Abstract
We have examined the lipids of three isolates, Romboutsia lituseburensis, Romboutsia ilealis, and Romboutsia sp. strain FRIFI, of the newly described genus Romboutsia by two-dimensional thin-layer chromatography (2D-TLC) and by liquid chromatography/mass spectrometry (LC/MS). We have found three phospholipids, phosphatidylglycerol (PG), cardiolipin and phosphatidic acid in all three species. A fourth phospholipid, lysyl-PG, was found in R. lituseburensis and strain FRIFI. Polyprenyl-phosphates were identified in the lipid extracts of all three species. Three glycolipids, mono-, di- and tri-hexosyldiacylglycerol, were common to all three species. An additional glycolipid, tetrahexosyl-diacylglycerol was identified in strain FRIFI. Acylated trihexosyldiacylglycerol and acyl-tetrahexosydiacylglycerol were also found in R. ilealis and strain FRIFI. Remarkably, no alk-1-enyl ether lipids (plasmalogens) were present in Romboutsia as distinct from bacteria of the related genus Clostridium in which these ether lipids are common. We have compared the lipidome of Romboutsia with that recently described for Clostridium difficile, which has plasmalogens, no lysyl-PG, and no tetrahexosyl-diacylglycerol. According to 16S rRNA gene sequencing, Romboutsia spp. and Cosmarium difficile are closely related (>95% sequence identity).
Keywords: Romboutsia, Glycolipid, Mass spectrometry, Phospholipid, Thin-layer chromatography
1. Introduction
A Gram-positive, rod-shaped, non-motile, spore-forming obligately anaerobic bacterium isolated from the rat gastro-intestinal tract was recently characterized and classified in a new genus, Romboutsia. On the basis of 16S rRNA gene sequencing and its low G + C content, 28.1 mol%, the new strain, named Romboutsia ilealis, was considered to be closely related to several species of the genus Clostridium, including Clostridium lituseburense (97.2%), Clostridium glycolicum (96.2%), Clostridium irregulare (95.5%) and two other species of clostridia. It was therefore decided to transfer C. lituseburense to this new genus and to reclassify C. glycolicum and C. irregulare into new genera [1]. Furthermore, strain FRIFI was recently isolated from human ileostoma effluent and was shown to represent a new species within the genus Romboutsia [2]. As part of this study, the polar lipids were examined by two-dimensional thin-layer chromatography (2D-TLC) with appropriate staining. Two of the major phospholipids were tentatively identified as cardiolipin (diphosphatidylglycerol) and phosphatidylglycerol (PG). Several other phospholipids and a number of glycolipids were detected, but not identified. It was noted that unlike species of Clostridium cluster I previously studied [3–9], there was no phosphatidylethanolamine. Since we have recently described the polar lipids of C. difficile which is related to the newly reclassified R. lituseburensis [10], we have carried out a 2D-TLC and mass spectrometric lipidomic analysis of three species in this new genus. We have identified PG, cardiolipin (CL) and phosphatidic acid (PA) in all three species. In addition we have identified lysyl-PG in two of the three species, Romboutsia sp. strain FRIFI and R. lituseburensis, but not in R. ilealis. Monohexosyldiacylglycerol (MHDAG), dihexosyldiacylglcyerol (DHDAG) and trihexosyldiacylglycerol (TriHDAG) were observed in all three species. Strain FRIFI contains a tetrahexosyldiacylglycerol (TetraHDAG) in addition to the other three glycosyldiacylglycerols. Acylated trihexosyldiacylglycerol and acyl-tetrahexosyldiacylglycerol were also identified in R. ilealis and strain FRIFI. Mass spectrometry revealed the presence of decaprenyl and/or nonaprenyl (C50 and C45)-P in all three species.
2. Methods
2.1. Bacterial strains and growth conditions
R. lituseburensis DSM 797T, R. ilealis CRIBT (DSM 25109) and Romboutsia sp. strain FRIFI (DSM 28814) were grown in liquid DSM medium 104b at 37 °C. This medium consisted of (per litre distilled water): 5 g trypticase peptone, 5 g peptone from meat (pepsin-digested), 10 g yeast extract, 5 glucose, 1 mg resazurin, 40 ml salt solution [11] and 0.5 g L-cysteine hydrochloride (pH 7). Cells were harvested in mid-exponential to end-exponential phase and lyophilized.
2.2. Lipid extraction and thin-layer chromatography (TLC)
Total lipids were extracted from the lyophilized cells after addition of 0.5 to 1.0 ml of water using chloroform-methanol [12] with minor modifications [13]. Two-dimensional TLC was carried out on silica gel 60, 10 × 10 cm, thin-layer plates using the following solvent systems: System A, chloroform/methanol/concentrated ammonia/water, 65:30:2.5:2.5 (by vol.) in the first dimension and System B, chloroform/methanol/acetic acid/water, 80:18:12:5 (by vol.) in the second dimension. Amine-containing lipids were detected with 0.3% ninhydrin in ethanol, followed by heating at 120 °C for 10 min. On the same plates, phospholipids were detected with 0.3% (w/v) molybdenum blue. After recording the results at each step, the lipids were charred by heating at 120 °C for 15 min. On separate plates, glycolipids were detected by α-naphthol staining [14]. All reagents were obtained from Sigma-Aldrich, St. Louis, MO.
Preparative TLC of strain FRIFI total lipids was performed on a 10 × 10 cm silica gel 60 TLC plate. The lipids were chromatographed in solvent system A and all but the left and right edges was scraped in 1 cm bands which were eluted as described above for extraction of cellular lipids. The remaining left edge was stained for glycolipids with α-naphthol and the remaining right edge was stained for phospholipids with molybdenum blue reagent and charred.
2.3. Liquid chromatography/tandem mass spectrometry (LC/ESI–MS/MS)
Methods for LC/ESI–MS/MS have been described [3,15]. Briefly, normal phase LC was performed on an Agilent 1200 Quaternary LC system equipped with an Ascentis Silica HPLC column, 5 µm, 25 cm × 2.1 mm (Sigma-Aldrich, St. Louis, MO) as described. The LC eluent (with a total flow rate of 300 µl/min) was introduced into the ESI source of a high resolution TripleTOF5600 mass spectrometer (Applied Biosystems, Foster City, CA). Instrumental settings for negative ion ESI and MS/MS analysis of lipid species were as follows: IS = −4500 V; CUR = 20 psi; GSI = 20 psi; DP = −55 V; and FP = −150 V. The MS/MS analysis used nitrogen as the collision gas. Data analysis was performed using Analyst TF1.5 software (Applied Biosystems, Foster City, CA).
3. Results
3.1. Identification of diacylglycerol and phospholipids
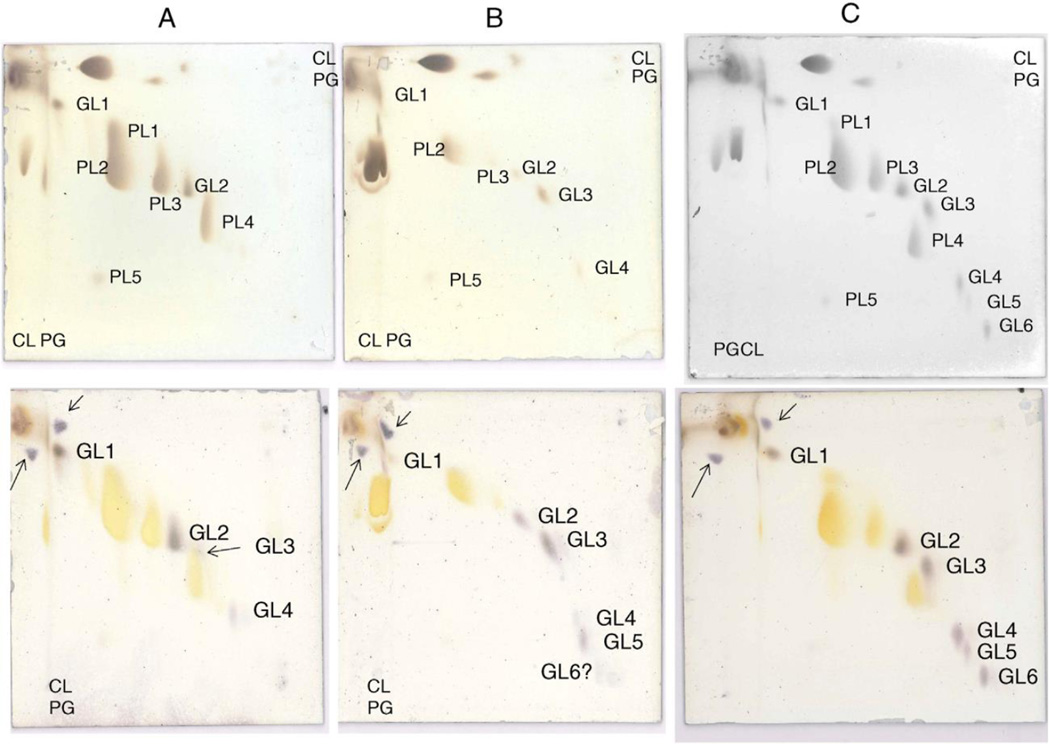
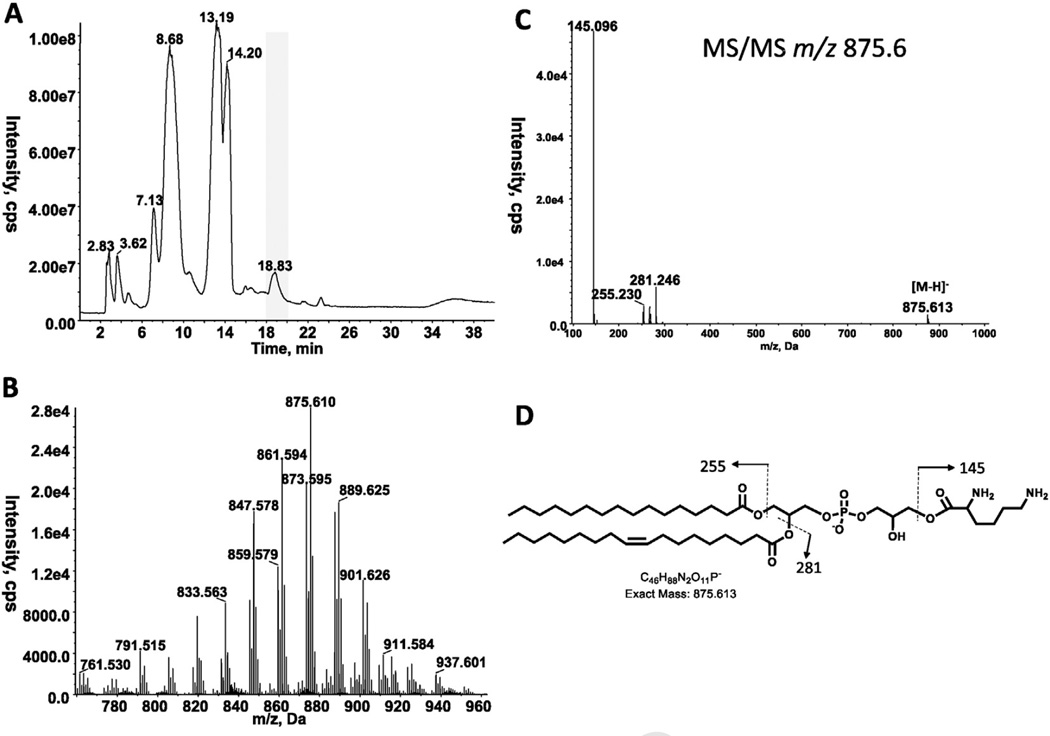
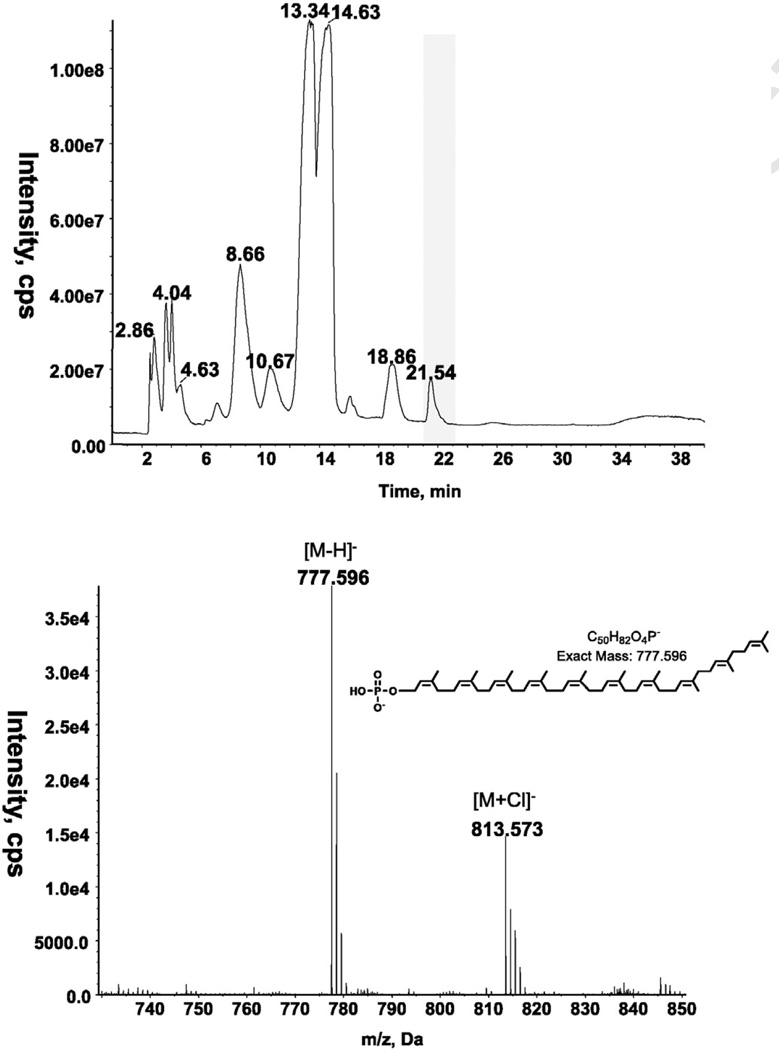
Diacylglycerol was identified by LC/MS in all three species of Romboutsia (Tables 1 and 2). As described previously, 2DTLC revealed the presence of several phospholipids and glycolipids in R. ilealis and R. lituseburensis, but some differences between species were noted [1]. We have now analyzed the polar lipids of three species, R. lituseburensis, R. ilealis and strain FRIFI, by 2D-TLC using solvent systems that differ from those used in the previous study. PG and CL were identified in all three species (Fig. 1) and confirmed by LC/MS (Table 2). A major ninhydrin and molybdate-positive spot was seen in R. lituseburensis and strain FRIFI, but not in R. ilealis. Its position relative to other lipids corresponds to that of lysyl-PG (Fig. 1) [7]. The presence of lysyl-PG was confirmed by LC/ESIMS and by MS/MS (Fig. 2). As found by Gerritsen et al. [1], no phosphatidylethanolamine is present in any of the three species. Fig. 1 also shows a spot (PL5) corresponding to a lipid that was previously identified as PA [7]. The presence of PA was confirmed by mass spectrometry of strain FRIFI and R. ilealis lipids (Table 2). Mass spectrometry revealed the presence of decaprenyl-P (C50-P) or nonaprenyl-P (C45-P) in all three species. The latter was predominant in R. lituseburensis. A representative spectrum of the lipids from strain FRIFI is shown in Fig. 3.
Table 1.
Romboutsia lipids identified by 2D-TLC and LC/MS/MS.
| Lipid | R. lituseburensis | R. ilealis | strain FRIFI |
|---|---|---|---|
| Diacylglycerol | + | + | + |
| Phosphatidylglycerol | + | + | + |
| Cardiolipin | + | + | + |
| Lysyl-PG | + | − | + |
| MHDAGa | + | + | + |
| DHDAGa | + | + | + |
| TriHDAGa | + | + | + |
| TetraHDAGa | − | tr | + |
| Acyl-TriHDAG | + | + | + |
| Acyl-TetraHDAG | − | + | + |
| Polyprenyl-P | + | + | + |
| Phosphatidic acid | + | + | + |
MHDAG, DHDAG, TriHDAG and TetraHDAG are mono-, di-, tri-, and tetra-hexosyldiacylglycerol, respectively.
Table 2.
Majormolecular species of Romboutsia diacylglycerol and phospholipids seen by negative ion ESI/MS.
| Major molecular speciesa | [M-H]−b | R. lituseburensis | R. ilealis | strain FRIFI |
|---|---|---|---|---|
| DAG, 31:1 | 587.443 | + | + | + |
| DAG, 32:1 | 601.459 | + | + | + |
| DAG, 33:1 | 615.474 | + | + | + |
| DAG, 34:1 | 629.490 | + | + | + |
| DAG, 35:1 | 643.505 | + | + | |
| PA, 31:0 | 633.449 | c | + | |
| PA 32:1 | 645.450 | c | + | + |
| PA 33:1 | 659.465 | c | + | |
| PA 33:0 | 661.481 | c | + | |
| PA 34:1 | 673.481 | c | + | + |
| PA 34:0 | 675.496 | c | + | |
| PA 35:1 | 687.497 | c | + | + |
| PG 32:1 | 719.488a | + | + | + |
| PG 33:1 | 733.503 | + | + | + |
| PG 34:1 | 747.518 | + | + | + |
| PG 35:1 | 761.533 | + | + | |
| CL 60:1 | 1293.886 | + | ||
| CL 61:1 | 1307.902 | + | ||
| CL 63:2 | 1333.916 | + | + | |
| CL 64:2 | 1347.933 | + | + | + |
| CL 65:2 | 1361.949 | + | + | + |
| CL 66:2 | 1375.964 | + | + | + |
| CL 67:2 | 1389.980 | + | ||
| CL 68:2 | 1403.995 | + | + | |
| CL 69:1 | 1418.011 | + | ||
| CL 69:3 | 1415.996 | + | ||
| Lysyl-PG 31:1 | 833.563 | + | ||
| Lysyl-PG 32:1 | 847.578 | + | + | |
| Lysyl-PG 33:1 | 861.540 | + | + | |
| Lysyl-PG 34:1 | 875.610 | + | + | |
| Lysyl-PG 35:2 | 887.611 | + | ||
| Lysyl-PG 35:1 | 889.625 | + | + |
DAG, diacylglycerol; PA, phosphatidic acid; PG, phosphatidylglycerol; CL, cardiolipin.
These m/z values are for strain FRIFI. The values for other species may vary slightly.
Although PA has been detected in R. lituseburensis nomass spectral data are available.
Fig. 1.
Analysis by 2D-TLC of lipids from: A) R. lituseburensis, B) R. ilealis, and C) strain FRIFI. The upper plates were stained with ninhydrin and after recording, with molybdenum blue followed by charring and the lower plates were stained with α-naphthol. The major lipids were identified by their staining properties and confirmed by LC/MS as follows: PL1, PG; PL2, CL; PL3, PG; PL4, lysyl-PG, and PL5, PA. The glycolipids were initially shown by staining with α-naphthol and their identities revealed by LC/MS as follows: GL1, MHDAG, GL2, DHDAG; GL3, acyl-TriHDAG, GL4, TriHDAG; GL5, acyl-TetraHDAG; and GL6, TetraHDAG. Lysyl-PG was ninhydrin-positive. Standards of PG and CL were applied to the left margin and in the upper right hand corner, upper plates and a standard of MHDAG was applied to the left margin and upper right hand corner of the lower plates stained with α-naphthol. The resulting spots are shown with arrows.
Fig. 2.
LC/MS/MS identification of lysyl-PG. A. Total ion chromatogram of lipids fromR. lituseburensis. B. Negative ion ESI/MS spectrum showing the [M-H]− ions of the lipid emerging at 18 to 20 min. C. MS/MS spectrum of [M-H]− at m/z 875.6. D. The product ion at m/z 145 corresponds to the lysine residue.
Fig. 3.
LC/MS identification of decaprenyl (C50)-P from strain FRIFI. A. Total ion chromatogram of lipids from strain FRIFI (top) and negative ion ESI/MS spectrum showing the [M-H]− and [M+Cl]− ions of the lipid emerging at 21 to 22.5min (bottom).
3.2. Identification of glycolipids
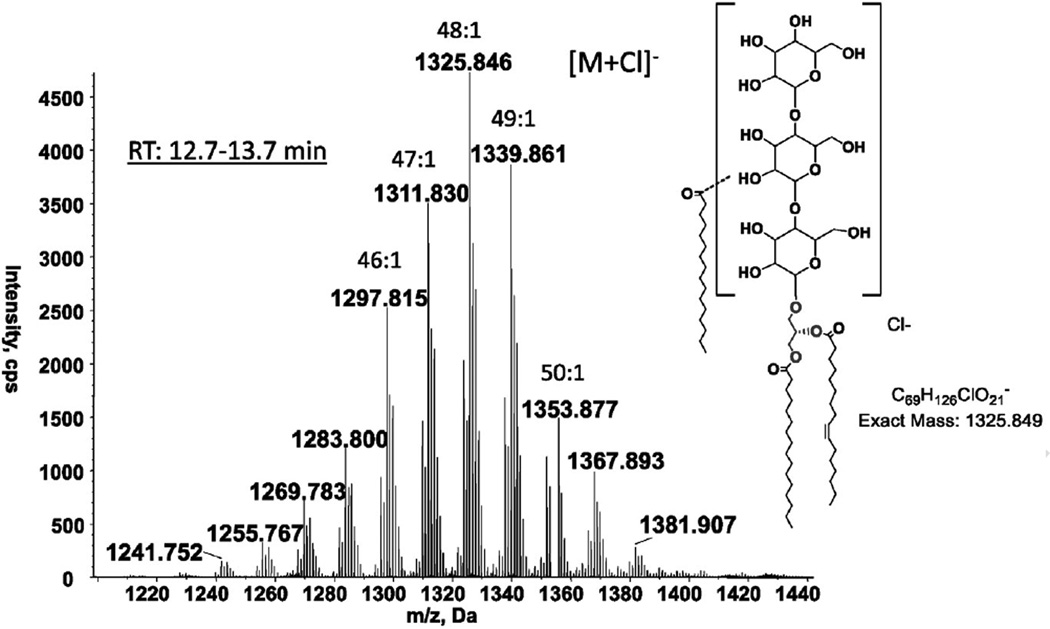
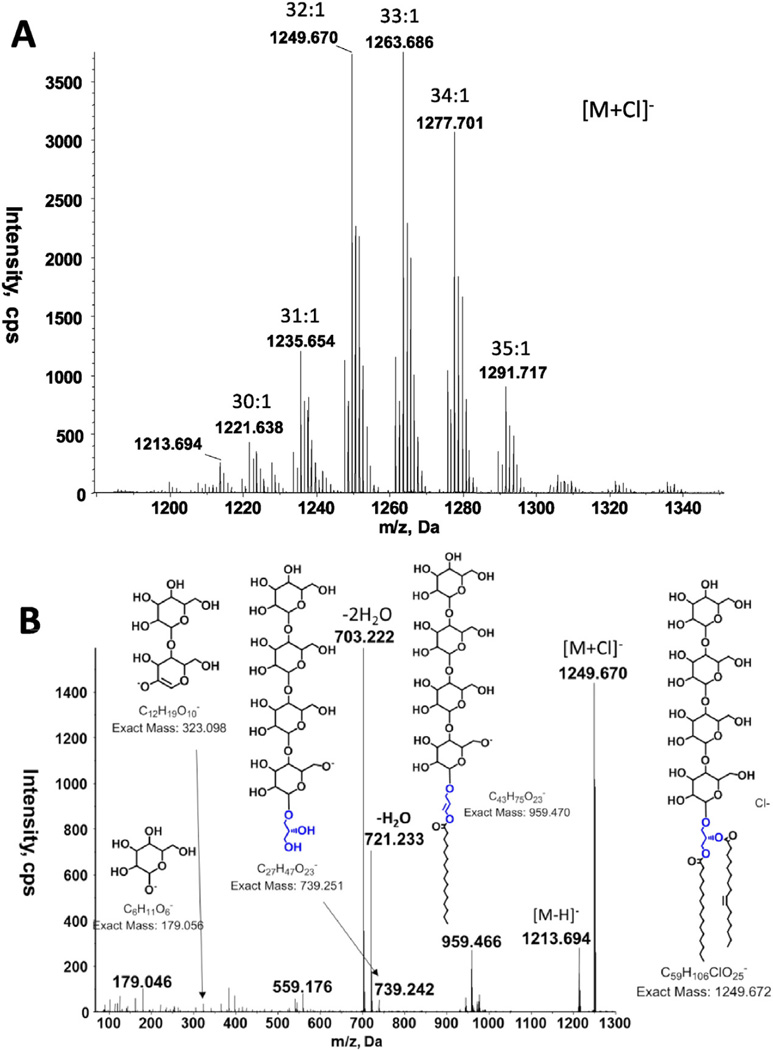
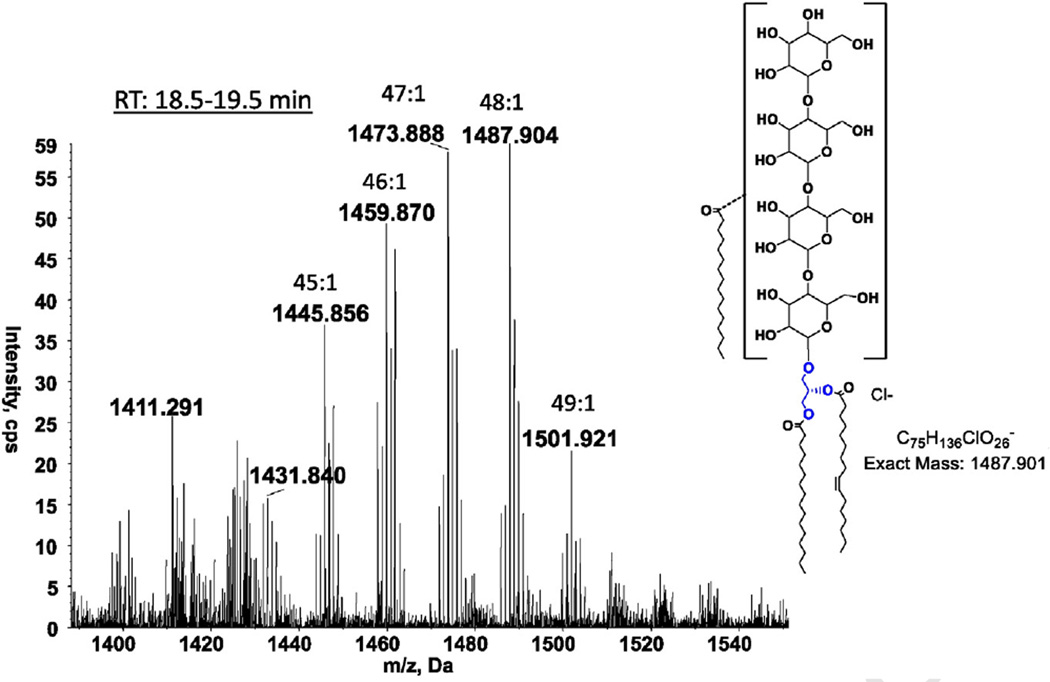
Three glycolipids, mono-, di- and tri-hexosyldiacylglycerol, were common to all three species of Romboutsia (Fig. 1, GL1, GL2 and GL4). All three species had another glycolipid designated as GL3 (Fig. 1), with only a faint hint of this lipid in R. lituseburensis visible after α-naphthol staining. Preparative TLC of strain FRIFI lipids followed by LC/ESI–MS revealed the presence of an acyl-TriHDAG in the band corresponding to GL3 (Fig. 4). After preparative TLC a tetrahexosyldiacylglycerol (TetraHDAG) was identified by accurate mass measurements and MS/MS (Fig. 5) in the fraction corresponding to GL6 in Fig. 1C. Lastly, a glycolipid isolated by preparative TLC in the band corresponding to GL5 (Fig. 1C) was identified as an acylated (primarily C16:0) derivative of TetraHDAG (Fig. 6) by accurate mass measurement. Some unacylated TetraHDAG also appears to be present in R. ilealis. The major molecular species of Romboutsia glycolipids are given in Table 3.
Fig. 4.
LC/MS identification of acyl-triHDAG from strain FRIFI. Negative ion ESI/MS showing the [M + Cl]− ions of the lipid emerging at 12.7 to 13.7 min. The structural drawings of carbohydrates are for the illustration of chemical formula, and are based on previous reports that bacterial glycolipids, at least majority of them, contain hexopyrnoses (glucose, galactose and mannose) [16].
Fig. 5.
LC/MS/MS identification of tetraHDAG from strain FRIFI. A. Negative ion ESI/MS showing the [M + Cl]− ions of the lipid emerging at 25.5 to 26.5 min. B. MS/MS spectrum of the [M + Cl]− at m/z 1249.670.
Fig. 6.
LC/MS identification of acyl-tetraHDAG from strain FRIFI. Negative ion ESI/MS showing the [M+ Cl]− ions of the lipid emerging at 18.5 to 19.5 min.
Table 3.
Major molecular species of Romboutsia glycolipids seen by negative ion ESI/MS.
| Major molecular speciesa | [M + Cl]−b | R. lituseburensis | R. ilealis | strain FRIFI |
|---|---|---|---|---|
| MHDAG 31:1 | 749.498 | + | + | + |
| MHDAG 32:1 | 763.513 | + | + | + |
| MHDAG 33:1 | 777.529 | + | + | + |
| MHDAG 34:1 | 791.544 | + | + | + |
| DHDAG 31:1 | 911.550 | + | + | + |
| DHDAG 32:1 | 925.566 | + | + | + |
| DHDAG 33:1 | 939.561 | + | + | + |
| DHDAG 34:1 | 953.596 | + | + | |
| TriHDAG 31:1 | 1073.602 | + | + | + |
| TriHDAG 32:1 | 1087.618 | + | + | + |
| TriHDAG 33:1 | 1101.634 | + | + | + |
| TriHDAG 34:1 | 1115.650 | + | + | |
| TriHDAG 35:1 | 1129.665 | + | + | |
| TetraHDAG 31:1 | 1235.654 | tr | + | |
| TetraHDAG 32:1 | 1249.670 | tr | + | |
| TetraHDAG 33:1 | 1263.686 | tr | + | |
| TetraHDAG 34:1 | 1277.701 | tr | + | |
| TetraHDAG 35:1 | 1291.717 | tr | + | |
| Acyl-TriHDAG 46:1 | 1297.815 | c | c | + |
| Acyl-TriHDAG 47:1 | 1311.830 | c | c | + |
| Acyl-TriHDAG 48:1 | 1325.846 | c | c | + |
| Acyl-TriHDAG 49:1 | 1339.861 | c | c | + |
| Acyl-TriHDAG 50:1 | 1353.877 | c | c | + |
| Acyl-TetraHDAG 45:1 | 1445.856 | c | + | |
| Acyl-TetraHDAG 46:1 | 1459.870 | c | + | |
| Acyl-TetraHDAG 47:1 | 1473.888 | c | + | |
| Acyl-TetraHDAG 48:1 | 1487.904 | c | + | |
| Acyl-TetraHDAG 49:1 | 1501.921 | c | + |
MHDAG, DHDAG, TriHDAG and TetraHDAG are mono-, di-, tri-, and tetra-hexosyldiacylglycerol, respectively.
These m/z values are for strain FRIFI. The values for other species may vary slightly.
Although these lipids have been detected in R. lituseburensis and R. ilealis, nomass spectral data are available.
4. Discussion
The lipids of currently known members of the new genus Romboutsia differ from those of most clostridia in the absence of plasmalogens and of phosphatidylethanolamine. Like clostridia, Romboutsia spp. have low DNA G + C content (27–28.1 mol%) [1]. R. lituseburensis, previously C. lituseburense, was placed in Clostridium Cluster XI and seen to be closely related to C. difficile by 16S rRNA gene sequencing [10]. Lawson and Rainey have recently proposed to restrict the genus Clostridium to Clostridium butyricum and related species in cluster I [17]. Our recent work showed that C. difficile has several phospholipids and glycolipids most of which were present as both diacyl and plasmalogen species [3]. These include PG, CL, MHDAG, DHDAG and TriHDAG. Previous 2D-TLC studies on the lipids of R. lituseburensis and R. ilealis showed the presence of a number of phospholipids, two of which were tentatively identified as PG and CL, and a number of unidentified glycolipids. In order to shed more light on the lipidome of Romboutsia, we undertook a 2D-TLC and ESI/MS examination of the lipids of three species. Our work confirms the presence of three phospholipids, PG, CL and PA in R. ilealis. An additional phospholipid, lysyl-PG, was identified in R. lituseburensis and strain FRIFI. Decaprenyl and/or nonaprenyl (C50 and C45)-P are detected in all three species. In comparison, undecaprenyl (C55)-P is found in Escherichia coli and Francisella novicida [18].
Three glycolipids, MHDAG, DHDAG and TriHDAG, were identified in all three species by LC/ESI–MS, and a fourth, TetraHDAG, was identified in strain FRIFI. A fifth glycolipid (Fig. 1, GL3) was observed in all three species by 2D-TLC and was identified in strain FRIFI, by preparative TLC followed by LC/ESI–MS as an acylated TriHDAG. A sixth glycolipid observed in both strain FRIFI and R. ilealis, was similarly identified as an acylated TetraHDAG. We have not studied the linkages of the acyl chains and the sugars in these glycolipids since that would require isolation of each lipid from large amounts of cells of each species followed by NMR and chemically-based linkage studies. Mass spectrometry alone, cannot determine these linkages.
Lysyl-PG has previously been identified in a variety of Gram-positive bacteria including aerobic and anaerobic species. Among clostridia containing this lipid are Clostridium novyi [7] and two strains of Clostridium botulinum Group III [6]. One of these strains, D1873, like C. novyi, also contains alanyl-PG. Clostridium perfringens also has both lysyl-PG and alanyl-PG [19,20]. The formation of amino-acyl PG is catalyzed by amino-acyl-PG synthetase (MprF), an enzyme that catalyzes the transfer of an amino acid from aminoacyl-tRNA to PG [21,22]. In C. perfringens two phylogenetically distinct MprF paralogs have been identified; MprF1 responsible for the formation of alanyl-PG and MprF2 responsible for the formation of lysyl-PG [23]. Genes encoding homologs of MprF2 in C. perfringens were identified in the genomes of R. lituseburensis and strain FRIFI, but not in R. ilealis, supporting the observation that lysyl-PG can be produced by the first two strains, but not the latter. The gene mprF (multi peptide resistance factor) confers resistance to cationic peptides. In tissue cultures and in mouse models of infection, Listeria monocytogenes mutants defective in mprF were found to be attenuated in virulence [24]. In a recent study mprF was found also to confer resistance to osmolytes such as urea, sodium chloride and sodium sulfate and to bile salts [25]. Thus, the presence of amino-acyl PG can provide protection from a variety of environmental insults encountered by both pathogenic and non-pathogenic species [26]. R. lituseburensis was originally isolated from mud [27], R. ilealis was isolated from ileal digesta of rats [1] and strain FRIFI was isolated from human ileostoma effluent [2]. Thus the multiple functions of amino-acyl PG could serve to protect these species in both the soil and within the mammalian intestine. PA is an intermediate in the biosynthesis of phospholipids in bacteria. It is usually either undetectable or present in trace amounts, as appears to be the case in the three species of Romboutsia we have investigated.
A large variety of glycolipids has been found in Gram-positive bacteria [16,28]. Recent studies of clostridia have demonstrated that these are major components of the lipidome in C. difficile [3], Clostridiumtetani [8], Clostridium sporogenes, some strains of C. botulinum Group I that are related to C. sporogenes [6], Clostridium psychrophilum [4], and Clostridium acetobutylicum [5,29,30]. We note that three of the glycolipids common to Romboutsia, MHDAG, DHDAG and TriHDAG, were also found in C. difficile. The unusual glycolipids TetraHDAG, and acylated forms of TriHDAG and TetraHDAG found in Romboutsia, were not seen in C. difficile. Acylated tri- and tetra-hexoses have been found as lipid anchors of the lipoteichoic acid of Lactobacillus casei [31] and Lactobacillus gasseri [32]. Acyl-TriHDAG and TetraHDAG were previously isolated as free lipids from L. casei [33]. Their potential role in the assembly of lipoteichoic acids has been considered [31]. The lipid composition of strain FRIFI has a striking resemblance to that of L. casei with the exception of free acyl-tetraHDAG, which was not found in L. casei.
It should be noted that our picture of the complex lipidome of Romboutsia is only a snapshot. We have only analyzed the lipidome of cells grown in a single medium and harvested at one time. The relative amounts of the lipids will undoubtedly change when cells are grown under different conditions and harvested at different stages of growth. Our lipidomic analysis of Romboutsia suggests that the major difference between C. difficile and Romboutsia is not in the compositions of phospholipids and glycolipids, but rather in the presence (C. difficile) or absence (Romboutsia) of plasmalogens. So far, all species of Clostridium that have been studied contain plasmalogens. These and previous studies on the lipidomes of Clostridium, including those species that may be considered to belong to new genera [10], indicate that they can shed light on bacterial taxonomy as an adjunct to the powerful technique of 16S rRNA gene sequencing.
Acknowledgments
The authors thank Aleksandr Umanetc for his help with growing the bacterial strains. The mass spectrometry facility in the Department of Biochemistry of Duke University Medical Center and ZG were supported by the Lipid Maps glue grant (GM-069338) and EY023666 from NIH. JG and HS were supported by a grant of SenterNovem (FND-07013).
Abbreviations
- CL
cardiolipin
- DHDRG
dihexosyldiacylglycerol
- MHDRG
monohexosyldiacylglycerol
- PG
phosphatidylglycerol
- THDRG
trihexosyldiacylglycerol
- TetraHDAG
tetrahexosyldiacylglycerol
References
- 1.Gerritsen J, Fuentes S, Grievink W, van Niftrik L, Tindall BJ, Timmerman HM, Rijkers GT, Smidt H. Characterization of Romboutsia ilealis gen. nov., sp. nov., isolated from the gastro-intestinal tract of a rat, and proposal for the reclassification of five closely related members of the genus Clostridium into the genera Romboutsia gen. nov., Intestinibacter gen. nov., Terrisporobacter gen. nov and Asaccharospora gen. nov. Int. J. Syst. Evol. Microbiol. 2014;64:1600–1616. doi: 10.1099/ijs.0.059543-0. [DOI] [PubMed] [Google Scholar]
- 2.Gerritsen J. The Genus Romboutsia — Genomic and Functional Characterization of Novel Bacteria Dedicated to Life in the Intestinal Tract. Wageningen University; 2015. [Google Scholar]
- 3.Guan Z, Katzianer D, Zhu J, Goldfine H. Clostridium difficile contains plasmalogen species of phospholipids and glycolipids. Biochim. Biophys. Acta. 2014;1841:1353–1359. doi: 10.1016/j.bbalip.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guan Z, Tian B, Perfumo A, Goldfine H. The polar lipids of Clostridium psychrophilum, an anaerobic psychrophile. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2013;1831:1108–1112. doi: 10.1016/j.bbalip.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian B, Guan Z, Goldfine H. An ethanolamine-phosphate modified glycolipid in Clostridium acetobutylicum that responds to membrane stress. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2013;1831:1185–1190. doi: 10.1016/j.bbalip.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan Z, Johnston NC, Raetz CRH, Johnson EA, Goldfine H. Lipid diversity among botulinum neurotoxin-producing clostridia. Microbiology. 2012;158:2577–2584. doi: 10.1099/mic.0.060707-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan Z, Johnston NC, Aygun-Sunar S, Daldal F, Raetz CR, Goldfine H. Structural characterization of the polar lipids of Clostridium novyi NT. Further evidence for a novel anaerobic biosynthetic pathway to plasmalogens. Biochim. Biophys. Acta. 1811;2011:186–193. doi: 10.1016/j.bbalip.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnston NC, Aygun-Sunar S, Guan Z, Ribeiro AA, Daldal F, Raetz CR, Goldfine H. A phosphoethanolamine-modified glycosyl diradylglycerol in the polar lipids of Clostridium tetani. J. Lipid Res. 2010;51:1953–1961. doi: 10.1194/jlr.M004788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldfine H, Johnston NC. Membrane lipids of Clostridia. In: Dürre P, editor. Hand- book on Clostridia. Boca Raton, FL: Taylor & Francis; 2005. pp. 297–310. [Google Scholar]
- 10.Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow JAE. The phylogeny of the genus Clostridium; proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 11.Holdeman LV, Cato EP, Moore WEC. Anaerobe Laboratory Manual. Blacksburg, VA: Virginia Polytechnic Institute and State University; 1977. [Google Scholar]
- 12.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 13.Kates M. Techniques of Lipidology. Isolation, Analysis and Identification of Lipids, Techniques of Lipidology. Isolation Analysis and Identification of Lipids. 1990;106 [Google Scholar]
- 14.Siakotos AN, Rouser G. Analytical separation of nonlipid water soluble substances and gangliosides from other lipids by dextran gel column chromatography. J. Am. Chem. Soc. 1965;42:913–919. doi: 10.1007/BF02632444. [DOI] [PubMed] [Google Scholar]
- 15.Goldfine H, Guan Z. Lipidomic analysis of bacteria by thin layer chromatography and liquid chromatography/mass spectrometry. In: McGenity TJ, Timmis KN, Nogales-Fernández B, editors. Hydrocarbon and Lipid Microbiology Protocols. Berlin Heidelberg: Springer Verlag; 2015. [Google Scholar]
- 16.Shaw N. Bacterial glycolipids. Bacteriol. Rev. 1970;34:365–377. doi: 10.1128/br.34.4.365-377.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawson PA, Rainey FA. Proposal to restrict the genus Clostridium (Prazmowski) to Clostridium butyricum and related species. Int. J. Syst. Evol. Microbiol. 2016 doi: 10.1099/ijsem.0.000824. [DOI] [PubMed] [Google Scholar]
- 18.Guan Z, Eichler J. Liquid chromatography/tandem mass spectrometry of dolichols and polyprenols, lipid sugar carriers across evolution. Biochim. Biophys. Acta. 2011;1811:800–806. doi: 10.1016/j.bbalip.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macfarlane MG. Characterization of lipoamino-acids as O-amino-acid esters of phosphatidylglycerol. Nature (London) 1962;196:136–138. [Google Scholar]
- 20.Johnston NC, Baker JK, Goldfine H. Phospholipids of Clostridium perfringens: a re- examination. FEMS Microbiol. Lett. 2004;233:65–68. doi: 10.1016/j.femsle.2004.01.048. [DOI] [PubMed] [Google Scholar]
- 21.Lennarz WJ, Bonsen PPM, Vandeene LL. Substrate specificity of O-L-Lysylphosphatidylglycerol synthetase. Enzymatic studies on structure of O-L-Lysylphosphatidylglycerol. Biochemistry. 1967;6 doi: 10.1021/bi00860a005. (2307-&) [DOI] [PubMed] [Google Scholar]
- 22.Lennarz WJ, Nesbitt JA, Reiss J. Biosynthesis of Lysyl phosphatidylglycerol. Fed. Proc. 1966;25 (521-&) [Google Scholar]
- 23.Roy H, Ibba M. RNA-dependent lipid remodeling by bacterial multiple peptide resistance factors. Proc. Natl. Acad. Sci. U. S. A. 2008;105:4667–4672. doi: 10.1073/pnas.0800006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thedieck K, Hain T, Mohamed W, Tindall BJ, Nimtz M, Chakraborty T, Wehland J, Jansch L. The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol. Microbiol. 2006;62:1325–1339. doi: 10.1111/j.1365-2958.2006.05452.x. [DOI] [PubMed] [Google Scholar]
- 25.Roy H, Dare K, Ibba M. Adaptation of the bacterial membrane to changing environments using aminoacylated phospholipids. Mol. Microbiol. 2009;71:547–550. doi: 10.1111/j.1365-2958.2008.06563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldfine H. Charge counter charge. Bacterial response to antimicrobial cationic peptides and more. Virulence. 2014;5:451–453. doi: 10.4161/viru.28662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prevot AR. Étude des bactéries anaerobes d'AFrique occidentale française (Senégal, Guinée, Côte d'Ivoire) Ann. Inst. Pasteur (Paris) 2015;74:157–170. [Google Scholar]
- 28.Kates M. Glyco-, phospho-, and sulfoglycoglycerolipids of bacteria. In: Kates M, editor. Handbook of Lipid Research, Glycolipids, Phosphoglycolipids, and Sulfoglycolipids. Vol. 6. New York: Plenum Press; 1990. pp. 1–122. [Google Scholar]
- 29.Oulevey J, Bahl H, Thiele OW. Novel alk-1-enyl ether lipids isolated from Clostridium acetobutylicum. Arch. Microbiol. 1986;144:166–168. [Google Scholar]
- 30.Johnston NC, Goldfine H. Replacement of the aliphatic chains of Clostridium acetobutylicum by exogenous fatty acids: regulation of phospholipid and glycolipid composition. J. Bacteriol. 1992;174:1848–1853. doi: 10.1128/jb.174.6.1848-1853.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakano M, Fischer W. Trihexosyldiacylglycerol and acyltrihexosyldiacylglycerol as lipid anchors of the lipoteichoic acid of Lactobacillus casei DSM20021. Hoppe Seylers Z. Physiol. Chem. 1978;359:1–11. doi: 10.1515/bchm.1978.359.1.1. [DOI] [PubMed] [Google Scholar]
- 32.Shiraishi T, Yokota S, Morita N, Fukiya S, Tomita S, Tanaka N, Okada S, Yokota A. Characterization of a Lactobacillus gasseri JCM1131(T) lipoteichoic acidwith a novel glycolipid anchor structure. Appl. Environ. Microbiol. 2013;79:3315–3318. doi: 10.1128/AEM.00243-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakano M, Fischer W. The glycolipids of Lactobacillus casei DSM 20021. Hoppe Seylers Z. Physiol. Chem. 1977;358:1439–1453. doi: 10.1515/bchm2.1977.358.2.1439. [DOI] [PubMed] [Google Scholar]