Reflections between 2.24–2.26 Å had elevated average intensities due to ice diffraction obscuring that region and were removed from the dataset, resulting in data with 95.0% completeness to 2.06 Å resolution. Data for Fab E03 were collected at APS beamline 23-ID-B and processed with HKL-2000 (57) resulting in data to 2.20 Å resolution with 99.7% completeness. Data for Fab A01 were collected at SSRL beamline 12-2, processed with XDS and were 98.2% complete to 2.00 Å resolution.
Structure solution and refinement
All structures were determined by molecular replacement, using Phaser (58) and BLV1H12 (4K3D) (2) as a model. Structures were refined with Phenix (59) and structural figures made with PyMOL (60). Fabs B11 and A01 each have one Fab molecule in the asymmetric unit. Fab E03 crystallized with four Fabs in the asymmetric unit. One of the four Fabs (chains EF) has very poor electron density due to fewer crystal contacts, and the Fab with the best electron density (chains AB) was used for the comparisons presented here.
Supplementary Material
Fig. S1. Electron density for bovine CDR H3 stem regions.
Fig. S2. Comparison of the bovine CDR H3 stem with corresponding region of human CDR H3s encoded by the IgH J6*03 gene.
Fig. S3. Wall-eyed stereoview of stabilizing contacts from light chain CDRs to the CDR H3 stalk.
Table S1. Cysteine Cα-Cα distances (Å) in bovine CDR H3 knobs.
Table S2. Intra-stem side-chain hydrogen bond partners in CDR H3.
Acknowledgments
We thank Yuanzi Hua and Henry Tien for excellent technical assistance, and Omar Bazirgan for providing antibody heavy chain vectors. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393) and the National Center for Research Resources (P41RR001209). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS, NCRR or NIH. Other portions of the research were carried out at GM/CA@APS, which has been funded in whole or in part with Federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. This is manuscript number 29310 from The Scripps Research Institute.
Funding: Supported in part by National Institutes of Health grants 5R01 GM105826 (V.V.S.), R56 AI117675 (I.A.W.) and R01 AI084817 (I.A.W.).
Footnotes
Author contributions: R.L.S. designed and performed experiments, analyzed the data, and wrote the manuscript. I.A.W. and V.V.S. directed the project, analyzed the data, and wrote the manuscript.
Competing interests: V.S. has equity in Sevion, Inc., which has interest in commercial development of cow antibodies.
Data and materials availability: The coordinates and structure factors for this study have been deposited to the Protein Databank with codes: 5IHU (Fab B11), 5IJV (Fab E03), and 5ILT (Fab A01).
References and Notes
- 1.Saini SS, Allore B, Jacobs RM, Kaushik A. Exceptionally long CDR3H region with multiple cysteine residues in functional bovine IgM antibodies. Eur J Immunol. 1999;29:2420–2426. doi: 10.1002/(SICI)1521-4141(199908)29:08<2420::AID-IMMU2420>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 2.Wang F, et al. Reshaping antibody diversity. Cell. 2013;153:1379–1393. doi: 10.1016/j.cell.2013.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zemlin M, et al. Regulation of repertoire development through genetic control of DH reading frame preference. J Immunol. 2008;181:8416–8424. doi: 10.4049/jimmunol.181.12.8416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffin LM, et al. Analysis of heavy and light chain sequences of conventional camelid antibodies from Camelus dromedarius and Camelus bactrianus species. J Immunol Meth. 2014;405:35–46. doi: 10.1016/j.jim.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Diaz M, Stanfield RL, Greenberg AS, Flajnik MF. Structural analysis, selection, and ontogeny of the shark new antigen receptor (IgNAR): identification of a new locus preferentially expressed in early development. Immunogenetics. 2002;54:501–512. doi: 10.1007/s00251-002-0479-z. [DOI] [PubMed] [Google Scholar]
- 6.Wu L, et al. Fundamental characteristics of the immunoglobulin VH repertoire of chickens in comparison with those of humans, mice, and camelids. J Immunol. 2012;188:322–333. doi: 10.4049/jimmunol.1102466. [DOI] [PubMed] [Google Scholar]
- 7.Almagro JC, et al. Analysis of the horse VH repertoire and comparison with the human IGHV germline genes, and sheep, cattle and pig VH sequences. Mol Immunol. 2006;43:1836–1845. doi: 10.1016/j.molimm.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 8.Johansson J, Aveskogh M, Munday B, Hellman L. Heavy chain V region diversity in the duck-billed platypus (Ornithorhynchus anatinus): long and highly variable complementarity-determining region 3 compensates for limited germline diversity. J Immunol. 2002;168:5155–5162. doi: 10.4049/jimmunol.168.10.5155. [DOI] [PubMed] [Google Scholar]
- 9.Walther S, Czerny CP, Diesterbeck US. Exceptionally long CDR3H are not isotype restricted in bovine immunoglobulins. PLoS One. 2013;8:e64234. doi: 10.1371/journal.pone.0064234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saini SS, Kaushik A. Extensive CDR3H length heterogeneity exists in bovine foetal VDJ rearrangements. Scand J Immunol. 2002;55:140–148. doi: 10.1046/j.1365-3083.2002.01028.x. [DOI] [PubMed] [Google Scholar]
- 11.Pasman Y, Kaushik A. In: Comparative Immunoglobulin Genetics. Kaushik A, Pasman Y, editors. Apple Academic Press, Inc; Toronto: 2014. pp. 187–221. [Google Scholar]
- 12.Aida Y, Takeshima S, Baldwin CL, Kaushik AK. In: The Genetics of Cattle. Garrick D, Ruvinsky A, editors. CAB International; 2015. pp. 153–191. [Google Scholar]
- 13.Koti M, Kataeva G, Kaushik AK. Novel atypical nucleotide insertions specifically at VH-DH junction generate exceptionally long CDR3H in cattle antibodies. Mol Immunol. 2010;47:2119–2128. doi: 10.1016/j.molimm.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 14.Saini SS, Farrugia W, Ramsland PA, Kaushik AK. Bovine IgM antibodies with exceptionally long complementarity-determining region 3 of the heavy chain share unique structural properties conferring restricted VH + Vλ pairings. Int Immunol. 2003;15:845–853. doi: 10.1093/intimm/dxg083. [DOI] [PubMed] [Google Scholar]
- 15.Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. Sequences of proteins of immunological interest. 5. Vol. 1 U.S. Department of Health and Human Services; 1991. [Google Scholar]
- 16.Chothia C, Lesk AM. Canonical structures for the hypervariable regions of immunoglobulins. J Mol Biol. 1987;196:901–917. doi: 10.1016/0022-2836(87)90412-8. [DOI] [PubMed] [Google Scholar]
- 17.Abhinandan KR, Martin AC. Analysis and improvements to Kabat and structurally correct numbering of antibody variable domains. Mol Immunol. 2008;45:3832–3839. doi: 10.1016/j.molimm.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 18.Honegger A, Pluckthun A. Yet another numbering scheme for immunoglobulin variable domains: an automatic modeling and analysis tool. J Mol Biol. 2001;309:657–670. doi: 10.1006/jmbi.2001.4662. [DOI] [PubMed] [Google Scholar]
- 19.Stanfield RL, Zemla A, Wilson IA, Rupp B. Antibody elbow angles are influenced by their light chain class. J Mol Biol. 2006;357:1566–1574. doi: 10.1016/j.jmb.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 20.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 21.Petersen MT, Jonson PH, Petersen SB. Amino acid neighbours and detailed conformational analysis of cysteines in proteins. Protein Eng. 1999;12:535–548. doi: 10.1093/protein/12.7.535. [DOI] [PubMed] [Google Scholar]
- 22.Mouquet H, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci USA. 2012;109:E3268–3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sok D, et al. The effects of somatic hypermutation on neutralization and binding in the PGT121 family of broadly neutralizing HIV antibodies. PLoS Pathog. 2013;9:e1003754. doi: 10.1371/journal.ppat.1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Julien JP, et al. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 2013;9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pejchal R, et al. A conformational switch in human immunodeficiency virus gp41 revealed by the structures of overlapping epitopes recognized by neutralizing antibodies. J Virol. 2009;83:8451–8462. doi: 10.1128/JVI.00685-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hashiguchi T, et al. Structural basis for Marburg virus neutralization by a cross-reactive human antibody. Cell. 2015;160:904–912. doi: 10.1016/j.cell.2015.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-Lazikani B, Lesk AM, Chothia C. Standard conformations for the canonical structures of immunoglobulins. J Mol Biol. 1997;273:927–948. doi: 10.1006/jmbi.1997.1354. [DOI] [PubMed] [Google Scholar]
- 28.Chothia C, et al. Conformations of immunoglobulin hypervariable regions. Nature. 1989;342:877–883. doi: 10.1038/342877a0. [DOI] [PubMed] [Google Scholar]
- 29.North B, Lehmann A, Dunbrack RL., Jr A new clustering of antibody CDR loop conformations. J Mol Biol. 2011;406:228–256. doi: 10.1016/j.jmb.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramsland PA, Kaushik A, Marchalonis JJ, Edmundson AB. Incorporation of long CDR3s into V domains: implications for the structural evolution of the antibody-combining site. Exp Clin Immunogenet. 2001;18:176–198. doi: 10.1159/000049197. [DOI] [PubMed] [Google Scholar]
- 31.Ekiert DC, et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 2012;489:526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 33.Daly NL, Clark RJ, Craik DJ. Disulfide folding pathways of cystine knot proteins. Tying the knot within the circular backbone of the cyclotides. J Biol Chem. 2003;278:6314–6322. doi: 10.1074/jbc.M210492200. [DOI] [PubMed] [Google Scholar]
- 34.Meiyappan M, Birrane G, Ladias JA. Structural basis for polyproline recognition by the FE65 WW domain. J Mol Biol. 2007;372:970–980. doi: 10.1016/j.jmb.2007.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, et al. Ligand-receptor binding revealed by the TNF family member TALL-1. Nature. 2003;423:49–56. doi: 10.1038/nature01543. [DOI] [PubMed] [Google Scholar]
- 36.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dreyfus C, et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson IA, Stanfield RL. Antibody-antigen interactions. Curr Opin Struct Biol. 1993;3:113–118. doi: 10.1016/0959-440x(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 39.Mian IS, Bradwell AR, Olson AJ. Structure, function and properties of antibody binding sites. J Mol Biol. 1991;217:133–151. doi: 10.1016/0022-2836(91)90617-f. [DOI] [PubMed] [Google Scholar]
- 40.Birtalan S, et al. The intrinsic contributions of tyrosine, serine, glycine and arginine to the affinity and specificity of antibodies. J Mol Biol. 2008;377:1518–1528. doi: 10.1016/j.jmb.2008.01.093. [DOI] [PubMed] [Google Scholar]
- 41.Koide S, Sidhu SS. The importance of being tyrosine: lessons in molecular recognition from minimalist synthetic binding proteins. ACS Chem Biol. 2009;4:325–334. doi: 10.1021/cb800314v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saini SS, Kaushik A, Basrur PK, Yamashiro S. Ultrastructural and immunologic characteristics of mouse x cattle xenogeneic hybridomas originating from bovine leukemia virus-infected cattle. Vet Pathol. 2003;40:460–464. doi: 10.1354/vp.40-4-460. [DOI] [PubMed] [Google Scholar]
- 43.Craik DJ, Simonsen S, Daly NL. The cyclotides: novel macrocyclic peptides as scaffolds in drug design. Curr Opin Drug Discov Devel. 2002;5:251–260. [PubMed] [Google Scholar]
- 44.Huang YH, et al. Design of substrate-based BCR-ABL kinase inhibitors using the cyclotide scaffold. Sci Rep. 2015;5:1–15. doi: 10.1038/srep12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ji Y, et al. In vivo activation of the p53 tumor suppressor pathway by an engineered cyclotide. J Am Chem Soc. 2013;135:11623–11633. doi: 10.1021/ja405108p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang CK, et al. Molecular grafting onto a stable framework yields novel cyclic peptides for the treatment of multiple sclerosis. ACS Chem Biol. 2014;9:156–163. doi: 10.1021/cb400548s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu T, et al. Rational design of CXCR4 specific antibodies with elongated CDRs. J Am Chem Soc. 2014;136:10557–10560. doi: 10.1021/ja5042447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu T, et al. Rational design of antibody protease inhibitors. J Am Chem Soc. 2015;137:4042–4045. doi: 10.1021/ja5130786. [DOI] [PubMed] [Google Scholar]
- 49.Liu T, et al. Functional human antibody CDR fusions as long-acting therapeutic endocrine agonists. Proc Natl Acad Sci USA. 2015;112:1356–1361. doi: 10.1073/pnas.1423668112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, et al. Functional antibody CDR3 fusion proteins with enhanced pharmacological properties. Angew Chem Int Ed Engl. 2013;52:8295–8298. doi: 10.1002/anie.201303656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, et al. An antibody CDR3-erythropoietin fusion protein. ACS Chem Biol. 2013;8:2117–2121. doi: 10.1021/cb4004749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, et al. An antibody with a variable-region coiled-coil “knob” domain. Angew Chem Int Ed Engl. 2014;53:132–135. doi: 10.1002/anie.201307939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, Liu Y, Wang Y, Schultz PG, Wang F. Rational design of humanized dual-agonist antibodies. J Am Chem Soc. 2015;137:38–41. doi: 10.1021/ja510519u. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, et al. Rational design of a humanized glucagon-like peptide-1 receptor agonist antibody. Angew Chem Int Ed Engl. 2015;54:2126–2130. doi: 10.1002/anie.201410049. [DOI] [PubMed] [Google Scholar]
- 55.Peng Y, et al. A general method for insertion of functional proteins within proteins via combinatorial selection of permissive junctions. Chem Biol. 2015;22:1134–1143. doi: 10.1016/j.chembiol.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 56.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol. 1997;276A:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 58.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.The PyMOL Molecular Graphics System, Version 1.7.4. Schrödinger, LLC; [Google Scholar]
- 61.Huber R, Deisenhofer J, Colman PM, Matsushima M, Palm W. Crystallographic structure studies of an IgG molecule and an Fc fragment. Nature. 1976;264:415–420. doi: 10.1038/264415a0. [DOI] [PubMed] [Google Scholar]
- 62.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Electron density for bovine CDR H3 stem regions.
Fig. S2. Comparison of the bovine CDR H3 stem with corresponding region of human CDR H3s encoded by the IgH J6*03 gene.
Fig. S3. Wall-eyed stereoview of stabilizing contacts from light chain CDRs to the CDR H3 stalk.
Table S1. Cysteine Cα-Cα distances (Å) in bovine CDR H3 knobs.
Table S2. Intra-stem side-chain hydrogen bond partners in CDR H3.