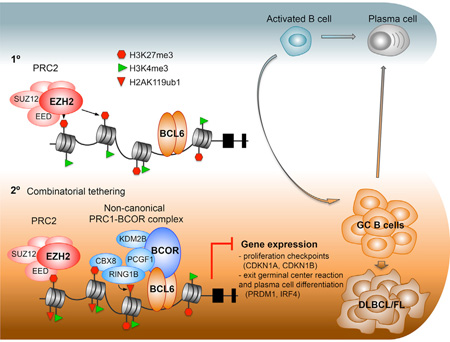
SUMMARY
The EZH2 histone methyltransferase mediates the humoral immune response and drives lymphomagenesis through formation of bivalent chromatin domains at critical germinal center (GC) B cell promoters. Herein we show that the actions of EZH2 in driving GC formation and lymphoma precursor lesions require site-specific binding by the BCL6 transcriptional repressor and the presence of a non-canonical PRC1-BCOR-CBX8 complex. The chromodomain protein CBX8 is induced in GC B cells, binds to H3K27me3 at bivalent promoters, and is required for stable association of the complex and the resulting histone modifications. Moreover, oncogenic BCL6 and EZH2 cooperate to accelerate diffuse large B cell lymphoma (DLBCL) development and combinatorial targeting of these repressors results in enhanced anti-lymphoma activity in DLBCLs.
Graphical Abstract
INTRODUCTION
During the humoral response, mature resting B cells are stimulated to differentiate into antibody-secreting plasma cells. Promoters for genes encoding key regulators of the plasma cell phenotype feature active chromatin marked by H3K4me3. However, a subset of B cells follows an alternative fate. They are able to suppress the plasma cell program and instead transiently become germinal center (GC) B cells, characterized by rapid proliferation and somatic hypermutation. Once GC B cells complete affinity maturation, they resume their normal path of plasma cell differentiation (Hatzi and Melnick, 2014). Hence, a salient feature of this process is the transient repression of the plasma cell transcriptional program and cell-cycle checkpoint genes. Importantly, a majority of B cell lymphomas arise from this inherently tumorigenic GC B cell phenotype.
GC B cells feature upregulation of EZH2 (Raaphorst et al., 2000; Velichutina et al., 2010), a core component of Polycomb repressive complex (PRC) 2 that methylates lysine 27 of histone 3 to generate H3K27me3, a histone mark associated with gene repression. Conditional deletion of EZH2 results in failure to form GCs. EZH2 enables GC formation at least in part by suppressing cell-cycle checkpoint genes like CDKN1A and possibly impairing DNA damage responses (Beguelin et al., 2013; Caganova et al., 2013). EZH2 also represses genes involved in plasma cell differentiation such as IRF4 and PRDM1, and is thus essential to maintain the GC phenotype. The EZH2 loss-of-function phenotype is strikingly similar to that of the BCL6 transcriptional repressor, a master regulator of the GC phenotype (reviewed in Hatzi and Melnick, 2014). This effect is dependent on its N-terminal BTB domain (Huang et al., 2013), which serves as the docking site for the BCL6 corepressor (BCOR) (Huynh et al., 2000). Notably, EZH2 and BCL6 share many target genes in GC B cells (Caganova et al., 2013; Velichutina et al., 2010).
The most common B cell lymphomas, follicular lymphoma (FL) and diffuse large B cell lymphoma (DLBCL), arise from B cells that have transited the GC reaction. EZH2 is often highly expressed in GC B cell-derived DLBCL and is required to maintain lymphoma cell proliferation and survival (Beguelin et al., 2013; Raaphorst et al., 2000; Velichutina et al., 2010). Moreover, in 30% of GCB-type DLBCLs and 27% of FLs, EZH2 is affected by heterozygous gain-of-function somatic mutations in its catalytic SET domain (Bodor et al., 2013; Morin et al., 2010). These mutations most typically affect the EZH2 Y641 residue, enhancing the efficiency of H3K27 trimethylation, and result in more pronounced repression of its target genes. Mice engineered to express mutant Ezh2Y641 in GC B cells develop GC hyperplasia and accumulate high levels of H3K27me3. Accordingly, patients with EZH2 overexpression or Y641 somatic mutation exhibit a characteristic gene expression signature featuring hyper-repression of genes involved in terminal differentiation and proliferation checkpoints (Beguelin et al., 2013). In a second parallel phenotype with EZH2, constitutive expression of BCL6 also results in GC hyperplasia and development of GC-derived lymphomas (Cattoretti et al., 2005). Drugs targeting BCL6 or EZH2 profoundly suppress the growth of human lymphoma cells (Cerchietti et al., 2010; McCabe et al., 2012; Knutson et al., 2012).
In embryonic and tissue-specific stem cells, EZH2 contributes to modifying gene promoters into a poised bivalent state characterized by overlapping H3K27me3 repressive mark with H3K4me3 activation mark (Bernstein et al., 2006). Bivalent chromatin maintains genes in a transiently repressed state from which they can become activated or stably repressed, depending on lineage commitment. Strikingly, in GC B cells, EZH2 mediates de novo generation of over 1,000 new bivalently marked promoters. Almost all of these domains originate from H3K4me3-only promoters in resting B cells (Beguelin et al., 2013). Many of these EZH2 target genes are specific to GC B cells and not embryonic stem cells, such as those involved in GC exit and plasma cell differentiation. Hence, in GC B cells, EZH2 mediates dynamic poising of genes involved in proliferation arrest and differentiation, and this effect is locked in through acquisition of EZH2 mutations.
The canonical mechanism by which EZH2 represses transcription is through recruitment of PRC1 complexes. However, GC centroblast B cells lack canonical core PRC1 components such as PCGF2/MEL18 and PCGF4/BMI1 (Raaphorst et al., 2000), raising the question of how EZH2 coordinates repression in this context. The critical dependency of GC B cells on EZH2 thus provides an opportunity to explore key determinants of its non-canonical and context-specific mechanisms of action. Various other modes of action of EZH2 have been proposed, including potential cooperation with sequence-specific transcription factors (Schuettengruber and Cavalli, 2009; Simon and Kingston, 2009). Along these lines, the parallels between EZH2 and BCL6 are especially intriguing (Cattoretti et al., 2005; Ci et al., 2008) and prompted us to explore whether and how these proteins might cooperate to control transcriptional repression and mediate the GC phenotype.
RESULTS
EZH2 Is Required for BCL6 to Drive GC Hyperplasia
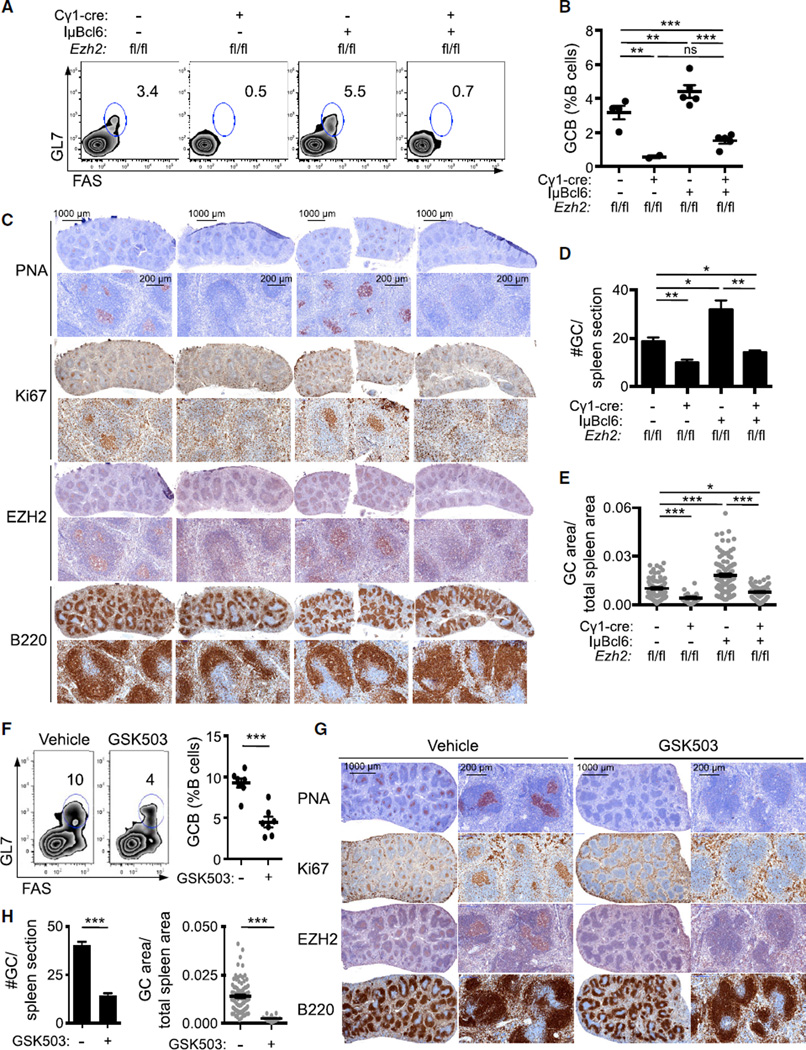
The similar effects of BCL6 and EZH2 on the GC phenotype prompted us to evaluate whether BCL6 and EZH2 cooperate in the development of GCs. To explore this question, we crossed conditional Ezh2fl/fl knockout mice (Su et al., 2003) with the Cγ1-cre strain, which expresses CRE recombinase in established GC B cells (Casola et al., 2006). These animals were crossed to IµBcl6 mice, which maintain constitutive BCL6 expression in GC B cells (Cattoretti et al., 2005). Ezh2fl/fl; Cγ1-cre, IµBcl6, Ezh2fl/fl;Cγ1-cre;IµBcl6, and Ezh2fl/fl control mice were immunized with T cell-dependent antigen sheep red blood cells (SRBC) to induce GC formation and sacrificed 10 days later, at which time the GC reaction is at its peak. As previously reported, Ezh2fl/fl;Cγ1-cre displayed defective formation of GCs (Beguelin et al., 2013; Caganova et al., 2013), whereas constitutive BCL6 expression induced GC hyperplasia (Cattoretti et al., 2005). CRE expression had no effect (Figures S1A–S1C). Notably, deletion of Ezh2 from GC B cells not only abrogated the BCL6-induced hyperplastic phenotype but also resulted in profound reduction in GC B cells (FAS+/GL7+/B220+, p < 0.001; Figures 1A and 1B). Immunohistochemical analysis using peanut agglutinin (a GC B cell marker) further revealed a reduction in the number and size (p < 0.05) of GCs in Ezh2fl/fl;Cγ1-cre;IµBcl6 versus Ezh2fl/fl controls (Figures 1C–1E). There was also marked reduction in Ki67-positive cells consistent with loss of the proliferative GC B cell compartment (Figure 1C). Residual GC B cells in Ezft2fl/fl;Cγ1-cre and Ezft2fl/fl;Cγ1-cre;IµBcl6 mice were EZH2 positive (Figures S1D–S1F), consistent with incomplete CRE-mediated excision of Ezh2. To determine if the requirement for EZH2 is dependent on its enzymatic function, we next immunized IµBcl6 mice with SRBC followed by daily treatment for 9 days with the EZH2 inhibitor GSK503 (Beguelin et al., 2013) or vehicle. GSK503 prevented GC hyperplasia in IµBcl6 mice after SRBC immunization, manifested by fewer GC B cells by flow cytometry (p < 0.001; Figure 1F) and reduced number and volume of GCs by immunohistochemistry (p < 0.001; Figures 1G and 1H). Collectively, these data show that constitutive expression of BCL6 is unable to drive GC hyperplasia in the absence of EZH2 protein or its catalytic activity.
Figure 1. EZH2 is Required for BCL6 to Drive GC Hyperplasia.
(A–E) Ezh2fl/fl, Ezh2fl/fl;Cγ1-cre, IµBcl6, and Ezh2fl/fl;Cγ1-cre;IµBcl6 mice (n = 5 per group) were immunized with SRBC to induce germinal center (GC) formation and sacrificed 10 days later. (A) Flow cytometry plot of one representative mouse spleen per group. The gated area shows the percentage of GC B cells (GL7+FAS+) within live B cells (B220+DAPI−). (B) Average of GC B populations of each group of mice quantified by flow cytometry. Each dot represents the percentage of GC B cells within splenic live B cells of one mouse. (C) Splenic tissue was stained for peanut agglutinin (PNA), Ki67, EZH2, and B220. (D and E) Quantification of PNA staining from (C). The #GC/spleen section is the count of all GCs per spleen section (D). The GC area/total spleen area is the quantified area of each individual GC divided by the total area of the spleen section (E).
(F–H) IµBcl6 mice were immunized with SRBC, treated daily with GSK503(150 mg/kg/day, n = 7) or vehicle (n = 7) and sacrificed 10 days after immunization. (F) Representative flow cytometry plot of splenic GC B cells (left) and quantification (right) as in (A) and (B). (G) Splenic tissue was stained for PNA, Ki67, EZH2, and B220. (H) Quantification of PNA staining from (G) as in (D) and (E).
Values in (B), (D), (E), (F), and (H) are shown as means ± SEM. t test, *p < 0.05, **p < 0.01, ***p < 0.001. See also Figure S1.
BCL6 Is Required for Mutant EZH2Y641 to Drive GC Hyperplasia
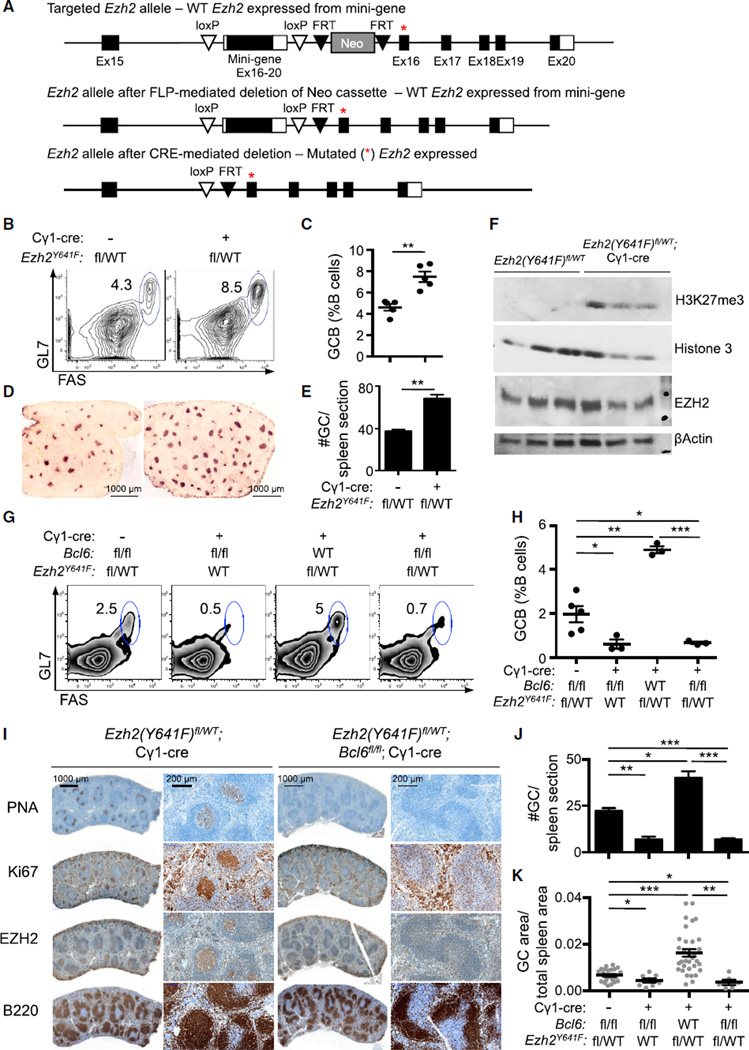
We performed the reciprocal experiment to determine whether BCL6 is required for hyperactive mutant EZH2Y641 to drive lymphoid hyperplasia. Because Bcl6 constitutive knockout has a complex and lethal phenotype (Ye et al., 1995), we first generated conditional Bcl6fl/fl;Cγ1-cre mice. As expected, conditional deletion of Bcl6 resulted in profound reduction in GC B cells, underlining that wild-type (WT) EZH2 alone is not sufficient to drive GC formation (Figures 2G, 2H, 2J, and 2K). To determine whether BCL6 was also required to support the function of hyperactive mutant EZH2Y641, we generated an additional conditional allele, Ezh2(Y641F)fl, that expresses Ezh2Y641F from the endogenous Ezh2 locus when activated by CRE (Figure 2A). Similar to Ezh2Y641N-ColA1 mice (Beguelin et al., 2013), Ezh2(Y641F)fl/WT;Cγ1-cre mice exhibited GC hyperplasia after immunization (Figures 2B–2E and S2A). The histologic appearance of the spleen and primary lymphoid follicles was otherwise normal, and marginal zone and follicular B cells were unaffected (Figures S2B–S2D). The phenotype was accompanied by a significant increase in the abundance of H3K27me3 in sorted GC B cells from Ezh2(Y641F)fl/WT;Cγ1-cre versus non-recombined Ezh2(Y641F)fl/WT mice (Figure 2F), analogous to what is observed in mutant EZH2 DLBCL cell lines (McCabe et al., 2012; Sneeringer et al., 2010).
Figure 2. BCL6 Is Required for Mutant EZH2Y641 to Drive GC Hyperplasia.
(A) Generation of Ezh2(Y641F)fl conditional mice. A mini-gene comprising exons 16–20 flanked by loxP sites and the pGK-Neo cassette flanked by FRT sites were inserted into intron 15–16 of the Ezh2 allele. The Neo cassette was removed by crossing to FLP deleter strain. Prior to Cre expression, the WT product is generated from the mini-gene. CRE-mediated deletion of the mini-gene results in expression of the mutant form (represented with a red asterisk).
(B–F) Ezh2(Y64lF)fl/WT crossed with Cγ1-cre positive (n = 5) or negative control mice (n = 5) were immunized with SRBC and sacrificed 10 days later. (B and C) Percentages of splenic GC B cells were measured by flow cytometry (B) and quantified (C) as in Figures 1A and 1B. (D) Splenic tissue was stained for PNA. (E) Quantification of PNA staining from (D). (F) Immunoblotting with anti-EZH2 and H3K27me3 antibodies was performed in sorted GC B splenocytes, using βactin and histone 3 as loading controls.
(G–K) Bcl6fl/fl, Bcl6fl/fl;Cγ1-cre, Ezh2(Y641F)fl/WT;Cγ1-cre and Bcl6fl/fl;Ezh2(Y641F)fl/WT;Cγ1-cre mice (n = 3 to 5 per group) were immunized with SRBC and sacrificed 10 days later. (G and H) Representative flow cytometry plot of splenic GC cells (G) and quantification (H) as shown in Figures 1A and 1B. (I) Splenic tissue was stained for PNA, Ki67, EZH2, and B220. (J and K) Quantification of GC number (J) and area (K) based on PNA staining in (I). Values in (C), (E), (H), (J) and (K) are shown as means ± SEM. t test, *p < 0.05, **p < 0.01, ***p < 0.001. See also Figure S2.
We next used the Ezh2(Y641F)fl allele to determine whether BCL6 is required to support mutant EZH2Y641. We assessed the GC reaction in the offspring of Bcl6 conditional KO mice crossed with Ezh2(Y641F)fl/WT;Cγ1-cre strain. Bcl6fl/fl; Ezh2(Y641F)fl/WT;Cγ1-cre mice failed not only to develop EZH2 driven GC hyperplasia but also exhibited profound reduction of GC B cells (p < 0.001 versus mutant EZH2 and p < 0.05 versus WT EZH2; Figures 2G and 2H). There was also significant reduction in the number and size of GCs as shown by immunohistochemistry (p < 0.001; Figures 2I–2K). Notably, homozygous Ezh2(Y641F)fl/fl;Cγ1-cre mice failed to form GCs upon SRBC immunization compared with Ezh2WT/WT and heterozygous Ezh2(Y641F)fl/WT;Cγ1-cre mice (p < 0.001; Figure S2E). These data are consistent with the fact that, whereas WT EZH2 preferentially mediates H3K27me1 and me2, the mutant form is more active at converting H3K27me2 into H3K27me3 (Sneeringer et al., 2010). Our results demonstrate that the phenotypic effects of mutant EZH2Y641 requires cooperation with the WT allele thus explaining why the gain-of-function mutations of EZH2 are always heterozygous in patients.
EZH2 and BCL6/BCOR Complexes Are Both Required to Repress Key De Novo GC B Cell Bivalent Promoters
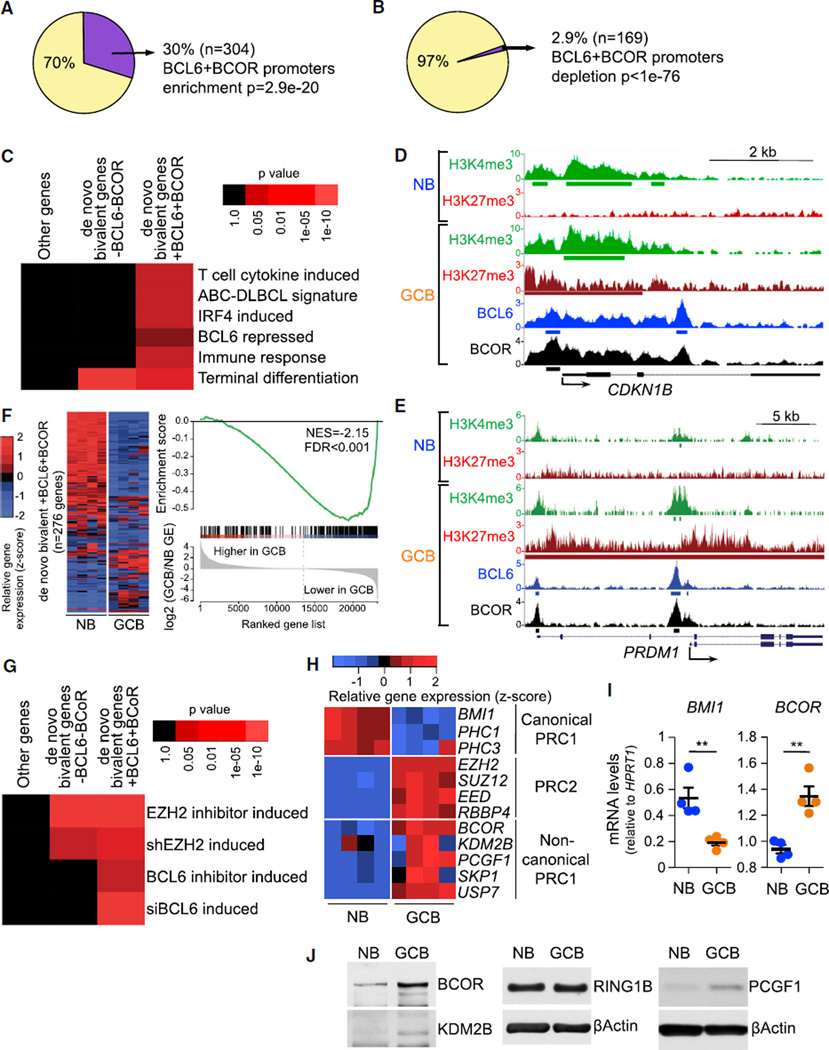
The above data suggest functional dependency between EZH2 and BCL6. EZH2 mediates its effects in GC B cells in part through de novo formation of bivalent promoters (Beguelin et al., 2013). BCL6 represses promoters mainly by recruiting the corepressor protein BCOR (Hatzi et al., 2013). To explore potential mechanistic links between EZH2 and BCL6, we examined the genomic distribution of H3K27me3, H3K4me3, BCL6, and BCOR in purified primary human naive B (NB) cells and GC B cells using chromatin immunoprecipitation sequencing (ChIP-seq) data (Beguelin et al., 2013; Hatzi et al., 2013). We observed significant overlap of GC de novo bivalent promoters with BCL6 and BCOR (hypergeometric test, p = 2.9 × 10−20; Figure 3A). In contrast, BCL6 and BCOR were excluded from monovalent H3K27me3 genes (depletion p < 1 × 10−76; Figure 3B and data not shown). BCL6/BCOR-occupied GC de novo bivalent genes were significantly enriched for pathways involved in GC exit and terminal differentiation, including genes induced by interferon regulatory factor 4 (IRF4) and T cell cytokines, genes highly expressed in activated B cell (ABC)-DLBCL signature compared with GCB-DLBCL (which includes GC exit genes), and genes associated with immune responses (Figure 3C). Among BCL6-BCOR-occupied de novo bivalent genes were key proliferation checkpoint (CDKN1A, CDKN1B) and B cell differentiation (PRDM1, IRF4) genes (as exemplified in Figures 3D and 3E). Bivalent genes without BCL6/BCOR complexes were not preferentially linked to these pathways. We next compared RNA-seq gene expression profiles of NB versus GC B cells (Beguelin et al., 2013) and found that de novo bivalent genes bound by BCL6 and BCOR at their promoters are significantly more repressed in GC B compared with NB cells (gene set enrichment analysis [GSEA] false discovery rate, q < 0.001; Figure 3F). To determine whether these BCL6-BCOR-bound de novo bivalent genes were actively repressed by EZH2 and BCL6, we examined RNA-seq profiles of GC-derived DLBCL cell lines treated with an EZH2 inhibitor or a BCL6 inhibitor that disrupts BCL6-BCOR interaction; or EZH2 small hairpin RNAs (shRNAs) (Beguelin et al., 2013) or BCL6 small interfering RNAs (siRNAs) (Hatzi et al., 2013). In all cases, we observed significant de-repression of BCL6-BCOR de novo bivalent genes. In contrast, de novo bivalent genes lacking BCL6/BCOR were only de-repressed by EZH2 shRNA or inhibitors (Figure 3G; for gene lists, see Table S1). These data suggest functional cooperation between EZH2 and BCL6, specifically at genes where BCL6 recruits BCOR, since EZH2 alone or BCL6 alone is not sufficient to maintain repression of these bivalent genes. Another 25% of de novo bivalent genes with enrichment for BCL6 and BCOR that did not reach peak threshold were also regulated in a similar way (Figure S3A and data not shown), suggesting even more widespread regulation of bivalent genes by BCL6-BCOR complexes.
Figure 3. EZH2 and BCL6/BCOR Complexes Are Both Required to Repress Key De Novo GC B Cell Bivalent Promoters.
(A and B) Percentage and number of de novo bivalent promoters (H3K27me3+H3K4me3, n = 1,011) (A) and H3K27me3 monovalent promoters (n = 5,798) (B) overlapping ChIP-seq peaks of BCL6 and BCOR in GC B cells.
(C) Heatmap of over-represented gene categories among genes with BCL6+BCOR-occupied de novo bivalent promoters compared with non-bivalent genes and de novo bivalent genes without BCL6 or BCOR. Enrichment was measured using hypergeometric p values.
(D and E) CDKN1B (D) and PRDM1 (E) gene loci showing H3K4me3, H3K27me3, BCL6, and BCOR ChIP-seq read density in naive B cells (NB) and germinal center B cells (GCB). Green, red, blue, and black bars: H3K4me3, H3K27me3, BCL6, and BCOR peaks, respectively.
(F) Heatmap of the gene expression level and GSEA of de novo bivalent genes with BCL6+BCOR in 4 NB and 4 GCB samples. NES, normalized enrichment score; FDR, false discovery rate.
(G) Heatmap of over-represented de novo bivalent genes with and without BCL6- and BCOR-occupied promoters for genes induced by EZH2 or BCL6 inhibitors (2 µM GSK343 for 5 days and 25 µM FX1 for 12 hr) or shRNAs for EZH2 (7 days) or siRNAs for BCL6 (2 days) in OCI-Ly1, OCI-Ly7, SUDHL5, SUDHL6, Farage, WSU-DLCL2, and Pfeiffer DLBCL cell lines. Enrichment measured using hypergeometric p values.
(H and I) Heatmap of the gene expression level (H) and RT-qPCR (I) of canonical and non-canonical PRC1 components in 4 NB and 4 GCB samples. Values in (I) are shown as means ± SEM. t test, **p < 0.01.
(J) Immunoblotting of whole-cell lysates from NB and GCB samples.
Transcriptional repression of H3K27me3 marked chromatin is mediated by PRC1 complexes. However, we found that genes for canonical PRC1 components BMI1 (PCGF4), PHC1, and PHC3 are repressed and downregulated in GC B cells compared with NB cells (Figure 3H). The canonical PRC1 component PCGF2 (MEL18) was absent in both GC B and NB cells (Figure S3B). However, BCOR forms an alternative non-canonical complex with certain PRC1 subunits (Gearhart et al., 2006; Gao et al., 2012). Non-canonical PRC1 genes BCOR, PCGF1, KDM2B, SKP1, and USP7 are upregulated in GC B cells, similar to PRC2 (Figure 3H). BMI1 downregulation and BCOR upregulation in GC B cells were confirmed by RT-qPCR (Figure 3I). Immunoblot analysis likewise revealed that GC B cells express higher protein levels of BCOR, KDM2B, and PCGF1 than NB cells (Figure 3J). Remarkably, the core canonical PRC1 component BMI1 is among the de novo bivalent genes bound and repressed by BCL6-BCOR complexes (Figure S3C). PCGF2 is highly repressed since its promoter is marked only by H3K27me3 in both GCB and NB cells (Figure S3D). We found almost complete overlap of BCOR and KDM2B at bivalent genes by ChIP-seq (Figure S3E). Taken together, these data indicate that the non-canonical PRC1-BCOR complex may represent the dominant PRC1 in GC B cells.
Mutant EZH2 Fails to Induce GC Hyperplasia in the Absence of BCOR in a BCL6-Dependent Manner
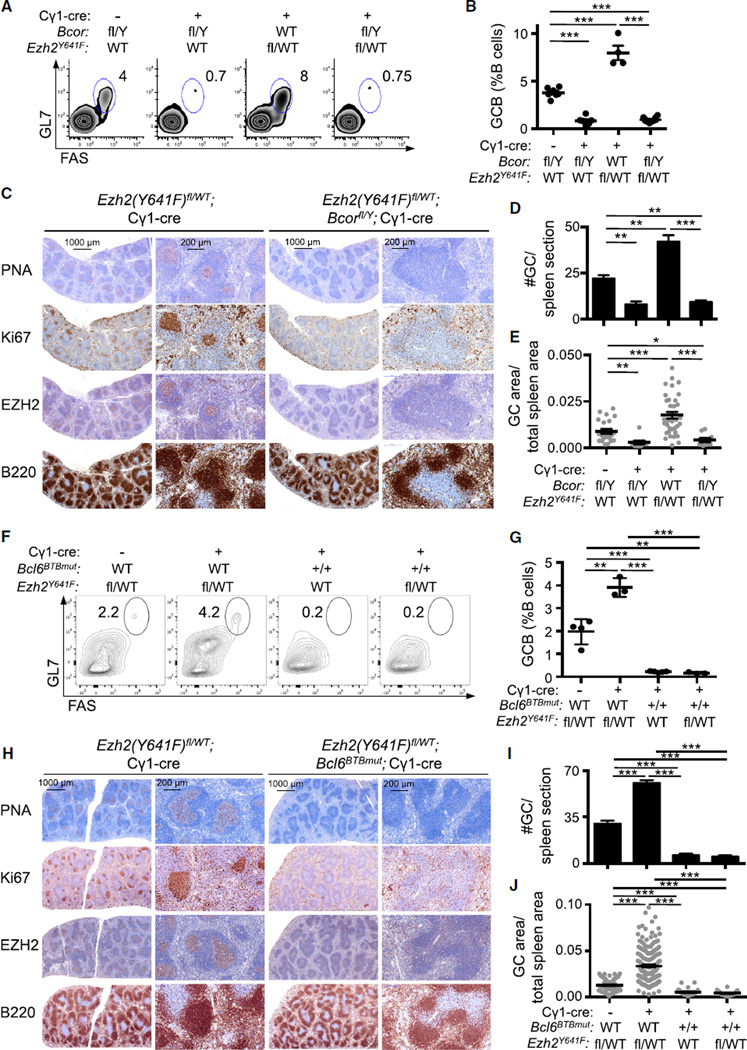
The above data suggest that BCOR, like BCL6, may be required for the transcriptional and biological effects of EZH2. Thus, to determine whether BCOR is required for GC formation, we used a conditional Bcor allele, Bcorfl (M.Y.H., C.M.C., M.D.G., and V.J.B, unpublished data) together with the Cγ1-cre allele. Bcorfl/Y;Cγ1-cre mice failed to form GCs after immunization, similar to the case of Ezh2 or Bcl6 deletion (Figures 4A–4E). To evaluate if mutant EZH2 can drive GC formation or hyperplasia in the absence of BCOR, we performed immunization experiments in mice bred for simultaneous conditional knockout of Bcor and conditional knockin of Ezh2(Y641F)fl/WT with the Cγ1-cre allele. Bcor deletion in Bcorfl/Y;Ezh2(Y641F)fl/WT;Cγ1-cre mice resulted in marked depletion in the number of GC B cells (p < 0.001; Figures 4A and 4B) and significant reduction in the number and volume of GCs by immunohistochemistry (p < 0.001; Figures 4C–4E). There was no effect on total B220+ B cells or marginal zone or follicular B cell numbers (Figure S4A).
Figure 4. Mutant EZH2 Fails to Induce GC Hyperplasia in the Absence of BCOR in a BCL6-Dependent Manner.
(A–E) Bcorfl/Y, Bcorfl/Y;Cγ1-cre, Ezh2(Y641F)fl/WT;Cγ1-cre, and Bcorfl/Y;Ezh2(Y641F)fl/WT;Cγ1-cre mice (n = 4 to 6 per group) were immunized with SRBC and sacrificed 10 days later. Note that Bcor is on the X chromosome; hence male mice have only one floxed Bcor allele and Y indicates the Y chromosome. (A) Representative flow cytometry plot of splenic GC B cells as in Figure 1A. (B) Quantification of GC B cells by flow cytometry as in Figure 1B. (C) Splenic tissue was stained for PNA, Ki67, EZH2, and B220. (D and E) Quantification of GC number (D) and area (E) based on PNA staining in (C). (F–J) Ezh2(Y641F)fl/WT, Ezh2(Y641F)fl/WT;Cγ1-cre, Bcl6BTBmut;Cγ1-cre and Bcl6BTBmut;Ezh2(Y641F)fl/WT;Cγ1-cre mice (n = 4 per group) were immunized with SRBC and sacrificed 10 days later.
(F) Representative flow cytometry plot of splenic GC B cells as in Figure 1A.
(G) Quantification of GC B cells by flow cytometry as in Figure 1B.
(H) Splenic tissue was stained for PNA, Ki67, EZH2, and B220.
(I and J) Quantification of GC number (I) and area (J) based on PNA staining in (H).
Values in (B), (D), (E), (G), (I), and (J) are shown as means ± SEM. t test, *p < 0.05, **p < 0.01, ***p < 0.001. See also Figure S4.
These data suggest that BCL6 and BCOR are each required for the actions of EZH2 in established GC B cells but do not address whether it is the interaction between BCL6 and BCOR that mediates this effect. To address this point, we used a Bcl6 allele that encodes a mutant BCL6 protein unable to bind to BCOR (Bcl6BTBmut) (Huang et al., 2013). As previously reported, homozygous Bcl6BTBmut mice were unable to form GCs (Figures 4F–4J). Bcl6BTBmut homozygous mice crossed with Ezh2(Y641F)fl/WT;Cγ1-cre rescued the GC hyperplasia phenotype and again abrogated GC formation, as shown by flow cytometry (p < 0.001; Figures 4F and 4G) and immunohistochemistry (p < 0.001; Figures 4H–4J). As an alternative approach to disrupting the BCL6-BCOR interaction, we used a small molecule that specifically blocks BCL6 binding to BCOR called FX1 (Cardenas et al., 2016). Ezh2(Y641F)fl/WT;Cγ1-cre mice were treated with FX1 or vehicle for 10 days after immunization. Reminiscent of Bcl6 or Bcor conditional deletion, FX1 prevented GC hyperplasia and induced a significant reduction in the number of GC B cells as opposed to vehicle-treated Ezh2(Y641F)fl/WT;Cγ1-cre mice (p < 0.01; Figure S4B). FX1 also significantly reduced the number and size of GCs compared with vehicle-treated Ezh2WT/WT and Ezh2(Y641F)fl/WT;Cγ1-cre mice (p < 0.05 and p < 0.001, respectively; Figures S4C and S4D). Collectively, these results demonstrate that WT and gain-of-function mutant EZH2 require a functional BCL6-BCOR complex to drive formation of GCs and GC hyperplasia, respectively.
PRC1-BCOR Complex Requires Both PRC2 and BCL6 for Stable Association and Repression of Bivalent Promoters
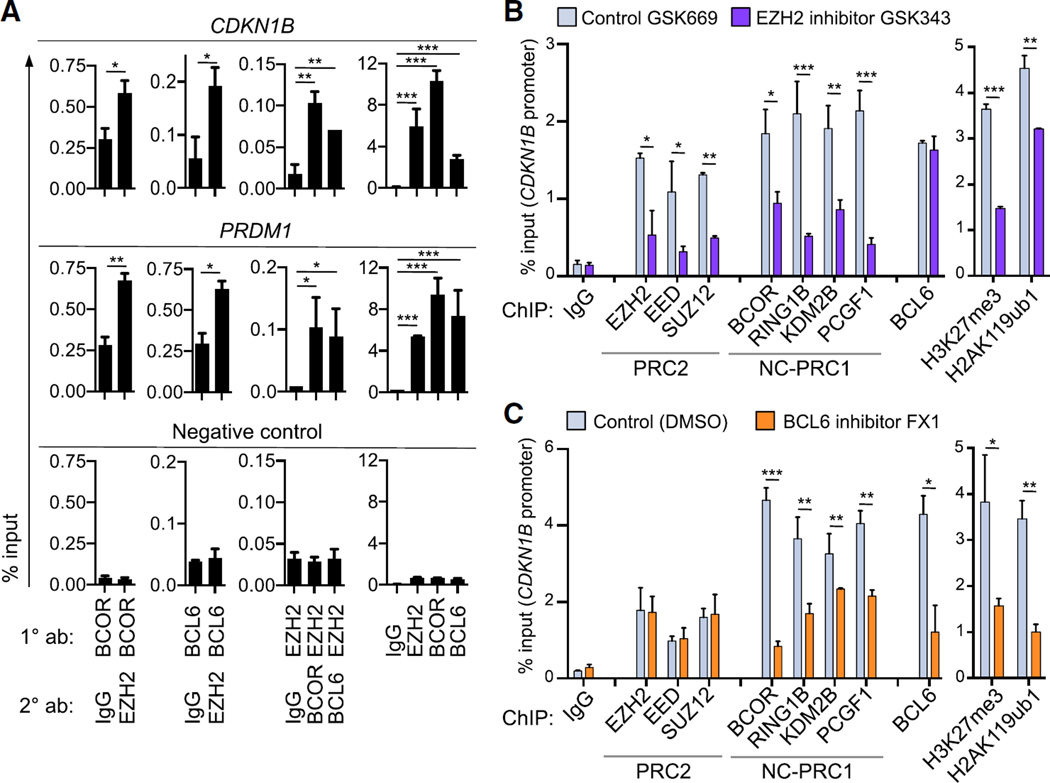
We next evaluated the mechanism through which EZH2, BCL6, and BCOR cooperate to mediate the GC phenotype. First we examined whether BCL6 could interact with EZH2. We were unable to detect any interaction either with endogenous or transfected proteins (data not shown). We then evaluated whether these proteins were bound to the same loci using sequential ChIP (ChIP re-ChIP) in GC-derived DLBCL cells. We found that EZH2 is co-recruited at the same bivalent promoters as BCOR and BCL6 as shown for the CDKN1B and PRDM1 loci (Figure 5A). Hence BCL6-BCOR and PRC2 complexes co-localize on chromatin without direct BCL6-EZH2 contact.
Figure 5. PRC1-BCOR Complex Requires Both PRC2 and BCL6 for Stable Association and Repression of Bivalent Promoters.
(A) ChIP re-ChIP in the bivalent promoters of CDKN1B and PRDM1 in OCI-Ly7 cells using the indicated antibodies for the first ChIP (1° ab) and the sequential ChIP (2° ab). As negative control, qPCR was performed using primers for a region in chromosome 6 where no BCOR, EZH2, or BCL6 enrichment was found by ChIP-seq read density in GC B cells.
(B and C) qChIP in CDKN1B promoter of SUDHL6 cells treated with (B) 2 µM EZH2 inhibitor GSK343 or control compound GSK669 for 72 hr, and (C) 25 µM BCL6 inhibitor FX1 or vehicle for 6 hr. NC-PRC1, non-canonical PRC1.
Values are shown as means of triplicates or quadruplicates ± SD. t test, *p < 0.05, **p < 0.01, ***p < 0.001. See also Figure S5.
To determine whether PRC2 and BCL6-BCOR functionally cooperate on chromatin, we treated GC-derived DLBCL cells with the EZH2 inhibitor GSK343 or the inactive compound GSK669. We then evaluated recruitment of PRC2 components EZH2, EED, and SUZ12; PRC1-BCOR complex components BCOR, RING1B, KDM2B, and PCGF1; BCL6; and the PRC2 histone mark H3K27me3, and the RING1B mark H2AK119ub. We performed qChIP for these proteins in four independent GC-derived DLBCL cell lines at six key bivalent promoters (CDKN1B, PRDM1, IRF4, CDKN1A, ARID3A, and ARID3B), as well as a negative control region. EZH2 inhibitor caused a significant reduction in both recruitment of PRC2 and BCOR complex components, along with concordant loss of H3K27me3 and H2AK119ub (Figures 5B, S5A, and S5B). In contrast, BCL6 binding was unaffected. This suggests that, even though EZH2 does not directly interact with the PRC1-BCOR complex, stable association of PRC1-BCOR complex with chromatin still requires PRC2 activity. BCL6 occupancy alone is not sufficient to maintain maximal PRC1-BCOR recruitment or transcriptional repression of these genes, and BCL6 recruitment does not require H3K27me3. In reciprocal experiments, we treated the same cell lines with FX1 to block the interaction between BCL6 and BCOR. In this case, qChIP revealed loss of PRC1-BCOR complex recruitment with no effect on PRC2 occupancy (Figures 5C and S5A–S5B). There was depletion of both H2AK119ub and H3K27me3, consistent with loss of BCOR complex as well as impairment of PRC2 function. We also observed reduced BCL6 binding at many loci (although there was no reduction in BCL6, PRC1, or PRC2 protein levels; Figure S5C). Collectively, these data suggest a model whereby BCL6 and EZH2 must cooperate to mediate the stable recruitment of the non-canonical PRC1-BCOR complex to bivalent promoters in GC B cells. Neither BCL6 nor PRC2 alone are sufficient to optimally tether this complex or fully repress expression of these target genes.
CBX8 Recruits BCOR Complex to H3K27me3 Marked Bivalent Genes and Is Required for the Biological Actions of EZH2
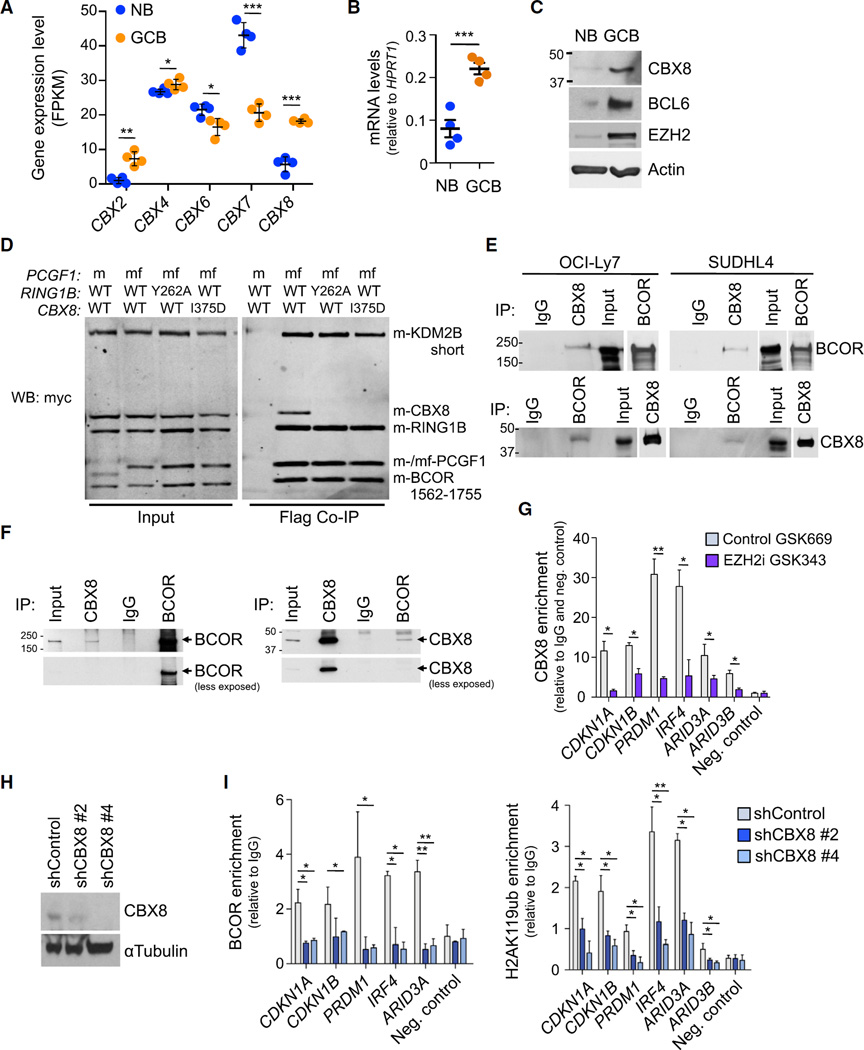
BCOR recruitment occurs through direct binding of BCOR to the BCL6 BTB domain (Ghetu et al., 2008; Huynh et al., 2000; Stogios et al., 2005). However, it is not known how BCOR is recruited to bivalent promoter regions through PRC2. Canonical PRC1 complexes contain chromobox homolog (CBX) histone reader proteins that bind to H3K27me3. Hence, we wondered whether CBX proteins might also mediate the PRC2-dependent recruitment of BCOR complex to bivalent promoters in the GC B cell context. We examined RNA-seq gene expression profiles to identify CBX family proteins potentially relevant to the GC B cell context (Figure 6A). Among these, CBX8 was the most differentially upregulated chromobox homolog family member in GC B cells. We confirmed CBX8 upregulation in purified GC B cells and NB cells using qPCR and immunoblots (Figures 6B and 6C).
Figure 6. CBX8 Recruits BCOR Complex to H3K27me3 Marked Bivalent Genes and Is Required for the Biological Actions of EZH2.
(A) Expression level in FPKM (fragments per kilobase of transcript per million mapped reads) of CBX genes in four naive B cells (NB) and four germinal center B cells (GCB) samples. Values are means ± SEM.
(B) RT-qPCR of CBX8 in four NB and four GCB samples. Values are means ± SEM.
(C) Immunoblotting of whole-cell lysates from NB and GCB cell samples.
(D) Myc immunoblotting of Flag IPs using whole-cell extracts from Sf9 insect cells transfected with expression vectors for the indicated proteins, m, amino terminal myc epitope tag; f, flag tag; mf, both tags.
(E) Immunoblotting of immunoprecipitation (IP) using whole-cell lysates from the indicated cell lines.
(F) Immunoblotting of IP using whole-cell lysates from primary GCB from human tonsils.
(G) CBX8 qChIP in OCI-Ly7 cells treated with 2 µM GSK343 or GSK669 for 72 hr. qPCR was performed using primers targeting the promoters of the indicated genes.
(H) Immunoblotting of whole-cell lysates from OCI-Ly7 cells expressing two independent CBX8 shRNAs or control.
(I) BCOR and H2AK119ub qChIP was done in cells from (H).
Values in (G) and (I) are shown as means of triplicates or quadruplicates ±SD. t test, *p < 0.05, **p < 0.01, ***p < 0.001. See also Figure S6, Tables S2, and S3.
To determine whether CBX8 might form part of the PRC1-BCOR complex, we performed BCOR tandem affinity purifications followed by mass spectrometry in HEK293 cells and identified CBX8 as a co-purifying protein (Table S2). We also observed CBX8 enrichment after PCFG1 (BCOR complex component) affinity purification (Table S3). PCGF1 was also reported enriched in CBX8 purifications in HeLa cells (Vandamme et al., 2011) and differentiating embryonic stem cells (Creppe et al., 2014), and CBX8 was also associated to a KDM2B-BCOR complex (Sanchez et al., 2007). To confirm these results, we developed an insect cell reconstitution system for BCOR complexes and showed that CBX8 can be incorporated into BCOR complex and immunoprecipitated with BCOR complex components (Figure 6D). A previously described co-structure of CBX7 and RING1B and associated mutational analysis (Wang et al., 2010) allowed us to identify residues that might inhibit the CBX8-RING1B interaction (Figure S6A). Mutation of these residues on RING1B (Y262A) or CBX8 (I375D) resulted in failure to incorporate CBX8 into the BCOR complex (Figure 6D). We confirmed this interaction between CBX8 and the BCOR complex in human cells transduced with tagged WT or mutant CBX8. Immunoprecipitation of WT CBX8 revealed a robust association with endogenous BCOR, as well as the endogenous BCOR complex component PCGF1 (Figure S6B). In contrast, there was no enrichment of BCOR or PCGF1 by mutant CBX8 as it cannot bind RING1B. RING1B binds PCGF1, which interacts directly with BCOR (Junco et al., 2013). To further investigate whether CBX8 associates with BCOR in GC B cells, we performed co-immunoprecipitation experiments for the endogenous CBX8 and BCOR proteins in two DLBCL cell lines. CBX8 antibody enriched for BCOR as did the reciprocal experiment with BCOR and CBX8 immunoblot (Figure 6E). BCOR-CBX8 co-precipitation remained stable in more stringent experiments using higher salt concentrations in the immunoprecipitation buffer (Figure S6C). Most importantly, we confirmed endogenous CBX8 association with BCOR in purified GC B cells from human tonsils (Figure 6F). CBX8 is thus an integral component of the PRC1-BCOR complex in GC B cells in a RING1B-dependent manner.
To evaluate the functional relevance of CBX8, we first examined whether it was recruited to bivalent promoters. CBX8 binding was observed using qChIP assays in four DLBCL cell lines at the same six bivalent promoters evaluated earlier, but not at a negative control locus (Figures 6G and S6D). To determine if CBX8 binding is linked to H3K27me3, we treated these cell lines with the EZH2 inhibitor GSK343 or inactive control GSK669. In all cases, EZH2 inhibition resulted in profound loss of CBX8 recruitment (Figures 6G and S6D). Next, to determine whether CBX8 was necessary for BCOR recruitment, we depleted CBX8 from GC-derived DLBCL cells using two independent shRNA or a control shRNA. Both shRNAs induced significant reduction of CBX8 protein (Figures 6H and S6E) and resulted in decreased BCOR recruitment to bivalent promoters, as well as reduction of the PRC1-BCOR complex catalyzed H2AK119ub histone mark (Figure 6I and S6F), without affecting the abundance of H3K27me3 (Figure S6G).
CBX8 Phenocopies the EZH2 Loss-of-Function Phenotype In Vitro and In Vivo
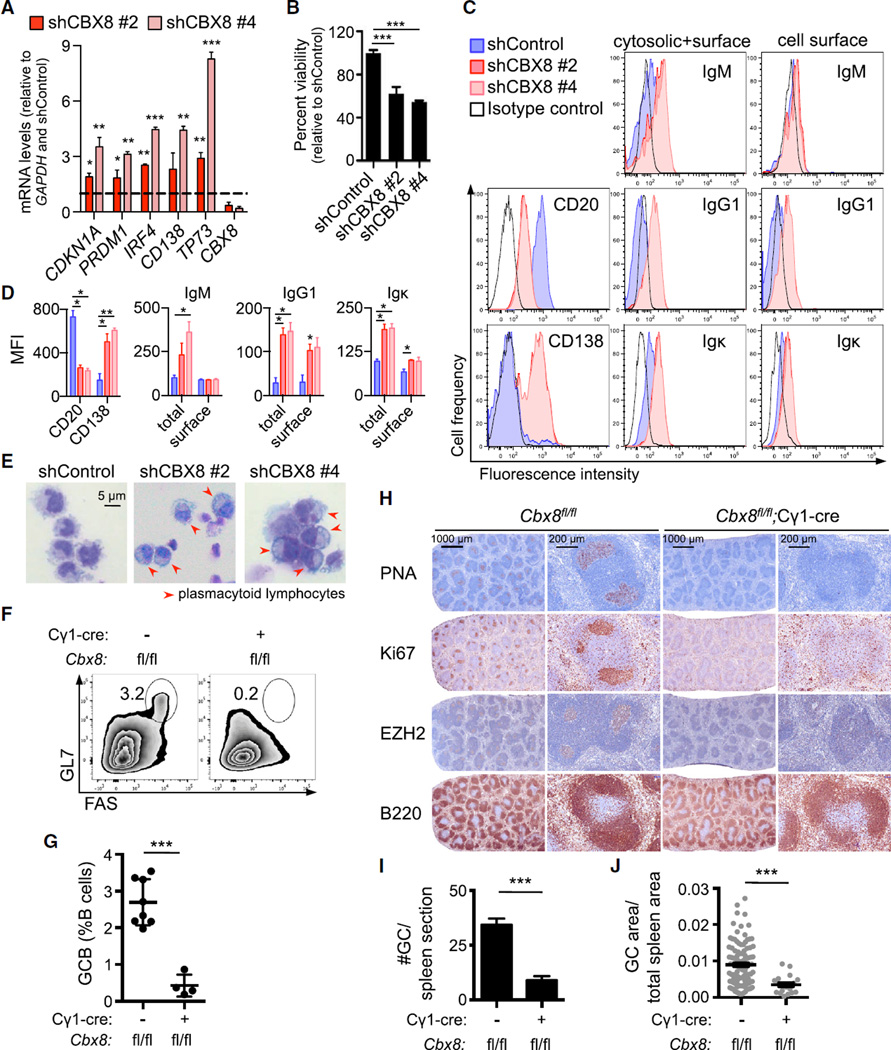
Depletion or inhibition of EZH2 causes growth suppression and plasma cell differentiation in GC-derived DLBCL cells (Beguelin et al., 2013). We observed that CBX8 was required for repression of key EZH2 bivalent genes including CDKN1A, PRDM1, and IRF4, since CBX8 shRNA resulted in their de-repression (Figures 7A and S7A). To determine whether loss of CBX8 mimics the effects of loss of EZH2, we examined the phenotype of DLBCL cells after CBX8 depletion relative to shRNA control in DLBCL cell lines. In all cases, CBX8 loss results in significant growth suppression (p < 0.001; Figures 7B and S7B). We also observed induction of the plasma cell genes PRDM1, TP73, and CD138 by qPCR (Figures 7A and S7A). Plasma cell differentiation was further demonstrated by decreased B cell surface marker CD20, increased plasma cell marker CD138, and surface expression of immunoglobulin heavy and light chains using flow cytometry (Figures 7C, 7D, S7C, and 7D). Morphologically, the DLBCL cells exhibited the characteristic features of plasma cell differentiation, including basophilic cytoplasm, eccentric more condensed nuclei, prominent Golgi apparatus, etc. (Figure 7E). A subset of cell lines also upregulated memory B cell marker CD27 (Figures S7C and S7D).
Figure 7. CBX8 Phenocopies the EZH2 Loss-of-Function Phenotype In Vitro and In Vivo.
(A) RT-qPCR of the indicated mRNAs from OCI-Ly7 cells expressing two independent CBX8 shRNAs or control used in Figures 6H and 6I. p Values shown are compared with the shControl.
(B) Viability of OCI-Ly7 cells from (A) was evaluated 7 days after infection using cell titer blue.
(C) WSU-DLCL2 cells were infected with two independent CBX8 shRNAs or control for 5 days, and CD20, CD138, and immunoglobulin (Ig) expression levels were examined by flow cytometry.
(D) Quantification of mean fluorescence intensity (MFI) from (C) (n = 3).
(E) Representative images of WSU-DLCL2 cells infected as in (C).
(F–J) Cbx8fl/fl (n = 8) and Cbx8fl/fl;Cγ1-cre mice (n = 4) were immunized with SRBC and sacrificed 10 days later. (F) Representative flow cytometry plot of splenic GC B cells as in Figure 1A. (G) Quantification of GC B cells by flow cytometry as in Figure 1B. (H) Splenic tissue was stained for PNA, Ki67, EZH2, and B220. (I and J) Quantification of GC number (I) and area (J) based on PNA staining in (H).
Values in (A) and (B) are shown as means of triplicates or quadruplicates ± SD. Values in (D), (G), (I), and (J) are means ± SEM. t test, *p < 0.05, **p< 0.01, ***p < 0.001. See also Figure S7.
These data suggested that CBX8 is a required component of the PRC1-BCOR complex in GC B cells. To confirm whether this is truly the case, we generated Gbx8fl/fl;Cγ1-cre mice (Tan et al., 2011) and performed immunization experiments to induce GCs. Cbx8 deletion resulted in marked depletion of GC B cells (p < 0.001; Figures 7F and 7G) and significant reduction in the number and volume of GCs (p < 0.001; Figures 7H–7J). Together these results indicate that CBX8 is the component of the PRC1-BCOR complex that tethers the complex to chromatin downstream of the actions of EZH2, thus enabling repression of bivalent promoters and mediating the actions of EZH2 on GC formation.
Mutant EZH2 and Constitutive BCL6 Cooperate to Induce Lymphomagenesis
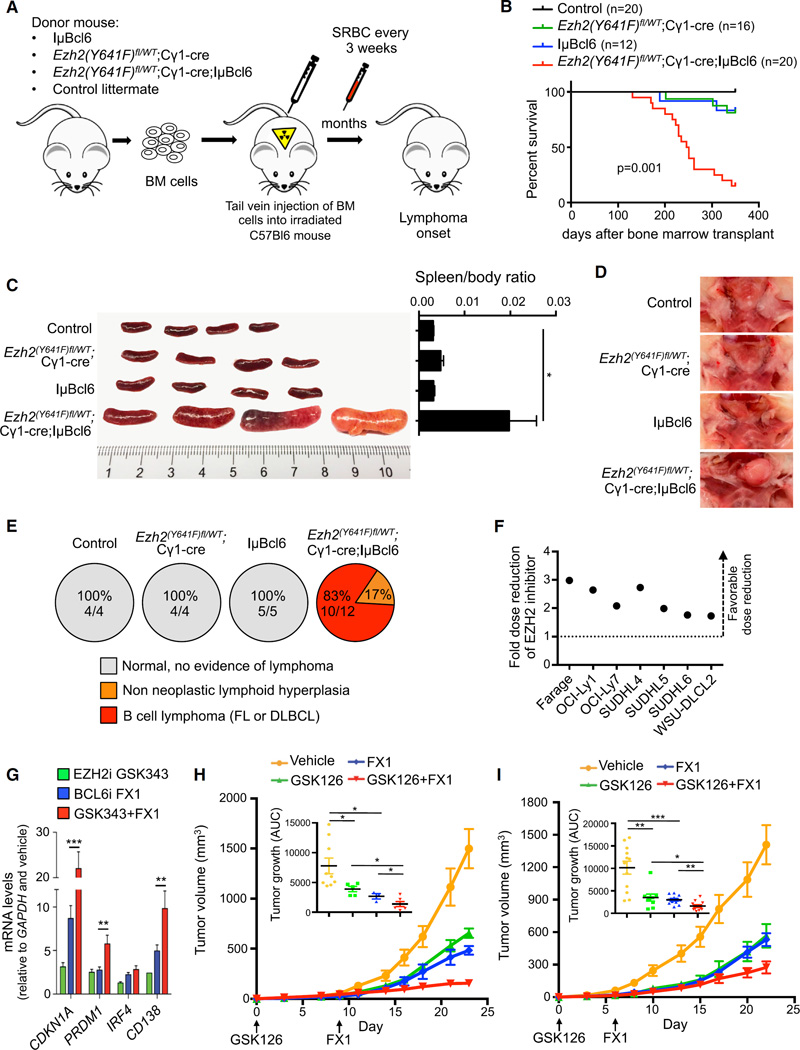
Both EZH2Y641 mutation and BCL6 constitutive expression induce GC hyperplasia. Having established the mechanistic basis for cooperation and interdependence of EZH2 and BCL6 in repressing critical GC B cell genes, we next examined whether their combined gain-of-function alleles might cooperate to drive the transformation of GC B cells to form DLBCLs. BCL6 is constitutively expressed in the GCB-DLBCLs in which EZH2 somatic mutations occur. Therefore, we crossed IµBcl6 with Ezh2(Y641F)fl/WT;Cγ1-cre mice to engineer BCL6 constitutive expression and mutant EZH2 activity in GC B cells. The breeding resulted in four different allele combinations: IµBcl6 alone, Ezh2(Y641F)fl/WT;Cγ1-cre alone, Ezh2(Y641F)fl/WT;Cγ1-cre;IµBcl6, and control littermates. The bone marrow of these four groups were next transplanted to lethally irradiated recipient mice (Figure 8A). Animals were immunized with SRBC every 3 weeks to ensure continuous formation of GCs and were observed for survival. Ezh2(Y641F)fl/WT;Cγ1-cre;IµBcl6 mice showed significant acceleration of lethality compared with IµBcl6 and Ezh2(Y641F)fl/WT;Cγ1-cre mice (p = 0.001; Figure 8B). A second cohort of mice (control n = 4, Ezh2(Y641F)fl/WT;Cγ1-cre n = 4, IµBcl6 n = 5, Ezh2(Y641F)fl/WT;Cγ1-cre;IµBcl6 n = 12) was euthanized 223 days after transplant for more detailed phenotypic analysis. Macroscopic examination of spleens and lymph nodes showed massive splenomegaly and submandibular lymph node enlargement in Ezh2(Y641F)fl/WT;Cγ1-cre;IµBcl6 versus the other groups (p< 0.05; Figure 8C and 8D). Histopathologic examination indicated that, whereas all Ezh2(Y641F)fl/WT; Cγ1-cre;IµBcl6 mice had developed a B cell lymphoma (FL or DLBCL n = 10/12) or pre-neoplastic lymphoid hyperplasia (n = 2/12), by contrast, none of the other groups showed either phenotype at this time point (Figure 8E). Ezh2(Y641F)fl/WT;Cγ1-cre;IµBcl6 mice displayed extensive tissue infiltration by neoplastic, proliferative B220+ B cells (Figures S8A and S8B, and data not shown). Spleens and lymph nodes exhibited effacement of normal architecture by sheets of lymphoma cells (Figure S8A).
Figure 8. Mutant EZH2 and Constitutive BCL6 Cooperate to Induce Lymphomagenesis, and Combinatorial Targeting of EZH2 and BCL6 Yields Enhanced Anti-Lymphoma Effect.
(A) Bone marrow transplantation was performed using IµBcl6, Ezh2(Y641F)fl/WT;Cγ1-cre, Ezh2(Y641F)fl/WT;Cγ1-cre;IµBcl6, and control negative littermate donor mice. BM, bone marrow.
(B) Survival curve of transplanted mice.
(C) Representative pictures of spleens from mice sacrificed 223 days after transplantation and quantification of the spleen weight (control, n=4; Ezh2(Y641F)fl/WT;Cγ1-cre, n = 4; IµBcl6, n = 5; Ezh2(Y641F)fl/WT;Cγ1-cre;IµBcl6, n = 12).
(D) Representative pictures of submandibular lymph nodes from mice sacrificed in (C).
(E) Percentage and number of mice from cohort used in (C) that did or did not develop lymphoma.
(F) Dose reduction plot for GSK343 at 90% growth inhibition after exposure of cells to increasing concentrations of GSK343 for 6 days and FX1 for 2 days. Data represent means of triplicate experiments.
(G) RT-qPCR of the indicated mRNAs from OCI-Ly7 treated with 2 µM GSK343 for 72 hr, 25 µM FX1 for 12 hr, or the combination. Values are means of triplicates ±SD.
(H and I) Tumor growth curves and area under the curve (AUC) for SUDHL6 (H) and WSU-DLCL2 (I) xenografted mice treated with vehicle (SUDHL6, n = 9; WSU-DLCL2, n = 11), GSK126 (80 mg/kg/day, SUDHL6, n = 5; WSU-DLCL2, n = 10), FX1 (12 mg/kg/day, SUDHL6, n = 4; WSU-DLCL2, n = 10), or the combination of GSK126 and FX1 (SUDHL6, n = 6; WSU-DLCL2, n = 12).
Values in (C), (H), and (I) are means ± SEM. t test, *p < 0.05, **p < 0.01, ***p < 0.001. See also Figure S8.
Similar results were obtained when we transduced bone marrow of IµBcl6 mice with retrovirus encoding EZH2Y641F or GFP alone (empty vector) and transplanted them into lethally irradiated recipients (Figure S8C). EZH2Y641F;IµBcl6 mice showed significant acceleration of lethality compared with IµBcl6 transduced mice (p < 0.0001; median survival EZH2Y641F;IµBcl6, 333 days; IµBcl6, 520 days; Figure S8D), with splenomegaly and lymph node enlargement (Figures S8E and S8F). Immunoglobulin gene rearrangement showed more prominent clonal patterning in the B220+ splenocytes of EZH2Y641F;IµBcl6 versus IµBcl6 mice (Figure S8G). Pathologic analysis of these mice yielded massive tissue infiltration by a B220+ B cell lymphoma (FL or DLBCL; Figures S8H and S8I). These data suggest that mutant EZH2 and BCL6 cooperate to induce and accelerate the development of DLBCL-like disease.
BCL6 and EZH2 Inhibitors Cooperate to Kill DLBCLs and Suppress Tumor Xenografts and Primary Human DLBCL Growth
Both BCL6 and EZH2 inhibitors are proposed as potential therapies for patients with B cell lymphomas. BCL6 and EZH2 are both constitutively expressed and required to maintain the growth of GCB-type DLBCLs regardless of whether EZH2 is mutated. Given that BCL6 and EZH2 cooperate to induce maximal repression of their key target promoters, we asked whether simultaneously targeting both proteins might yield enhanced anti-lymphoma activity. We exposed a panel of GCB-DLBCL cells to increasing concentrations of GSK343 in combination with FX1 (along with their respective controls). In almost every case, the concentration of GSK343 required to yield 50% growth inhibition was reduced when cells were concomitantly treated with FX1 (Figures 8F and S8J). This was not due to unexpected toxicity as the combination had no effect on control cell lines (Figure S8K). We also observed that EZH2-BCL6 bivalent target genes were significantly further de-repressed by combining GSK343 and FX1 versus the single drugs (p < 0.01; Figures 8G and S8L). Hence targeting both arms of PRC1-BCOR tethering through EZH2 and BCL6 results in more powerful target gene de-repression with corresponding greater biological activity against lymphoma cells.
To determine the impact of combinatorial BCL6-EZH2 targeted therapy in a preclinical model, we evaluated the action of the EZH2 inhibitor GSK126 and FX1 alone or in combination at submaximal doses in mice bearing established human DLBCL cell line (SUDHL6 and WSU-DLCL2) xenografts. Although both EZH2 and BCL6 inhibitors inhibited tumor growth alone, the combination more potently and significantly suppressed lymphoma growth in vivo as demonstrated by growth curves and tumor weight (Figures 8H, 8I, S8M, and S8N). The combination of BCL6 and EZH2 inhibitors was well tolerated, and there was no significant difference in the gross body weight of animals in the different treatment arms (Figures S8M and S8N). These data were replicated in vitro and in vivo using the structurally distinct peptidomimetic BCL6 inhibitor RI-BPI, and also using the alternative in vivo EZH2 inhibitor GSK503 (Figures S8O–S8Q). Finally, four primary GCB-type DLBCLs were exposed to GSK343 and RI-BPI. The combination of BCL6 and EZH2 inhibitors yielded significantly greater anti-lymphoma activity against these primary specimens (p < 0.01; Figure S8R). Collectively, targeting BCL6 and EZH2 together may provide the basis for rational combinatorial therapies for GC-derived B cell lymphomas.
DISCUSSION
Herein, we show that the GC phenotype and lymphomagenesis are mediated through cooperative and mutually interdependent actions of EZH2 together with the transcriptional repressor BCL6. Our data suggest a scenario whereby, within early GC B cells, BCL6, EZH2, CBX8, BCOR, and other non-canonical PRC1 components are upregulated while canonical PRC1 components are repressed. This allows the formation of a PRC1-BCOR complex containing CBX8. BCL6, a sequence-specific transcription factor, binds to gene promoters mostly through direct binding of its cognate DNA consensus site. At the same time, EZH2 is recruited by still ill-defined mechanisms to a subset of the promoters of genes bound by BCL6, specifically those linked to cell-cycle checkpoints and plasma cell differentiation. EZH2 mediates H3K27 methylation of nucleosomes that were previously marked as active with H3K4me3 in mature resting B cells (Beguelin et al., 2013). EZH2 and BCL6 appear to arrive at these genes independently and do not physically interact. What ensues is a form of combinatorial tethering, whereby the presence of BCL6 and H3K27me3 at bivalent chromatin formed by EZH2 is required for the stable recruitment of the BCOR-CBX8 non-canonical PRC1 complex. However, neither BCL6 binding to BCOR nor CBX8 binding to H3K27me3 alone is sufficient to maintain the association of the BCOR complex. Notably, binding of BCOR to BCL6 occurs with surprisingly low affinity (~20 µM) given the extended binding surface between these two proteins. Among CBX proteins, CBX8 has relatively lower binding affinity for H3K27me3 (Kaustov et al., 2011) and may only result in meta-stable binding. Apparently both of these independent protein interactions are required to sustain and stabilize the association of BCOR with this particular set of key GC B cell promoters. Thus engaged, the PRC1-BCOR-CBX8 complex then mediates PRC1 functions including H2AK119 ubiquitylation, which in turn may contribute to repression of transcription at these loci. Notably, CBX8 has not been previously implicated in B cell biology or lymphomagenesis. The fact that CBX8 loss of function recapitulates the effects of EZH2 loss in normal and malignant GC B cells speaks to its critical function in this cellular context.
Our proposed model of combinatorial tethering conceptually links the proposed instructive versus sampling modes of action for PRC2 functionality (Klose et al., 2013). In this instance, neither instructive (transcription factor directed) nor sampling (PRC2 directed) modes are sufficient to direct PRC1 recruitment and instead must cooperate for cell-context-specific gene repression. Thus PRC2 mediated formation of bivalent chromatin at specific promoters in B cells provides a required link to support a stoichiometrically weak interaction between transcription factors and their corepressors. By the same token, BCL6 binding to BCOR is insufficient to sustain and stabilize its activity and requires the independent action of EZH2. Combinatorial tethering may also help to explain certain puzzling aspects of transcriptional repression. For example, BCL6 is known to bind to many promoters, yet only represses the subset of these where it forms a complex with BCOR (Hatzi et al., 2013). Perhaps the combined actions of BCL6 and H3K27me3 together represent a combinatorial code that limits the formation of competent repression complexes only to sites relevant to GC B cells. Indeed, the genes where this combinatorial mechanism occurs are critical for the GC phenotype, such as CDKN1A, CDKN1B, IRF4, PRDM1, etc. In contrast, BCL6 and BCOR are mostly excluded from monovalent H3K27me3 domains, and hence are not involved in repressing these regions.
It remains possible that PRC1, PRC2, and BCL6 function to mutually sustain and stabilize their respective binding in bivalent promoters. Along these lines, Kalb et al. (2014) identified a positive feedback loop in which H2Aub promotes PRC2 binding and H3K27 trimethylation, and H3K27me3, in turn, promotes binding of canonical PRC1. H2A ubiquitylation mediated by non-canonical complexes was shown to facilitate recruitment of PRC2 in embryonic stem cells engineered to contain a Polycomb tethering sequence (Blackledge et al., 2014). Recruitment of PRC2 was also reported to occur through canonical PRC1-mediated H2A ubiquitylation (Cooper et al., 2014). Indeed, our data hint at additional aspects of Polycomb functionality. For example, the finding that disruption of the BCL6-BCOR interaction reduces EZH2-mediated H3K27me3 without loss of PRC2 complex binding could be linked to EZH2 requirement for H2A ubiquitylation; or to impaired PRC2 function due to increasing H3K36 methylation because of loss of KDM2B (Yuan et al., 2011). Indeed, H3K36me2 was increased after treating three different GCB-DLBCL cell lines with GSK343 or after disrupting BCL6-BCOR interaction with FX1 (Figure S8S). Collectively, the combinatorial tethering model expands notions on how transcription factors and Polycomb complexes can work integratively to direct gene-specific repression. It is important to also underline that these results do not explain all the effects of BCL6, EZH2, and BCOR in B cells. For example, BCOR binds to many promoters independent of the presence of BCL6. It was recently shown in embryonic stem cells that KDM2B, a subunit of the PRC1-BCOR complex, can contribute to recruitment of non-canonical PRC1 complexes through binding of its CXXC motif to unmethylated CpG islands (Farcas et al., 2012; Wu et al., 2013). Our analysis of BCOR distribution in GC B cells suggests that a similar mechanism may be at play at different sets of target genes independent of the BCL6-EZH2 mechanism described herein (Hatzi et al., 2013).
Constitutive expression of BCL6 as well as somatic mutation of EZH2 can prevent the resolution of the GC phenotype, potentially explaining how they induce lymphomas. Here, we show that this is at least in part linked to their common action in the combinatorial tethering of the non-canonical PRC1-BCOR complex to bivalent chromatin domains formed during the humoral immune response. The enhanced anti-lymphoma activity observed by combining EZH2 with BCL6 inhibitors is likely at least in part due to more profound disruption of bivalent gene repression, since the combination results further increased expression of these transcripts. Administration of BCL6 and EZH2 inhibitors may thus constitute a mechanism-oriented rational combinatorial therapy by disabling both arms of the PRC1-BCOR tethering mechanism.
EXPERIMENTAL PROCEDURES
A more detailed description of the experimental procedures and reagents used in this study can be found in the Supplemental Experimental Procedures.
The Research Animal Resource Center of the Weill Cornell Medical College of Medicine approved all mouse procedures. The use of human tissue was approved by the research ethics board of the Vancouver Cancer Center/University of British Columbia and the Weill Cornell Medical Center.
Generation of Conditional Ezh2Y641F Knockin Mice
Ezh2Y641F mice were generated by inGenious Targeting Laboratory using a mini-gene approach. A mini-gene comprising exons 16–20 and flanked by loxP sites was inserted into intron 15–16 of Ezh2 endogenous allele. The point mutation (TAC→TTC; aa, T > P) was engineered in exon 16 as indicated in the diagram in Figure 2A. The FRT-flanked Neo cassette followed by a LoxP site was inserted immediately downstream of the mini-gene. The mini-gene is composed of a LoxP site and 3.9 kb genomic sequence spanning from 3′ intron 15 to exon 20 including 200–300 bps of intronic sequence and the 3′UTR to ensure correct splicing and processing of the transcript. The mini-gene was inserted into intron 15 and is 492 bp upstream of exon 16. Prior to cre expression, the WT product is generated from the mini-gene. CRE-mediated deletion of the mini-gene results in expression of the mutant form.
Statistics
Pairwise comparisons of cell numbers, GC and tumor phenotypes, qPCR, and qChIP were assessed using Student’s t test. Enrichment of ChIP-seq marks in promoters and pathway enrichment were determined using the hypergeometric test. Gene set enrichment was assessed using the GSEA algorithm, a computational method based on the Kolmogorov-Smirnov test. Significance of survival probability was determined using the Kaplan-Meier estimator and Cox proportional hazards test.
Supplementary Material
In Brief.
Béguelin et al. show that EZH2 and BCL6 cooperate to recruit a non-canonical PRC1/BCOR complex containing CBX8 to repress differentiation gene expression in germinal center B cells and promote lymphomagenesis. Targeting both BCL6 and EZH2 elicits strong anti-lymphoma activity in diffuse large B cell lymphoma.
Highlights.
EZH2 and BCL6 mediate combinatorial tethering of non-canonical PRC1-BCOR complex
CBX8 binding to bivalent promoters enables GC B cell-specific PRC1-BCOR recruitment
BCOR tethering by EZH2 activity and BCL6 is required for GC and drives GC hyperplasia
Combinatorial targeting of EZH2 and BCL6 yields enhanced anti-lymphoma effect
Significance.
The humoral immune response requires transient silencing of B cell differentiation and cell-cycle checkpoint genes to permit immunoglobulin affinity maturation. Aberrant persistence of this state causes malignant transformation. We show that silencing depends on a combinatorial tethering mechanism that enables recruitment of the non-canonical PRC1-BCORCBX8 complex to bivalent gene promoters methylated by EZH2. CBX8 anchors these proteins to bivalent promoter H3K27me3, while the transcriptional repressor BCL6 directly binds to DNA and BCOR and stabilizes PRC1-BCOR complex in an EZH2-independent manner. Combinatorial tethering explains how transcription factors, PRC2, and non-canonical PRC1 intersect on chromatin to mediate context-specific transient silencing of active promoters. It also provides the basis for improved lymphoma therapy through rational combination of BCL6 and EZH2 inhibitors.
Acknowledgments
We thank Dr. Alexander Tarakhovsky (Rockefeller University) for sharing the Ezh2 conditional knockout mouse strain, Dr. Ricardo Dalla-Favera (Columbia University) for the IµBcl6 strain, Dr. Haruhiko Koseki (Riken Center, Japan) for sharing the Cbx8 conditional knockout mouse, and Dr. Sebastien Monette for the mouse pathology reports. We thank GlaxoSmithKline for GSK compounds and Fengtian Xue (University of Maryland) for the synthesis of FX1 compound. W.B. was supported by a Charles Revson Senior Fellowship in Biomedical Science and an American Society of Hematology Scholar Award. M.Y.H was supported by NIH 5F30HL093996, NIH 5T32GM008244, AHA 0810194Z, and the University of Minnesota Wetzel Fund. V.J.B. is supported by NCI R01 CA071540 and funds from the Minnesota Masonic Charities, and the University of Minnesota Medical School and Office of the Vice President for Research. A.M. is supported by NCI R01 CA187109 and LLS TRP 6141-14. A.M. is a consultant to Epizyme, and receives research funding (unrelated to the topic of this manuscript) from Janssen, Eli Lilly, GSK, and Roche.
Footnotes
ACCESSION NUMBERS
The Gene Expression Omnibus accession numbers for the ChIP-seq, RNA-seq, and the expression microarray data reported in this paper are GEO: GSE45982, GSE23501, and GSE81705.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, eight figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ccell.2016.07.006.
AUTHOR CONTRIBUTIONS
Conceptualization, W.B. and A.M.M.; Methodology, W.B., M.D.G., V.J.B., and A.M.M.; Software, Formal Analysis, and Data Curation, M.T. and O.E.; Investigation, W.B., M.D.G., M.T.C.F., R.L.G., K.H., M.R., H.S., C.M.C., M.Y.H., O.A.W., and R.S.; Resources, M.G.C., R.D.G., R.L.L., O.A.W., and J.D.L.; Writing – Original Draft, W.B. and A.M.M.; Writing – Review & Editing, W.B., M.D.G., V.J.B., and A.M.M.; Visualization, W.B. and M.T.; Funding Acquisition, V.J.B. and A.M.M.
REFERENCES
- Beguelin W, Popovic R, Teater M, Jiang Y, Bunting KL, Rosen M, Shen H, Yang SN, Wang L, Ezponda T, et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell. 2013;23:677–692. doi: 10.1016/j.ccr.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LL, Ito S, Cooper S, Kondo K, Koseki Y, et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157:1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor C, Grossmann V, Popov N, Okosun J, O’Riain C, Tan K, Marzec J, Araf S, Wang J, Lee AM, et al. EZH2 mutations are frequent and represent an early event in follicular lymphoma. Blood. 2013;122:3165–3168. doi: 10.1182/blood-2013-04-496893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caganova M, Carrisi C, Varano G, Mainoldi F, Zanardi F, Germain PL, George L, Alberghini F, Ferrarini L, Talukder AK, et al. Germinal center dysregulation by histone methyltransferase EZH2 promotes lymphomagenesis. J. Clin. Invest. 2013;123:5009–5022. doi: 10.1172/JCI70626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas MG, Yu W, Beguelin W, Teater M, Geng H, Goldstein RL, Oswald E, Hatzi K, Yang S, Cohen J, et al. Therapeutic targeting of GCB- and ABC-DLBCLs by rationally designed BCL6 inhibitors. J. Clin. Invest. 2016;126 doi: 10.1172/JCI85795. http://dx.doi.org/10.1172/JCI85795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola S, Cattoretti G, Uyttersprot N, Koralov SB, Seagal J, Hao Z, Waisman A, Egert A, Ghitza D, Rajewsky K. Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc. Natl. Acad. Sci. USA. 2006;103:7396–7401. doi: 10.1073/pnas.0602353103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoretti G, Pasqualucci L, Ballon G, Tam W, Nandula SV, Shen Q, Mo T, Murty VV, Dalla-Favera R. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7:445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Cerchietti LC, Ghetu AF, Zhu X, Da Silva GF, Zhong S, Matthews M, Bunting KL, Polo JM, Fares C, Arrowsmith CH, et al. A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell. 2010;17:400–411. doi: 10.1016/j.ccr.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ci W, Polo JM, Melnick A. B-cell lymphoma 6 and the molecular pathogenesis of diffuse large B-cell lymphoma. Curr. Opin. Hematol. 2008;15:381–390. doi: 10.1097/MOH.0b013e328302c7df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S, Dienstbier M, Hassan R, Schermelleh L, Sharif J, Blackledge NP, De Marco V, Elderkin S, Koseki H, Klose R, et al. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep. 2014;7:1456–1470. doi: 10.1016/j.celrep.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creppe C, Palau A, Malinverni R, Valero V, Buschbeck M. A Cbx8-containing polycomb complex facilitates the transition to gene activation during ES cell differentiation. PLoS Genet. 2014;10:e1004851. doi: 10.1371/journal.pgen.1004851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farcas AM, Blackledge NP, Sudbery I, Long HK, McGouran JF, Rose NR, Lee S, Sims D, Cerase A, Sheahan TW, et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife. 2012;1:e00205. doi: 10.7554/eLife.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearhart MD, Corcoran CM, Wamstad JA, Bardwell VJ. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol. Cell Biol. 2006;26:6880–6889. doi: 10.1128/MCB.00630-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetu AF, Corcoran CM, Cerchietti L, Bardwell VJ, Melnick A, Prive GG. Structure of a BCOR corepressor peptide in complex with the BCL6 BTB domain dimer. Mol. Cell. 2008;29:384–391. doi: 10.1016/j.molcel.2007.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzi K, Melnick A. Breaking bad in the germinal center: how deregulation of BCL6 contributes to lymphomagenesis. Trends Mol. Med. 2014;20:343–352. doi: 10.1016/j.molmed.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzi K, Jiang Y, Huang C, Garrett-Bakelman F, Gearhart MD, Giannopoulou EG, Zumbo P, Kirouac K, Bhaskara S, Polo JM, et al. A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Rep. 2013;4:578–588. doi: 10.1016/j.celrep.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Hatzi K, Melnick A. Lineage-specific functions of Bcl-6 in immunity and inflammation are mediated by distinct biochemical mechanisms. Nat. Immunol. 2013;14:380–388. doi: 10.1038/ni.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh KD, Fischle W, Verdin E, Bardwell VJ. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev. 2000;14:1810–1823. [PMC free article] [PubMed] [Google Scholar]
- Junco SE, Wang R, Gaipa JC, Taylor AB, Schirf V, Gearhart MD, Bardwell VJ, Demeler B, Hart PJ, Kim CA. Structure of the polycomb group protein PCGF1 in complex with BCOR reveals basis for binding selectivity of PCGF homologs. Structure. 2013;21:665–671. doi: 10.1016/j.str.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb R, Latwiel S, Baymaz HI, Jansen PW, Muller CW, Vermeulen M, Muller J. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol. 2014;21:569–571. doi: 10.1038/nsmb.2833. [DOI] [PubMed] [Google Scholar]
- Kaustov L, Ouyang H, Amaya M, Lemak A, Nady N, Duan S, Wasney GA, Li Z, Vedadi M, Schapira M, et al. Recognition and specificity determinants of the human cbx chromodomains. J. Biol. Chem. 2011;286:521–529. doi: 10.1074/jbc.M110.191411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Cooper S, Farcas AM, Blackledge NP, Brockdorff N. Chromatin sampling-an emerging perspective on targeting polycomb repressor proteins. PLoS Genet. 2013;9:e1003717. doi: 10.1371/journal.pgen.1003717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, Sacks JD, Raimondi A, Majer CR, Song J, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat. Chem. Biol. 2012;8:890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A, 3rd, Diaz E, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaphorst FM, van Kemenade FJ, Fieret E, Hamer KM, Satijn DP, Otte AP, Meijer CJ. Cutting edge: polycomb gene expression patterns reflect distinct B cell differentiation stages in human germinal centers. J. Immunol. 2000;164:1–4. doi: 10.4049/jimmunol.164.1.1. [DOI] [PubMed] [Google Scholar]
- Sanchez C, Sanchez I, Demmers JA, Rodriguez P, Strouboulis J, Vidal M. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol. Cell Proteomics. 2007;6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, Copeland RA. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc. Natl. Acad. Sci. USA. 2010;107:20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol. 2003;4:124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- Tan J, Jones M, Koseki H, Nakayama M, Muntean AG, Maillard I, Hess JL. CBX8, a polycomb group protein, is essential for MLL-AF9-induced leukemogenesis. Cancer Cell. 2011;20:563–575. doi: 10.1016/j.ccr.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme J, Volkel P, Rosnoblet C, Le Faou P, Angrand PO. Interaction proteomics analysis of polycomb proteins defines distinct PRC1 complexes in mammalian cells. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.002642. M110.002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velichutina I, Shaknovich R, Geng H, Johnson NA, Gascoyne RD, Melnick AM, Elemento O. EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood. 2010;116:5247–5255. doi: 10.1182/blood-2010-04-280149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Taylor AB, Leal BZ, Chadwell LV, Ilangovan U, Robinson AK, Schirf V, Hart PJ, Lafer EM, Demeler B, et al. Polycomb group targeting through different binding partners of RING1B C-terminal domain. Structure. 2010;18:966–975. doi: 10.1016/j.str.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Johansen JV, Helin K. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol. Cell. 2013;49:1134–1146. doi: 10.1016/j.molcel.2013.01.016. [DOI] [PubMed] [Google Scholar]
- Ye BH, Chaganti S, Chang CC, Niu H, Corradini P, Chaganti RS, Dalla-Favera R. Chromosomal translocations cause deregulated BCL6 expression by promoter substitution in B cell lymphoma. EMBO J. 1995;14:6209–6217. doi: 10.1002/j.1460-2075.1995.tb00311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 2011;286:7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.