Abstract
Brucellosis, caused by a facultative intracellular pathogen Brucella, is one of the most prevalent zoonosis worldwide. Host infection relies on several uncanonical virulence factors. A recent research hotpot is the links between carbon metabolism and bacterial virulence. In this study, we found that a carbon metabolism-related pyruvate kinase (Pyk) encoded by pyk gene (locus tag BAB_RS24320) was associated with Brucella virulence. Determination of bacterial growth curves and resistance to environmental stress factors showed that Pyk plays an important role in B. abortus growth, especially under the conditions of nutrition deprivation, and resistance to oxidative stress. Additionally, cell infection assay showed that Pyk is necessary for B. abortus survival and evading fusion with lysosomes within RAW264.7 cells. Moreover, animal experiments exhibited that the Pyk deletion significantly reduced B. abortus virulence in a mouse infection model. Our results elucidated the role of the Pyk in B. abortus virulence and provided information for further investigation of Brucella virulence associated carbon metabolism.
Introduction
Brucellosis, which is characterized by a febrile disease and infectious abortion in animals, is caused by Brucella spp, and remains one of the most common zoonotic diseases worldwide [1]. As the threat of Brucella to human health and animal husbandry has continued to increase in recent years, this disease has attracted ever more attention from scientific community and governments in affected areas throughout the world [2]. Moreover, Brucella strains have the potential to be used in biological warfare [3]. Therefore, in-depth research of Brucella virulence is truly pressing and meaningful.
Brucella, a genus of gram-negative bacteria, can grow in vitro and multiply in both phagocytic cells and non-phagocytic cells. Besides, it has evolved an amazing ability to evade host immunity and establish chronic infection. However, Brucella has no classic virulence factors, such as exotoxins, cytolysins, capsules, fimbria, and endotoxic lipopolysaccharide (LPS) [4]. Up to date, several virulence-associated factors have been identified, which are indispensable for the survival of Brucella in host cells, including a type IV secretion system, a two-component regulatory system composed of regulatory (BvrR) and sensory (BvrS) proteins, cyclic β-1,2-glucans, superoxide dismutase, catalase and urease [4]. Recently, much progress has been made in the study of possible links between carbon metabolism and intracellular bacterial virulence, especially in model intracellular pathogens, such as Listeria monocytogenes, Shigella flexneri, Salmonella entericaserovar typhimurium and Mycobacterium tuberculosis [5–8].
After entering the host cells, intracellular pathogens have to adjust their metabolism to the environmental conditions encountered in its intracellular replicative niche, including low oxygen and nutrient levels, acidic pH and so on [9]. In this process, the regulation of carbon metabolism may directly or indirectly influence the expression of the virulence genes within the host cell and, hence, pathogen virulence [5]. In Brucella, many genes reportedly associated with carbon metabolism are necessary for virulence, such as 6-phosphogluconate dehydrogenase (gnd), phosphoglucose isomerase (pgi), pyruvate carboxylase (pyc) and some erythritol catabolism genes (e.g., eryB, eryC) [9–11]. According to the previous reports, it has been accepted that glucose is mainly catabolized in Brucella through the pentose phosphate pathway in conjunction with the tricarboxylic acid (TCA) cycle [12, 13]. Of these, pyruvate is one of the more important substances that connects the glucose catabolism pathway with TCA cycle, in which several related genes have been shown to be necessary for Brucella virulence. Pyruvate phosphate dikinase (Ppdk), which is involved in classical gluconeogenesis, is required for full virulence in B. abortus [14]. Pyc is an enzyme of the ligase class that catalyzes the irreversible carboxylation of pyruvate to oxaloacetate, which was identified as a virulence-related gene by random mutagenesis [9]. It has been suggested that pyruvate catabolism plays an essential role in the full virulence of Brucella.
Our recent study found that pyruvate kinase, which is encoded by the pyk gene (gene locus BAB_RS24320) is associated with B. abortus virulence by PCR-based on signature-tagged mutagenesis (data unpublished). Pyk catalyzes the synthesis of pyruvate from phosphoenolpyruvate (PEP), that is, adenosine diphosphate + phosphoenolpyruvate = adenosine triphosphate + pyruvate, which is required for glucose catabolism through the glycolysis pathway. In this study, we further investigated the role of Pyk on B abortus virulence and found that Pyk plays important roles on the bacterial resistance to oxidative stress, escaping from fusion with lysosome within macrophages, and establishing infection in BALB/c mouse.
Materials and methods
Ethic statement
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Institutional Animal Care and Use Committee guidelines set by Shanghai Veterinary Research Institute, the Chinese Academy of Agricultural Sciences (CAAS). Mice (SLAC Experimental Animal Inc., Shanghai, China) were housed in cages with water and food ad libitum under biosafety conditions. Animal handling and procedures were approved by the Committee on the Ethics of Animal Experiments of Shanghai Veterinary Research Institute, CAAS (permit number: SHVRI-mo-0175).
Bacterial strains and growth conditions
B. abortus strain S2308 was obtained from the Chinese Veterinary Culture Collection Center (Beijing, China) and routinely grown in tryptic soy broth (TSB) (Difco™, BD BioSciences, Franklin Lakes, NJ, USA) or tryptic soy agar (TSA). Escherichia coli strain DH5α was grown on Luria–Bertani medium. When appropriate, 100 μg/mL of ampicillin or 20 μg/mL of chloramphenicol (Sigma–Aldrich Corporation, St. Louis, MO, USA) respectively, were added. All strains and plasmids used in the study are listed in Table 1.
Table 1.
Strains and plasmids used in the study
| Strains and plasmids | Characteristics | Source |
|---|---|---|
| Brucella abortus | ||
| S2308 | Wild type strain; Smooth phenotype | ATCC |
| ∆pyk | pyk gene deletion mutant strain; Smooth phenotype | This study |
| ∆pyk(Pyk-3 × Flag) | Cmr; complementation strain; ∆pyk carrying the complementation plasmid pBBR-pyk-3 × Flag; Smooth phenotype | This study |
| Escherichia coli | ||
| DH5α | F− φ80lacZ∆M15 ∆(lacZYA-argF)U169 recA1 endA1 hsdR17(r−k, m+k) phoA supE44 thi-1 gyrA96 relA1 λ− | Invitrogen |
| Plasmids | ||
| pBBR1MCS1 | Cmr; Broad-host-range cloning vector | [17] |
| pSC | Ampr; pUC19 plasmid containing SacB gene | [16] |
| pSC-∆pyk | Ampr; pSC plasmid containing the ∆pyk fragment; used to construct deletion strain | This study |
| p3 × Flag-CMV-14 | Ampr; eukaryotic expression plasmid | Sigma-Aldrich |
| pBBR-pyk-3 × Flag | Cmr; pBBR1MCS1 containing the pyk gene flanked by its upstream and downstream regions containing a C-terminal 3 × flag tag. | This study |
Construction of suicide and complementation plasmids
Suicide plasmids were constructed using an overlap PCR assay, as previously reported [15]. Briefly, efficient primers were designed for amplification of a 947-bp upstream fragment and a 995-bp downstream fragment of the pyk gene by a first round of PCR. After purification by gel extraction, the recovered products containing joined flanking sequences were used as templates for a second round of overlap PCR. Then, the PCR product was gel purified, digested with XbaI, and cloned into the XbaI-digested pSC plasmid [15, 16]. The recombinant suicide plasmid pSC-Δpyk was transformed into competent DH5α cells (Invitrogen Corporation, Carlsbad, CA, USA) for propagation and then extracted to construct the mutants.
In order to construct the complementation plasmid, the pyk gene was amplified by PCR using the primer pair Cpyk-F/Cpyk-R, the product was recovered, digested with the restriction enzymes KpnI and BamHI, and inserted into the p3 × Flag-CMV-14 plasmid (Sigma-Aldrich) to construct the recombinant plasmid p3 × Flag-pyk, and then the inserted pyk fragment with a 3 × Flag tag fragment was amplified by PCR using the primers Cpyk-F and 3 × Flag-R from the p3 × Flag-pyk plasmid, the fragment was digested with KpnI and XbaI, and inserted into the pBBR1-MCS1 plasmid [17]. The complementation plasmid was designated as pBBR-pyk-3 × Flag.
Construction of the pyk mutant and the complementation strain
The pyk mutant was constructed by allelic replacement using a two-step strategy, as previously reported [18]. Briefly, competent B. abortus strain S2308 cells were prepared through two washes with ice-cold sterile water and suicide plasmid (0.5–1.0 μg) was transformed into competent cells by electroporation. The single exchanged recombinants were selected by plating on TSA containing ampicillin, and then colonies were inoculated into TSB without antibiotics. The second exchanged recombinants were selected by plating on TSA containing 5% sucrose. All colonies were selected and verified by PCR amplification.
The complementation strain was constructed by transforming the plasmid pBBR-pyk-3 × Flag by electroporation, as described above, and the recombinant strain was designated as Δpyk(Pyk-3 × Flag).
Western blotting analysis
Western blotting analysis was performed to verify the complementation strain Δpyk(Pyk-3 × Flag). The fresh cultural Δpyk(Pyk-3 × Flag) and S2308 strains were harvested, respectively, by centrifugation 8000 × g for 5 min, the pellets were suspended in a mixture of deionized water and 2 × SDS-PAGE loading buffer (Beyotime Institute of Biotechnology, Shanghai, China), and boiled for 5 min. Proteins were separated on a 12.5% SDS polyacrylamide gel and transferred onto nitrocellulose membrane (Whatman®; Sigma-Aldrich). The membrane was blocked for 2 h at room temperature with phosphate-buffered saline (PBS; hyclone) contained 5% skim milk, and then incubated with mouse anti-Flag monoclonal antibody (Sigma-Aldrich) in PBST (PBS containing 0.2% tween-20) overnight at 4 °C. After washing five times with PBST, the membrane was incubated with IRDye 680RD-conjugated donkey anti-mouse polyclonal antibody (LI-COR Biosciences, Lincoln, NE, USA) in PBST for 1 h at room temperature. After washing five times with PBST, protein bands were detected using the Odyssey Infrared Imaging System (LI-COR Biosciences).
RNA isolation and real-time qRT-PCR
To verify the transcriptional level of the pyk upstream gene bab_RS24315 and the downstream gene bab_RS24325, total RNA was extracted from both S2308 and the pyk mutant by TRlzol reagent (invitrogen), and the bacterial genomic contamination was removed by the TURBO DNA-free kit (Ambion®; Invitrogen). The RNA was quantified by absorption at 260 nm using a NanoVue™ plus spectrophotometer (GE Healthcare Life Sciences, Logan, UT, USA). Then, qRT-PCR was carried out using GoTaq qPCR Master Mix (Promega, Fitchburg, WI, USA) and the Mastercycler ep realplex Real-Time PCR system (Eppendorf, Hamburg, Germany). For each gene, PCRs were performed in triplicate and relative transcription levels were determined by the 2−∆∆Ct method using glyceraldehyde phosphate dehydrogenase (gapdh) as an internal control for data normalization. All primers used for real-time qRT-PCR are listed in Table 2.
Table 2.
Primers used in the study
| Primers | Oligonucleotide sequence (5′ to 3′) | Target genes | Products (bp) |
|---|---|---|---|
| Cpyk-F | GGGGTACCTTGTCCAATATAAAGCGATGAC KpnI, underlined | The pyk containing the promoter region | 1934 |
| Cpyk-R | CGGGATCCAATGCCGGATTTTCCGTCAGCG BamHI, underlined | ||
| 3 × Flag-R | GCTCTAGACAGGGATGCCACCCGGGATC XbaI, underlined | The 3 × Flag tag of p3 × Flag-CMV-14 | |
| pyk-UF | CGGGATCCCGGGGGTTATGGAAAGCAACT | The upstream fragment of pyk | 947 |
| pyk-UR | TGACGACGCAATGCAGGCTCGAGGGTGAAGGTCTGG | ||
| pyk -DF | CCAGACCTTCACCCTCGAGCCTGCATTGCGTCGTCA | The downstream fragment of pyk | 995 |
| pyk- DR | CGGGATCCCGTACGGGTGCGGGTGTTTC | ||
| In-pyk-F | ATGCCGTGCTGAAGGAAGAG | The inside fragment of pyk gene | 493 |
| In-pyk-R | GCGTCAATGATGGTCGAATAGG | ||
| Out-pyk-F | GGGGTACCCCGACGGTGGGAAGGCAAAG | The outside fragment of pyk gene | 1885 |
| Out-pyk-R | CGGGATCCCGGAGCGGCTCCAGAAATCG | ||
| RT-pyk-F | AAAACTGCATCTGGTGGCTG | pyk | 207 |
| RT-pyk-R | GGGCGCTGGATAAAGGAAAG | ||
| RT-upstream-F | ATGACATCAATCGCACGCTG | bab_RS24315 | 161 |
| RT-upstream-R | GAAATTCTTTTGGGCGTCGC | ||
| RT-downstream-F | AAGCTGCAAAACCCTGATCG | bab_RS24325 | 209 |
| RT-downstream-R | AGCTTGATTGTTCCCCGGTA | ||
| RT-GAPDH-F | GACATTCAGGTCGTCGCCATCA | gapdh | 188 |
| RT-GAPDH-R | TCTTCCTTCCACGGCAGTTCGG |
Growth curve in TSB and minimal medium
Bacterial growth was measured at an optical density at 600 nm (OD600). For growth curve analysis, bacterial strains were incubated in TSB for 24 h, and then diluted with TSB to an OD600 value of 0.01 and cultured in a rotary shaker (200 rpm) at 37 °C. Cultures were taken at appropriate interval and OD600 values were recorded.
Bacterial growth in minimal medium was measured as described above except minimal medium was used as the diluting solution. Components of the minimal medium were glucose (10 g/L), yeast extract (1 g/L), (NH4)2SO4 (13.2 g/L), Na2S2O3·5H2O (0.1 g/L), MgSO4 (10 mg/L), MnSO4 (0.1 mg/L), NaCl (5 g/L) and KH2PO4 (3 g/L). The pH was adjusted to 6.8 to 7 [19]. In order to further evaluate the effect of metabolic product (sodium pyruvate) by Pyk catalysis, sodium pyruvate was added into the minimal medium with concentrations of 1, 5 or 10 mM or glucose was replaced by pyruvate as sole carbon source at final concentration of 200 mM in minimal medium.
Cell infection assay
RAW 264.7 macrophages were used to assess the ability of the pyk mutant to survive intracellularly. The experiment was performed as previously reported [20]. Briefly, the cells were seed in 24-well plates and grown in Dulbecco’s Modified Eagle Medium (DMEM) (Hyclone™; GE Healthcare) supplemented with 10% fetal bovine serum (FBS) (Gibco®; Invitrogen) at 37 °C with 5% CO2 for 24 h. The cell monolayer was washed twice with DMEM and infected with B. abortus S2308 or the pyk mutant at a multiplicity of infection of 100. Bacteria were centrifuged onto the cells at 400 × g for 10 min and the cells were then incubated at 37 °C for 1 h. Non-adherent bacteria were removed by rinsing the wells twice with PBS. To kill extra-cellular bacteria, the cells were incubated with DMEM containing gentamicin (100 μg/mL) for an additional 1 h, washed twice with PBS and the medium was replaced with DMEM containing 2% FBS and 20 μg/mL gentamicin. At 2, 8, 24 and 48 h post-infection, the macrophages were lysed with 0.2% Triton X-100 in sterile water and the live bacteria were enumerated by plating on TSA plate. All assays were performed in triplicate and repeated at least three times and the results are the averages from at triplicate infection samples.
Immunofluorescence assay
RAW264.7 cells cultured on glass coverslips (Thermo Fisher Scientific, Waltham, MA, USA) were infected with Brucella at a multiplicity of infection of 100. The infected cells were fixed with 3.7% (w/v) paraformaldehyde at 4 °C for 24 h post-infection. The immunofluorescence assay was performed as previously described [15] using rabbit anti-Brucella serum (diluted 1:1000) and Rat anti-LAMP-1 monoclonal antibody [1D4B] (diluted 1:500, Abcam, Cambridge, MA, USA) as the primary antibody, and Alexa Fluor 488-conjugated goat anti-rabbit IgG (diluted 1:1000) and Alexa Fluor 555-conjugated goat anti-rat lgG (diluted 1:1000) (Invitrogen) as the secondary antibody. The coverslips were mounted onto glass slides using Eukitt quick-hardening mounting medium for microscopy (Sigma-Aldrich) and the cells were observed under a Nikon Eclipse 80i microscope (Nikon Corporation, Tokyo, Japan) with 100× oil immersion objective. Images were saved in TIFF format and imported to Adobe Photoshop CS4 (Adobe Systems Incorporated, San Jose, CA, USA), where they were merged using RGB format. To determine the percentage of bacteria positive for the lysosome marker LAMP-1, 100 intracellular bacteria were counted randomly. The assays were performed in triplicate.
Stress resistance assay
The S2308 strain and the pyk mutant were cultured to mid-logarithmic phase (the value of OD600 ≈ 1.0) at 37 °C in TSB medium, and then the bacterial suspension was diluted with PBS and adjusted to a concentration to 4 × 105 colony-forming units (CFU)/mL. Afterward, 50 μL of bacterial suspension was mixed with 50 μL of the appropriate reagent. The effects of various stress factors were tested as follows. H2O2 was used to determine sensitivity to oxidative stress and added at final concentrations of 0.5, 1 or 1.5 mM. Polymyxin B at concentrations of 50, 100 or 200 μg/mL was used to test sensitivity to cationic bactericidal peptides. Bovine serum and heat-inactivated bovine serum were used to assess resistance to natural serum killing with a bactericidal activity assay. In all tested groups, a negative-control group was introduced by adding 50 μL of PBS to the same bacterial suspension. After exposure for 1 h at 37 °C, the mixtures were rapidly diluted and plated on TSA plates to determine viability. Results are expressed as the mean percentage of the negative control from independent triplicate samples.
To determine the ability of the pyk mutant to resist acidic pH, an acid tolerance assay was performed as previously described with some modifications [20]. Briefly, the bacterial suspension of the S2308 strain and the pyk mutant were cultured and diluted to 2 × 107 CFU/mL in TSB with pH of 7.3, 5.5 or 4.5. After 1 h of incubation at 37 °C, cells were serially diluted and plated on TSA to determine the number of bacterial CFU. The percentage of surviving bacteria was calculated with respect to CFU obtained from bacteria incubated in TSB at pH 7.3 (100% survival).
Infection of mice
Virulence assay using BALB/c mice was performed as reported previously with some modifications [16]. Briefly, 6-week-old female BALB/c mice (n = 6 per group) were intraperitoneally inoculated with 0.1 mL of suspension containing 1 × 106 CFU of the S2308 strain, the pyk mutant or the complementation strain. The survival of the bacteria in mice was evaluated by bacterial enumeration in the spleens at different time points post-infection. At 1 or 5 weeks post-infection, mice were sacrificed by cervical dislocation. The spleens were harvested, and homogenized in 5 mL of PBS-0.2% Triton X-100. After that, serial dilutions of the homogenates were made and plated on TSA plates to determine the bacterial loads. The data are expressed as the log10 CFU per spleen. Besides, spleen weight was measured to evaluate splenomegaly.
Statistical analysis
Statistical analysis was performed using GraphPad Prism software 5.0 (GraphPad Software Inc., La Jolla, CA, USA). Statistical significance was determined by either an unpaired or two-tailed Student’s t test, or in the case of groups, a one-way analysis of variance followed by the Tukey’s test. A probability (p) values of ≤0.05 were considered significant.
Results
The pyk mutant and the complementation strain were successfully constructed
The pyk mutant was verified by PCR amplification of the inside and outside fragments using respective primers (Figure 1A). The results indicated that a 493-bp inside fragment and a 1885-bp outside fragment were amplified from the S2308 strain, respectively. However, the inside fragment was not amplified from the pyk mutant and the size of outside fragment was obviously shorter than that of S2308 due to deletion of the pyk gene (Figure 1B). In order to further identify the mutant and exclude the possible polar effects of pyk deletion, qRT-PCR was performed to quantify the expression of pyk and its flanking genes at the transcriptional level. The results showed that the pyk gene was not transcribed in the pyk mutant, and transcription of the upstream and downstream genes was not affected (Figure 1C). In addition, the complementation strain Δpyk(Pyk-3 × Flag) was verified by western blotting, as shown in Figure 1D, Pyk-3 × Flag was successfully expressed in the pyk mutant.
Figure 1.
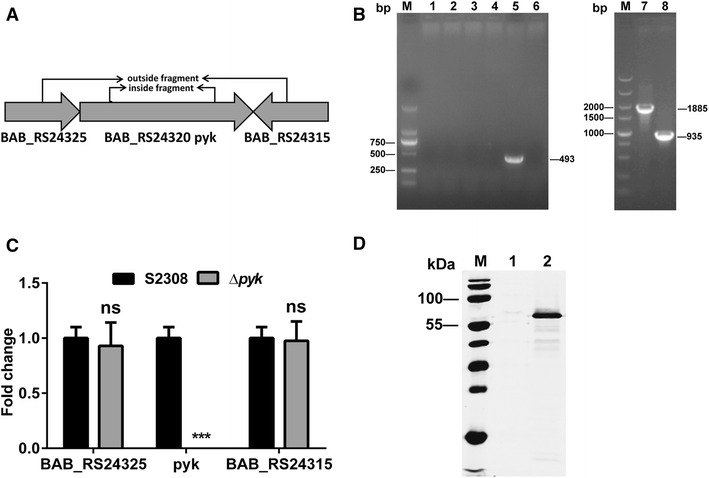
Identification of the pyk mutant and the complementation strain. A A schematic of the pyk gene and its flanking genes. The inside and outside fragments amplified using respective primers are indicated. B PCR amplification confirmed deletion of the pyk gene in the pyk mutant. Lane M: DNA marker DL2000 (Takara Bio, Inc., Shiga, Japan). Lanes 1–6: amplification of the inside fragment. Lanes 1–4: four different clones of the pyk mutant, no fragment was amplified. Lane 5: the S2308 strain (positive control), a 493-bp fragment was amplified; Lane 6: sterile water (negative control). Lanes 7 and 8: amplification of the outside fragment. Lane 7: the S2308 strain (positive control), a 1885-bp fragment was amplified; Lane 8: the pyk mutant, a 935-bp fragment was amplified. C Real-time qRT-PCR confirmed the deletion of the pyk gene at the transcriptional level. The pyk deletion did not affect expression of the flanking genes. The gene transcriptional level of the pyk mutant was compared with that of the S2308 strain. ***p ≤ 0.001, ns: no significant difference. D Western blotting confirmed expression of pyk in the complementation strain Δpyk(Pyk-3 × Flag). Lane M: prestained protein ladder (Thermo Fisher Scientific); Lane 1: the S2308 strain (negative control), no flag expression was detected; Lane 2: the complementation strain Δpyk(Pyk-3 × Flag), about 55 kDa of the flag expression product was shown.
Pyk is required for intracellular replication and trafficking of B. abortus
To determine the effect of the pyk gene in intracellular survival, the ability of the pyk mutant to survive within macrophages was assessed at 2, 8, 24 and 48 h post-infection. As shown in Figure 2 there was no significant difference in intracellular survival between the S2308 strain and the pyk mutant at 2 and 8 h post-infection, indicating a similar ability of both strains to invade macrophages. However, a marked decrease in bacterial recovery from RAW264.7 cells infected with the pyk mutant, as compared with that of S2308 infected cells at 24 and 48 h post-infection (Figure 2). These results encouraged us to determine if the difference observed in the intracellular viable counts was a consequence of increased degradation or inactivation of the mutant. To this end, we determined the number of LAMP-1-positive Brucella-containing vacuoles (BCVs) at 4 and 24 h post-infection of RAW264.7 cells with the pyk mutant and the S2308 strain. As shown in Figure 3, the mutant showed a reduced capacity to exclude the lysosome marker LAMP-1 at 24 h post-infection (about 45% co-localization), as compared with the S2308 strain (about 20% co-localization), indicating that deletion of pyk decreased the ability of the Brucella to avoid the fusion of the BCV with lysosome.
Figure 2.
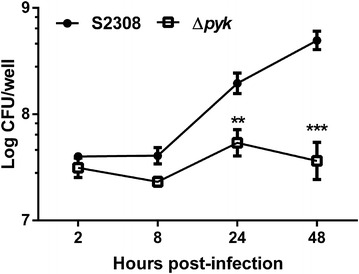
Intracellular survival within RAW 264.7 macrophages of the S2308 strain and the pyk mutant. Values are means ± standard errors for triplicate infection samples, and the experiments were repeated three times with similar results. **p ≤ 0.01, ***p ≤ 0.001.
Figure 3.
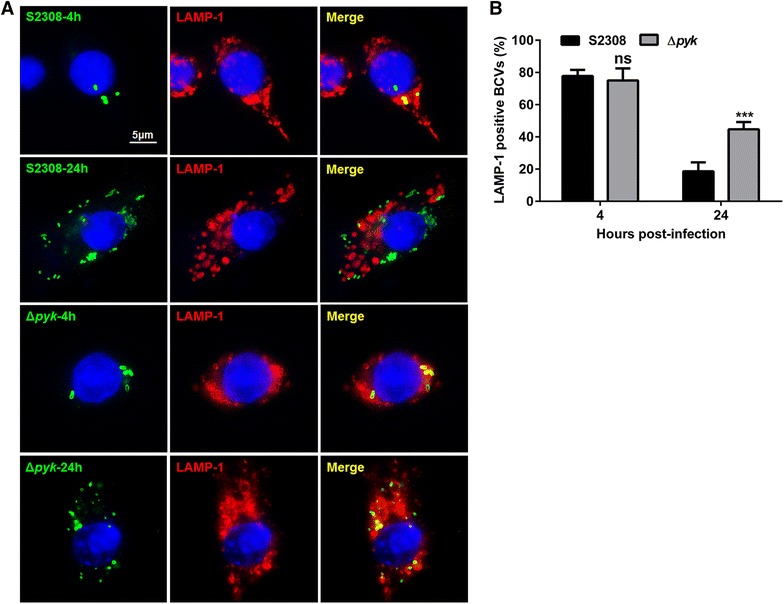
The pyk mutant was not able to efficiently exclude lysosome-associated membrane protein-1 within RAW264.7 cells. A Representative images of LAMP-1-positive and -negative BCVs. B Determination of the LAMP-1 positive BCVs at 4 and 24 h post-infection of the S2308 strain and the pyk mutant. The percentage of positive BCVs per 100 random BCVs is shown. ***p ≤ 0.001. ns: no significant difference.
Deletion of pyk in Brucella impaired its growth in TSB and in minimal medium
The Pyk is an important protein in glycolysis, and catalyzation of PEP to pyruvate, accompanied by the release of ATP. To further confirm the role of pyruvate in Brucella growth, we compared the bacterial growth of the pyk mutant with that of the S2308 strain in rich TSB and in minimal medium. As shown in Figure 4A, growth of the pyk mutant was slightly reduced at the logarithmic phase, but reached a similar stationary phase in rich TSB in comparison with the S2308 strain. However, in minimal medium the pyk mutant exhibited a significant growth defect throughout the growth stage, as compared with the S2308 strain (Figure 4B), suggesting that Pyk plays an important role in Brucella growth, especially under the condition of nutrition deprivation. To further assess the effect of pyruvate synthesis on Brucella growth, we assessed the ability of the Brucella to growth in minimal medium containing different concentrations of pyruvate. As shown in Figure 4B, growth of the pyk mutant restored to a similar level of the S2308 strain supplemented with pyruvate at final concentration of 1 mM, when supplemented with 5 or 10 mM of pyruvate, the growth of the mutant was faster than that of the S2308 strain. To further confirm the role of pyruvate in Brucella growth, we replaced glucose by pyruvate as sole carbon source in minimal medium at the concentration of 200 mM, no significant difference was found between the growth of the mutant and the S2308 strain (Figure 3C). These results revealed that the Pyk is necessary for Brucella growth, especially under the condition of nutrition deprivation, and the growth defect was due to the loss of pyruvate synthesis catalyzed by Pyk.
Figure 4.
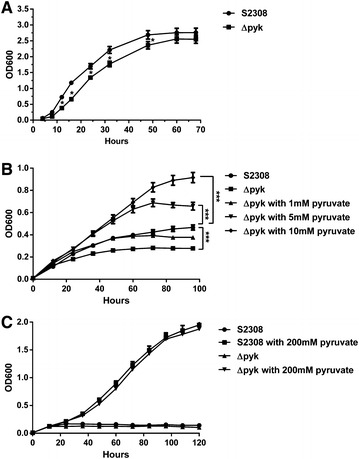
Bacterial growth was affected by pyk deletion and recovered by supplementation with sodium pyruvate. A Growth curves in TSB. B Growth curves in minimal medium with or without sodium pyruvate. C Growth curves in minimal medium without glucose or replaced glucose by pyruvate as sole carbon source at concentration of 200 mM. Each point represents the mean for triplicate samples (error bars are within the size of the symbols). The experiment was repeated three times with similar results. *p ≤ 0.05; ***p ≤ 0.001.
The pyk mutant exhibited greater sensitivity to H2O2
For Brucella spp. to successfully infect and establish chronic infection in their preferred host, the bacteria must have the ability to resist host bactericidal activity from the innate immune response, such as reactive oxygen and nitrogen species, bactericidal peptides, and low PH within macrophages. To this end, we assessed the ability of the pyk mutant to resist environmental stress factors. The results showed that the pyk mutant displayed higher sensitivity to different concentrations of H2O2, as compared with the S2308 strain. At 1.5 mM H2O2 exposure for 1 h, the mutant exhibited about 50% survival in comparison with 80% survival of the S2308 strain, and the mutant harboring the complementation plasmid pBBR-pyk-3 × Flag recovered resistance to H2O2 (Figure 5A). However, the mutant and the S2308 strain showed similar sensitivity to polymyxin B, bovine serum and low PH (Figures 5B–D).
Figure 5.
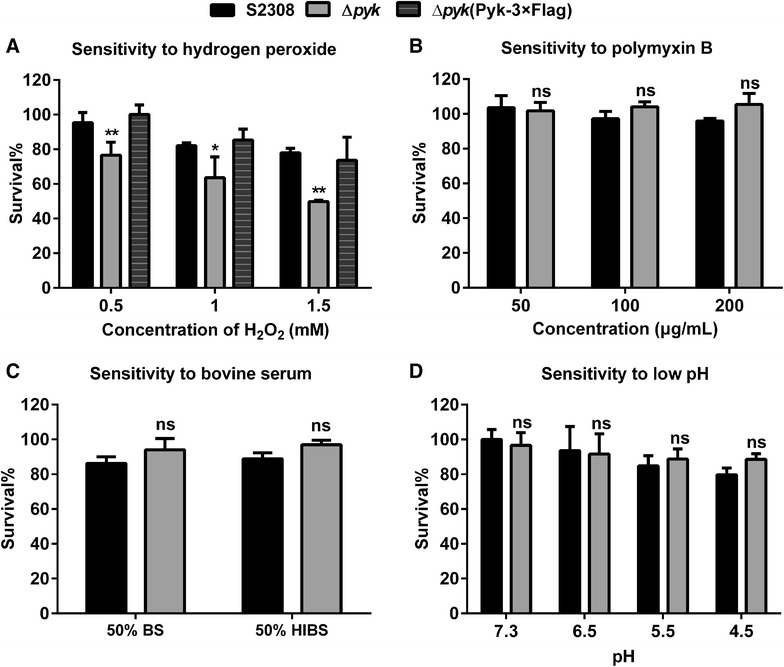
Stress resistance assays. A Sensitivity to H2O2 at concentrations of 0.5, 1 or 1.5 mM. B Sensitivity to polymyxin B at concentrations of 50, 100 or 200 μg/mL. C Sensitivity to bovine serum (BS) and heat inactivated bovine serum (HIBS). D Resistance to low pH. Values are means ± standard errors for triplicate samples. * p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ns: no significant difference.
The pyk mutant was attenuated in mice
The main manifestation of Brucella virulence is the ability to establish chronic infection in the host, therefore, we investigated the capacity of the pyk mutant to establish chronic infection in BALB/c mice. As shown in Figure 6A, the mutant exhibited significantly reduced level of spleenic colonization at 1 and 5 week post-infection. Similarly, the splenomegaly in mice infected with the mutant was less severe than that infected with the S2308 strain (Figure 6B). Besides, when harboring the pBBR1-pyk-3 × Flag plasmid, the pyk complementation strain restored the ability of splenic colonization and induced splenomegaly in infected BALB/c mice at 1 week post-infection (Figures 6C and D). These data suggested that the Pyk plays an important role in Brucella virulence.
Figure 6.
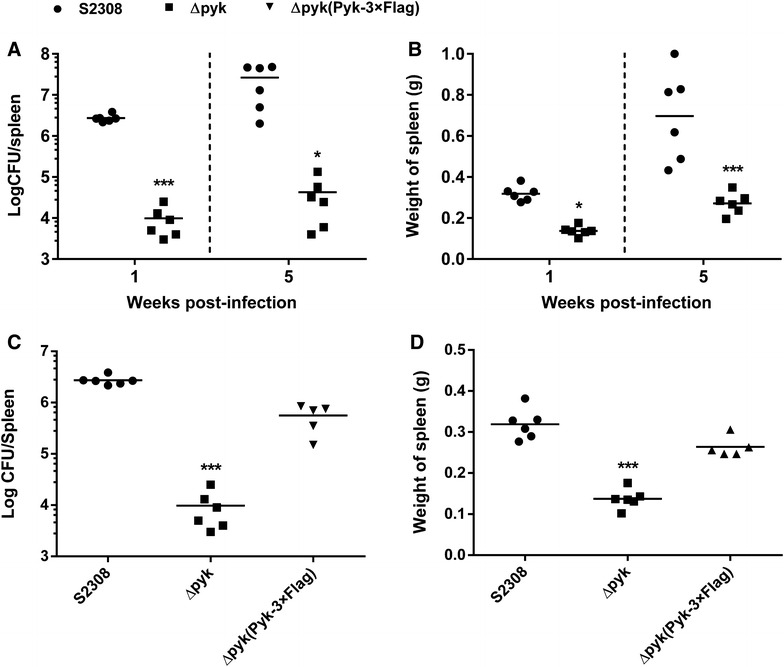
Pyk is crucial for the virulence of B. abortus in mouse model of infection. A Bacterial loads of the S2308 strain, the pyk mutant in the spleens of BALB/c mouse at 1 and 5 weeks post-infection. B Spleen weight of the S2308 strain and the pyk mutant infected mice at 1 and 5 weeks post-infection. C Bacterial loads of the S2308 strain, the pyk mutant and the complementation strain in the spleens of BALB/c mouse at 1 week post-infection. D Spleen weight of the S2308 strain, the pyk mutant and the complementation strain infected mice at 1 week post-infection *p ≤ 0.05; ***p ≤ 0.001.
Discussion
Pyk in the glycolysis pathway converts PEP to pyruvate and releases one molecule of ATP. In minimal medium, the pyk mutant was not able to reach a similar level at stationary phase like the S2308 strain, and showed a marked growth defect (Figure 4B). However, in TSB, the mutant exhibited a slight growth defect at the exponential phase, but finally achieved a similar level at the stationary phase (Figure 4A). The growth of the pyk mutant was restored when pyruvate was added, which suggested that the loss of the metabolic product pyruvate is the main reason for defective Brucella growth. It was suggested that Pyk plays an important role in Brucella growth, especially under the condition of nutrition deprivation. In Brucella, it has been reported that gluconeogenesis pathway is very important for full virulence of Brucella, and the classical genes involved in this pathway, ppdk and mae, are responsible for the markedly reduced multiplication of the mutant within macrophages and virulence in mouse model [14]. These observations confirmed the deduction that amino acids could be the preferred carbon source in vivo, which requires a gluconeogenic mechanism [21]. However, some studies suggested the availability of sugars in the replicative niche of intracellular Brucella. Xavier et al. observed that glucose uptake is crucial for increased replication of B. abortus in alternatively activated macrophages and for chronic infection in a mouse model [22]. These results indicated that the glycolytic pathway may also play an important role in metabolism and virulence of intracellular Brucella. Pyk, as an important enzyme for the pyruvate-TCA cycle node, catalyzes PEP to pyruvate irreversibly as the last step of the glycolytic pathway, which plays a significant role in Brucella virulence. Our results are consistent with those reported for other pathogens, such as the enteric pathogen Yersinia pseudotuberculosis, as Pyk resulted in significantly reduced virulence of a Yersinia mutant in a mouse infection model [23].
In order to further explain the attenuated mechanism of the pyk mutant, we analyzed other biological properties of the mutant and found that the pyk mutant was more sensitive to H2O2 than the S2308 strain. In a further study, we used real-time qRT-PCR to analyze transcription levels of genes associated with antioxidant mechanisms, including katA, sodC and oxyR, but found no significant difference in expression levels between the pyk mutant and the S2308 strain (data not shown), suggesting that reduced resistance to oxidative stress of the pyk mutant was barely attributable to the conventional antioxidant mechanism. The change in the outer membrane was also shown to affect sensitivity to oxidative stress [24–26], so the liposaccharide integrity of the mutant was confirmed by silver-staining, but no difference was found between the mutant and the S2308 strain (data not shown). The role of Pyk in B. abortus to resist oxidative stress warrants further investigation. Another manifestation of Brucella virulence is the ability of intracellular survival within target cells. Once taken up, Brucella resides in a vacuole, designated as BCV. BCVs initially fuse with endosome, and then fuse with lysosome to some extent, acquire the lysosome marker LAMP-1. Meanwhile, the BCVs are acidified by lysosome, which is a key step the Brucella needs to express the important virulence components such as Type IV secretion system, to help it to escape the fusion of BCV and lysosome, and then the BCVs re-localize to endoplasmic reticulum, forming permissive replicative Brucella vacuole. Co-localization of BCV and LAMP-1 is a key marker for determining the efficiency of BCV excluding lysosome [27]. At late stage of Brucella trafficking within host cell, the pyk mutant barely excluded the lysosomal marker LAMP-1 (Figure 3), which explained the reason why the mutant was not able to replicate efficiently within macrophages. Therefore, we speculated the mechanism of virulence attenuation for the pyk mutant as defected growth ability under a condition of nutrition deprivation, reduced resistance to oxidative response, and enhanced fusion efficiency with lysosome within host cells.
In summary, this study illuminated that the carbon metabolism gene pyk plays important roles on B. abortus growth, especially under the condition of nutrition deprivation, intracellular survival, and establishment of chronic infection in mice. This study provides further insight into the role of Pyk on B. abortus virulence.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JG and MT participated in design of the study, analyzed the data and prepared the manuscript. JG, MT, YB, PL, JL, CD, SW and TL carried out the experiments. SY designed the study, revised the manuscript and coordinated the research. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by the Scientific and Technical Innovation Project of the Chinese Academy of Agricultural Science (SHVRI-ASTIP-2014-8), the First-class General Financial Grant from the China Postdoctoral Science Foundation (2015M570184), Shanghai Sailing Program (16YF1414600) and National Basic Fund for Research Institutes, which is supported by Chinese Academy of Agricultural Sciences (2016JB06).
Footnotes
Jianpeng Gao and Mingxing Tian contributed equally to this work
Contributor Information
Jianpeng Gao, Email: gjp_veterinary@163.com.
Mingxing Tian, Email: tianmx530@126.com.
Yanqing Bao, Email: hunterzap@sina.com.
Peng Li, Email: 1067276714@qq.com.
Jiameng Liu, Email: 792900163@qq.com.
Chan Ding, Email: shoveldeen@shvri.ac.cn.
Shaohui Wang, Email: shwang@shvri.ac.cn.
Tao Li, Email: litao@shvri.ac.cn.
Shengqing Yu, Email: yus@shvri.ac.cn.
References
- 1.Corbel MJ. Brucellosis: an overview. Emerg Infect Dis. 1997;3:213–221. doi: 10.3201/eid0302.970219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ratushna VG, Sturgill DM, Ramamoorthy S, Reichow SA, He Y, Lathigra R, Sriranganathan N, Halling SM, Boyle SM, Gibas CJ. Molecular targets for rapid identification of Brucella spp. BMC Microbiol. 2006;6:13. doi: 10.1186/1471-2180-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moreno E, Moriyon I. Brucella melitensis: a nasty bug with hidden credentials for virulence. Proc Natl Acad Sci U S A. 2002;99:1–3. doi: 10.1073/pnas.022622699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seleem MN, Boyle SM, Sriranganathan N. Brucella: a pathogen without classic virulence genes. Vet Microbiol. 2008;129:1–14. doi: 10.1016/j.vetmic.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 5.Eisenreich W, Dandekar T, Heesemann J, Goebel W. Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat Rev Microbiol. 2010;8:401–412. doi: 10.1038/nrmicro2351. [DOI] [PubMed] [Google Scholar]
- 6.Eylert E, Schar J, Mertins S, Stoll R, Bacher A, Goebel W, Eisenreich W. Carbon metabolism of Listeria monocytogenes growing inside macrophages. Mol Microbiol. 2008;69:1008–1017. doi: 10.1111/j.1365-2958.2008.06337.x. [DOI] [PubMed] [Google Scholar]
- 7.Munoz-Elias EJ, McKinney JD. Carbon metabolism of intracellular bacteria. Cell Microbiol. 2006;8:10–22. doi: 10.1111/j.1462-5822.2005.00648.x. [DOI] [PubMed] [Google Scholar]
- 8.Ray K, Marteyn B, Sansonetti PJ, Tang CM. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol. 2009;7:333–340. doi: 10.1038/nrmicro2112. [DOI] [PubMed] [Google Scholar]
- 9.Kohler S, Foulongne V, Ouahrani-Bettache S, Bourg G, Teyssier J, Ramuz M, Liautard JP. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc Natl Acad Sci U S A. 2002;99:15711–15716. doi: 10.1073/pnas.232454299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foulongne V, Bourg G, Cazevieille C, Michaux-Charachon S, O’Callaghan D. Identification of Brucella suis genes affecting intracellular survival in an in vitro human macrophage infection model by signature-tagged transposon mutagenesis. Infect Immun. 2000;68:1297–1303. doi: 10.1128/IAI.68.3.1297-1303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim S, Watarai M, Kondo Y, Erdenebaatar J, Si Makino, Shirahata T. Isolation and characterization of mini-Tn5Km2 insertion mutants of Brucella abortus deficient in internalization and intracellular growth in HeLa cells. Infect Immun. 2003;71:3020–3027. doi: 10.1128/IAI.71.6.3020-3027.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robertson DC, McCullough WG. The glucose catabolism of the genus Brucella. I. Evaluation of pathways. Arch Biochem Biophys. 1968;127:263–273. doi: 10.1016/0003-9861(68)90225-7. [DOI] [PubMed] [Google Scholar]
- 13.Robertson DC, McCullough WG. The glucose catabolism of the genus Brucella. II. Cell-free studies with B. abortus (S-19) Arch Biochem Biophys. 1968;127:445–456. doi: 10.1016/0003-9861(68)90249-X. [DOI] [PubMed] [Google Scholar]
- 14.Zuniga-Ripa A, Barbier T, Conde-Alvarez R, Martinez-Gomez E, Palacios-Chaves L, Gil-Ramirez Y, Grillo MJ, Letesson JJ, Iriarte M, Moriyon I. Brucella abortusdepends on pyruvate phosphate dikinase and malic enzyme but not on Fbp and GlpX fructose-1,6-bisphosphatases for full virulence in laboratory models. J Bacteriol. 2014;196:3045–3057. doi: 10.1128/JB.01663-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian M, Qu J, Han X, Zhang M, Ding C, Ding J, Chen G, Yu S. Microarray-based identification of differentially expressed genes in intracellular Brucella abortus within RAW264.7 cells. PLoS One. 2013;8:e67014. doi: 10.1371/journal.pone.0067014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M, Han X, Liu H, Tian M, Ding C, Song J, Sun X, Liu Z, Yu S. Inactivation of the ABC transporter ATPase gene in Brucella abortus strain 2308 attenuated the virulence of the bacteria. Vet Microbiol. 2013;164:322–329. doi: 10.1016/j.vetmic.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 17.Kovach ME, Phillips RW, Elzer PH, Roop RM, 2nd, Peterson KM. pBBR1MCS: a broad-host-range cloning vector. Biotechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 18.Kahl-McDonagh MM, Ficht TA. Evaluation of protection afforded by Brucella abortus and Brucella melitensis unmarked deletion mutants exhibiting different rates of clearance in BALB/c mice. Infect Immun. 2006;74:4048–4057. doi: 10.1128/IAI.01787-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gourley CR (2012) Virulence effects and signaling partners modulated by Brucella melitensis light-sensing histidine kinase. PHD thesis, University of Wisconsin-Madison, Department of Pathobiological Sciences, Madison
- 20.Tian M, Qu J, Han X, Ding C, Wang S, Peng D, Yu S. Mechanism of Asp24 upregulation in Brucella abortusrough mutant with a disrupted O-antigen export system and effect of Asp24 in bacterial intracellular survival. Infect Immun. 2014;82:2840–2850. doi: 10.1128/IAI.01765-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamontagne J, Forest A, Marazzo E, Denis F, Butler H, Michaud JF, Boucher L, Pedro I, Villeneuve A, Sitnikov D, Trudel K, Nassif N, Boudjelti D, Tomaki F, Chaves-Olarte E, Guzman-Verri C, Brunet S, Cote-Martin A, Hunter J, Moreno E, Paramithiotis E. Intracellular adaptation of Brucella abortus. J Proteome Res. 2009;8:1594–1609. doi: 10.1021/pr800978p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xavier MN, Winter MG, Spees AM, den Hartigh AB, Nguyen K, Roux CM, Silva TM, Atluri VL, Kerrinnes T, Keestra AM, Monack DM, Luciw PA, Eigenheer RA, Baumler AJ, Santos RL, Tsolis RM. PPARgamma-mediated increase in glucose availability sustains chronic Brucella abortus infection in alternatively activated macrophages. Cell Host Microbe. 2013;14:159–170. doi: 10.1016/j.chom.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bucker R, Heroven AK, Becker J, Dersch P, Wittmann C. The pyruvate-tricarboxylic acid cycle node: a focal point of virulence control in the enteric pathogen Yersinia pseudotuberculosis. J Biol Chem. 2014;289:30114–30132. doi: 10.1074/jbc.M114.581348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corbeil LB, Blau K, Inzana TJ, Nielsen KH, Jacobson RH, Corbeil RR, Winter AJ. Killing of Brucella abortus by bovine serum. Infect Immun. 1988;56:3251–3261. doi: 10.1128/iai.56.12.3251-3261.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eisenschenk FC, Houle JJ, Hoffmann EM. Serum sensitivity of field isolates and laboratory strains of Brucella abortus. Am J Vet Res. 1995;56:1592–1598. [PubMed] [Google Scholar]
- 26.Allen CA, Adams LG, Ficht TA. Transposon-derived Brucella abortus rough mutants are attenuated and exhibit reduced intracellular survival. Infect Immun. 1998;66:1008–1016. doi: 10.1128/iai.66.3.1008-1016.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Bargen K, Gorvel JP, Salcedo SP. Internal affairs: investigating the Brucella intracellular lifestyle. FEMS Microbiol Rev. 2012;36:533–562. doi: 10.1111/j.1574-6976.2012.00334.x. [DOI] [PubMed] [Google Scholar]


