Abstract
Ossification of the ligamentum flavum (OLF) is a disorder of heterotopic ossification of spinal ligaments and is the main cause of thoracic spinal canal stenosis. Previous studies suggested that miR-132-3p negatively regulates osteoblast differentiation. However, whether miR-132-3p is involved in the process of OLF has not been investigated. In this study, we investigated the effect of miR-132-3p and its target genes forkhead box O1 (FOXO1), growth differentiation factor 5 (GDF5) and SRY-box 6 (SOX6) on the osteogenic differentiation of ligamentum flavum (LF) cells. We demonstrated that miR-132-3p was down-regulated during the osteogenic differentiation of LF cells and negatively regulated the osteoblast differentiation. Further, miR-132-3p targeted FOXO1, GDF5 and SOX6 and down-regulated the protein expression of these genes. Meanwhile, FOXO1, GDF5 and SOX6 were up-regulated after osteogenic differentiation and the down-regulation of endogenous FOXO1, GDF5 or SOX6 suppressed the osteogenic differentiation of LF cells. In addition, we also found FOXO1, GDF5 and SOX6 expression in the ossification front of OLF samples. Overall, these results suggest that miR-132-3p inhibits the osteogenic differentiation of LF cells by targeting FOXO1, GDF5 and SOX6.
Keywords: ossification of the ligamentum flavum, miR-132-3p, FOXO1, GDF5, SOX6, osteogenic differentiation
1. Introduction
Ossification of the ligamentum flavum (OLF) is a rare disorder of heterotopic ossification of spinal ligaments that is almost exclusively reported in Eastern Asian countries. OLF primarily occurs in the thoracolumbar spine and is the main cause of thoracic spinal canal stenosis and myelopathy [1,2]. In the past several years, many studies have examined OLF development and progression at both histopathological and cellular levels. While these studies identified potential contributing factors, such as mechanical [3,4,5,6], metabolic [7,8], degenerative [9] and genetic factors [10,11], OLF development and progression continues to be inadequately understood.
One potential way that osteogenic differentiation is modulated is through microRNAs (miRNAs). MiRNAs, which comprise a substantial family of small (18–24 nucleotides), single-stranded non-coding RNAs, function in the regulation of mammalian cell gene expression. miRNAs regulate a target mRNA by binding its 3′-untranslated region (UTR) and subsequently mediating its degradation via the RNA-induced silencing complex (RISC) [12]. MiRNAs regulate a variety of physiological and pathological processes, with previous studies showing that particular miRNAs have the potential to positively or negatively regulate cells’ osteogenic differentiation and bone development [13,14]. For instance, miR-29b [15], miR-21 [16], miR-548d-5p [17], and miR-22 [18] were found to promote osteogenic differentiation and miR-92a [19], miR-214 [20], miR-30 [21], and miR-103a [22] were discovered to suppress osteogenesis, while miR-34a may have dimorphic effects [12,23]. Further, as reviewed by Huang C, studies on the miRNA revealed that miRNAs regulate osteogenesis by targeting osteoblast-related genes, particularly runt related transcription factor 2 (RUNX2) and Osterix, and targeting signaling pathways such as Wnt/β-catenin, transforming growth factor beta (TGF-β)/bone morphogenetic protein (BMP) and Notch [24].
The miR-212/132 family is highly conserved in vertebrates [25]. miR-132 and miR-212 are found in an intergenic region, exhibit similar mature sequences and share the same seed region, but the expression levels and the physiological functions of miR-132 and miR-212 are largely different [26]. Previous studies suggested that miR-132-3p can inhibit osteoblast differentiation and participate in the regulation of bone loss [27,28]. In addition, miR-132-3p was also found to regulate osteosarcoma [29,30,31,32], an osteogenic tumor that has the capability of osteoblast differentiation. However, whether miR-132-3p is involved in the process of OLF has not been investigated.
To explore the important role of miR-132-3p in thoracic ossification of the ligamentum flavum (TOLF), the present study investigated the effect of miR-132-3p and its target genes on the osteogenic differentiation of ligamentum flavum (LF) cells. Our results implied that miR-132-3p, which is down-regulated during osteogenic differentiation of LF cells, inhibits the differentiation process by targeting forkhead box O1 (FOXO1), growth differentiation factor 5 (GDF5) and SRY-box 6 (SOX6).
2. Results
2.1. MiR-132-3p Inhibits the Osteogenic Differentiation of Ligamentum Flavum (LF) Cells
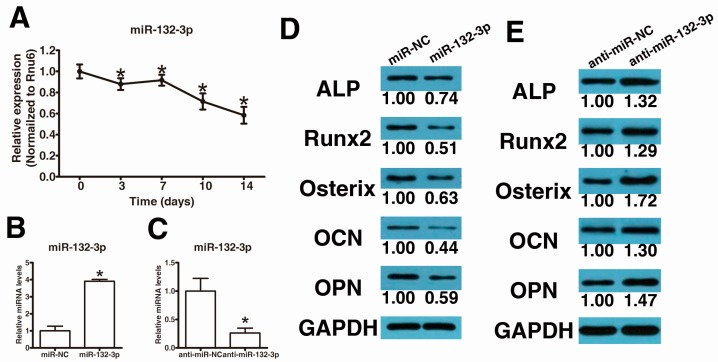
We first determined the temporospatial expression pattern of miR-132-3p in human LF cells cultured in osteogenic medium by qPCR to investigate the potential roles of miR-132-3p in the osteogenic differentiation of LF cells. Expression of miR-132-3p decreased at day 3 compared with that at day 0 and continuously decreased to day 14 (Figure 1A). This result suggests that miR-132-3p might negatively regulate the osteogenic differentiation of LF cells.
Figure 1.
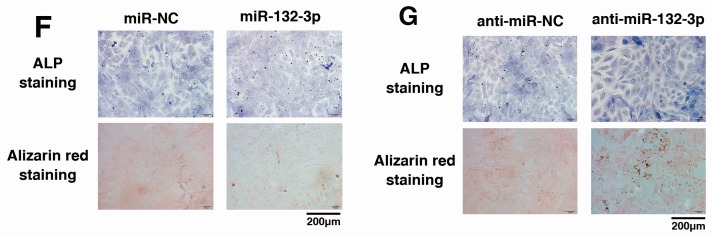
MiR-132-3p inhibits the osteogenic differentiation of ligamentum flavum cells. (A) Endogenous miR-132-3p expression levels were measured via quantitative real-time polymerase chain reaction (qRT-PCR) at different time points during osteogenic differentiation of ligamentum flavum cells; * p < 0.05 compared with day 0; (B) miR-132-3p expression was assessed via qRT-PCR in ligamentum flavum cells transfected with miRNA mimics; * p < 0.05 compared with nonspecific microRNA (miR-NC) group; (C) miR-132-3p expression was assessed via qRT-PCR in ligamentum flavum cells transfected with miRNA inhibitors; * p < 0.05 compared with anti-miR-NC group; (D,E) Western blot analysis of osteogenic marker protein expression after miR-132-3p overexpression (D) and reduction (E) at day 14; (F,G) alkaline phosphatase (ALP) staining and Alizarin Red staining at day 14 showed ALP activity and calcification after miR-132-3p overexpression (F) and reduction (G). Scale bar represents 200 µm and is fitted for every figure.
To further elucidate the role of miR-132-3p in the regulation of osteogenic differentiation of LF cells, synthetic mimics of miR-132-3p and inhibitors were transfected into LF cells, and the osteogenic capacity was examined. Intracellular miR-132-3p levels were markedly up-regulated by miR-132-3p mimics (Figure 1B) and substantially down-regulated by miR-132-3p inhibitors (Figure 1C). Furthermore, osteogenic differentiation was significantly inhibited after over-expression of miR-132-3p (Figure 1D,E) and significantly promoted after reduction of miR-132-3p (Figure 1F,G), as indicated by the expression changes of the osteogenic transcription factors, RUNX2 and Osterix, and osteoblastic markers, alkaline phosphatase (ALP), osteopontin (OPN) and osteocalcin (OCN), as well as ALP and Alizarin Red staining.
2.2. MiR-132-3p Directly Targets FOXO1, GDF5 and SOX6
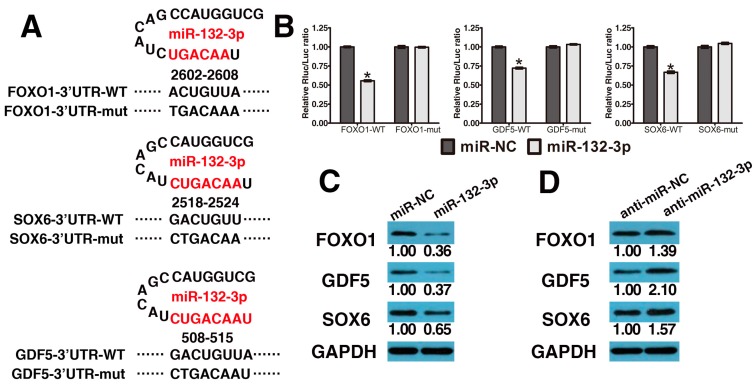
To reveal the molecular mechanism by which miR-132-3p regulates the osteogenic differentiation of LF cells, TargetScan (http://www.targetscan.org/vert_71/) was utilized to forecast potential miR-132-3p targets. Among the candidates, we found that three osteogenesis-related genes, FOXO1, GDF5 and SOX6, contain miR-132-3p binding sites in their 3′-UTRs. Next, we constructed luciferase reporters for each gene that contained either a wild-type (WT) 3′-UTR or a mutant (mut) 3′-UTR with mutant sequences of the miR-132-3p binding site (Figure 2A). The results showed that miR-132-3p repressed the luciferase activity of the 3′-UTR of each gene when compared to the nonspecific microRNA (miR-NC) control group, respectively. Additionally, no statistically significant alteration in luciferase activity was observed in the presence of the mutated 3′-UTR site (Figure 2B).
Figure 2.
MiR-132-3p directly targets FOXO1, GDF5 and SOX6. (A) The wild-type and mutated type miR-132-3p binding sites in the FOXO1, GDF5 and SOX6 3′-UTR. The sequences in red font showed the blinding sites; (B) The wild-type (WT) 3′-UTR or mutant (MUT) 3′-UTR reporter plasmids of the three genes were co-transfected into HEK293T cells with either miR-132-3p or miR-NC and fluorescence was quantified; * p < 0.05 compared with miR-NC group; (C,D) FOXO1, GDF5 and SOX6 protein expression levels were examined via Western blot following miR-132-3p mimics (C) and inhibitors (anti-miR-132-3p) (D) transfection in ligamentum flavum cells.
Next, we detected the gene expression of FOXO1, GDF5 and SOX6 after transfecting LF cells with the mimics of miR-132-3p and the inhibitor. We confirm that overexpression of miR-132-3p resulted in down-regulation of FOXO1, GDF5 and SOX6 in LF cells based on Western blot analysis (Figure 2C) and the reduction of miR-132-3p resulted in the opposite effects (Figure 2D). These results suggested that the effect of miR-132-3p during the osteogenic differentiation of LF cells is mediated by targeting these osteogenesis-related genes.
2.3. FOXO1, GDF5 and SOX6 Knockdown Inhibits Osteogenic Differentiation in LF Cells
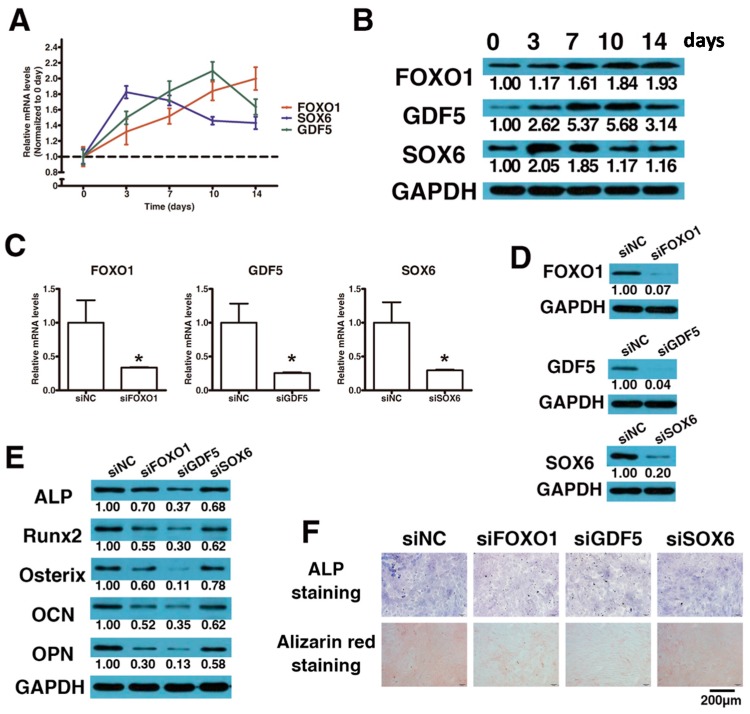
The mRNA and protein expression levels of FOXO1, GDF5 and SOX6 were determined by quantitative real-time polymerase chain reaction (qRT-PCR) (Figure 3A) and Western blot analyses (Figure 3B). All three were up-regulated in expression after osteogenic differentiation compared with day 0. Specifically, FOXO1 was continually increased to day 14, while GDF5 and SOX6 were decreased (still higher than day 0) after day 10 and 7, respectively.
Figure 3.
FOXO1, GDF5 and SOX6 knockdown inhibits osteogenic differentiation of ligamentum flavum cells. (A,B) FOXO1, GDF5 and SOX6 mRNA and protein expression levels examined via qRT-PCR and Western blot at different time points during osteogenic differentiation of ligamentum flavum cells; (C,D) FOXO1, GDF5 and SOX6 mRNA and protein expression level examined via qRT-PCR and Western blot following siRNAs transfection in ligamentum flavum cells; * p < 0.05 compared with non-targeting siRNA (si-NC) group; (E) Osteogenic marker protein expression examined via Western blot at day 14 after FOXO1, GDF5 or SOX6 knockdown; (F) ALP staining and Alizarin Red staining at day 14 showed inhibited ALP activity and calcification following FOXO1, GDF5 or SOX6 knockdown when compared with siNC group. Scale bar represents 200 µm and is fitted for every figure.
To examine the functional effects of FOXO1, GDF5 and SOX6 on the osteogenic differentiation of LF cells, siRNA-induced mRNA knockdown for each gene was employed and it significantly reduced both mRNA and protein expression of FOXO1, GDF5 and SOX6 (Figure 3C,D). Furthermore, FOXO1, GDF5 or SOX6 knockdown inhibited the osteogenic differentiation of LF cells, as indicated by reduced RUNX2, Osterix, ALP, OPN and OCN expression (Figure 3E) and reduced ALP and Alizarin Red staining (Figure 3F).
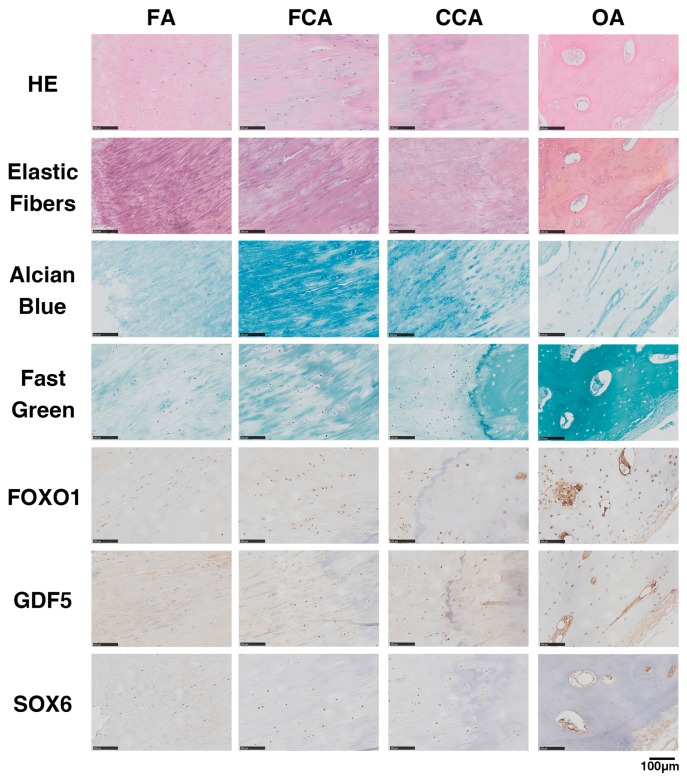
2.4. FOXO1, GDF5 and SOX6 Protein Expression in OLF Samples
Structurally, pathological specimens of endochondral ossification exhibit an ossification front, including a fibrocartilage area (FCA) and calcified cartilage area (CCA), between the ossified area (OA) and ligamentous fiber area (FA). The HE staining, tissue-specific staining and immunohistochemical (IHC) staining in each area are shown in Figure 4. In FA, the Elastic Fibers staining showed positive and regular expression, and the IHC for FOXO1, GDF5 and SOX6 was negative. In FCA and CCA, the expressions of Elastic Fibers staining were decreased and irregular, and the Alcian Blue staining was positive. The IHC results suggested that FOXO1, GDF5 and SOX6 were positive in the round cells of the FCA and CCA. Further, in OA, the Fast Green staining was positive. IHC results showed that FOXO1 and GDF5 were strongly stained in the nuclei of osteoblasts, but the nuclei of osteoblasts showed no nuclear reactivity for SOX6.
Figure 4.
Representative hematoxylin-eosin staining, tissue-specific staining and immunohistochemical staining for FOXO1, GDF5 or SOX6 in OLF samples. Scale bar represents 100 µm and is fitted for every figure.
3. Discussion
miR-132, which is located on chromosome 17p13.3, has been widely researched in recent years and is mainly implicated in neuropsychiatric disorders [25,26,33,34] and tumor progression in tumors, especially in osteosarcoma [29,30,31,32,35,36]. Recent evidence suggested that miRNA-132-3p inhibits osteoblast differentiation in simulated microgravity [27] and Type 2 Diabetes Mellitus-induced osteoporosis [28]. Furthermore, in a recent integrated microRNA-mRNA study, miR-132-3p was found to be one of the top 10 down-regulated miRNAs in ossified posterior longitudinal ligament (PLL) cells compared with normal PLL cells [37], thus suggesting that miR-132-3p may be involved in the process of ligament ossification. The present study provides the first evidence that miR-132-3p suppresses the osteogenic differentiation of LF cells. Our results showed that miR-132-3p was down-regulated during osteogenic differentiation. Further, inhibition of the expression of miR-132-3p promoted osteogenic differentiation, while miR-132-3p overexpression inhibited osteogenic differentiation. These findings suggest that miR-132-3p acts as a negative regulator of the osteogenic differentiation of LF cells.
It is well known that one miroRNA can regulate multiple target genes. Among the potential miR-132-3p targets predicted using Targetscan, three osteogenesis-related genes, FOXO1, GDF5 and SOX6, were selected in this study. The dual luciferase reporter assays identified these three genes as direct targets of miR-132-3p. Furthermore, miR-132-3p overexpression resulted in their down-regulation at the protein level, whereas inhibition of miR-132-3p led to their up-regulation, suggesting that FOXO1, GDF5 and SOX6 are regulated by miR-132-3p during osteogenic differentiation. Though the regulatory connection between miR-132-3p and FOXO1 has already been reported in gastric cancer cells [38], the direct targeting of miR-132-3p with GDF5 and SOX6 was newly discovered. Besides, miR-132-3p regulated osteoblast differentiation in osteoporosis by targeting EP300 [27] and SIRT1 [28]. Whether these two genes were involved in osteogenic differentiation of LF cells requires further study.
The pathological process of OLF involves the differentiation of fibroblasts into osteoblasts. Many cytokines, including transcription factors [39], growth factors [40] and inflammatory cytokines [41], have been reported to be involved in the ossification process. In the present study, FOXO1, GDF5 and SOX6 were all found to encourage the osteogenic differentiation of LF cells. We demonstrated that down-regulation of endogenous FOXO1, GDF5 or SOX6 suppressed osteogenesis to varying degrees, respectively. In addition, during the osteogenic differentiation of LF cells, the three osteogenesis-related genes were up-regulated, and the highest value of each gene expression was found at different times. The results indicated that these three genes may take effect in different stages of osteogenic differentiation and also suggested that miR-132-3p may regulate different genes in different stages of osteogenic differentiation. Location expressions of FOXO1, GDF5 and SOX6 were visually observed by immunohistochemistry. From the perspective of histopathology, the progression of OLF is viewed as a process of endochondral ossification. FOXO1 and GDF5 were positively expressed in both the cartilage and ossified area, while SOX6 was only positively expressed in the cartilage area. The results were in agreement with the cell experiment.
FOXO1 is the main member of the orkhead box O (FoxO) family expressed in bone and is a positive regulator of osteoblast differentiation [42,43,44]. GDF5, also called bone morphogenetic protein (BMP)-14 and cartilage-derived morphogenetic protein (CDMP)-1, is a member of the transforming growth factor β (TGF-β) superfamily and plays critical roles in organ development processes including bone, cartilage, ligament, and joint formation [45]. SOX6, a member of the D subfamily of sex determining region y-box (SRY-box)-related transcription factors, is a crucial transcription factor that regulates chondrogenesis and endochondral ossification [46]. A previous study showed that rhGDF-5 induces the osteogenic differentiation of LF cells through the activation of ERK1/2 and p38 mitogen-activated protein kinase (MAPK) [40]. However, the molecular mechanisms of FOXO1 and SOX6 in the osteogenic differentiation of LF cells were unclear. More research is required to understand the detailed mechanism of these osteogenesis-related genes in LF cells.
4. Materials and Methods
4.1. Patient Specimens
All experimental protocol involving humans (including the acquisition, processing and detection of the specimens) were approved by the Medical Scientific Research Ethics Committee of Peking University Third Hospital (PUTH-REC-SOP-06-3.0-A27, 2014003). All the methods were performed in accordance with relevant guidelines and regulations. TOLF patients who visited the orthopedic clinic and provided written informed consent for the study were utilized. Specialists diagnosed TOLF based on clinical symptoms and radiological examination as previously described [47]. Ligamentum flavum samples were obtained from TOLF patients during spinal surgery via en bloc resection of the lamina and ligamentum flavum as previously described [48].
4.2. Cell Cultures and Osteogenic Differentiation
Ligaments (approximately 0.5–1 cm2) were aseptically harvested from patients during surgery and rinsed with phosphate-buffered saline (PBS), while surrounding tissues were removed under a dissecting microscope to avoid possible osteogenic cell contamination. The collected ligaments were minced into approximately 0.5 mm3 pieces and digested using 0.25% trypsin, followed by 250 U/mL type I collagenase (Sigma-Aldrich, St. Louis, MO, USA). The specimen were washed with serum-containing medium and placed in 100-mm culturing dishes containing Dulbecco’s Modified Eagle′s medium (DMEM; GIBCO, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (GIBCO), 100 U/mL penicillin G sodium and 100 mg/mL streptomycin sulfate in a humidified atmosphere with 5% CO2 at 37 °C. Explant-derived cells derived were harvested using 0.25% trypsin for further passaging, with passages (P) 2 and 3 used for subsequent experimentation. To induce osteogenic differentiation, cells were cultured in osteogenic medium consisting of DMEM supplemented with 50 µM ascorbic acid (Sigma-Aldrich), 10 mM β-glycerophosphate (Sigma-Aldrich) and 10−8 M dexamethasone (Sigma-Aldrich).
4.3. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis
Total RNA was isolated using Trizol (Invitrogen, Carlsbad, CA, USA). Reverse transcription and qPCR for miR-132-3p were performed using a miDETECTA Track™ miRNA qRT-PCR Starter kit (RiboBio, Guangzhou, China) according to the manufacturer’s instructions on a BioRad IQ5 system. Each value was normalized to that of RnU6. Reverse transcription and qPCR for the mRNA levels of FOXO1, GDF5 and SOX6 were carried out as described previously [49]. Expression levels were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and relative gene expression levels were calculated using the 2-ΔΔCt method. All experiments were performed in triplicate. The primers were described in Table 1.
Table 1.
Primer sequences for quantitative real-time polymerase chain reaction (qRT-PCR).
| Gene | Primer (5′–3′) |
|---|---|
| FOXO1 | Fw: AAGCTCCCAAGTGACTTGGATG Rv: CTGCTCACTAACCCTCAGCCTGA |
| GDF5 | Fw: AAAAGGACAGCTTCCCGGAG Rv: GCCTCCCTTTCTGTCAGCAT |
| SOX6 | Fw: TCAACATGTGGCCTCCCATC Rv: GATGACAGAACGCTGTCCCA |
| GAPDH | Fw: TCAAGGCTGAGAACGGGAAG Rv: TGGACTCCACGACGTACTCA |
| miR-132-3p | RT: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCGACCATG Fw: GCGCGCGTAACAGTCTACAGC Rv: GTCGTATCCAGTGCAGGGTCC |
| U6 | RT: AAAATATGGAACGCTTCACGAATTTG Fw: CTCGCTTCGGCAGCACATATACT Rv: CGCTTCACGAATTTGCGTGT |
4.4. Western Blot Analysis
Cell lysates were obtained using RIPA lysis buffer (Beyotime, Shanghai, China) containing 10 mM phenylmethylsulphonylfluoride as a protease inhibitor (PMSF; Beyotime) and 50 µg of total protein was separated in a Bis-Tris polyacrylamide gel and transferred onto a nitrocellulose membrane. The membrane was then incubated in 5% bovine serum albumin (BSA) containing primary rabbit-anti-human polyclonal antibodies at 4 °C overnight. After incubating with horseradish peroxidase (HRP)-conjugated goat-anti-rabbit antibody at room for 1 h, protein was detected using electrochemiluminescence (ECL; Millipore, Darmstadt, Germany). The following primary rabbit-anti-human antibodies were used: anti-FOXO1 (1:1000; ab39670, Abcam, Cambridge, MA, USA); anti-GDF5 (1:1000; ab93855, Abcam); anti-SOX6 (1:1000; ab30455, Abcam); anti-Runx2 (1:1000; ab23981, Abcam); anti-Sp7/Osterix (1:2000; ab22552, Abcam); anti-ALP (1:2000; ab95462, Abcam); anti-OCN (1:500; ab93876, Abcam); anti-OPN (1:1000; ab8448, Abcam); and anti-GAPDH (1:2500; ab9485, Abcam). The results of Western blots were quantified using image-J software (http://imagej.net) analysis.
4.5. Alkaline Phosphatase (ALP) Activity Assay and Alizarin Red Staining
Cells were seeded in six-well plates at a density of 1 × 105 cells/well and cultured in osteogenic medium for 14 days. ALP activity was determined using an ALP activity staining kit (DE0004; Leagene Biotech, Beijing, China) and mineralization was assessed using an Alizarin Red S kit (DS0002; Leagene Biotech).
4.6. miRNA/siRNA Transfection
Synthetic miRNA and siRNA were purchased from RiboBio. ligamentum flavum cells were transfected with miRNA/siRNA using Lipofectamine® 2000 Transfection Reagent (Life Technologies, New York, NY, USA), according to the manufacturer′s instructions.
MiR-132-3p mimics or miR-132-3p inhibitor (anti-miR-132-3p) were transfected into LF cells at a concentration of 20 nM with nonspecific microRNA (miR-NC) or nonspecific microRNA inhibitor (anti-miR-NC) used as negative controls. SiRNA targeting FOXO1, GDF5 or SOX6 were transfected at a concentration of 50 nM with non-targeting siRNA (siNC) used as negative control.
4.7. Luciferase Constructs and Reporter Assay
The DNA sequences of FOXO1, GDF5 or SOX6 3′-UTR were amplified by PCR using HEK293T genomic DNA as a template. The amplified DNA sequences were inserted into pmiR-RB-REPORT™ vectors (RiboBio) to generate wild type (WT) FOXO1, GDF5 or SOX6 3′-UTR, with a mutated (mut) FOXO1, GDF5 or SOX6 3′-UTR luciferase vector generated using site-directed mutagenesis. For the reporter assay, HEK293T cells were cultured in a 96-well plate with 1.5 × 104 cells/well in 100 μL of culture medium/well for 24 h. Cells were then co-transfected with 50 nM miR-132-3p mimic or miR-NC and 100 ng of vector per well and cultured in fresh medium for an additional 48 h. The luciferase reporter assay was carried out using the Dual-Glo® Luciferase Assay System (Promega, Madison, WI, USA) according to the manufacturer’s instructions and luminescence was quantified using a Veritas™ 9100-002 luminometer (Promega).
4.8. Hematoxylin-Eosin (HE) Staining, Tissue-Specific Staining and Immunohistochemical (IHC) Analysis
Serial 5-mm-thick sections were prepared from paraffin-embedded specimens for staining.
Hematoxylin-eosin staining was performed in an autostainer machine (ST5010 XL; Leica Microsystems, Mannheim, Germany) using standard procedures.
Elastic Fibers staining kit for ligament, Alcian Blue staining kit for cartilage and Fast Green staining kit for bone were purchased from Leagene Biotech (DC0066; DB0060; DZ0046).
Sections for immunohistochemical staining were carried out as described previously [49]. The primary rabbit anti-human antibodies were: anti-FOXO1 (1:200; ab39670, Abcam); anti-GDF5 (1:200; ab93855, Abcam); anti-SOX6 (1:200; ab30455, Abcam).
5. Conclusions
Overall, our study provides a comprehensive profiling of miR-132-3p as a novel regulator during the osteogenic differentiation of LF cells. The findings presented herein show that miR-132-3p can suppress osteogenic differentiation of LF cells by targeting FOXO1, GDF5 and SOX6. Further, we demonstrated that FOXO1, GDF5 and SOX6 are expressed in LF cells and ossified ligamentum flavum tissues, and these osteogenesis-related genes may take effect at different stages of the osteogenic differentiation of LF cells. Our findings reveal a new mechanism of the TOLF pathological process and suggest that miR-132-3p and its target genes could possibly be viable therapeutic targets for TOLF and other skeletal disorders.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant numbers 81272031 and 81071505). We would like to extend a special thanks to all of the patients who provided specimens and thank Guangzhou RiboBio Technology Co. Ltd. (http://www.ribobio.com/) for their assistance in constructing the luciferase vectors.
Author Contributions
Xiaochen Qu designed the study, performed the majority of the laboratory work, statistics and data analysis, and drafted the manuscript. Zhongqiang Chen designed the study, secured funding, analyzed and interpreted data, and approved the final submitted manuscript. Dongwei Fan designed the study and performed some of the laboratory work, statistics, and data analysis. Chuiguo Sun and Yan Zeng specimen and clinical data acquisition. All authors have read and approved the final submitted manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Hou X., Sun C., Liu X., Liu Z., Qi Q., Guo Z., Li W., Zeng Y., Chen Z. Clinical features of thoracic spinal stenosis-associated myelopathy: A retrospective analysis of 427 cases. Clin. Spine Surg. 2016;29:86–89. doi: 10.1097/BSD.0000000000000081. [DOI] [PubMed] [Google Scholar]
- 2.Feng F.B., Sun C.G., Chen Z.Q. Progress on clinical characteristics and identification of location of thoracic ossification of the ligamentum flavum. Orthop. Surg. 2015;7:87–96. doi: 10.1111/os.12165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan D., Chen Z., Wang D., Guo Z., Qiang Q., Shang Y. Osterix is a key target for mechanical signals in human thoracic ligament flavum cells. J. Cell. Physiol. 2007;211:577–584. doi: 10.1002/jcp.21016. [DOI] [PubMed] [Google Scholar]
- 4.Chen J., Wang X., Wang C., Yuan W. Rotational stress: role in development of ossification of posterior longitudinal ligament and ligamentum flavum. Med. Hypotheses. 2011;76:73–76. doi: 10.1016/j.mehy.2010.08.034. [DOI] [PubMed] [Google Scholar]
- 5.Xu P., Wang C., He H., Xu G., Ye X. Spinal balance failure: A potential cause of spinal ligament ossification. Med. Hypotheses. 2011;76:908–910. doi: 10.1016/j.mehy.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Cai H.X., Yayama T., Uchida K., Nakajima H., Sugita D., Guerrero A.R., Yoshida A., Baba H. Cyclic tensile strain facilitates the ossification of ligamentum flavum through β-catenin signaling pathway: In vitro analysis. Spine. 2012;37:E639–E646. doi: 10.1097/BRS.0b013e318242a132. [DOI] [PubMed] [Google Scholar]
- 7.Fan D., Chen Z., Chen Y., Shang Y. Mechanistic roles of leptin in osteogenic stimulation in thoracic ligament flavum cells. J. Biol. Chem. 2007;282:29958–29966. doi: 10.1074/jbc.M611779200. [DOI] [PubMed] [Google Scholar]
- 8.Sohn S., Yoon J.W., Chung C.K. Increased bone mineral density in patients with ossification of the ligamentum flavum: A case-control study. J. Clin. Densitom. 2014;17:195–199. doi: 10.1016/j.jocd.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Lang N., Yuan H.S., Wang H.L., Liao J., Li M., Guo F.X., Shi S., Chen Z.Q. Epidemiological survey of ossification of the ligamentum flavum in thoracic spine: CT imaging observation of 993 cases. Eur. Spine J. 2013;22:857–862. doi: 10.1007/s00586-012-2492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong Q., Ma X., Li F., Guo Z., Qi Q., Li W., Yuan H., Wang Z., Chen Z. COL6A1 polymorphisms associated with ossification of the ligamentum flavum and ossification of the posterior longitudinal ligament. Spine. 2007;32:2834–2838. doi: 10.1097/BRS.0b013e31815b761c. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y., Zhao Y., Chen Y., Shi G., Yuan W. RUNX2 polymorphisms associated with OPLL and OLF in the Han population. Clin. Orthop. Relat. Res. 2010;468:3333–3341. doi: 10.1007/s11999-010-1511-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L., Holmstrom K., Qiu W., Ditzel N., Shi K., Hokland L., Kassem M. MicroRNA-34a inhibits osteoblast differentiation and in vivo bone formation of human stromal stem cells. Stem Cells. 2014;32:902–912. doi: 10.1002/stem.1615. [DOI] [PubMed] [Google Scholar]
- 13.Peng S., Gao D., Gao C., Wei P., Niu M., Shuai C. MicroRNAs regulate signaling pathways in osteogenic differentiation of mesenchymal stem cells. Mol. Med. Rep. 2016;14:623–629. doi: 10.3892/mmr.2016.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang S., Deng Y., Gu P., Fan X. MicroRNAs regulate bone development and regeneration. Int. J. Mol. Sci. 2015;16:8227–8253. doi: 10.3390/ijms16048227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan T., Song W., Gao H., Li T., Cao X., Zhong S., Wang Y. miR-29b-loaded gold nanoparticles targeting to the endoplasmic reticulum for synergistic promotion of osteogenic differentiation. ACS Appl. Mater. Interfaces. 2016;8:19217–19227. doi: 10.1021/acsami.6b02969. [DOI] [PubMed] [Google Scholar]
- 16.Song Q., Zhong L., Chen C., Tang Z., Liu H., Zhou Y., Tang M., Zhou L., Zuo G., Luo J., et al. miR-21 synergizes with BMP9 in osteogenic differentiation by activating the BMP9/smad signaling pathway in murine multilineage cells. Int. J. Mol. Med. 2015;36:1497–1506. doi: 10.3892/ijmm.2015.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang W., Zhang L., Zhou Y., Ji X., Liu J., Liu D., Yin P., Peng Y., Hao M., Zhang L., et al. Synergistic effects of BMP9 and miR-548d-5p on promoting osteogenic differentiation of mesenchymal stem cells. BioMed Res. Int. 2015;2015:309747. doi: 10.1155/2015/309747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang S., Wang S., Bian C., Yang Z., Zhou H., Zeng Y., Li H., Han Q., Zhao R.C. Upregulation of miR-22 promotes osteogenic differentiation and inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells by repressing HDAC6 protein expression. Stem Cells Dev. 2012;21:2531–2540. doi: 10.1089/scd.2012.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murata K., Ito H., Yoshitomi H., Yamamoto K., Fukuda A., Yoshikawa J., Furu M., Ishikawa M., Shibuya H., Matsuda S. Inhibition of miR-92a enhances fracture healing via promoting angiogenesis in a model of stabilized fracture in young mice. J. Bone Miner. Res. 2014;29:316–326. doi: 10.1002/jbmr.2040. [DOI] [PubMed] [Google Scholar]
- 20.Wang X., Guo B., Li Q., Peng J., Yang Z., Wang A., Li D., Hou Z., Lv K., Kan G., et al. miR-214 targets ATF4 to inhibit bone formation. Nat. Med. 2013;19:93–100. doi: 10.1038/nm.3026. [DOI] [PubMed] [Google Scholar]
- 21.Wu T., Zhou H., Hong Y., Li J., Jiang X., Huang H. miR-30 family members negatively regulate osteoblast differentiation. J. Biol. Chem. 2012;287:7503–7511. doi: 10.1074/jbc.M111.292722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuo B., Zhu J., Li J., Wang C., Zhao X., Cai G., Li Z., Peng J., Wang P., Shen C., et al. microRNA-103a functions as a mechanosensitive microRNA to inhibit bone formation through targeting Runx2. J. Bone Miner. Res. 2015;30:330–345. doi: 10.1002/jbmr.2352. [DOI] [PubMed] [Google Scholar]
- 23.Fan C., Jia L., Zheng Y., Jin C., Liu Y., Liu H., Zhou Y. miR-34a promotes osteogenic differentiation of human adipose-derived stem cells via the RBP2/notch1/cyclin D1 coregulatory network. Stem Cell Rep. 2016;7:236–248. doi: 10.1016/j.stemcr.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang C., Geng J., Jiang S. MicroRNAs in regulation of osteogenic differentiation of mesenchymal stem cells. Cell Tissue Res. 2016 doi: 10.1007/s00441-016-2462-2. [DOI] [PubMed] [Google Scholar]
- 25.Bicker S., Lackinger M., Weiss K., Schratt G. MicroRNA-132, -134, and -138: A microRNA troika rules in neuronal dendrites. Cell. Mol. Life Sci. 2014;71:3987–4005. doi: 10.1007/s00018-014-1671-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wanet A., Tacheny A., Arnould T., Renard P. miR-212/132 expression and functions: Within and beyond the neuronal compartment. Nucleic Acids Res. 2012;40:4742–4753. doi: 10.1093/nar/gks151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Z., Wang Y., Sun Z., Wang H., Zhou H., Zhang L., Zhang S., Cao X. miRNA-132-3p inhibits osteoblast differentiation by targeting Ep300 in simulated microgravity. Sci. Rep. 2015;5:18655. doi: 10.1038/srep18655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gong K., Qu B., Liao D., Liu D., Wang C., Zhou J., Pan X. miR-132 regulates osteogenic differentiation via downregulating Sirtuin1 in a peroxisome proliferator-activated receptor β/δ-dependent manner. Biochem. Biophys. Res. Commun. 2016;478:260–267. doi: 10.1016/j.bbrc.2016.07.057. [DOI] [PubMed] [Google Scholar]
- 29.Hou B., Cui X., Liu Y., Zhang W., Liu M., Sun Y.E., Ma Z., Gu X. Positive feedback regulation between microRNA-132 and CREB in spinal cord contributes to bone cancer pain in mice. Eur. J. Pain. 2016;20:1299–1308. doi: 10.1002/ejp.854. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y., Li Y., Liu J., Wu Y., Zhu Q. MicroRNA-132 inhibits cell growth and metastasis in osteosarcoma cell lines possibly by targeting Sox4. Int. J. Oncol. 2015;47:1672–1684. doi: 10.3892/ijo.2015.3147. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Wang J., Xu G., Shen F., Kang Y. miR-132 targeting cyclin E1 suppresses cell proliferation in osteosarcoma cells. Tumour Biol. 2014;35:4859–4865. doi: 10.1007/s13277-014-1637-2. [DOI] [PubMed] [Google Scholar]
- 32.Yang J., Gao T., Tang J., Cai H., Lin L., Fu S. Loss of microRNA-132 predicts poor prognosis in patients with primary osteosarcoma. Mol. Cell. Biochem. 2013;381:9–15. doi: 10.1007/s11010-013-1677-8. [DOI] [PubMed] [Google Scholar]
- 33.Hernandez-Rapp J., Rainone S., Hebert S.S. MicroRNAs underlying memory deficits in neurodegenerative disorders. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2016 doi: 10.1016/j.pnpbp.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 34.Tognini P., Pizzorusso T. MicroRNA212/132 family: Molecular transducer of neuronal function and plasticity. Int. J. Biochem. Cell Biol. 2012;44:6–10. doi: 10.1016/j.biocel.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 35.Ouyang L., Liu P., Yang S., Ye S., Xu W., Liu X. A three-plasma miRNA signature serves as novel biomarkers for osteosarcoma. Med. Oncol. 2013;30:340. doi: 10.1007/s12032-012-0340-7. [DOI] [PubMed] [Google Scholar]
- 36.Gougelet A., Pissaloux D., Besse A., Perez J., Duc A., Dutour A., Blay J.Y., Alberti L. Micro-RNA profiles in osteosarcoma as a predictive tool for ifosfamide response. Int. J. Cancer. 2011;129:680–690. doi: 10.1002/ijc.25715. [DOI] [PubMed] [Google Scholar]
- 37.Xu C., Chen Y., Zhang H., Chen Y., Shen X., Shi C., Liu Y., Yuan W. Integrated microRNA-mRNA analyses reveal OPLL specific microRNA regulatory network using high-throughput sequencing. Sci. Rep. 2016;6:21580. doi: 10.1038/srep21580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li W., Zhang J., Chen T., Yin P., Yang J., Cao Y. miR-132 upregulation promotes gastric cancer cell growth through suppression of FoxO1 translation. Tumour Biol. 2015 doi: 10.1007/s13277-015-3924-y. [DOI] [PubMed] [Google Scholar]
- 39.Uchida K., Yayama T., Cai H.X., Nakajima H., Sugita D., Guerrero A.R., Kobayashi S., Yoshida A., Chen K.B., Baba H. Ossification process involving the human thoracic ligamentum flavum: Role of transcription factors. Arthritis Res. Ther. 2011;13:R144. doi: 10.1186/ar3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong Z.M., Chen J.T., Zhang Y., Zha D.S., Lin Z.S., Zhao C.Y., Xu J.C., Li T., Xu Z.X. Growth/differentiation factor-5 induces osteogenic differentiation of human ligamentum flavum cells through activation of ERK1/2 and p38 MAPK. Cell. Physiol. Biochem. 2010;26:179–186. doi: 10.1159/000320526. [DOI] [PubMed] [Google Scholar]
- 41.Park J.O., Lee B.H., Kang Y.M., Kim T.H., Yoon J.Y., Kim H., Kwon U.H., Lee K.I., Lee H.M., Moon S.H. Inflammatory cytokines induce fibrosis and ossification of human ligamentum flavum cells. J. Spinal Disord. Tech. 2013;26:E6–E12. doi: 10.1097/BSD.0b013e3182698501. [DOI] [PubMed] [Google Scholar]
- 42.Rached M.T., Kode A., Xu L., Yoshikawa Y., Paik J.H., Depinho R.A., Kousteni S. FoxO1 is a positive regulator of bone formation by favoring protein synthesis and resistance to oxidative stress in osteoblasts. Cell Metab. 2010;11:147–160. doi: 10.1016/j.cmet.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siqueira M.F., Flowers S., Bhattacharya R., Faibish D., Behl Y., Kotton D.N., Gerstenfeld L., Moran E., Graves D.T. FOXO1 modulates osteoblast differentiation. Bone. 2011;48:1043–1051. doi: 10.1016/j.bone.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teixeira C.C., Liu Y., Thant L.M., Pang J., Palmer G., Alikhani M. Foxo1, a novel regulator of osteoblast differentiation and skeletogenesis. J. Biol. Chem. 2010;285:31055–31065. doi: 10.1074/jbc.M109.079962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin L., Li X. Growth differentiation factor 5 regulation in bone regeneration. Curr. Pharm. Des. 2013;19:3364–3373. doi: 10.2174/1381612811319190003. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y., Yang T.L., Li X., Guo Y. Functional analyses reveal the essential role of SOX6 and RUNX2 in the communication of chondrocyte and osteoblast. Osteoporos. Int. 2015;26:553–561. doi: 10.1007/s00198-014-2882-3. [DOI] [PubMed] [Google Scholar]
- 47.Chen Z.Q., Sun C.G. Clinical guideline for treatment of symptomatic thoracic spinal stenosis. Orthop. Surg. 2015;7:208–212. doi: 10.1111/os.12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun X., Sun C., Liu X., Liu Z., Qi Q., Guo Z., Leng H., Chen Z. The frequency and treatment of dural tears and cerebrospinal fluid leakage in 266 patients with thoracic myelopathy caused by ossification of the ligamentum flavum. Spine. 2012;37:E702–E707. doi: 10.1097/BRS.0b013e31824586a8. [DOI] [PubMed] [Google Scholar]
- 49.Qu X., Chen Z., Fan D., Sun C., Zeng Y., Hou X., Ning S. Notch signaling pathways in human thoracic ossification of the ligamentum flavum. J. Orthop. Res. 2016 doi: 10.1002/jor.23303. [DOI] [PubMed] [Google Scholar]