Abstract
Background
The intracellular bacteria Anaplasma phagocytophilum are emerging zoonotic pathogens affecting human and animal health, and a good model for the study of tick-host-pathogen interactions. This tick-borne pathogen is transmitted by Ixodes scapularis in the United States where it causes human granulocytic anaplasmosis. Tick midguts and salivary glands play a major role during tick feeding and development, and in pathogen acquisition, multiplication and transmission. Vertebrate host proteins are found in tick midguts after feeding and have been described in the salivary glands of fed and unfed ticks, suggesting a role for these proteins during tick feeding and development. Furthermore, recent results suggested the hypothesis that pathogen infection affects tick metabolic processes to modify host protein digestion and persistence in the tick with possible implications for tick physiology and pathogen life-cycle.
Methods
To address this hypothesis, herein we used I. scapularis female ticks fed on uninfected and A. phagocytophilum-infected sheep to characterize host protein content in midguts and salivary glands by proteomic analysis of tick tissues.
Results
The results evidenced a clear difference in the host protein content between tick midguts and salivary glands in response to infection suggesting that A. phagocytophilum selectively manipulates the levels of vertebrate host proteins in ticks in a tissue-specific manner to facilitate pathogen infection, multiplication and transmission while preserving tick feeding and development. The mechanisms by which A. phagocytophilum manipulates the levels of vertebrate host proteins are not known, but the results obtained here suggested that it might include the modification of proteolytic pathways.
Conclusions
The results of this study provided evidence to support that A. phagocytophilum affect tick proteolytic pathways to selectively manipulate the levels of vertebrate host proteins in a tissue-specific manner to increase tick vector capacity. Investigating the biological relevance of host proteins in tick biology and pathogen infection and the mechanisms used by A. phagocytophilum to manipulate host protein content is essential to advance our knowledge of tick-host-pathogen molecular interactions. These results have implications for the identification of new targets for the development of vaccines for the control of tick-borne diseases.
Electronic supplementary material
The online version of this article (doi:10.1186/s13071-016-1747-3) contains supplementary material, which is available to authorized users.
Keywords: Anaplasma, Tick, Ixodes, Proteomics, Hemoglobin, Immunology
Background
Infectious diseases transmitted by arthropod vectors such as ticks constitute a growing burden for human and animal health worldwide [1–4]. Anaplasma phagocytophilum (Rickettsiales: Anaplasmataceae) are tick-borne intracellular bacteria that infect vertebrate host granulocytes causing human, canine and equine granulocytic anaplasmosis and tick-borne fever of ruminants [5–8]. Human granulocytic anaplasmosis (HGA) is the second most common tick-borne disease in the United States, and tick-borne fever is an established and economically important disease of sheep in Europe [6, 9]. This emerging zoonotic pathogen is transmitted by Ixodes ticks of which the major vector species are I. scapularis in the United States and I. ricinus in Europe [2]. In ticks, A. phagocytophilum infects and multiply in different tissues including midguts [10] and salivary glands [11].
Anaplasma phagocytophilum is a good model for the study of tick-host-pathogen interactions because recent results have shown that infection affects gene expression and protein levels in vertebrate hosts and ticks [8, 12]. Recently, we proposed that the evolution of I. scapularis-host-A. phagocytophilum molecular interactions involving genetic traits of all parts resulted in conflict and cooperation between them, with mutual beneficial effects for ticks, hosts and pathogens [13]. Furthermore, A. phagocytophilum evolved common strategies for infection of vertebrate host and tick cells that include but are not limited to remodeling of the cytoskeleton, inhibition of cell apoptosis, manipulation of the immune response and control of host cell epigenetics [14].
Tick midguts are the tissue where blood digestion occurs while salivary glands produce and secrete proteins and other molecules that modulate host defenses to enhance blood feeding [15–21]. Additionally, tick midguts and salivary glands play a major role during pathogen acquisition, multiplication and transmission [22, 23]. The functional role of these tissues during tick feeding and pathogen infection is reflected at the transcriptome and proteome levels, with tissue-specific differences between midguts and salivary glands [12, 20, 21].
Vertebrate host proteins are found in tick midguts after feeding and have been described in the salivary glands and saliva of fed ticks [24–30]. Additionally, vertebrate host proteins and particularly alpha and beta-globin chains of hemoglobin have been identified in unfed I. scapularis and Amblyomma americanum nymphs [31] and Rhipicephalus sanguineus adult ticks [32]. Wickramasekara et al. [31] suggested that because blood meal digestion in ticks occurs gradually within midgut cells after endocytosis [19], the combination of slow assimilation and uptake of some host proteins into tick hemolymph might explain the persistence of host blood proteins for months after feeding and molting. In fact, Francischetti et al. [33] did not identify vertebrate host proteins in salivary glands from fed soft ticks Ornithodoros coriaceus, probably associated with the fact that hard ticks feed for several days while soft ticks feed for less than 1 hour, therefore decreasing the probability for host proteins to persist. Nevertheless, Diaz-Martin et al. [26] did find host proteins in the saliva of the soft ticks Ornithodoros moubata.
These results suggest a mechanism by which host proteins remain in the tick after blood digestion. For example, host proteins persisting in the tick after molting may serve as a reserve for nutrients until the next infestation and feeding cycle are completed. However, preliminary results in questing R. sanguineus infected with Rickettsia conorii suggested that pathogen infection modify tick digestion processes, thus provoking an increase in the concentration of some host proteins such as hemoglobin ingested with blood meal in infected ticks when compared to uninfected controls [32]. These results led to the hypothesis that pathogen infection affect tick metabolic processes to modify host protein digestion and persistence in the tick, with possible implications for tick physiology and pathogen life-cycle.
To address this hypothesis, in this study we used I. scapularis female ticks fed on uninfected and A. phagocytophilum-infected sheep to characterize host proteins present in midguts and salivary glands by mass spectrometry (MS) analysis of the proteome. The results evidenced a clear difference in the sheep host protein content between tick midguts and salivary glands in response to infection and provided evidence to support that A. phagocytophilum selectively manipulates the levels of vertebrate host proteins in the tick vector I. scapularis.
Methods
Ethics statement
Animals were housed and experiments conducted with the approval and supervision of the Oklahoma State University Institutional Animal Care and Use Committee (Animal Care and Use Protocol, ACUP No. VM1026).
Ticks and sample preparation
Ticks and sample preparation were previously described [12]. Briefly, I. scapularis ticks were obtained from the laboratory colony maintained at the Oklahoma State University Tick Rearing Facility [34]. Adult female I. scapularis were infected with A. phagocytophilum by feeding on a sheep inoculated intravenously with approximately 1 × 107A. phagocytophilum (NY18 isolate)-infected HL-60 cells (90–100 % infected cells). In this model, over 85 % of the ticks become infected with A. phagocytophilum in nymphs, midguts and salivary glands. One hundred adult female ticks were removed from the sheep 7 days after infestation, held in the humidity chamber for 4 days and dissected for DNA, RNA and protein extraction from midguts and salivary glands using the AllPrep DNA/RNA/Protein Mini Kit (Qiagen, Valencia, CA, USA). Midguts and salivary glands were washed in PBS after collection to remove hemolymph-related cells. Uninfected ticks were prepared in a similar way but feeding on an uninfected sheep. Two independent samples were collected and processed for proteomics analysis for each tick tissue. Ten individual female ticks were dissected and samples collected and processed as described above to characterize A. phagocytophilum infection and the mRNA or protein levels of selected genes/proteins after RNA sequencing (RNAseq) or proteomics analyses.
Proteomics data collection and analysis
Proteins were digested using the filter aided sample preparation (FASP) protocol [35]. The FASP method allows processing total SDS lysates of essentially any class of protein from biological samples of any origin, thus solving the long-standing problem of efficient and unbiased solubilization of all cellular proteins irrespective of their subcellular localization and molecular weight. Samples were dissolved in 50 mM Tris-HCl pH8.5, 4 % SDS and 50 mM DTT, boiled for 10 min and centrifuged. Protein concentration in the supernatant was measured by the Direct Detect system (Millipore, Billerica, MA, USA). About 150 μg of protein were diluted in 8 M urea in 0.1 M Tris-HCl (pH 8.5) (UA), and loaded onto 30 kDa centrifugal filter devices (FASP Protein Digestion Kit, Expedeon, TN, USA). With this method, the sample is solubilized in 4 % SDS, then retained and concentrated into microliter volumes in an ultrafiltration device. The filter unit then acts as a ‘proteomic reactor’ for detergent removal, buffer exchange, chemical modification and protein digestion. Notably, during peptide elution, the filter retains high-molecular-weight substances that would otherwise interfere with subsequent peptide separation [35]. The denaturation buffer was replaced by washing three times with UA. Proteins were later alkylated using 50 mM iodoacetamide in UA for 20 min in the dark, and the excess of alkylation reagents were eliminated by washing three times with UA and three additional times with 50 mM ammonium bicarbonate. Proteins were digested overnight at 37 °C with modified trypsin (Promega, Madison, WI, USA) in 50 mM ammonium bicarbonate at 40:1 protein:trypsin (w/w) ratio. The resulting peptides were eluted by centrifugation with 50 mM ammonium bicarbonate (twice) and 0.5 M sodium chloride. Trifluoroacetic acid (TFA) was added to a final concentration of 1 % and the peptides were finally desalted onto C18 Oasis-HLB cartridges and dried-down for further analysis. For stable isobaric labeling, the resulting tryptic peptides were dissolved in Triethylammonium bicarbonate (TEAB) buffer and labeled using the 4-plex iTRAQ Reagents Multiplex Kit (Applied Biosystems, Foster City, CA, USA) according to manufacturer’s protocol. Briefly, each peptide solution was independently labeled at room temperature for 1 h with one iTRAQ reagent vial (mass tag 114, 115, 116 or 117) previously reconstituted with 70 μl of ethanol. Reaction was stopped after incubation at room temperature for 15 min with diluted TFA, and peptides were combined. Samples were evaporated in a Speed Vac, desalted onto C18 Oasis-HLB cartridges and dried-down for further analysis. Labeled peptides were loaded into the liquid chromatography (LC)-MS/MS system for on-line desalting onto C18 cartridges and analyzing by LC-MS/MS using a C-18 reversed phase nano-column (75 μm I.D. × 50 cm, 2 μm particle size, Acclaim PepMap 100 C18; Thermo Fisher Scientific, Waltham, MA, USA) in a continuous acetonitrile gradient consisting of 0–30 % B in 145 min, 30–43 % B in 5 min and 43–90 % B in 1 min (A = 0.5 % formic acid; B = 90 % acetonitrile, 0.5 % formic acid). A flow rate of c.200 nl/min was used to elute peptides from the reverse phase nano-column to an emitter nanospray needle for real time ionization and peptide fragmentation on a QExactive mass spectrometer (Thermo Fisher Scientific). For increasing proteome coverage, iTRAQ-labeled samples were run at least twice. For peptide identification, all spectra were analyzed with Proteome Discoverer (version 1.4.0.29, Thermo Fisher Scientific) using a Uniprot databases containing all sequences from Ruminantia and Ixodida (April 14, 2014). For database searching, parameters were selected as follows: trypsin digestion with 2 maximum missed cleavage sites, precursor and fragment mass tolerances of 2 Da and 0.02 Da, carbamidomethyl cysteine as fixed modification and methionine oxidation as dynamic modifications. For iTRAQ labeled peptides, N-terminal and Lys iTRAQ modification was added as a fixed modification. Peptide identification was validated using the probability ratio method [36] and false discovery rate (FDR) was calculated using inverted databases and the refined method [37] with an additional filtering for precursor mass tolerance of 12 ppm. Only peptides with a confidence of at least 99 % were used to quantify the relative abundance of each peptide determined as described previously [38]. Protein quantification from reporter ion intensities and statistical analysis of quantitative data were performed as described previously using QuiXoT [39, 40]. The intensity of the reporter peaks was used to calculate the fitting weight of each spectrum in the statistical model as described previously [39]. Outliers at the scan and peptide levels and significant protein-abundance changes were detected from the Z-values (the standardized variable used by the model that expresses the quantitative values in units of standard deviation) by using a FDR threshold of 1 % as described previously [39]. Results are the mean of two replicates. The gene ontology (GO) analysis was performed using Uniprot (http://www.uniprot.org) annotations.
Characterization of the digestion of sheep host hemoglobins in tick midguts and salivary glands
Sheep hemoglobin alpha 1/2 (P68240) and beta (P02075) peptides detected by MS analysis with 1 % FDR in midguts and salivary glands from uninfected and A. phagocytophilum-infected ticks were used for analysis. For this analysis, a new database search of MS spectra was performed with the same parameters described above but selecting “no enzyme” instead of “trypsin” digestion to identify the non-tryptic peptides present in the samples. The preferred cleavage sites for hemoglobinolytic enzymes Trypsin, Leucine aminopeptidase, Legumain, Cathepsin B, Cathepsin C, Serine carboxypeptidase were determined by searching against the MEROPS Peptidase Database (https://merops.sanger.ac.uk, release 10.0) (see in Additional file 1: Dataset S1).
Characterization of the I. scapularis proteolytic and heme transport pathways mRNA and protein levels in response to A. phagocytophilum infection
The quantitative transcriptomics and proteomics data for midguts and salivary glands from uninfected and A. phagocytophilum-infected I. scapularis were obtained from previously published results and deposited at the Dryad Digital Repository database with the dataset identifier http://dx.doi.org//10.5061/dryad.50kt0 [12]. The analysis of the tick proteolytic pathway included the genes/proteins annotated as protease, proteinase, peptidase, and its inhibitors [41].
Determination of hemoglobin protein levels by ELISA
Proteins were extracted from midguts and salivary glands dissected from individual uninfected and A. phagocytophilum-infected I. scapularis female ticks using the AllPrep DNA/RNA/Protein Mini Kit (Qiagen, Inc. Valencia, CA, USA) according to manufacturer instructions. Extracted proteins were resuspended in PBS with 0.5 % Triton X-100 and protein concentration was determined with the Pierce BCA Protein Assay Kit (Thermo Scientific, San Jose, CA, USA) using bovine serum albumin (BSA) as standard. Hemoglobin protein levels were determined by ELISA (Cloud-Clone Corp., Houston, TX, USA) following manufacturer instructions. Optical density values were converted to μg/ml hemoglobin using the assay standard curve and regression analysis. Hemoglobin values were compared between groups by one-tailed Student’s t-test for samples with unequal variance (P < 0.05; n = 2 biological replicates).
Determination of tick mRNA levels by real-time RT-PCR
The expression of selected genes was characterized using total RNA extracted from individual I. scapularis female midguts and salivary glands obtained from uninfected and A. phagocytophilum-infected samples as previously described [12]. All ticks were confirmed as infected or uninfected by real-time PCR analysis of A. phagocytophilum msp4 DNA. Real-time RT-PCR was performed on RNA samples with gene-specific oligonucleotide primers (see Additional file 2: Table S1) using the iScript One-Step RT-PCR Kit with SYBR Green and the iQ5 thermal cycler (Bio-Rad, Hercules, CA, USA) following the manufacturer’s recommendations. A dissociation curve was run at the end of the reaction to ensure that only one amplicon was formed and that the amplicons denatured consistently in the same temperature range for every sample. The mRNA levels were normalized against tick cyclophilin and ribosomal protein S4 as described previously using the genNorm method (ddCT method as implemented by Bio-Rad iQ5 Standard Edition, Version 2.0) [12]. Normalized Ct values were compared between infected and uninfected tick samples by Student’s t-test with unequal variance (P < 0.05; n = 3–17 biological replicates).
Immunofluorescence assay (IFA)
Female ticks fed on A. phagocytophilum-infected and uninfected sheep and fixed with 4 % paraformaldehyde in 0.2 M sodium cacodylate buffer were embedded in paraffin and used to prepare sections on glass slides as previously described [12]. The paraffin was removed from the sections through two washes in xylene and the sections were hydrated by successive 5 min washes with a graded series of 100 %, 96 and 65 % ethanol and finally with distilled water. Next, the slides were treated with Proteinase K (Dako, Barcelona, Spain) for 7 min, washed with 0.1 % PBS-Tween 20 (Sigma-Aldrich, St. Louis, MI, USA) and blocked with 2 % bovine serum albumin (BSA; Sigma-Aldrich) in PBS-Tween 20 during 1 h at room temperature. The slides were then incubated overnight at 4 °C with rabbit anti-Cathepsin L (mature region No. pab0213-0; Covalab, Villeurbanne, France) antibodies diluted 1:1000 in 2 % BSA/PBS-Tween 20. This antibody was previously shown to recognize tick Cathepsin L by Western blot [42]. After 3 washes with PBS-Tween 20, the slides were incubated for 1 h with goat-anti-rabbit IgG conjugated with FITC (Sigma-Aldrich) diluted 1:160 in 2 % BSA/PBS-Tween 20. Finally, after two washes with PBS the slides were mounted on ProLong Diamond Antifade Mountant with DAPI reagent (Thermo Scientific™, Madrid, Spain). The sections were examined using a Leica SP2 laser scanning confocal microscope (Leica, Wetzlar, Germany) and IgGs from rabbit preimmune serum were used as controls.
Proteomics data
Data are available via Peptide Atlas (http://www.peptideatlas.org) with identifier PASS00854.
Results
The vertebrate host protein content differs between tick midguts and salivary glands in response to A. phagocytophilum infection
Infection with A. phagocytophilum affects gene expression and protein production in ticks in a tissue-specific manner, but the effect on host protein content in different tick tissues has not been characterized. To address this question, sheep host proteins present in midguts and salivary glands were characterized in uninfected and A. phagocytophilum-infected I. scapularis female ticks. A total of 1,753 sheep host proteins were identified in fed adult female ticks, of which 473 were identified with more than one peptide per protein in at least one of the samples (see Additional file 2: Table S1). Of these, 1,151 (364 identified with more than one peptide per protein) and 1,282 (414 identified with more than one peptide per protein) proteins were identified in tick midguts and salivary glands, respectively (see Additional file 3: Dataset S2). Of the host proteins identified with more than one peptide per protein, 388 proteins were found in both midguts and salivary glands (see Additional file 3: Dataset S2).
Sheep host proteins showing statistically significant protein abundance changes on the basis of Zq, or standardized log2-ratio of A. phagocytophilum-infected versus non-infected samples (Zq > 2, Zq < −2), were selected among proteins identified with more than one peptide in at least one of the samples [12, 39]. A total of 48 (6 underrepresented and 42 overrepresented) and 50 (36 underrepresented and 14 overrepresented) differentially represented sheep host proteins were found in tick midguts and salivary glands, respectively (see Additional file 3: Dataset S2). Of them, only 8 proteins were found in both midguts and salivary glands.
The GO analysis of differentially represented sheep host proteins showed that most biological processes (BPs) were found in both tick midguts and salivary glands (Fig. 1a, b). However, immune response, other and oxygen transport BPs contained 68 % of the differentially represented sheep host proteins in tick midguts while in salivary glands the most represented BPs were other, unknown, oxygen transport and translation, containing 64 % of the differentially represented sheep host proteins (Fig. 1a, b). These results evidenced a clear difference in the host protein content between tick midguts and salivary glands in response to A. phagocytophilum infection. Additionally, although the total number of differentially represented sheep host proteins was similar between tick tissues, 88 % of the differentially represented host proteins in midguts were overrepresented while in salivary glands only 28 % of the differentially represented proteins were overrepresented in infected ticks when compared to uninfected controls (Fig. 1c, d). Furthermore, while sheep host stress response and transcription/DNA replication proteins were overrepresented in midguts and salivary glands, proteins in the lipid metabolism, oxygen transport and immune response BPs were overrepresented in midguts and underrepresented in salivary glands in response to A. phagocytophilum infection (Fig. 1c, d).
Fig. 1.
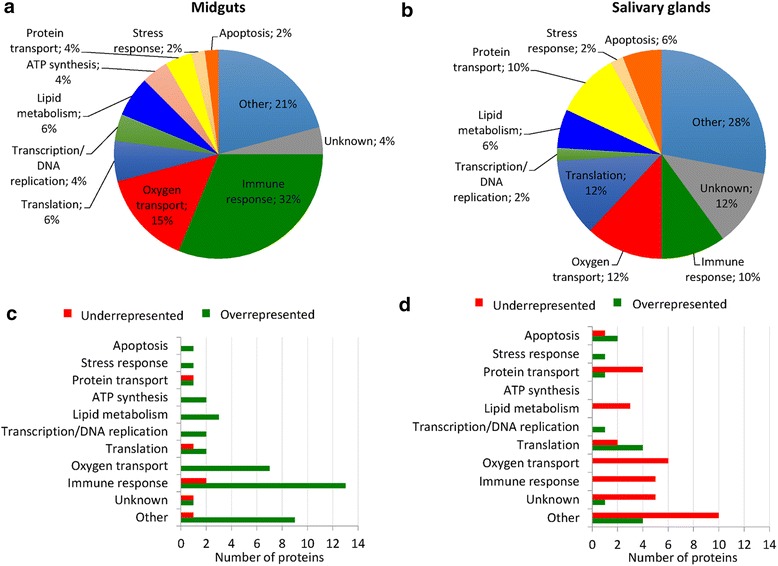
Tissue-specific effect of A. phagocytophilum infection on sheep host proteins represented in ticks. The results demonstrated a clear difference in the sheep host protein content between tick midguts and salivary glands in response to A. phagocytophilum infection. a Biological processes of differentially represented sheep host proteins in infected female tick midguts. b Biological processes of differentially represented sheep host proteins in infected female tick salivary glands. c Number of underrepresented and overrepresented sheep host proteins in different biological processes in infected female tick midguts when compared to uninfected controls. d Number of underrepresented and overrepresented sheep host proteins in different biological processes in infected female tick salivary glands when compared to uninfected controls
Sheep host heat shock and chromatin-related proteins are overrepresented in response to A. phagocytophilum infection in tick midguts and salivary glands
To characterize the putative physiological role of the host proteins differentially represented in tick midguts and salivary glands in response to A. phagocytophilum infection, we first focused on sheep host stress response and transcription/DNA replication proteins that were overrepresented in both tick tissues (Fig. 1c, d). The results showed that two sheep heat shock proteins (HSPs), HSP60 and HSP70, were overrepresented in infected tick midguts and salivary glands, respectively when compared to uninfected controls (Table 1). In the transcription/DNA replication BP, three host proteins involved in chromatin structure and function were overrepresented in response to A. phagocytophilum infection in tick midguts and salivary glands (Table 1).
Table 1.
Sheep host stress response, transcription/DNA replication, lipid metabolism, and immune response proteins differentially represented in tick midguts and salivary glands in response to A. phagocytophilum infection
| ID | Description | Log2 (infected/uninfected) fold change | Function | |
|---|---|---|---|---|
| Midguts | Salivary glands | |||
| Stress response proteins | ||||
| P31081 | HSP60 | +2.9 | ns | Response to cold |
| P0CB32 | HSP70 | ns | +1.4 | Heat shock response |
| Transcription/DNA replication | ||||
| P62803 | Histone H4 | +3.1 | ns | Chromatin structure |
| P68432 | Histone H3.1 | +3.1 | ns | Chromatin structure |
| F1MN93 | TOP1 uncharacterized protein | ns | +2.7 | Chromatin binding |
| Lipid metabolism | ||||
| W5QHX9 | Phospholipase B | +2.3 | -1.3 | Lipid absorption |
| Q9GL30 | Phospholipase B | +1.9 | ns | Lipid absorption |
| P15497 | Apolipoprotein A-I | +1.9 | ns | Cholesterol transport |
| Q32PF2 | ATP-citrate synthase | ns | -1.5 | Lipid synthesis |
| Q9TTS3 | Acetyl-CoA carboxylase 1 | ns | -1.6 | Lipid synthesis |
| Immune response | ||||
| W5NQK9 | S100A8 | +3.8 | ns | Innate immunity |
| W5NQJ0 | S100A12 | +3.2 | ns | Innate immunity |
| W5NQH6 | S100A9 | +3.1 | ns | Innate immunity |
| P28783 | S100A9 | +2.6 | ns | Innate immunity |
| D8X187 | Serpin peptidase inhibitor clade B ovalbumin member 1 | +2.1 | ns | Innate immunity |
| P62808 | Histone H2B type 1 | +3.4 | ns | Adaptive immunity |
| W5PGJ7 | PYD and CARD Domain-Containing uncharacterized protein | +2.9 | ns | Adaptive immunity |
| P49928 | Cathelin-related peptide SC5 | +3.1 | ns | Anti-bacterial immunity |
| P82018 | Cathelicidin-2 | +2.7 | ns | Anti-bacterial immunity |
| P50415 | Cathelicidin-3 | +2.3 | ns | Anti-bacterial immunity |
| P79360 | Myeloid antimicrobial peptide | +2.6 | ns | Anti-bacterial immunity |
| W5P7S6 | Alpha-1-acid glycoprotein | +2.6 | nf | Regulation of the immune response |
| W5PLV3 | RAB5B uncharacterized protein | +2.4 | nf | Antigen processing and presentation |
| W5PSQ7 | Ig-like uncharacterized protein | ns | -1.3 | Adaptive immunity |
| G5E513 | Ig-like uncharacterized protein | ns | -1.8 | Adaptive immunity |
| G5E5T5 | Ig-like uncharacterized protein | ns | -2.7 | Adaptive immunity |
| F1MQF6 | Apoptosis-associated speck-like protein-containing a CARD | nf | -1.5 | Innate immunity |
| W5PGJ7 | LOC101105208 uncharacterized protein | ns | -1.6 | Anti-bacterial immunity |
Abbreviations: ID protein (Uniprot; http://www.uniprot.org) accession numbers; +, overrepresented proteins in infected vs uninfected ticks; -, underrepresented proteins in infected vs uninfected ticks; nf, not found; ns, not significant
Sheep host proteins involved in lipid metabolism and immune response are overrepresented in tick midguts but underrepresented in salivary glands in response to A. phagocytophilum infection
To characterize further the putative physiological role of the host proteins differentially represented in response to A. phagocytophilum infection, we then focused on sheep host proteins in the lipid metabolism and immune response BPs that were overrepresented in midguts and underrepresented in salivary glands in response to A. phagocytophilum infection (Fig. 1c, d). The host lipid metabolism proteins overrepresented in A. phagocytophilum-infected tick midguts when compared to uninfected controls included proteins involved in lipid absorption, transport and excretion (Table 1). In tick salivary glands, sheep host proteins involved in lipid synthesis were underrepresented in infected ticks when compared to uninfected controls (Table 1).
In tick midguts, sheep host immune response proteins that were overrepresented in response to A. phagocytophilum infection included proteins involved in innate immunity (including several S100 proteins), adaptive immunity, anti-bacterial immunity, regulation of the immune response, and antigen processing and presentation (Table 1). In tick salivary glands, sheep host immunoglobulin (Ig)-like proteins and proteins involved in innate and anti-bacterial immunity were underrepresented in response to A. phagocytophilum infection (Table 1).
To confirm the origin of selected differentially represented proteins (Table 1), all of the peptides used to identify the proteins sharing tryptic peptides with I. scapularis proteins were revised to show the sequence of the peptides that are exclusive for host-derived proteins (Table 2).
Table 2.
Identification of host-derived proteins with identical tryptic peptides to I. scapularis tick homologues
| ID | Description | Unique host-derived peptides |
|---|---|---|
| P31081 | HSP60 | ALMLQGVDLLADAVAVTMGPK |
| VGGTSDVEVNEK | ||
| VGGTSDVEVNEKKDR | ||
| P0CB32 | HSP70 | FDLTGIPPAPR |
| RKELEQVCNPIITK | ||
| P68432 | Histone H3.1 | RVTIMPKDIQLAR |
| SAPATGGVK | ||
| SAPATGGVKKPHRYRPGTVALR | ||
| F1MN93 | TOP1 uncharacterized protein | AGNEKEEGETADTVGCCSLR |
| HLQDLMEGLTAK | ||
| P62808 | Histone H2B type 1 | AMGIMNSFVNDIFER |
| EIQTAVRLLLPGELAK | ||
| EIQTAVR | ||
| ESYSVYVYK | ||
| SRKESYSVYVYK | ||
| STITSREIQTAVRLLLPGELAK | ||
| STITSREIQTAVR | ||
| VLKQVHPDTGISSK | ||
| W5PLV3 | RAB5B uncharacterized protein | TAMNVNDLFLAIAK |
| P62803 | Histone H4 | None |
| Q9TTS3 | Acetyl-CoA carboxylase 1 | None |
To confirm the origin for selected differentially represented proteins (Table 1), all of the peptides used to identify the proteins sharing tryptic peptides with I. scapularis proteins were revised. The peptides unique for host-derived proteins are shown. For protein P62803, we could not define the origin due to 100 % homology between sheep and tick proteins. For protein Q9TTS3, all peptides used for identification were identical in both host and tick-derived proteins
Anaplasma phagocytophilum infection impacts on vertebrate host hemoglobin content in tick midguts and salivary glands
The identification of differentially represented sheep host proteins in A. phagocytophilum-infected I. scapularis midguts and salivary glands suggested the question about the origin of these proteins. The analysis of cell compartment GO showed that over 50 % of the proteins were extracellular or associated with blood cells (Fig. 2a; see Additional file 3: Dataset S2). Nevertheless, other proteins were localized in the cell cytoplasm (Fig. 2a; see Additional file 3: Dataset S2), probably associated with host blood cells ingested by ticks during feeding. Most of the host proteins in the hemoglobin complex and blood microparticle classification were sheep hemoglobins in the oxygen transport BP represented in both tick midguts and salivary glands (Fig. 2b). These hemoglobins were overrepresented in midguts and underrepresented in salivary glands of A. phagocytophilum-infected ticks when compared to uninfected controls (Fig. 2c), a result that was corroborated by an independent analysis using a specific ELISA test (Fig. 2d).
Fig. 2.
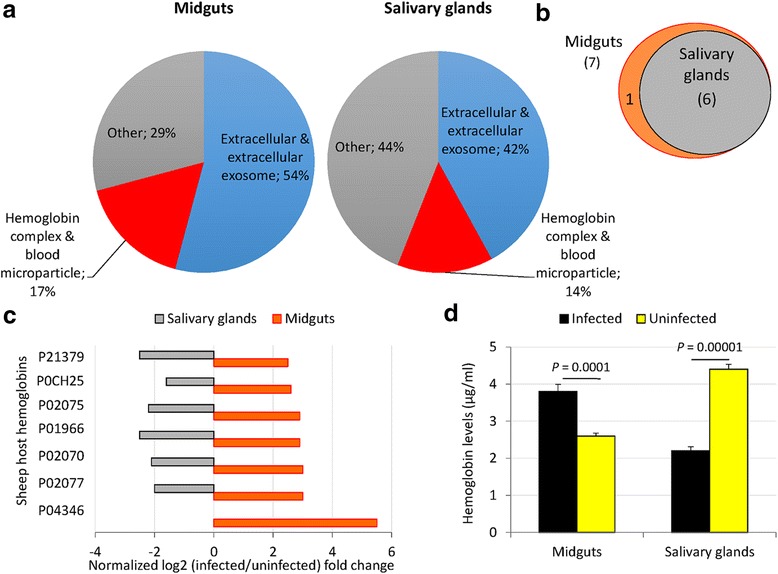
Sheep host hemoglobin levels vary in a tissue-specific manner in response to A. phagocytophilum infection in ticks. a Cell compartment classification of differentially represented sheep host proteins in infected female tick midguts and salivary glands. b Venn diagram of the sheep host hemoglobin differentially represented in infected vs uninfected tick tissues. c Differential host hemoglobin protein representation in response to A. phagocytophilum infection in tick midguts and salivary glands. d Hemoglobin levels in tick midguts and salivary glands from A. phagocytophilum-infected and uninfected ticks determined by ELISA in individual tick protein extracts, represented as the mean + standard deviation (SD) and compared between samples from infected and uninfected ticks by Student’s t-test with unequal variance (P < 0.05; 2 biological replicates)
Anaplasma phagocytophilum manipulates host protein content through modification of tick proteolytic pathways
To aid in the probable mechanism responsible for the differential representation of host proteins in midguts and salivary glands of A. phagocytophilum-infected ticks when compared to uninfected controls, the tissue-specific effect of infection was characterized on tick hemoglobinolytic enzymes and other proteases. The transcriptomics and proteomics data used in this study was previously validated by real-time RT-PCR and Western blot or immunofluorescence for selected genes and proteins, respectively [12, 43]. Nevertheless, 5 selected genes coding for hemoglobin digesting enzymes differentially regulated in response to A. phagocytophilum infection were used for analysis by real-time RT-PCR in individual tick midguts and salivary glands (see Additional file 2: Figure S1). As previously discussed [12], the differences observed between the results of both analyses that were evident in tick midguts considering the absence of transcriptomics data for some genes in salivary glands, could be attributed to intrinsic variation in gene expression and the fact that approximately 85 % of the ticks used for RNAseq were infected [44], while for real-time RT-PCR all ticks were confirmed uninfected or infected with A. phagocytophilum before analysis. At the protein level, an antibody recognizing tick Cathepsin L was used to corroborate proteomics results by IFA. Similar to proteomics analysis (Table 3), the results showed protein underrepresentation in the salivary glands of A. phagocytophilum-infected ticks when compared to uninfected controls (Fig. 3). Furthermore, although proteomics data were not available, Cathepsin L was overrepresented in tick midguts in response to infection (Fig. 3). Therefore, considering these results, the analysis of the differential expression/representation of tick hemoglobinolytic enzymes in response to A. phagocytophilum infection was presented by pondering mRNA (transcriptomics RNAseq and real-time RT-PCR) and protein (proteomics) data (see Additional file 2: Figure S2).
Table 3.
Differential expression/representation of enzymes involved in tick hemoglobinolytic and heme transport pathways in response to A. phagocytophilum infection
| ID | Description | Log2 (infected/uninfected) fold change (mRNA/protein) | Role in hemoglobin digestion | |
|---|---|---|---|---|
| Midguts | Salivary glands | |||
| Tick hemoglobinolytic pathway | ||||
| EF428204 | Cathepsin D | nf/ns | ns/nf | Primary cleavage |
| A4GTA5 | ||||
| HQ615697 | Cathepsin D2 | nf/ns | ns/nf | Primary cleavage |
| E7E820 | ||||
| ISCW000202 | Legumain | -0.6/ns | ns/ns | Primary cleavage |
| B7P6S9 | ||||
| ISCW015983 | Legumain | -1.3/nf | nf/nf | Primary cleavage |
| B7P2C6 | ||||
| ISCW000076 | Cathepsin L | -7.3/nf | nf/nf | Primary and secondary cleavage |
| B7P3N8 | ||||
| JX502821 | Cathepsin L | nf/nf | nf/-2.9 | Primary and secondary cleavage |
| J9QSA1 | ||||
| ISCW000080 | Cathepsin B | +0.5/ns | nf/-2.1 | Secondary and tertiary cleavage |
| B7P3P1 | ||||
| EU551624 | Cathepsin B | nf/ns | nf/-2.0 | Secondary and tertiary cleavage |
| B7SP39 | ||||
| ISCW013346 | Cathepsin B | -1.6/ns | ns/-2.5 | Secondary and tertiary cleavage |
| B7QCU7 | ||||
| ISCW000078 | Cathepsin B | +0.2/ns | -4.3/-1.8 | Secondary and tertiary cleavage |
| B7P3P0 | ||||
| ISCW003494 | Cathepsin C | +0.3/ns | -2.6/+2.2 | Tertiary cleavage |
| B7PEB4 | ||||
| ISCW001779 | Leucine aminopeptidase | +2.2/ns | ns/nf | Tertiary cleavage |
| B7P2N4 | ||||
| ISCW023735 | Leucine aminopeptidase | +2.0/ns | +1.1/ns | Tertiary cleavage |
| B7QLQ7 | ||||
| ISCW001780 | Leucine aminopeptidase | +1.4/ns | +0.5/ns | Tertiary cleavage |
| B7P2N5 | ||||
| ISCW013904 | Serine carboxipeptidase | +1.0/ns | ns/ns | Tertiary cleavage |
| B7QLB7 | ||||
| ISCW024536 | Serine carboxipeptidase | -0.5/ns | ns/nf | Tertiary cleavage |
| B7Q049 | ||||
| ISCW024751 | Serine carboxipeptidase | -0.7/ns | ns/ns | Tertiary cleavage |
| B7QD81 | ||||
| ISCW024883 | Serine carboxipeptidase | -4.5/nf | -2.4/nf | Tertiary cleavage |
| B7QK83 | ||||
| ISCW007492 | Serine carboxipeptidase | -1.7/ns | ns/ns | Tertiary cleavage |
| B7PTE5 | ||||
| ISCW003059 | Serine carboxipeptidase | -1.2/nf | ns/nf | Tertiary cleavage |
| B7PC00 | ||||
| Tick heme transport pathway | ||||
| ISCW001847 | Heme-responsive gene 1 (HRG1) | -0.8/nf | ns/nf | Heme transporter |
| B7P8M4 | ||||
| ISCW021709 | Heme-binding lipoprotein (HELP) | +3.3/ns | -0.2/ns | Heme transporter |
| B7Q406 | ||||
| ISCW013727 | Vitellogenin 1 (VG1) | ns/ns | ns/nf | Heme transporter |
| B7QJ67 | ||||
| ISCW021228 | Vitellogenin 2 (VG2) | -1.1/nf | ns/nf | Heme transporter |
| B7Q7E5 | ||||
Transcriptomics RNAseq and proteomics data from A. phagocytophilum-infected and uninfected tick samples were obtained from Ayllón et al. [12]. Except for Cathepsins D and D2, which were included to show that these proteins were identified in the proteomics analysis but were not significantly different between infected and uninfected samples, only genes/proteins with statistically significant differences in at least one of the analyses (transcriptomics or proteomics) and samples (midguts or salivary glands) were included. Abbreviations: ID, gene (GenBank; http://www.ncbi.nlm.nih.gov) and protein (Uniprot; http://www.uniprot.org) accession numbers
+, upregulated/overrepresented genes/proteins in infected vs uninfected ticks; -, downregulated/underrepresented genes/proteins in infected vs uninfected ticks; nf, not found; ns, not significant
Fig. 3.
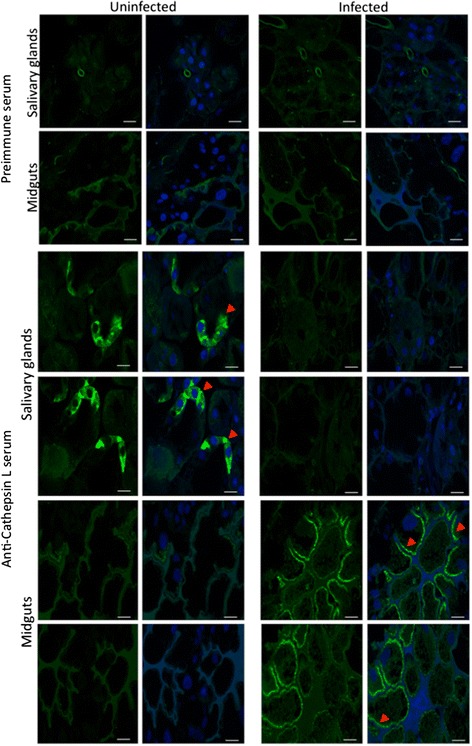
Characterization of Cathepsin L protein levels by IFA. Representative images of IFA of midguts and salivary glands of uninfected and A. phagocytophilum-infected adult female I. scapularis. Tick tissues were stained with rabbit anti-Cathepsin L (mature region No. pab0213-0; Covalab, Villeurbanne, France) antibodies (green, FITC) or DAPI (blue), and images were superimposed after staining (right panels). Preimmune control serum-treated samples showed similar results for uninfected and infected ticks. Uninfected and infected samples stained with anti-Cathepsin L antibodies showed higher protein levels in infected midguts while Cathepsin L was underrepresented in infected salivary glands when compared to uninfected controls (arrowheads). Scale-bars: 10 μm
The results suggested that in midguts from A. phagocytophilum-infected ticks when compared to uninfected controls, the hemoglobin primary cleavage was inhibited after Legumain underrepresentation while hemoglobin secondary and tertiary cleavages were probably not affected (Fig. 4 and Table 3). The hemoglobinolytic enzymes were also found in tick salivary glands, suggesting a role in hemoglobin digestion in this tissue (Table 3). In the salivary glands of infected ticks when compared to uninfected controls, the results suggested that hemoglobin primary and secondary cleavages were inhibited because Cathepsins L and B were underrepresented in response to infection while the hemoglobin tertiary cleavage was probably not affected (Fig. 4 and Table 3). The analysis of sheep hemoglobin alpha 1/2 (P68240) and beta (P02075) peptides identified by MS in tick midguts and salivary glands showed the presence of potential cleavage sites for trypsin (used in protein digestion for MS analysis), Legumain, Cathepsin B, Cathepsin C, Leucine aminopeptidase and Serine carboxipeptidase, therefore providing additional support for the activity of these enzymes in both tick tissues (Fig. 5a; see Additional file 1: Dataset S1).
Fig. 4.
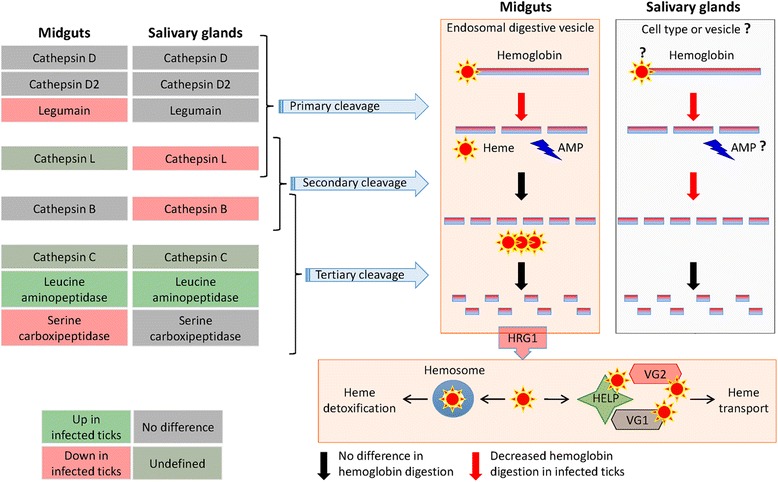
The levels of enzymes involved in the tick hemoglobinolytic pathway vary in a tissue-specific manner in response to A. phagocytophilum infection. Differential expression/representation of tick hemoglobinolytic enzymes in response to A. phagocytophilum infection was obtained from Ayllón et al. [12] and represented by pondering mRNA (transcriptomics RNAseq and real-time RT-PCR) and protein (proteomics) data. In tick midguts, the hemoglobinolytic pathway operating in the endosomal digestive vesicle was revised by Sojka et al. [19]. In tick salivary glands, these enzymes are also produced and may function under different conditions. Abbreviations: AMP, hemoglobin-derived antimicrobial peptides (Hemocidins and other); HRG1, Heme-responsive gene 1; HELP, Heme-binding lipoprotein; VG1, Vitellogenin 1; VG2, Vitellogenin 2
Fig. 5.
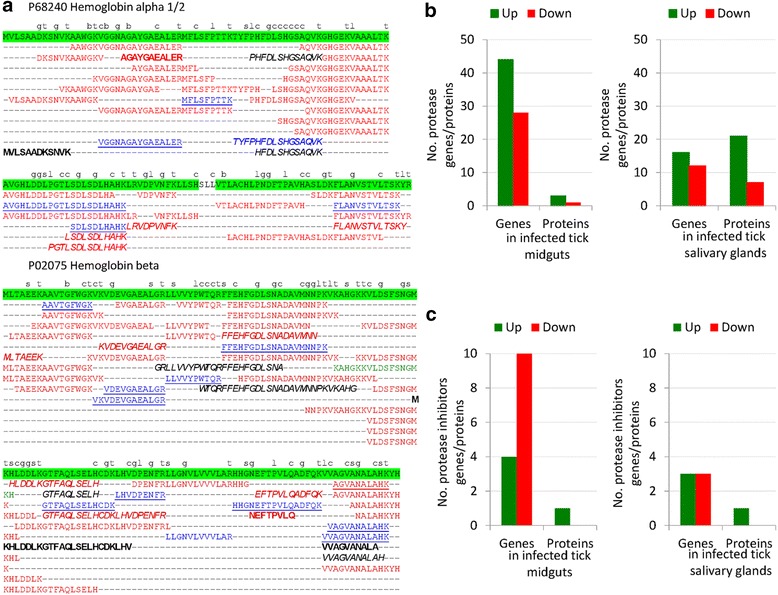
The digestion of sheep host hemoglobin varies between tick midguts and salivary glands in a tissue-specific manner in response to A. phagocytophilum infection. a Sheep hemoglobin alpha 1/2 (P68240) and beta (P02075) peptides detected by MS analysis with 1 % FDR in midguts and salivary glands from uninfected and A. phagocytophilum-infected ticks. Peptides detected in uninfected and infected (blue), infected (red), or uninfected (green) tick midguts and in uninfected and infected (underlined), infected (bold), or uninfected (italics) tick salivary glands are shown. Hemoglobin protein coverage by detected peptides is highlighted in green. The preferred cleavage sites for Trypsin and hemoglobinolytic enzymes are shown over P1 amino acid for Trypsin (t), Leucine aminopeptidase (l), Legumain (g), Cathepsin B (b), Cathepsin C (c), and Serine carboxypeptidase (s) (see Additional file 3: Dataset S2). b The number of protease genes/proteins different from hemoglobinolytic enzymes and differentially expressed/represented in response to A. phagocytophilum infection in tick midguts and salivary glands were extracted from transcriptomics and proteomics data [12]. c The number of protease inhibitor genes/proteins differentially expressed/represented in response to A. phagocytophilum infection in tick midguts and salivary glands were extracted from transcriptomics and proteomics data [12]
In the midguts of ticks infected with A. phagocytophilum, Heme-responsive gene 1 protein (HRG1) was underrepresented, while heme transport proteins Heme-binding lipoprotein (HELP) and Vitellogenin 2 (VG2) but not Vitellogenin 1 (VG1) were overrepresented and underrepresented, respectively, in response to infection (Fig. 4 and Table 3). Furthermore, sheep host blood coagulation factors Annexin A3 (Q3SWX7; overrepresented in infected tick midguts and involved in blood anti-coagulation as a Phospholipase 2 inhibitor), and Fibrinogen gamma-B, and uncharacterized protein APOH (P12799 and W5Q268; underrepresented in infected tick salivary glands and involved in blood coagulation) were differentially represented in infected ticks when compared to uninfected controls (see Additional file 3: Dataset S2), resulting in the inhibition of blood coagulation in both tick tissues.
In addition to hemoglobinolytic enzymes, other tick proteases were upregulated/overrepresented while protease inhibitors were down-regulated or did not change in midguts and salivary glands of A. phagocytophilum-infected ticks when compared to uninfected controls (Fig. 5b, c). However, as shown for the hemoglobinolytic enzymes (Fig. 3), the tick proteases differentially regulated in response to infection were predominantly different between midguts and salivary glands (see Additional file 2: Table S2).
Discussion
The characterization of sheep host proteins in the midguts and salivary glands of uninfected and A. phagocytophilum-infected I. scapularis female ticks showed tissue-specific differences in response to infection. Vertebrate host proteins in the transcription, lipid metabolism, immune response and oxygen transport (hemoglobins) were previously found to be highly abundant in the saliva of engorged I. scapularis ticks [24, 30]. The authors suggested that ticks have evolved mechanisms to selectively secrete host proteins in the saliva to aid in the feeding process [30]. Furthermore, anti-microbial peptides (AMP) such as S100 proteins [45] highly abundant in the saliva of engorged I. scapularis were proposed to function in clearing microbes from the feeding site to preserve ticks [30]. In A. phagocytophilum-infected ticks, host proteins from some of these pathways such as stress response and transcription were overrepresented in tick midguts and salivary glands, supporting the existence of a mechanism to facilitate tick feeding that was enhanced in response to infection. However, other host proteins overrepresented in A. phagocytophilum-infected tick midguts probably reflected the host response to infection. For example, proteins in the immune response BP are upregulated at the transcriptional level in sheep infected with A. phagocytophilum [46]. Nevertheless, immune response proteins were underrepresented in infected tick salivary glands when compared to uninfected controls, suggesting that A. phagocytophilum selectively manipulates the levels of host proteins to facilitate pathogen infection, multiplication and transmission.
The infection with A. phagocytophilum modulates lipid metabolism in vertebrate host cells and bacteria incorporate host cholesterol for survival [47–49]. In tick cells, A. phagocytophilum infection inhibits lipid metabolism through down-representation of tick proteins [50]. The overrepresentation of sheep host proteins involved in lipid absorption, transport and secretion in midguts and the underrepresentation of lipid synthesis proteins in salivary glands of infected ticks when compared to uninfected controls may constitute an additional mechanism by which A. phagocytophilum selectively manipulates lipid metabolism to enhance infection and multiplication in tick tissues.
Although ticks contain genes that encode heme synthesis enzymes, recent results demonstrate that they do not synthesize heme but obtain heme from the vertebrate host hemoglobin in the midgut and from tick heme transporters HELP/VG1/VG2 in other tissues [50–52]. Recently, Hajdusek et al. [50] proposed that the heme produced after host hemoglobin digestion is transported outside the endosomal digestive vesicle by HRG1 and subsequently detoxified in the hemosome or transported by HELP, VG1 and VG2 to other tick tissues such as salivary glands. However, as shown here and in previous reports [30], the presence of active tick hemoglobinolytic enzymes in the salivary glands and secreted in the saliva of engorged I. scapularis suggests the possibility that host hemoglobin may be also digested under different conditions to provide heme in the salivary glands. Although heme may not contribute to the cellular iron pool in ticks [52], the results reported here suggested that A. phagocytophilum affects hemoglobin primary cleavage in tick midguts and salivary glands, probably to reduce the production of hemoglobin-derived AMP to facilitate pathogen multiplication [50]. Furthermore, although A. phagocytophilum infection did not affect most of the enzymes involved in hemoglobin secondary and tertiary cleavage in tick midguts, the underrepresentation of HGR1 suggested a mechanism to reduce heme release into the cytoplasm of midgut cells (Fig. 4). This mechanism is probably manipulated by A. phagocytophilum to facilitate infection through reduction of the antimicrobial oxidative burden caused by reactive oxygen species (ROS) generated after heme release [19, 53, 54]. Furthermore, the inhibition of blood coagulation may be a mechanism driven by tick and/or A. phagocytophilum to facilitate tick feeding and pathogen multiplication. As recently proposed [13], these mechanisms may have evolved to guarantee A. phagocytophilum infection, multiplication and transmission while preserving tick life cycle.
Once shown that A. phagocytophilum selectively manipulates the levels of vertebrate host proteins in ticks in a tissue-specific manner, the next question was why these proteins were selected among all host proteins ingested by ticks during blood feeding? Some of these host proteins such as hemoglobins, S100 and Ig-like proteins that were overrepresented in tick midguts and underrepresented in salivary glands have a crucial role during tick feeding and pathogen infection. In feeding ticks, host hemoglobins are the source of heme and AMP, which together with other immune system proteins such as S100 and Ig-like proteins may be essential for tick feeding and antimicrobial response to control microbe levels in tick tissues [19, 30, 45, 50, 53, 54]. Additionally, most of these proteins are highly conserved among major domestic and natural vertebrate hosts for A. phagocytophilum and I. scapularis (see Additional file 2: Table S3). Therefore, these results suggested that the mechanisms responsible for the selective manipulation of vertebrate host proteins by A. phagocytophilum infection in tick tissues are evolutionary conserved.
The physiological significance of these findings was addressed by responding to the questions recently proposed by Sojka et al. [19] for a better understanding of how ticks handle the blood meal. Among these questions they proposed to address if the same tick enzyme machinery process hemoglobin and other vertebrate host proteins and the role of blood digestion and chemical reduction-oxidation reaction balance on pathogen infection and transmission in the tick midgut. The results of our study showed that tick hemoglobinolytic enzymes are present and active in both midguts and salivary glands of fed ticks and therefore may be involved in the digestion of hemoglobin and other host proteins. Although the effect of hemoglobin digestion and ROS production on pathogen infection and transmission was not directly addressed in our study, the results suggested that A. phagocytophilum selectively manipulate these and other processes to facilitate pathogen infection, multiplication and transmission.
The results reported here suggested that the mechanism used by A. phagocytophilum to selectively manipulate the levels of vertebrate host proteins in a tissue-specific manner is through modification of tick proteolytic pathways. How A. phagocytophilum modify tick proteolytic pathways is not known, but may include the regulation of gene expression through epigenetic mechanisms recently shown to be affected by pathogen infection in I. scapularis [43]. These epigenetic mechanisms are probably controlled by secreted bacterial effectors [55–58]. However, future experiments should address the physiological significance of tick proteolytic pathways during A. phagocytophilum infection and multiplication in midguts and salivary glands.
Conclusions
In summary, the results of this study corroborated that vertebrate host proteins are present in the midguts and salivary glands of fed female I. scapularis. To our knowledge, the results presented here showed for the first time that A. phagocytophilum selectively manipulates the levels of vertebrate host proteins in the tick vector to facilitate pathogen infection, multiplication and transmission while preserving tick feeding and development (Fig. 6). The mechanisms by which A. phagocytophilum manipulates the levels of vertebrate host proteins are not known, but may include modification of proteolytic pathways by affecting tick epigenetics and other biological processes.
Fig. 6.
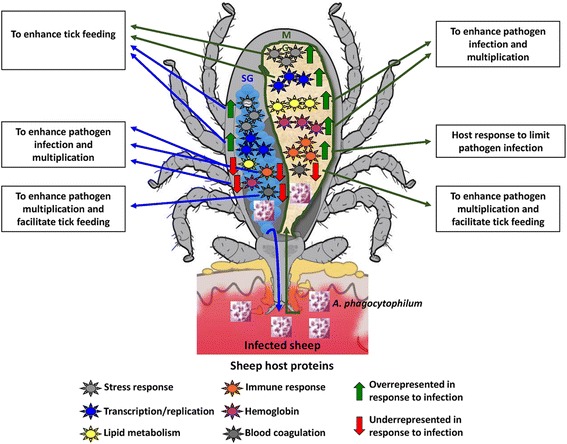
Proposed functional significance for the host proteins selectively manipulated by A. phagocytophilum in tick midguts and salivary glands. These results suggested that A. phagocytophilum selectively manipulates the levels of vertebrate host proteins in the tick midguts (MG) and salivary glands (SG) to facilitate pathogen infection, multiplication and transmission while preserving tick feeding and development
Despite the growing burden that A. phagocytophilum and other tick-borne pathogens represent for human and animal health worldwide, effective control measures have not been developed [59]. Investigating the biological relevance of host proteins in tick biology and pathogen infection and the mechanisms used by A. phagocytophilum to manipulate host protein content is essential to advance our knowledge of tick-host-pathogen molecular interactions. These results have implications for the identification of new targets for the development of vaccines for the control of tick-borne diseases.
Acknowledgements
We would like to acknowledge José Ramón Marín Tébar (University of Castilla - La Mancha, Spain) for technical assistance with immunofluorescence.
Funding
This research was supported by the Ministerio de Economia y Competitividad (Spain) grant BFU2011-23896 and the European Union (EU) Seventh Framework Programme (FP7) ANTIGONE project number 278976. NA was funded by Ministerio de Economía y Competitividad, Spain. MV was supported by the Research Plan of the University of Castilla - La Mancha, Spain. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Availability of data and material
The quantitative transcriptomics and proteomics data for midguts and salivary glands from uninfected and A. phagocytophilum-infected I. scapularis were obtained from previously published results and deposited at the Dryad Digital Repository database with the dataset identifier http://dx.doi.org//10.5061/dryad.50kt0 [12]. Proteomics data for sheep host proteins are available via Peptide Atlas (http://www.peptideatlas.org) with identifier PASS00854.
Authors’ contributions
JF and MV conceived and supervised the study. VL, NA, ACC, JAL, JV and PA performed the experiments. MV and JF wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Animals were housed and experiments conducted with the approval and supervision of the Oklahoma State University Institutional Animal Care and Use Committee (Animal Care and Use Protocol, ACUP No. VM1026).
Abbreviations
- HGA
human granulocytic anaplasmosis
- MS
mass spectrometry
- RNAseq
RNA sequencing
- FASP
filter aided sample preparation
- TFA
trifluoroacetic acid
- TEAB
TriethyLammonium bicarbonate
- LC
liquid chromatography
- FDR
false discovery rate
- GO
gene ontology
- IFA
immunofluorescence assay
- BSA
bovine serum albumin
- BP
biological process
- HSP
heat shock protein
- Ig
immunoglobulin
- HRG1
Heme-responsive gene 1 protein
- HELP
Heme-binding lipoprotein
- VG1
Vitellogenin 1
- VG2
Vitellogenin 2
- ROS
reactive oxygen species
- AMP
anti-microbial peptides
Additional files
Digestion of sheep host hemoglobin in tick midguts and salivary glands. (XLS 55 kb)
Analysis of RNAseq results by real-time RTPCR. Figure S2. Pondering mRNA and protein data. Table S1. Genes and oligonucleotide primers selected for gene expression analysis by real-time RT-PCR. Table S2. Differential expression of highly differentially regulated tick protease genes in response to A. phagocytophilum infection. Table S3. Percent homology for selected differentially represented host proteins in tick midguts and salivary glands in response to A. phagocytophilum infection. (PDF 337 kb)
Proteomics results for sheep host proteins in tick tissues. (XLSX 208 kb)
Contributor Information
Margarita Villar, Email: margaritam.villar@uclm.es.
Vladimir López, Email: vladimirlopez6@gmail.com.
Nieves Ayllón, Email: nieves.ayllon@uclm.es.
Alejandro Cabezas-Cruz, Email: cabezasalejandrocruz@gmail.com.
Juan A. López, Email: juan.lopez@cnic.es
Jesús Vázquez, Email: jesus.vazquez@cnic.es.
Pilar Alberdi, Email: maria.alberdi@uclm.es.
José de la Fuente, Email: jose_delafuente@yahoo.com.
References
- 1.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszac P. Global trends in emerging infectious diseases. Nature. 2008;451:990–994. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de la Fuente J, Estrada-Peña A, Venzal JM, Kocan KM, Sonenshine DE. Overview: Ticks as vectors of pathogens that cause disease in humans and animals. Front Biosci. 2008;13:6938–6946. doi: 10.2741/3200. [DOI] [PubMed] [Google Scholar]
- 3.Estrada-Peña A, Ostfeld RS, Peterson AT, Poulin R, de la Fuente J. Effects of environmental change on zoonotic disease risk: an ecological primer. Trends Parasitol. 2014;30:205–214. doi: 10.1016/j.pt.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Gortazar C, Reperant LA, Kuiken T, de la Fuente J, Boadella M, Martínez-Lopez B, et al. Crossing the interspecies barrier: Opening the door to zoonotic pathogens. PLoS Pathog. 2014;10:e1004129. doi: 10.1371/journal.ppat.1004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dumler JS, Barbet AC, Bekker CPJ, Dasch GA, Palmer GH, Ray SC, et al. Reorganization of the genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions subjective synonyms of Ehrlichia phagocytophila. Int J Syst Evol Microbiol. 2001;51:2145–2165. doi: 10.1099/00207713-51-6-2145. [DOI] [PubMed] [Google Scholar]
- 6.Stuen S. Anaplasma phagocytophilum - the most widespread tick-borne infection in animals in Europe. Vet Res Commun. 2010;31:79–84. doi: 10.1007/s11259-007-0071-y. [DOI] [PubMed] [Google Scholar]
- 7.Rikihisa Y. Molecular pathogensis of Anaplasma phagocytophilum. Current Microbiol Rev. 2011;24:469–489. doi: 10.1128/CMR.00064-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Severo MS, Pedra JHF, Ayllón N, Kocan KM, de la Fuente J. Anaplasma. In: Tang YW, Sussman M, Liu D, Poxton I, Schwartzman J, editors. Molecular Medical Microbiology. 2. London: Academic Press-Elsevier; 2015. pp. 2033–2042. [Google Scholar]
- 9.Dahlgren FS, Mandel EJ, Krebs JW, Massung RF, McQuiston JH. Increasing incidence of Ehrlichia chaffeensis and Anaplasma phagocytophilum in the United States, 2000–2007. Am J Trop Med Hyg. 2011;85:124–131. doi: 10.4269/ajtmh.2011.10-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reichard MV, Manzano-Roman R, Kocan KM, Blouin EF, de la Fuente J, Snider TA, et al. Inoculation of white-tailed deer (Odocoileus virginianus) with Ap-V1 or NY-18 strains of Anaplasma phagocytophilum and microscopic demonstration of Ap-V1 in Ixodes scapularis adults that acquired infection from deer as nymphs. Vector Borne Zoonotic Dis. 2009;9:565–568. doi: 10.1089/vbz.2008.0106. [DOI] [PubMed] [Google Scholar]
- 11.Sukumaran B, Narasimham S, Anderson JF, DePonte K, Marcantonio K, Krishnan MN, et al. An Ixodes scapularis protein required for survival of Anaplasma phagocytophilum in tick salivary glands. J Exp Med. 2006;6:1507–1517. doi: 10.1084/jem.20060208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ayllón N, Villar M, Galindo RC, Kocan KM, Šíma R, López JA, et al. Systems biology of tissue-specific response to Anaplasma phagocytophilum reveals differentiated apoptosis in the tick vector Ixodes scapularis. PLoS Genet. 2015;11. [DOI] [PMC free article] [PubMed]
- 13.de la Fuente J, Villar M, Cabezas-Cruz A, Estrada-Peña A, Ayllón N, Alberdi P. Tick-host-pathogen interactions: conflict and cooperation. PLoS Pathog. 2016;12:e1005488. doi: 10.1371/journal.ppat.1005488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de la Fuente J, Estrada-Peña A, Cabezas-Cruz A, Kocan KM. Anaplasma phagocytophilum uses common strategies for infection of ticks and vertebrate hosts. Trends Microbiol. 2016;3:173–180. doi: 10.1016/j.tim.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Ribeiro JM. Role of saliva in blood-feeding by arthropods. Annu Rev Entomol. 1987;32:463–478. doi: 10.1146/annurev.en.32.010187.002335. [DOI] [PubMed] [Google Scholar]
- 16.Anderson JM, Sonenshine DE, Valenzuela JG. Exploring the mialome of ticks: an annotated catalogue of midgut transcripts from the hard tick, Dermacentor variabilis (Acari: Ixodidae) BMC Genomics. 2008;9:552. doi: 10.1186/1471-2164-9-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francischetti IM, Sa-Nunes A, Mans BJ, Santos IM, Ribeiro JM. The role of saliva in tick feeding. Front Biosci. 2009;14:2051–2088. doi: 10.2741/3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliveira CJ, Sá-Nunes A, Francischetti IM, Carregaro V, Anatriello E, Silva JS, et al. Deconstructing tick saliva: Non-protein molecules with potent immunomodulatory properties. J Biol Chem. 2011;286:10960–10969. doi: 10.1074/jbc.M110.205047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sojka D, Franta Z, Horn M, Caffrey CR, Mareš M, Kopáček P. New insights into the machinery of blood digestion by ticks. Trends Parasitol. 2013;29:276–285. doi: 10.1016/j.pt.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Kotsyfakis M, Schwarz A, Erhart J, Ribeiro JM. Tissue- and time-dependent transcription in Ixodes ricinus salivary glands and midguts when blood feeding on the vertebrate host. Sci Rep. 2015;5:9103. doi: 10.1038/srep09103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chmelař J, Kotál J, Karim S, Kopacek P, Francischetti IM, Pedra JH, Kotsyfakis M. Sialomes and mialomes: A systems-biology view of tick tissues and tick-host interactions. Trends Parasitol. 2015;32:242–254. doi: 10.1016/j.pt.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hajdušek O, Šíma R, Ayllón N, Jalovecká M, Perner J, de la Fuente J, Kopáček P. Interaction of the tick immune system with transmitted pathogens. Front Cell Infect Microbiol. 2013;3:26. doi: 10.3389/fcimb.2013.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mastronunzio JE, Kurscheid S, Fikrig E. Postgenomic analyses reveal development of infectious Anaplasma phagocytophilum during transmission from ticks to mice. J Bacteriol. 2012;194:2238–2247. doi: 10.1128/JB.06791-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valenzuela JG, Francischetti IM, Pham VM, Garfield MK, Mather TN, Ribeiro JM. Exploring the sialome of the tick Ixodes scapularis. J Exp Biol. 2002;205:2843–2864. doi: 10.1242/jeb.205.18.2843. [DOI] [PubMed] [Google Scholar]
- 25.Madden RD, Sauer JR, Dillwith JW. A proteomics approach to characterizing tick salivary secretions. Exp Appl Acarol. 2002;28:77–87. doi: 10.1023/A:1025342015065. [DOI] [PubMed] [Google Scholar]
- 26.Diaz-Martin V, Manzano-Roman R, Valero L, Oleaga A, Encinas-Grandes A, Perez-Sanchez R. An insight into the proteome of the saliva of the argasid tick Ornithodoros moubata reveals important differences in saliva protein composition between the sexes. J Proteomics. 2013;80:216–235. doi: 10.1016/j.jprot.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 27.Oliveira CJ, Anatriello E, de Miranda-Santos IK, Francischetti IM, Sá-Nunes A, Ferreira BR, Ribeiro JM. Proteome of Rhipicephalus sanguineus tick saliva induced by the secretagogues pilocarpine and dopamine. Ticks Tick Borne Dis. 2013;4:469–477. doi: 10.1016/j.ttbdis.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tirloni L, Reck J, Terra RM, Martins JR, Mulenga A, Sherman NE, et al. Proteomic analysis of cattle tick Rhipicephalus (Boophilus) microplus saliva: a comparison between partially and fully engorged females. PLoS One. 2014;9:e94831. doi: 10.1371/journal.pone.0094831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tirloni L, Islam MS, Kim TK, Diedrich JK, Yates JR, 3rd, Pinto AF, Mulenga A, et al. Saliva from nymph and adult females of Haemaphysalis longicornis: a proteomic study. Parasit Vectors. 2015;8:338. doi: 10.1186/s13071-015-0918-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim TK, Tirloni L, Pinto AF, Moresco J, Yates JR, 3rd, DA Silva Vaz I, Jr, Mulenga A. Ixodes scapularis tick saliva proteins sequentially secreted every 24 h during blood feeding. PLoS Negl Trop Dis. 2016;10:e0004323. doi: 10.1371/journal.pntd.0004323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wickramasekara S, Bunikis J, Wysocki V, Barbour AG. Identification of residual blood proteins in ticks by mass spectrometry proteomics. Emerg Infect Dis. 2008;14:1273–1275. doi: 10.3201/eid1408.080227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villar M, Torina A, Nuñez Y, Zivkovic Z, Marina A, Alongi A, et al. Application of highly sensitive saturation labeling to the analysis of differential protein expression in infected ticks from limited samples. Proteome Sci. 2010;8:43. doi: 10.1186/1477-5956-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Francischetti IM, Meng Z, Mans BJ, Gudderra N, Hall M, Veenstra TD, et al. An insight into the salivary transcriptome and proteome of the soft tick and vector of epizootic bovine abortion. Ornithodoros Coriaceus J Proteomics. 2008;71:493–512. doi: 10.1016/j.jprot.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kocan KM, de la Fuente J, Coburn LA. Insights into the development of Ixodes scapularis: A resource for research on a medically important tick species. Parasit Vectors. 2015;8:592. doi: 10.1186/s13071-015-1185-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wisniewski J, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Meth. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 36.Martínez-Bartolomé S, Navarro P, Martín-Maroto F, López-Ferrer D, Ramos-Fernández A, Villar M, et al. Properties of average score distributions of SEQUEST: the probability ratio method. Mol Cell Proteomics. 2008;7:1135–1145. doi: 10.1074/mcp.M700239-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Navarro P, Vázquez J. A refined method to calculate false discovery rates for peptide identification using decoy databases. J Proteome Res. 2009;8:1792–1796. doi: 10.1021/pr800362h. [DOI] [PubMed] [Google Scholar]
- 38.Bonzon-Kulichenko E, Martínez-Martínez S, Trevisan-Herraz M, Navarro P, Redondo JM, Vázquez J. Quantitative in-depth analysis of the dynamic secretome of activated Jurkat T-cells. J Proteomics. 2011;75:561–571. doi: 10.1016/j.jprot.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 39.Navarro P, Trevisan-Herraz M, Bonzon-Kulichenko E, Núñez E, Martínez-Acedo P, Pérez-Hernández D, et al. General Statistical framework for quantitative proteomics by stable isotope labeling. J Proteome Res. 2014;13:1234–1247. doi: 10.1021/pr4006958. [DOI] [PubMed] [Google Scholar]
- 40.Jorge I, Navarro P, Martinez-Acedo P, Nunez E, Serrano H, Alfranca A, et al. Statistical model to analyze quantitative proteomics data obtained by 18O/16O labeling and linear ion trap mass spectrometry: application to the study of vascular endothelial growth factor-induced angiogenesis in endothelial cells. Mol Cell Proteomics. 2009;8:1130–1149. doi: 10.1074/mcp.M800260-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barrett AJ, McDonald JK. Nomenclature: protease, proteinase and peptidase. Biochem J. 1986;237:935. doi: 10.1042/bj2370935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Popara M, Villar M, Mateos-Hernández L, Fernández De Mera IG, Marina A, Del Valle M, et al. Lesser protein degradation machinery correlates with higher BM86 tick vaccine efficacy in Rhipicephalus annulatus when compared to R. microplus. Vaccine. 2013;31:4728–4735. doi: 10.1016/j.vaccine.2013.08.031. [DOI] [PubMed] [Google Scholar]
- 43.Cabezas-Cruz A, Alberdi P, Ayllón N, Valdés JJ, Pierce R, Villar M, de la Fuente J. Anaplasma phagocytophilum increases the levels of histone modifying enzymes to inhibit cell apoptosis and facilitate pathogen infection in the tick vector, Ixodes scapularis. Epigenetics. 2016;11:303–319. doi: 10.1080/15592294.2016.1163460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kocan KM, Busby AT, Allison RW, Breshears MA, Coburn L, Galindo RC, et al. Sheep experimentally-infected with a human isolate of Anaplasma phagocytophilum serve as a host for infection of Ixodes scapularis. Ticks Tick-Borne Dis. 2012;3:147–153. doi: 10.1016/j.ttbdis.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 45.Donato R, Cannon BR, Sorci G, Riuzzi F, Hsu K, Weber DJ, Geczy CL. Functions of S100 Proteins. Curr Mol Med. 2013;13:24–57. doi: 10.2174/156652413804486214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galindo RC, Ayoubi P, García-Pérez AL, Naranjo V, Kocan KM, Gortazar C, de la Fuente J. Differential expression of inflammatory and immune response genes in sheep infected with Anaplasma phagocytophilum. Vet Immunol Immunopathol. 2008;126:27–34. doi: 10.1016/j.vetimm.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 47.Lin M, Rikihisa Y. Ehrlichia chaffeensis and Anaplasma phagocytophilum lack genes for lipid A biosynthesis and incorporate cholesterol for their survival. Infect Immun. 2003;71:5324–5331. doi: 10.1128/IAI.71.9.5324-5331.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manzano-Roman R, Almazán C, Naranjo V, Blouin EF, Kocan KM, de la Fuente J. Expression of perilipin in human promyelocytic cells in response to Anaplasma phagocytophilum infection results in modified lipid metabolism. J Med Microbiol. 2008;57:159–163. doi: 10.1099/jmm.0.47504-0. [DOI] [PubMed] [Google Scholar]
- 49.Severo MS, Stephens KD, Kotsyfakis M, Pedra JH. Anaplasma phagocytophilum: deceptively simple or simply deceptive? Future Microbiol. 2012;7:719–731. doi: 10.2217/fmb.12.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Villar M, Ayllón N, Alberdi P, Moreno A, Moreno M, Tobes R, et al. Integrated metabolomics, transcriptomics and proteomics identifies metabolic pathways affected by Anaplasma phagocytophilum infection in tick cells. Mol Cell Proteomics. 2015;14:3154–3172. doi: 10.1074/mcp.M115.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gulia-Nuss M, Nuss AB, Meyer JM, Sonenshine DE, Roe RM, Waterhouse RM, et al. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat Commun. 2016;7:10507. doi: 10.1038/ncomms10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perner J, Sobotka R, Sima R, Konvickova J, Sojka D, Oliveira PL, et al. Acquisition of exogenous haem is essential for tick reproduction. eLife. 2016;5:e12318. doi: 10.7554/eLife.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Graça-Souza AV, Maya-Monteiro C, Paiva-Silva GO, Braz GR, Paes MC, Sorgine MH, et al. Adaptations against heme toxicity in blood-feeding arthropods. Insect Biochem Mol Biol. 2006;36:322–335. doi: 10.1016/j.ibmb.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 54.Toh SQ, Glanfield A, Gobert GN, Jones MK. Heme and blood-feeding parasites: friends or foes? Parasit Vectors. 2010;3:108. doi: 10.1186/1756-3305-3-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang B, Troese MJ, Howe D, Ye S, Sims JT, Heinzen RA, et al. Anaplasma phagocytophilum APH_0032 is expressed late during infection and localizes to the pathogen-occupied vacuolar membrane. Microb Pathog. 2010;49:273–284. doi: 10.1016/j.micpath.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rikihisa Y, Lin M, Niu H. Type IV secretion in the obligatory intracellular bacterium Anaplasma phagocytophilum. Cell Microbiol. 2010;12:1213–1221. doi: 10.1111/j.1462-5822.2010.01500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rennoll-Bankert KE, Garcia-Garcia JC, Sinclair SH, Dumler JS. Chromatin bound bacterial effector AnkA recruits HDAC1 and modifies host gene expression. Cell Microbiol. 2015;17:1640–1652. doi: 10.1111/cmi.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sinclair SH, Yegnasubramanian S, Dumler JS. Global DNA methylation changes and differential gene expression in Anaplasma phagocytophilum-infected human neutrophils. Clin Epigenetics. 2015;7:77. doi: 10.1186/s13148-015-0105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de la Fuente J, Contreras M. Tick vaccines: current status and future directions. Exp Rev Vaccines. 2015;14:1367–1376. doi: 10.1586/14760584.2015.1076339. [DOI] [PubMed] [Google Scholar]


