Abstract
Dysfunctional spermatozoa maturation is the main reason for the decrease in sperm motility and morphology in infertile men. Ejaculated spermatozoa from healthy fertile men were separated into four fractions using three-layer density gradient. Proteins were extracted and bands were digested on a LTQ-Orbitrap Elite hybrid mass spectrometer system. Functional annotations of proteins were obtained using bioinformatics tools and pathway databases. Western blotting was performed to verify the expression levels of the proteins of interest. 1469 proteins were identified in four fractions of spermatozoa. The number of detected proteins decreased according to the maturation level of spermatozoa. During spermatozoa maturation, proteins involved in gamete generation, cell motility, energy metabolism and oxidative phosphorylation processes showed increasing expression levels and those involved in protein biosynthesis, protein transport, protein ubiquitination, and response to oxidative stress processes showed decreasing expression levels. We validated four proteins (HSP 70 1A, clusterin, tektin 2 and tektin 3) by Western blotting. The study shows protein markers that may provide insight into the ejaculated spermatozoa proteins in different stages of sperm maturation that may be altered or modified in infertile men.
Keywords: bioinformatics, protein validation, proteins, proteomics, spermatozoa maturation
INTRODUCTION
Male infertility is a multi-factorial condition caused by gene mutations, chromosomal abnormalities, infectious diseases, varicocele, recurrent duct occlusion, radiation, and chemotherapy.1,2 However, in 50% of the cases, there is no identifiable cause of infertility and this condition is termed as idiopathic.3,4 Maturation of spermatozoa is a highly complex process that involves the initial process of spermatogenesis occurring in the testis. The latter process of sperm maturation occurs in the epididymis. It involves membrane and nuclear remodeling that result in sperm differentiation and acquisition of sperm motility.5 Spermatozoa shed the excess cytoplasm before they are released in the epididymis.6
If these events are not completed in the testes, immature sperm that are destined to undergo apoptosis escape the elimination process in the testis and appear in the ejaculate.7,8,9 Abnormalities related to epididymal maturation may lead to teratozoospermia and asthenozoospermia.5,10,11 Immature spermatozoa in the ejaculate exhibit cytoplasmic retention, increased production of reactive oxygen species (ROS) and lipid peroxidation, in addition to the metabolic alterations reported in immature spermatozoa.6,12 Cell-to-cell variation in ROS production in spermatozoa prepared on a three-layer density gradient has been reported in both fertile and infertile men.6 Lowest levels of ROS were reported in mature, morphologically-normal spermatozoa from fertile donors compared to abnormal spermatozoa in fertile men and in fertile men with normal and abnormal semen parameters. Similarly, DNA damage was correlated with immature germ cell concentration and percentage of abnormal sperm in different subsets. Highest levels of DNA fragmentation were reported in immature spermatozoa with abnormal morphology.12
There are a number of studies that have characterized the proteome of isolated germ cell populations in animal models and provided invaluable insights into the mechanism of spermatozoa maturation. However, these studies can only provide us indirect evidence for human spermatozoa maturation.13,14,15,16 Human spermatozoa development is significantly different from that of these animal models. Compared to many of the animal models, spermatogenesis in human testis is very inefficient, being only 25% efficient. This is related to the number of maturation steps being significantly reduced to 8 and of these only 2 are involved in nuclear remodeling.5,6,17 Human epididymal anatomy is unique when compared with that of other mammalian species: the caput epididymal segment is formed by a branched vas deferens; the corpus is not as thin as in other mammalian species; and the cauda epididymis does not possess the usual bulbous appearance.18 The protein profile from the human testis is quite different from that of the rat testis.19 Spermatozoa are terminally differentiated, highly specialized cells. They are transcriptionally and translationally inactive. As they traverse the epididymis, post-translational modifications occur during epididymal maturation. However, the biochemical processes driving these events are still not well understood as are the mechanisms associated with the pathological conditions that lead to defective sperm function and male infertility. Post-translational modifications (PTMs) are major components of the post-testicular events, involved in sperm development. PTMs involve both canonical PTMs such as phosphorylation or non-canonical pathways involving ROS.20,21,22
Proteomic tools such as two-dimensional electrophoresis (2-DE) have allowed the generation of large inventories of proteins present in the spermatozoa. In addition, liquid chromatography mass spectrometry (LC-MS) also provides information on the peptide changes occurring in a given pathway.23,24,25,26
Currently, research is geared toward identifying protein markers from semen samples that can be helpful in explaining underlying problems with male infertility. Some studies in male infertility have employed proteomic techniques, such as 2D-polyacrylamide gel electrophoresis (2D-PAGE), 2D-differential in gel electrophoresis (2D-DIGE), and two-dimensional liquid chromatography-tandem mass spectrometry (LC-MS/MS). These tools have allowed the identification of a large number of sperm-specific proteins.27 Proteomic analysis also provides us with a new method to find novel markers in the study of infertility, especially idiopathic infertile men who have normal semen parameters but are unable to father children. Frapsauce et al. used 2D-DIGE and detected two proteins, laminin receptor LR 67 and L-xylulose reductase (P34H) that may serve as biomarkers to distinguish idiopathic infertility from normal fertile men because of their important roles in gamete interaction.28 These approaches have increased our understanding of protein function involved in sperm processes such as motility, capacitation, acrosome reaction, and fertilization.29 We have reported a number of proteins that may be important in infertility patients exhibiting sperm dysfunction as a result of high levels of ROS.20,21,24,25,26,27,28,29
The density gradient separation method is the most common method used for obtaining highly motile, morphologically normal sperm for use in intrauterine insemination or assisted reproductive techniques (ARTs).17 Sperm preparation by density gradient centrifugation separates sperm cells based on their density. Morphologically normal and abnormal spermatozoa have different densities. A mature morphologically normal spermatozoon has a slightly higher density of 1.10 g ml−1 whereas an immature and morphologically abnormal spermatozoon has a lower density between 1.06 and 1.09 g ml−1. At the end of centrifugation, each spermatozoon is situated at the gradient level that matches its density.17 In our previous study, we demonstrated that by using different density gradients, spermatozoa in the human seminal ejaculate can be separated into four fractions of sperm undergoing varying levels of maturation.6 As expected, with increases in motility and normal morphology in the four fractions, a decrease in the ROS levels and DNA fragmentation was observed,6,30,31 demonstrating that spermatozoa in the whole ejaculate can be separated into four stages of maturation according to their density.
In the present study, the goal was to identify proteins that were differentially expressed in immature and mature ejaculated spermatozoa obtained from ejaculated spermatozoa from fertile men after density gradient separation. The in-depth investigation of the maturation process in ejaculated spermatozoa will help us understand the heterogeneity that is present in the ejaculated sperm population. By comparing the protein profiles among the immature and mature fractions, we may be able to identify proteins that are important in spermatozoa maturation and fertilization. This may improve our understanding of the key processes of spermatozoa maturation that may become dysfunctional or altered in ejaculated spermatozoa in infertile men with various clinical diagnoses.
MATERIALS AND METHODS
Study subjects
Following the approval of the study by the Institutional Review Board of Cleveland Clinic, semen samples were collected from 12 proven fertile men. All fertile men provided written consent to be enrolled in the study.
Inclusion/exclusion criteria
Fertile healthy men with normal semen parameters who had fathered at least one healthy child within 2 years without assisted reproductive measures; aged 20–40 years. Each fertile subject selected in the study was asked to fill out a general questionnaire regarding their lifestyle habits, i.e., smoking, alcohol, recreational drugs, fever, medication, and prior history of illness. The selected fertile subjects had no adverse lifestyle conditions. Fertile men presenting with recurring fever in the 90-day period prior to semen analysis and specimens with >1 × 106 white blood cells ml−1 of seminal ejaculate were excluded to avoid the interference of leukocytes in semen samples.
Sample collection and semen analysis
Semen samples were examined according to 2010 World Health Organization (WHO, 2010) criteria.32 All specimens were collected by masturbation at the Andrology Center after a period of sexual abstinence of 48–72 h and were allowed to liquefy completely for 15–20 min at 37°C before further processing. Following liquefaction, manual semen analysis was performed using a MicroCell counting chamber (Vitrolife, San Diego, CA, USA) to determine sperm concentration, percent motility, and the presence of round cells. Viability was determined by Eosin-Nigrosin stain. Smears of the semen were prepared, air-dried and were stained with a Diff-Quik kit (Baxter Healthcare Corporation, Inc., McGaw Park, IL, USA) for assessment of sperm morphology. Concentration of round cells in the wet smear was examined and if >1 × 106 ml−1 round cells were present; they were examined for the presence of white blood cells.
Leukocytospermia was confirmed by the peroxidase or the Endtz test.2 In brief, 20 μl of seminal ejaculate was mixed with 20 μl of phosphate buffered saline (PBS) and 40 μl of working Endtz solution in an amber colored Eppendorf tube and incubated for 5 min. The presence of white blood cells was done by examining the dark purple colored cells using a Makler chamber. Samples with concentration >1 × 106 white blood cells ml−1 of the semen was an indication of leukocytospermia and the sample was excluded.
Separation of immature and mature spermatozoa
For separating immature and mature sperm, a three-layer density gradient consisting of 2 ml of 40%, 60%, and 80% of the Upper Layer, Intermediate Layer and Lower Layer was prepared from the stock (100%) solution of the gradient with the SpermRinse medium. The gradient is antibiotic-free bicarbonate and HEPES buffered medium containing silane-coated, colloid silica particles, whereas the Sperm Rinse medium is bicarbonate and HEPES buffered medium containing human serum albumin and gentamycin as an antibiotic (Vitrolife, San Diego, CA, USA). This is a slight modification from the two-layer density gradient method routinely used for preparing sperm for ART techniques, especially intrauterine insemination.17 Briefly, 1–2 ml of liquefied semen sample was carefully loaded on the 40% gradient and centrifuged at 300g for 20 min. The resulting interfaces between the seminal plasma and 40% (fraction 1); 40%–60% (fraction 2); 60% and 80% (fraction 3) and the 80% pellet (fraction 4, mature fraction) were carefully aspirated (Figure 1a), resuspended in human tubal fluid media (HTF, Irvine Scientific, Santa Ana, CA, USA) and centrifuged at 300 ×g for 7 min. The pellets of each fraction were resuspended in 0.5–1 ml HTF and the total sperm count, motility, and morphology were checked again.
Figure 1.
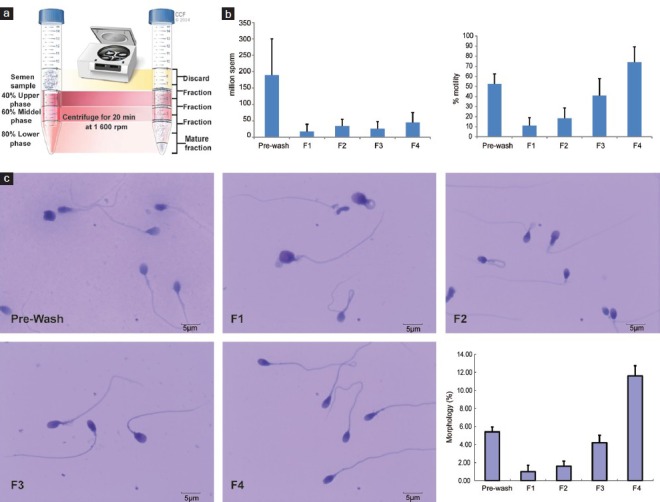
Separation of immature and mature spermatozoa (a) after a three-layer density gradient. (b) Sperm count, percentage of motility. Error bars represent standard deviations values for each parameter. (c) Sperm morphology before (Prewash) and after density gradient in each fraction. Error bars represent standard deviations values for each parameter. For 1c: Sperm morphology in various fractions is shown as: Prewash; Fraction 1 (F1); Fraction 2 (F2); Fraction 3 (F3); Fraction 4 (F4); and percentage of normal sperm morphology in each fractions. Error bars were shown as s.d. values of each parameter.
Preparation of samples for proteomic analysis
Protein extraction
Spermatozoa from each fraction were centrifuged (300 ×g, 7 min) and washed with PBS for 3 times. Sperm count was calculated and samples from each fraction were pooled such that each sample contributed equally to the sperm pool. The spermatozoa from pooled or individual samples were solubilized in RIPA lysis buffer (Sigma-Aldrich, Santa Louis, MO, USA) containing the proteinase inhibitor cocktail (Roche, Indianapolis, IN, USA). The spermatozoa samples were stored overnight at 4°C to allow for complete lysis of the spermatozoa including the cell membranes. After centrifugation at 13 000 ×g for 30 min, the supernatant was aspirated and the protein concentration was determined by using a BCA kit (Thermo, Rockford, IL, USA).
Proteomic analysis
A 25 μg aliquot of each sample mixed with SDS page buffer was boiled and a standard sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was run on a 12.5% Tris-HCl gel (Bio-Rad, Hercules, CA, USA) with constant voltage of 150 V for 35 min. The gel was fixed for 30 min, washed with water thoroughly and stained with GelCode Blue (Thermo, Rockford, IL, USA). For the protein digestion, the bands were cut to minimize excess polyacrylamide and divided into six smaller pieces. The gel pieces were then washed with water and dehydrated in acetonitrile. The proteins were reduced with dithiothreitol (DTT) and alkylated with iodoacetamide, and digested in-gel using trypsin, by adding 5 μl 10 ng μl−1 trypsin in 50 mmol l−1 ammonium bicarbonate and incubated overnight at room temperature to achieve complete digestion. The peptides that were formed were extracted from the polyacrylamide in two aliquots of 30 μl 50% acetonitrile with 5% formic acid. The extracts were combined and evaporated to <10 μl in Speedvac and then resuspended in 1% acetic acid to make up a final volume of ~30 μl for LC-MS analysis.
Liquid chromotography mass spectrometer analysis (LC-MS)
The LC-MS system was a Finnigan LTQ-Orbitrap Elite hybrid mass spectrometer system (Thermo, Rockford, IL, USA). The HPLC column was a Dione × 15 cm × 75 μm internal diameter Acclaim PepMap C18, 2 μm, 100 Å reversed phase capillary chromatography column (Thermo, Rockford, IL, USA). Five micro liters of the extract was injected and the peptides eluted from the column by an acetonitrile 0.1% formic acid gradient at a flow rate of 0.25 μl/min were introduced into the source of the mass spectrometer on-line. The microelectrospray ion source was operated at 2.5 kV. The digest was analyzed using the data dependent multitask capability of the instrument acquiring full scan mass spectra to determine peptide molecular weights and production spectra to determine amino acid sequence in successive instrument scans. Each sample was run 3 times to give the average of the errors.
Database searching
Tandem mass spectra were extracted by Proteome Discoverer version 1.4.1.288 (Thermo Fisher Scientific, San Jose, CA, USA). Charge state deconvolution and de-isotoping were not performed. All MS/MS samples were analyzed using Mascot (Matrix Science, London, UK; version 2.3.02), Sequest (Thermo Fisher Scientific, San Jose, CA, USA; version 1.4.0.288) and X! Tandem (The GPM, thegpm.org; version CYCLONE (2010.12.01.1)). Mascot, Sequest and X! Tandem were set up to search the human reference with the database (33292 entries) assuming the digestion enzyme trypsin. These searches were performed with a fragment ion mass tolerance of 1.0 Da, and parent ion tolerance of 10 parts per million (PPM). Carbamidomethylation of cysteine was specified as a fixed modification, and oxidation of methionine was specified as variable modifications.
To validate MS/MS-based peptide and protein identifications Scaffold (version Scaffold 4.0.6.1, Proteome Software Inc., Portland, OR, USA) was used. Peptide identifications were accepted if they could be established at >95.0% probability by the PeptideProphet algorithm with Scaffold delta-mass correction.33 Protein identifications were accepted if they could be established at >99.0% probability to achieve a false detection rate (FDR) <1.0% and contained at least 2 identified peptides. Protein probabilities were assigned by the ProteinProphet algorithm.34 Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Quantitation of the relative abundance of protein in sperm
For proteomic analysis, the relative quantity of the proteins was determined by comparing the number of spectra, termed spectral counts, used to identify each protein. The selection of peptide ions for MS/MS analysis is based on the abundance of the peptides, therefore, the more abundant the protein in the sample, the more often peptides from this protein is selected for MS/MS analysis. The relative quantity of these proteins was determined by comparing the number of spectra, termed spectral counts, used to identify each protein. The numerical values used in the quantitation correspond to the normalized spectral abundance factor (NSAF = SC/(ΣSC × protein length). The error observed for the SC measurements is greater for lower abundant proteins compared to higher abundant proteins. Due to this, different filtering criteria were used to determine if proteins are differentially present based on the overall abundance. NSAF approach was applied prior to relative protein quantification.35,36
Identification of differentially expressed proteins
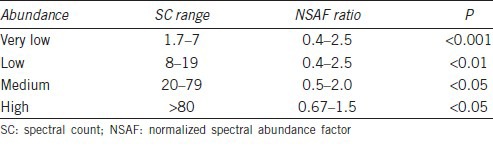
Appropriate filters were used to identify DEP that were dependent on the overall abundance of the proteins. It has been reported that accurate quantification and determination of real biological change is dependent on the number of SpCs and hence different constraints have to be applied to SpC levels in order to circumvent the biases and maintain a constant FPR for all proteins.37 The categorization of overall abundance along with the filtering criteria used for differential expression analysis is summarized in Table 1.
Table 1.
Criteria used to determine if proteins are differentially present in sperm samples
Bioinformatic analysis
Functional bioinformatics analysis was performed using publicly available software packages such as the Gene Ontology (GO) Term Finder38 UNIPROT, STRAP,39 and Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.niaid.nih.gov) along with the proprietary pathway database, Ingenuity Pathway Analysis (IPA) from Ingenuity® Systems, to identify the differentially affected processes, pathways, interactions and cellular distribution of the proteins in the four study fractions. Protein-protein interaction networks were generated using the IPA network generation algorithm. This algorithm generates sub-networks (and enlists the core functions associated with them) by identifying the interconnections between these significant proteins relative to all molecules they are connected to in the Ingenuity Knowledge database.
Western blotting analysis
The differentially expressed proteins of interest were verified in four individual samples using Western blotting analysis. For Western blotting, 20 μg protein of each sample was loaded to a 4%–15% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Bio-Rad, Hercules, CA, USA). Proteins in the gel were transferred to polyvinylidene fluoride (PVDF) membrane (Thermo, Rockford, IL, USA) at 18 volts for 30 min on Trans-Blot® SD Semi-Dry Electrophoretic Transfer Cell (Bio-Rad, Hercules, CA, USA). First antibodies were incubated at 4°C overnight, then the membranes were washed using Tris-buffered saline tween-20 (TBST) for 3 times 5 min each, second antibodies were incubated at room temperature for 1 h, another wash with TBST for 3 times 5 min each. Signal was developed using ECL Western blotting detection reagent (GE Healthcare, Life Sciences, Chicago, IL, USA) on X-ray film (Thermo, Rockford, IL). The expression of protein was compared with an internal reference of β-actin and compared between each research group and quantified using Image Lab™ software (Bio-Rad Inc., Hercules, CA, USA). Mouse β-actin monoclonal antibody (sc-4778), rabbit clusterin polyclonal antibody (sc-8354), goat anti-rabbit lgG-HRP (sc-2004), goat anti rabbit lgG-HRP (sc-2005), and donkey anti goat lgG-HRP (sc-2005) were obtained from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA). Rabbit HSP 1A1 polyclonal antibody (H00003305-D01), mouse TEKT2 polyclonal antibody (H00027285-B01), and mouse TEKT2 polyclonal antibody (H00064518-B01) were obtained from Abnova Inc. (Walnut, CA, USA).
Statistical analysis
The results were expressed as mean and standard deviations. To compare the differences from fraction 1 to fraction 4, we used Jonckheere–Terpstra test or the Jonckheere trend test. It is similar to the Kruskal–Wallis test where the null hypothesis is that several independent samples are from the same population. However, there is no priori ordering of the populations from which the samples are drawn. When there is a priori ordering, the Jonckheere test has more power than the Kruskal–Wallis test. In this test, there is no issue of normality and does not require log transformation of the data. The statistical analysis was performed using the Statistical Package for the Social Sciences (SPSS) Version 16.0 (SPSS, Chicago, IL, USA). Similarly, Jonckheere–Terpstra test was used in Western blot analysis of four proteins of interest. For proteomic analysis, we compared the DEP taking F4 as the baseline. Therefore, each fraction was statistically compared with F4 independently using t-test. A difference was considered significant at P < 0.05. For IPA, P < 0.05 was used using the right-tailed Fisher's exact test. This is inbuilt into the IPA software.
RESULTS
Motility and morphology changes in four fractions after density gradient
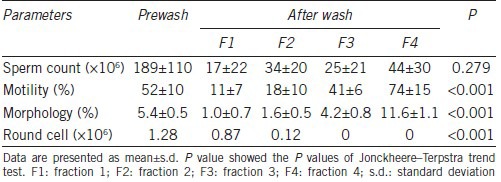
Twelve fertile men participated in this study. Semen parameters of these donors before separation on a three-layer density gradient were within normal range according to the WHO 2010 criteria. Results of sperm count, motility and morphology of different sperm fractions obtained before and after density gradient centrifugation are shown in Table 2 and Figure 1b. Motility and normal sperm morphology percentages were significantly increased from fraction 1 to fraction 4 (P < 0.01). Sperm morphology in various fractions is shown in Figure 1c. Compared to Prewash sample, Fraction 1 was enriched with round cells and immature spermatozoa with abnormal head and cytoplasmic retention (F1, Figure 1c). Fraction 2 contained immature spermatozoa with cytoplasmic retention and coiled tails (F2, Figure 1c). Fraction 3 contained a mixture of morphologically normal and abnormal spermatozoa. Most of the abnormal spermatozoa in this fraction had abnormal necks (F3, Figure 1c). Fraction 4 had the most number of morphologically normal spermatozoa. Most of the abnormal spermatozoa in this fraction had minor abnormalities in their heads or necks (F4, Figure 1c). The progressive improvement in sperm morphology from F1 to F4 is shown in the last panel of Figure 1c.
Table 2.
Sperm parameters before and after density gradient (n=12)
Global protein profiling for protein abundance, peptide coverage, and overall protein abundance
Pooling of samples is common in proteomic studies since the availability of semen sample is a major limitation that limits the use of an adequate number of individual samples. Therefore, protein normalization is very important to avoid sampling errors. To do this, the protein contribution must be from equal number of spermatozoa from each sample. Of the 12 recruited donors, 4 of them had total sperm count in each fraction of more than 10 × 106. We pooled together the spermatozoa from each fraction of these four donors. In addition, we used two individual samples to validate the results of the pooled samples.
The proteomic study showed a total of 1469 proteins in the pooled samples, whereas 1856 and 1650 proteins were found in two individual samples (Figure 2a). Of these proteins, 1207 proteins were found in both pooled and individual samples (Figure 2a). Of the 525 proteins found only in individual sample 1 but not in pooled sample, 443 of them (84.4%) had spectral counts <8.0 in all of the four fractions, indicating that their expression levels are very low, and 345 of them (65.7%) could be found in only one of the four fractions. Of the 399 proteins found only in individual sample 2 but not in pooled sample, 320 of them (80.2%) had spectral count values <8.0 (very low abundance), and 213 of them (53.4%) could be found in one of the four fractions only. Most of the proteins found in the two individual samples (84.4% and 80.2%) had very low expression levels (SC <8.0), which means that these proteins were not detected in pooled sample. This is because pooling may filter (exclude) those proteins with very low abundance. This is clearly one of the advantages of pooling samples in proteomic assay – which is to avoid biological variation of individual samples.
Figure 2.
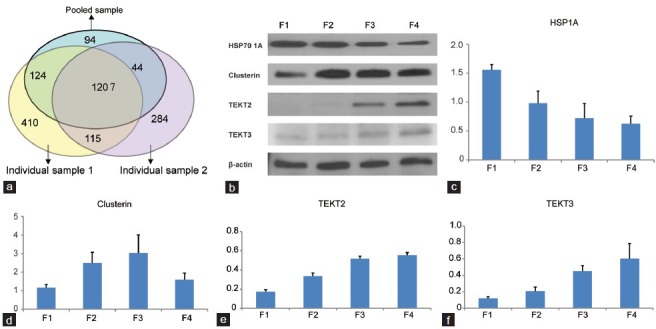
Venn diagram showing a total of 1469 proteins in the pooled samples. 1856 and 1650 proteins were found in two individual samples respectively (a). Of these proteins, 1207 proteins were found in both pooled and individual samples. Of the 525 proteins found only in individual sample 1 but not in pooled sample, (b) Western blotting showing HSP 70 1A, Clusterin, Tektin 2, and Tektin 3 β-actin in four fractions (n = 4), (c–f) trend in protein expression in four fractions. Error bars represent standard deviations values for each parameter.
Of the 1469 proteins detected in pooled fractions, 1169, 1106, 1091, and 963 proteins were found in fraction F1, F2, F3, and F4, respectively. In the three replicates, the number of peptides identified in all the replicates was 73%, 76%, 73%, and 74% in fractions F1, F2, F3, and F4, respectively. The average number of peptides in the three replicates of each fraction was as follows F1 = 2–73, F2 = 2–65, F3 = 2–73, and F4 = 2–78. The peptide coverage was set at >40% for differentially expressed proteins and ranged from F1 = 40%–85%, F2 = 40%–80%, F3 = 40%–84%, and F4 = 40%–86%. Similarly, the cutoff for spectral count set as >7 for low abundance protein was 2456 (F1), 1911 (F2), 132 (F3), and 3515 (F4). Total spectral count was 39 445 in F1, 39 239 in F2, 9361 in F3, and 40 049 in F4, respectively. The names of the proteins are listed in the Supplementary Table 1 (306KB, pdf) . The number of detected proteins decreased with the maturation of spermatozoa. A similar trend was seen in the two individual samples.
Proteins detected in four fractions of fertile men ejaculated semen
Besides the decrease in protein biosynthesis during spermatozoa maturation, proteins participating in proteolysis and cell cycle also decreased from fraction 1 to fraction 4, indicating that in fraction 1, during spermatozoal head condensation stage, protein ubiquitination was dramatic, and then, along with the progressive maturation, protein ubiquitination decreased gradually, and ubiquitination was almost complete in the most mature spermatozoa. This phenomenon could further explain the decrease in protein numbers in mature spermatozoa.
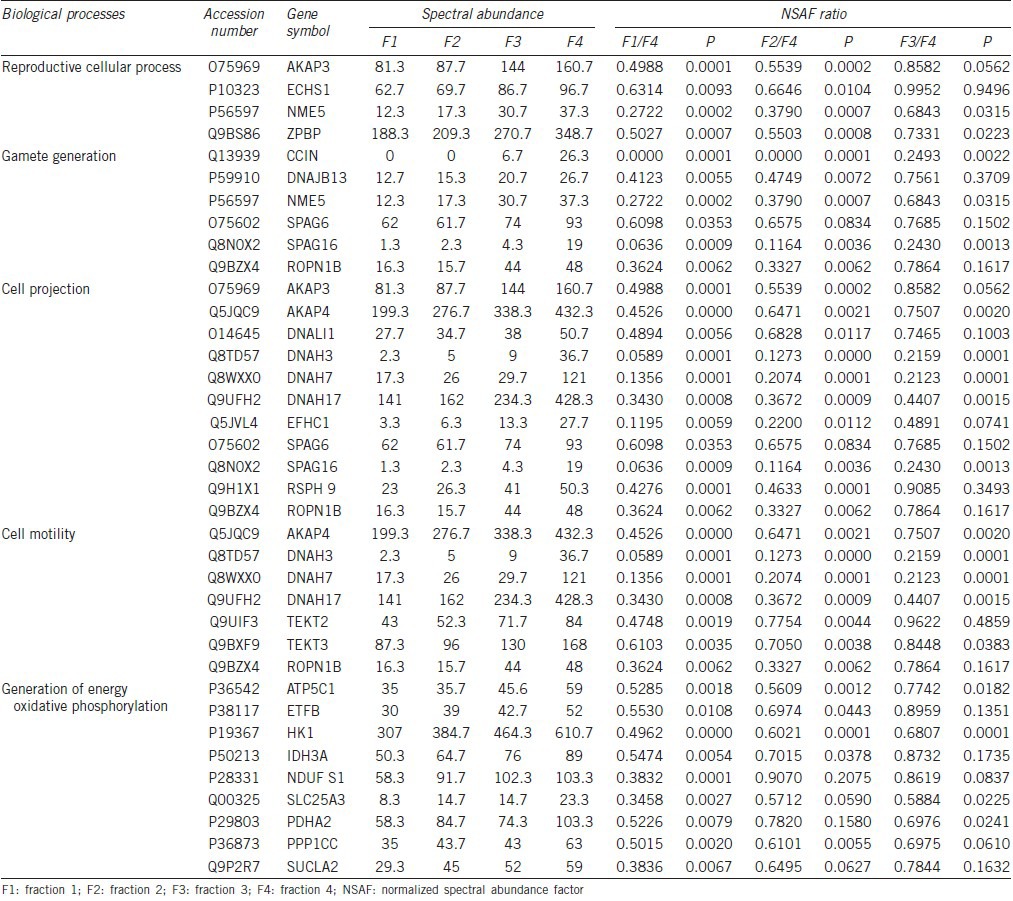
Similarly, of the 1469 detected proteins, 98 of them showed an increasing trend in expression levels. Functional bioinformatics annotation analysis showed these proteins take part in the reproductive cellular process, gamete generation, cell projection, cell motility, and oxidative phosphorylation, all of which are related to the acquisition of the reproductive ability. Key biological processes associated with proteins that show increasing trend during spermatozoa maturation were analyzed by DAVID (Table 3) showed that some proteins, such as A-kinase anchor proteins (AKAPs), dynein heavy chain proteins, and ropporin were equally present in these processes, indicating that these processes are linked together during the spermatozoa maturation process. These processes coincide with the general process of spermatozoa maturation.
Table 3.
Key biological processes associated with proteins that show increasing trend during spermatozoa maturation
Top pathways and networks for proteins that increase or decrease during the sperm maturation process
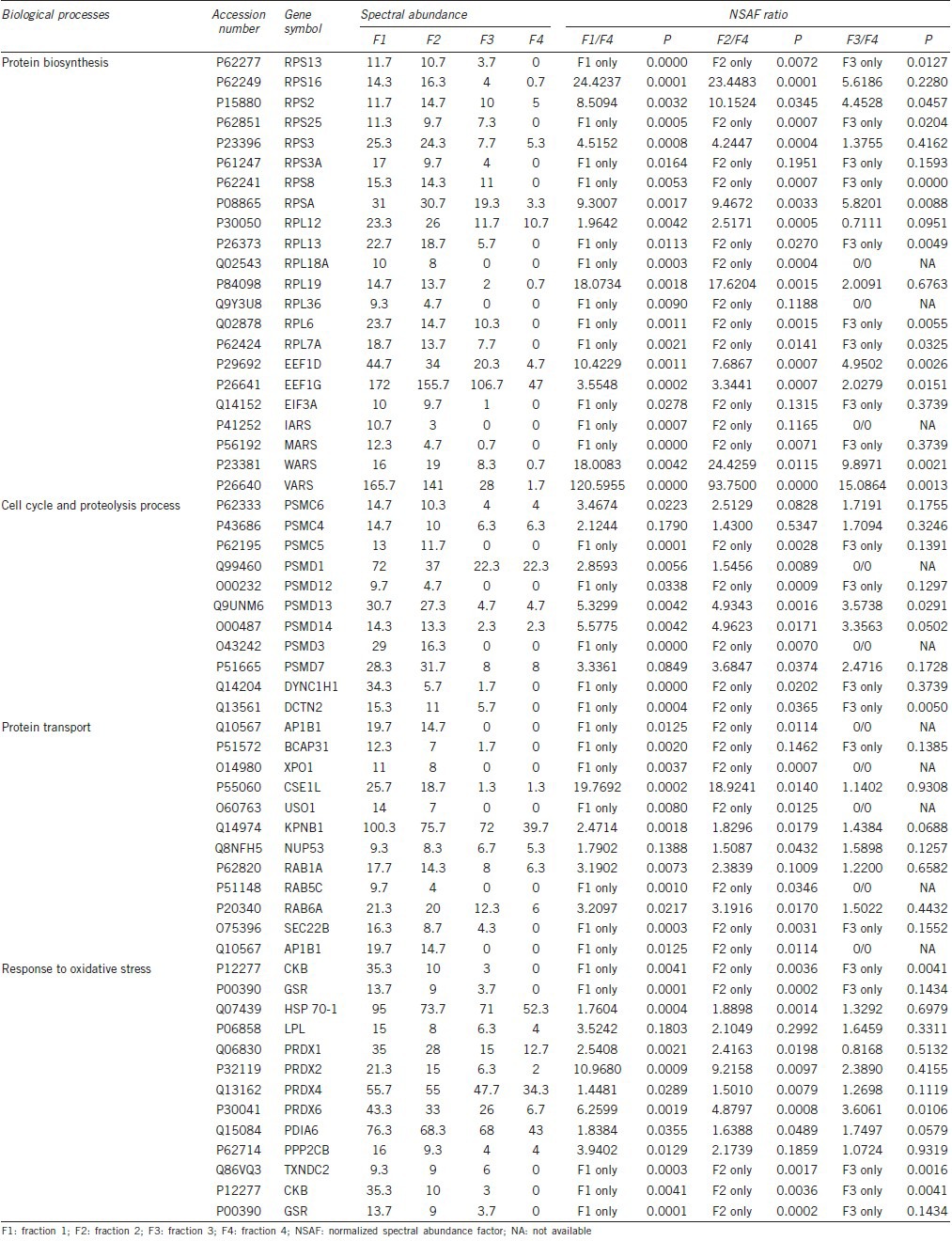
A majority of proteins with increasing trend during the process of spermatozoa maturation were observed to be involved in the reproductive cellular process, gamete generation, cell projection, cell motility, energy metabolism, and oxidative phosphorylation. In parallel, most of the proteins with decreasing trends during the spermatozoa maturation process took part in protein biosynthesis, protein transport, protein ubiquitination, and response to oxidative stress. Proteins with increasing and decreasing trend associated with each process are listed in Tables 3 and 4, respectively.
Table 4.
Key biological processes associated with proteins that show decreasing trend during spermatozoa maturation
To gain insights into the relationship among these proteins that show either increasing or decreasing trends in expression during the spermatozoa maturation process, protein-protein interaction networks were generated using the IPA network generation algorithm. From a list of top networks generated using IPA, we selected the sub-networks associated with the functions that were more pertinent to the maturation process. For the proteins exhibiting increasing trends, IPA analysis showed that these proteins mainly participate in three networks. These were: (1) 12 proteins participating in free radical scavenging, molecular transport, developmental disorder; (2) 9 proteins participating in cellular development, embryonic development and organ development; and (3) 1 protein involved in cell morphology, organ morphology, reproductive system development and function. We selected the 2nd sub-network for further analysis because cellular development is one of the key processes during sperm maturation. Therefore for the proteins with increasing levels of expression as spermatozoa mature, 9 proteins were identified in this network (Supplementary Figure 1a (306KB, pdf) ).
The cellular development and cell motility functions associated network identified by IPA network generation algorithm (a). The proteins marked in “blue” are the ones from our dataset while the remaining ones in “black” are the proteins in the Ingenuity Knowledge Base (IKB) that interact with ours. The nine proteins in blue show increasing trends expression during spermatozoa maturation process. ACR: acrosin; AKAP3: a kinase anchor protein 3; AKAP4: a kinase anchor protein 3; ALAT: dihydrolipoamids S-acetyltransferase; PDHA2: pyruvate dehydrogenase (lipoamide) alpha 2; PRSS12: protease, serine, 12; SLC 25A3: solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3; APAG6: sperm associated antigen 6; SPAG 16: sperm associated antigen 16. (b) Molecular transport and cellular assembly functions the seven proteins in blue showed significant decreasing trends in spermatozoa maturation. CSE1L: CSE1 chromosome segregation 1-like; PRDX1: peroxiredoxin 1; PRDX2: peroxiredoxin 2; PRDX4: peroxiredoxin 4: RANBP1: RAN binding protein 1; XPO1: exportin 1; KPNB1: karyopherin (importin) beta 1.
The interaction network was associated with reproductive processes such as ACR: acrosin; AKAP3: a kinase anchor protein 3; AKAP4: a kinase anchor protein 3; ALAT: dihydrolipoamids S-acetyltransferase; PDHA2: pyruvate dehydrogenase (lipoamide) alpha 2; PRSS12: protease, serine, 12; SLC 25A3: solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3; SPAG6: sperm associated antigen 6; SPAG 16: sperm associated antigen 16. These are associated with cellular processes, gamete generation, cell projection, cell motility, and generation of energy via oxidative phosphorylation.
Similarly, for the proteins exhibiting a decreasing trend, IPA analysis showed these proteins to participate in three networks. These are: (1) 23 proteins participating in cellular assembly and organization, cellular function and maintenance, cellular movement; (2) 18 proteins involved in infectious disease, cellular movement, hair and skin development and function; and (3) 7 proteins that were involved in molecular transport, protein trafficking, cellular assembly and organization. We selected the 3rd sub-network for further analysis because the molecular and protein synthesis and transport are gradually paused or becomes inactive during sperm maturation as spermatozoa become fully mature and transcriptionally inactive. Therefore for the seven proteins with decreased expression identified during sperm maturation, molecular transport and cellular assembly functions network were exportin-1 (XPO1), exporting-2 (CSE1L), importin subunit beta 1 (KPNB1), cytoplasmic proteins counteracting oxidative stress such as the peroxiredoxin 1 (PRDX1), peroxiredoxin 2 isoform a (PRDX2), and peroxiredoxin-4 precursor (PRDX4) and RAN binding protein 1 (RANBP1) that forms a complex with Ras-related nuclear protein (Ran) and metabolizes guanoside triphosphate (Supplementary Figure 1b (306KB, pdf) ).
Validation of protein expression by Western blotting
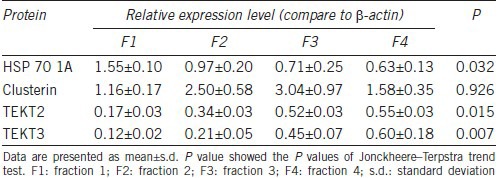
From the list of proteins that showed increasing or decreasing trends during the spermatozoa maturation, we selected 4 that are involved in key processes such as spermatozoa maturation and those that have been corroborated by other studies, for Western blotting validation. HSP 70 1A showed decreasing trends during spermatozoa maturation and was shown to take part in the response to oxidative stress. Tektin 2 and tektin 3 showed progressively increasing expression during spermatozoa maturation and were demonstrated to take part in motility acquisition. In addition, we selected a protein, clusterin, which did not show significant difference among four fractions in proteomic analysis, but was previously demonstrated by many other studies as an important protein for spermatozoa maturation.
Western blotting results confirmed and validated our findings from the proteomic analysis. Further, these results showed decreased expression levels of HSP 70 1A, while tektin 2 and tektin 3 expression levels were increased during spermatozoa maturation (Table 5). However, clusterin showed increasing trend from fraction 1 to fraction 3, and decreased from fraction 3 to fraction 4 (Figure 2b–2f).
Table 5.
Relative protein expression levels in four fractions of spermatozoa obtained from four individual samples from fertile donors by Western blotting assay
DISCUSSION
Obtaining human germ cells at different maturation stages is a big challenge. The ideal situation would be to conduct a proteomic study using different stages of sperm maturation such as gonocyte, spermatocyte, spermatid, and spermatozoa. However, this is not feasible without the use of invasive techniques. That is why no work has been done in this area. Even in animal studies, density gradient has been used for separating spermatozoa with different development stages in testis. Therefore, a proteomic study of human spermatozoa in different stages of maturation is necessary for a complete understanding of the process of spermatozoa maturation.
During spermatozoa maturation, nuclear histones are replaced by protamine, spermatozoa head is condensed, and extra cytoplasm is excluded gradually.5 Spermatozoa density increases proportionately with the maturation process, where the most mature and morphologically normal spermatozoa or the highest-quality spermatozoa display very high motility and are concentrated at the bottom of the sperm preparation media with the highest density. While it is true that spermatozoa in each fraction are not pure, and cell sorting technology or antibody labeled magnetic beads to enrich cells with different maturation stages would be ideal for sorting and separation of immature and mature cells, both the above techniques are not feasible in the present study. This is because of the fact that antibody availability for each stage of spermatozoa maturation is not available, and more importantly after sorting, cell numbers in each fraction would be too small for extraction of the proteins. Many of the studies, including our two previous studies6,12 have demonstrated that density gradient can separate spermatozoa at different maturation stages.6,12,17,30,31 Obtaining mature, morphologically normal, motile sperm with minimum DNA damage by sperm preparation on a density gradient is the most common accepted procedure in assisted reproductive laboratories for intrauterine insemination and in vitro fertilization.
We did not measure ROS levels in this study, as we have already reported the cell-to-cell variation in ROS levels in four fractions of sperm cells separated by a three-layer density gradient from samples obtained from fertile men as well as infertile men with normal and abnormal semen parameters and leukocytospermia.6 Similarly, we had also demonstrated chromatin damage measured by sperm chromatin structure assay in the same study groups in our earlier work.12 Sample limitation prevented us from repeating this again in our present study as this restricted the availability of the semen sample for proteomic study and Western blot analysis.
Proteomic study of pooled samples showed results similar to individual samples. 82.2% of the proteins could be found in both pooled and individual samples. Most of the proteins found only in the individual but not in pooled sample showed very low expression levels and could only be found in one of the four fractions. These data demonstrate that pooled samples are a good representative of the abundant proteins observed in individual samples. In proteomic studies, pooling is an important strategy. There are numerous studies in the literature that highlight the advantages of pooling samples when it is not feasible to analyze individual samples either due to limitations of the sample or the study design. It has been shown that pooling can reduce biological variations in studies such as two-dimensional differential in gel electrophoresis (2D-DIGE).40,41 A detailed evaluation of the benefits of pooling of samples was conducted by Diz et al.42 The authors concluded pooling of samples can provide similar results when compared to an individual analysis. These studies showed that although some information from the individual samples may be missed, pooled samples have the advantage of reducing biological variance and resource constraints.
In our proteomic study, we have shown that fraction 1 had the most abundant proteins. The numbers of detected proteins decreased proportionally to the maturation stage of spermatozoa. This finding was confirmed by bioinformatics analysis, that the expression levels of proteins involved in protein biosynthesis showed decreased expression according to the maturation stage of the spermatozoa. Our results showed that many of the ribosomal proteins showed decreasing trends. We believe that the decrease of ribosomal proteins could be due to both the loss of cytoplasm and the decrease of its expression. The loss of cytoplasm (along with the ribosomes) could further exacerbate the decrease of protein synthesis.
In fraction 4, some of the ribosome protein expression levels were decreased to very low or even non-existent levels. Many studies have shown that during spermatozoa maturation, transcription and translation are decreased because of post-meiotic DNA packaging and removal of the excessive cytoplasm.5,43 Our present results confirm that the number of expressed proteins is significantly decreased in mature spermatozoa. Our results of DAVID analysis demonstrated 11 proteins that take part in the decreasing trends of cell cycle and the proteolysis process (as shown in Table 4) that shows key biological processes associated with proteins that display a decreasing trend during spermatozoa maturation. In this table, we have demonstrated that 9 of the 11 proteins are regulatory subunits of the 26S proteasome.
Most of the proteins involved in this process were 26S protease regulatory proteins. 26S proteasome, in collaboration with ubiquitin, operate the energy-dependent and well-regulated proteolytic degradation of a variety of oncoproteins, transcription factors, cell cycle specific cyclins, cyclin-dependent kinase inhibitors, ornithine decarboxylase, and other key regulatory cellular proteins in eukaryotic cells.44,45 Studies have highlighted the importance of the 26S proteasome in various intractable diseases, such as cancer and heart disease.46 However, the role of 26S protease regulatory proteins in spermatozoa maturation, or in sperm function is not reported. Since oxidative stress and DNA fragmentation are higher in immature spermatozoa than in mature spermatozoa,6,30 and the 26S proteasome complex takes part in multiple cellular events including apoptosis, immune response, and DNA damage response,45 we deduce that besides protein ubiquitination and cell cycle arrest, these 26S proteosomal proteins might also take part in oxidative stress response and apoptosis of the immature spermatozoa. However, the exact function of 26S protease regulatory proteins in spermatozoa maturation needs further exploration.
Proteins such as importin, exportin, and ras-related proteins (Rab-1A isoform 1a; Rab-5C isoform b and ras–related Rab-6A isoform a, that take part in protein transport were observed to decrease as the spermatozoa matures. Our results show that during sperm maturation, along with decreased protein biosynthesis and the establishment of sperm homeostasis during spermatogenesis, protein transport declines as the sperm prepare for fertilization. During spermatogenesis, frequent nucleocytoplasmic transport has been reported. However, this process is almost paused or stopped in mature sperm, and can be activated after fertilization.47,48 Importin beta 1 (KPNB1) was also found to be a key point for the molecular transport and cellular assembly functions network.
Response to oxidative stress during spermatozoa maturation has been well-studied. It is well-known that immature spermatozoa especially those with the presence of excessive residual cytoplasm demonstrate higher ROS level than mature spermatozoa.6,12,30,31,49 The present study also found that many proteins were decreased along with the decrease in ROS level during the process of spermatozoa maturation. Of these proteins, we selected a well-studied protein, HSP 70 1A for the validation of our proteomic results using Western blotting. Various isoforms of HSP 70 have been known to be present in stage-specific and developmentally-regulated manner during spermatogenesis and spermatozoa maturation in mouse, rat, claw crayfish, and human.50,51,52,53,54 All these results show that HSP 70 has a higher expression level in immature spermatozoa than in mature spermatozoa. Both our proteomic results and Western blotting results demonstrate that HSP 70 1A had similar trend in expression as reported in the other studies.53,54
Flagellum development is one of the most important events in spermatozoa maturation. The principle piece of flagellum, which makes up about three-fourth of the length of flagellum, is comprised of the fibrous sheath. Nearly half of the proteins in fibrous sheaths isolated from mouse sperm include AKAP4.55 AKAP4 and AKAP3 have anchoring sites for cAMP-dependent protein kinase.56,57 AKAP3 also anchors ropporin, a spermatogenic cell-specific protein that is linked through rhophilin to the small GTPase Rho.58 AKAPs and their anchoring proteins are thought to be serine-threonine phosphorylation in response to cAMP signaling pathway which regulates the sperm maturation processes of motility, capacitation and hyperactivation.55 In an earlier study, we also demonstrated that AKAPs were decreased in ROS positive semen samples which exhibit lower motility.29 Our finding of the increase in AKAPs and its anchoring protein, ropporin during spermatozoa maturation corroborated with previous findings that show that AKAPs and their anchoring proteins are important for the development of sperm fibrous sheath.
Axonemal dynein heavy chain was found to have a higher expression level in fertile men than in asthenozoospermia patients and the mutation of axonemal dynein heavy chain gene can lead to male infertility.59 Tektins are evolutionarily conserved flagellar (and ciliary) filamentous proteins present in the axoneme and peri-axonemal structures in diverse metazoan species.60 Five tektins have been reported in human sperm flagella and all of them are thought to be essential for progressive sperm motility.61,62,63,64 Gene knockout for tektins in mouse models have shown normal epididymal sperm counts with reduced or almost absence of sperm motility and hyperactivated motility.62 In the present study, tektin 2 and tektin 3 were found to be significantly increased from fraction 1 to fraction 4 both in proteomic results and in Western blotting results. Our study findings demonstrate the importance of tektin 2 and tektin 3 in the maturation of spermatozoa.
For validation by Western blot, we selected HSP 70 1A which showed a decreasing trend and 2 proteins (tektin 2 and tektin 3) that showed an increasing trend from immature (fraction F1) to mature sperm (fraction F4). These 3 proteins have been well-studied for their functions during spermatozoa maturation. The expression levels changed in our proteomic study and this was confirmed by Western blotting study. Besides these three significant proteins mentioned above, we also studied another protein – clusterin that was reported to be important in the maturation of spermatozoa.65,66,67 It is secreted by the epididymis and seminal vesicle and is found to have a positive relationship with DNA fragmentation and morphological abnormalities of the spermatozoa.68 Higher expression level of clusterin has been reported in infertile patients than in fertile men.69 However, one recent study showed that clusterin can also be self-synthesized by spermatozoa during the later stages of spermatogenesis, but not in spermatogonia.70
Interestingly, in our proteomic results, clusterin was found to increase from fraction 1 to fraction 3, but decreased in fraction 4 as shown by the change in SC values from fraction 1 to fraction 4 were 153.0, 167.3, 192.3, and 127.7, respectively. Western blotting demonstrated similar trend in the expression of clusterin. These results show that clusterin did have a higher expression level in immature spermatozoa than in mature spermatozoa. However, in the immature spermatozoa, the expression level was increased along with the maturation process.
Clusterin is a major glycoprotein in mammalian semen. It is synthesized and secreted by the testis, epididymis, and seminal vesicle.71,72 Regardless of whether or not semen is of poor quality, clusterin mainly exists on the surface of immature, low motile or morphologically abnormal spermatozoa.71,73 Clusterin in the semen and on abnormal spermatozoa belongs to the secretory form of clusterin (sCLU). As the spermatozoa move through the rete testis and the efferent duct, testis-derived clusterin is replaced by the clusterin from the epididymis or seminal vesicles.72,74,75 Therefore, clusterin on the abnormal spermatozoa is implicated in decreased sperm motility, sperm aggregation and infertility.73,76 It is thus regarded as a marker of pathological spermatozoa. In humans, clusterin has also been reported in normal spermatozoa; however, this is a novel form of clusterin that is distinct from the heterodimeric sCLU.70
We hypothesize that, at the later stage of spermatogenesis (round spermatids to long spermatids), clusterin was synthesized by spermatozoa. Then during the epididymis maturation, spermatozoal - synthesized clusterin was gradually replaced by epididymis-secreted clusterin which then bind to the morphologically abnormal spermatozoa during maturation. Along with the morphological maturation of spermatozoa, clusterin may be proteolyzed gradually by proteinase, therefore, was expressed at very low levels in mature spermatozoa.
Male molecular causes of failures in IVF were identified by Frapsauce et al.28 They used sperm prepared by density gradient and utilized 2D-DIGE. They identified 17 proteins that were differentially expressed between fertilizing and nonfertilizing spermatozoa (mature and immature spermatozoa). Similarly, Dorin and Barratt reported the role of β-defensins in male infertility.77 In addition to the established role in innate immunity; these authors have demonstrated the role of β-defensins on sperm function. DEFB126 (β-defensins 126) was shown to be present all along the length of the sperm. DEFB126 mutations are believed to have a role in men with unexplained infertility and reduced ability to fertilize.77
There were some limitations of our study: (1) the number of healthy fertile subjects that were enrolled in our proteomic study was only 4 because of the strict exclusion criteria (2) although proteomic study provides us with significant information, we only analyzed those proteins with increasing and decreasing trends - from most immature to mature spermatozoa fraction. We did not analyze proteins that were unique, (3) although we have demonstrated many proteins that may be potential markers of spermatozoa maturation; we validated only three markers by Western blotting and (4) the fact that there is a statistically significant contamination with round cells in fractions 1 and 2 (Table 1) raises the possibility that some of the differences detected and reported here in the protein's relative abundance in fractions 1 and 2 could be due to this differential contamination with round cells. Examining a signal network that includes all of these significant proteins regulating spermatozoa maturation should be examined.
CONCLUSION
Our study provides important information about the proteins that are critical to spermatozoa maturation, motility, and fertilization capacity. It provides insight into the biological processes that are activated or suppressed during spermatozoa maturation. Future studies should be conducted in a larger patient population to examine the distribution of proteins in various stages of maturation in ejaculated spermatozoa obtained from infertile men with various clinical diagnoses. This will further help in understanding the underlying pathology of male infertility.
AUTHOR CONTRIBUTION
ZC conducted the study and helped with the data collection and management of this study. RS helped with the writing, reviewing, and editing of the manuscript. AA conceived the idea, supervised the study and edited the article for submission. All authors read and approved the final manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing interests.
ACKNOWLEDGMENTS
This study was supported by funds from the American Center for Reproductive Medicine. The Orbitrap Elite mass spectrometer used in this study was purchased with funds from an NIH shared instrument grant 1S10RR031537-01 to Belinda Willard, Lerner Research Institute. Zhihong Cui received a fellowship from the Chinese Government and the Institute of Toxicology, Third Military Medical University, Chongqing, People's Republic of China.
The authors are grateful to the Andrology Center technologists for scheduling the study subjects and Jeff Hammel for statistical analysis.
Supplementary information is linked to the online version of the paper on the Asian Journal of Andrology website.
REFERENCES
- 1.Wiser HJ, Sandlow J, Köhler TS. Causes of male infertility. In: Parekattil SJ, Agarwal A, editors. Male Infertility: Contemporary Clinical Approaches, Andrology, ART and Antioxidants. New York: Springer Science+Bussiness Media; 2012. [Google Scholar]
- 2.Agarwal A, Tvrda E, Sharma R. Relationship amongst teratozoospermia, seminal oxidative stress and male infertility. Reprod Biol Endocrinol. 2014;12:45. doi: 10.1186/1477-7827-12-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gudeloglu A, Brahmbhatt JV, Parekattil SJ. Medical management of male infertility in the absence of a specific etiology. Semin Reprod Med. 2014;32:313–8. doi: 10.1055/s-0034-1375184. [DOI] [PubMed] [Google Scholar]
- 4.Sabanegh E, Agarwal A. Male infertility. In: Wein AJ, editor. Campbell's Urology. Philadelphia, PA, USA: Elsevier Saunders; 2012. [Google Scholar]
- 5.Sharma R, Agarwal A. Spermatogenesis: an overview. In: Zini A, Agarwal A, editors. Sperm Chromatin: Biological and Clinical Applications in Male Infertility and Assisted Reproduction. New York, NY: Springer Science+Business Media; 2011. pp. 305–28. [Google Scholar]
- 6.Gil-Guzman E, Ollero M, Lopez MC, Sharma RK, Alvarez JG, et al. Differential production of reactive oxygen species by subsets of human spermatozoa at different stages of maturation. Hum Reprod. 2001;16:1922–30. doi: 10.1093/humrep/16.9.1922. [DOI] [PubMed] [Google Scholar]
- 7.Sakkas D, Mariethoz E, Manicardi G, Bizzaro D, Bianchi PG, et al. Origin of DNA damage in ejaculated human spermatozoa. Rev Reprod. 1999;4:31–7. doi: 10.1530/ror.0.0040031. [DOI] [PubMed] [Google Scholar]
- 8.Sakkas D, Seli E, Bizzaro D, Tarozzi N, Manicardi GC. Abnormal spermatozoa in the ejaculate: abortive apoptosis and faulty nuclear remodelling during spermatogenesis. Reprod Biomed Online. 2003;7:428–32. doi: 10.1016/s1472-6483(10)61886-x. [DOI] [PubMed] [Google Scholar]
- 9.Sakkas D, Seli E, Manicardi GC, Nijs M, Ombelet W, et al. The presence of abnormal spermatozoa in the ejaculate: did apoptosis fail? Hum Fertil (Camb) 2004;7:99–103. doi: 10.1080/14647270410001720464. [DOI] [PubMed] [Google Scholar]
- 10.Dun MD, Aitken RJ, Nixon B. The role of molecular chaperones in spermatogenesis and the post-testicular maturation of mammalian spermatozoa. Hum Reprod Update. 2012;18:420–35. doi: 10.1093/humupd/dms009. [DOI] [PubMed] [Google Scholar]
- 11.Lin C, Tholen E, Jennen D, Ponsuksili S, Schellander K, et al. Evidence for effects of testis and epididymis expressed genes on sperm quality and boar fertility traits. Reprod Domest Anim. 2006;41:538–43. doi: 10.1111/j.1439-0531.2006.00710.x. [DOI] [PubMed] [Google Scholar]
- 12.Ollero M, Gil-Guzman E, Lopez MC, Sharma RK, Agarwal A, et al. Characterization of subsets of human spermatozoa at different stages of maturation: implications in the diagnosis and treatment of male infertility. Hum Reprod. 2001;16:1912–21. doi: 10.1093/humrep/16.9.1912. [DOI] [PubMed] [Google Scholar]
- 13.Park YJ, Kim J, You YA, Pang MG. Proteomic revolution to improve tools for evaluating male fertility in animals. J Proteome Res. 2013;12:4738–47. doi: 10.1021/pr400639x. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Liu F, Liu X, Liu J, Zhu P, et al. Mapping of the human testicular proteome and its relationship with that of the epididymis and spermatozoa. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.004630. doi:10.1074/mcp.M110.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon WS, Rahman MS, Lee JS, Yoon SJ, Park YJ, et al. Discovery of predictive biomarkers for litter size in boar spermatozoa. Mol Cell Proteomics. 2015;14:1230–40. doi: 10.1074/mcp.M114.045369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guyonnet B, Zabet-Moghaddam M, SanFrancisco S, Cornwall GA. Isolation and proteomic characterization of the mouse sperm acrosomal matrix. Mol Cell Proteomics. 2012;11:758–74. doi: 10.1074/mcp.M112.020339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beydola T, Sharma R, Agarwal A. Sperm preparation and selection techniques. In: Rizk B, Aziz N, Agarwal A, Sabanegh E, editors. Male Infertility Practice. New Delhi: Jaypee Brothers Medical Publishers; 2013. pp. 244–51. [Google Scholar]
- 18.Cornwall GA. New insights into epididymal biology and function. Hum Reprod Update. 2009;15:213–27. doi: 10.1093/humupd/dmn055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macleod G, Varmuza S. The application of proteomic approaches to the study of mammalian spermatogenesis and sperm function. FEBS J. 2013;280:5635–51. doi: 10.1111/febs.12461. [DOI] [PubMed] [Google Scholar]
- 20.Aitken RJ, Buckingham DW, West KM. Reactive oxygen species and human spermatozoa: analysis of the cellular mechanisms involved in luminol- and lucigenin-dependent chemiluminescence. J Cell Physiol. 1992;151:466–77. doi: 10.1002/jcp.1041510305. [DOI] [PubMed] [Google Scholar]
- 21.Baker MA, Krutskikh A, Aitken RJ. Biochemical entities involved in reactive oxygen species generation by human spermatozoa. Protoplasma. 2003;221:145–51. doi: 10.1007/s00709-002-0057-0. [DOI] [PubMed] [Google Scholar]
- 22.Baker MA, Hetherington L, Weinberg A, Naumovski N, Velkov T, et al. Analysis of phosphopeptide changes as spermatozoa acquire functional competence in the epididymis demonstrates changes in the post-translational modification of Izumo1. J Proteome Res. 2012;11:5252–64. doi: 10.1021/pr300468m. [DOI] [PubMed] [Google Scholar]
- 23.Baker MA, Hetherington L, Reeves GM, Aitken RJ. The mouse sperm proteome characterized via IPG strip prefractionation and LC-MS/MS identification. Proteomics. 2008;8:1720–30. doi: 10.1002/pmic.200701020. [DOI] [PubMed] [Google Scholar]
- 24.Sharma R, Agarwal A, Mohanty G, Du Plessis SS, Gopalan B, et al. Proteomic analysis of seminal fluid from men exhibiting oxidative stress. Reprod Biol Endocrinol. 2013;11:85. doi: 10.1186/1477-7827-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma R, Agarwal A, Mohanty G, Hamada AJ, Gopalan B, et al. Proteomic analysis of human spermatozoa proteins with oxidative stress. Reprod Biol Endocrinol. 2013;11:48. doi: 10.1186/1477-7827-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma R, Agarwal A, Mohanty G, Jesudasan R, Gopalan B, et al. Functional proteomic analysis of seminal plasma proteins in men with various semen parameters. Reprod Biol Endocrinol. 2013;11:38. doi: 10.1186/1477-7827-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliva R, De Mateo S, Estanyol JM. Sperm cell proteomics. Proteomics. 2009;9:1004–17. doi: 10.1002/pmic.200800588. [DOI] [PubMed] [Google Scholar]
- 28.Frapsauce C, Pionneau C, Bouley J, Delarouziere V, Berthaut I, et al. Proteomic identification of target proteins in normal but nonfertilizing sperm. Fertil Steril. 2014;102:372–80. doi: 10.1016/j.fertnstert.2014.04.039. [DOI] [PubMed] [Google Scholar]
- 29.Hamada A, Sharma R, Du Plessis SS, Willard B, Yadav SP, et al. Two-dimensional differential in-gel electrophoresis-based proteomics of male gametes in relation to oxidative stress. Fertil Steril. 2013;99:1216–26. doi: 10.1016/j.fertnstert.2012.11.046. [DOI] [PubMed] [Google Scholar]
- 30.Brahem S, Mehdi M, Elghezal H, Saad A. Semen processing by density gradient centrifugation is useful in selecting sperm with higher double-strand DNA integrity. Andrologia. 2011;43:196–202. doi: 10.1111/j.1439-0272.2010.01050.x. [DOI] [PubMed] [Google Scholar]
- 31.Castillo J, Simon L, De Mateo S, Lewis S, Oliva R. Protamine/DNA ratios and DNA damage in native and density gradient centrifuged sperm from infertile patients. J Androl. 2011;32:324–32. doi: 10.2164/jandrol.110.011015. [DOI] [PubMed] [Google Scholar]
- 32.WHO. World Health Organization Laboratory Manual for the Examination and Processing of Human Semen. 5th ed. Geneva: World Health Organization Press; 2010. [Google Scholar]
- 33.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 34.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 35.Zybailov B, Coleman MK, Florens L, Washburn MP. Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal Chem. 2005;77:6218–24. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Wen Z, Washburn MP, Florens L. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal Chem. 2010;82:2272–81. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
- 37.Gokce E, Shuford CM, Franck WL, Dean RA, Muddiman DC. Evaluation of normalization methods on GeLC-MS/MS label-free spectral counting data to correct for variation during proteomic workflows. J Am Soc Mass Spectrom. 2011;22:2199–208. doi: 10.1007/s13361-011-0237-2. [DOI] [PubMed] [Google Scholar]
- 38.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, et al. GO: TermFinder – Open source software for accessing Gene Ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–5. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhatia VN, Perlman DH, Costello CE, McComb ME. Software tool for researching annotations of proteins: open-source protein annotation software with data visualization. Anal Chem. 2009;81:9819–23. doi: 10.1021/ac901335x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karp NA, Lilley KS. Investigating sample pooling strategies for DIGE experiments to address biological variability. Proteomics. 2009;9:388–97. doi: 10.1002/pmic.200800485. [DOI] [PubMed] [Google Scholar]
- 41.Chao HC, Chung CL, Pan HA, Liao PC, Kuo PL, et al. Protein tyrosine phosphatase non-receptor type 14 is a novel sperm-motility biomarker. J Assist Reprod Genet. 2011;28:851–61. doi: 10.1007/s10815-011-9602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diz AP, Truebano M, Skibinski DO. The consequences of sample pooling in proteomics: an empirical study. Electrophoresis. 2009;30:2967–75. doi: 10.1002/elps.200900210. [DOI] [PubMed] [Google Scholar]
- 43.Govin J, Gaucher J, Ferro M, Debernardi A, Garin J, et al. Proteomic strategy for the identification of critical actors in reorganization of the post-meiotic male genome. Mol Hum Reprod. 2012;18:1–13. doi: 10.1093/molehr/gar063. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka K, Mizushima T, Saeki Y. The proteasome: molecular machinery and pathophysiological roles. Biol Chem. 2012;393:217–34. doi: 10.1515/hsz-2011-0285. [DOI] [PubMed] [Google Scholar]
- 45.Frankland-Searby S, Bhaumik SR. The 26S proteasome complex: an attractive target for cancer therapy. Biochim Biophys Acta. 2012;1825:64–76. doi: 10.1016/j.bbcan.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crawford LJ, Walker B, Irvine AE. Proteasome inhibitors in cancer therapy. J Cell Commun Signal. 2011;5:101–10. doi: 10.1007/s12079-011-0121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Itman C, Miyamoto Y, Young J, Jans DA, Loveland KL. Nucleocytoplasmic transport as a driver of mammalian gametogenesis. Semin Cell Dev Biol. 2009;20:607–19. doi: 10.1016/j.semcdb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 48.Miyamoto Y, Boag PR, Hime GR, Loveland KL. Regulated nucleocytoplasmic transport during gametogenesis. Biochim Biophys Acta. 2012;1819:616–30. doi: 10.1016/j.bbagrm.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 49.Moustafa MH, Sharma RK, Thornton J, Mascha E, Abdel-Hafez MA, et al. Relationship between ROS production, apoptosis and DNA denaturation in spermatozoa from patients examined for infertility. Hum Reprod. 2004;19:129–38. doi: 10.1093/humrep/deh024. [DOI] [PubMed] [Google Scholar]
- 50.Angelier N, Moreau N, Rodriguez-Martin ML, Penrad-Mobayed M, Prudhomme C. Does the chaperone heat shock protein hsp70 play a role in the control of developmental processes? Int J Dev Biol. 1996;40:521–9. [PubMed] [Google Scholar]
- 51.Chan SH, Wang LL, Chang KF, Ou CC, Chan JY. Altered temporal profile of heat shock factor 1 phosphorylation and heat shock protein 70 expression induced by heat shock in nucleus tractus solitarii of spontaneously hypertensive rats. Circulation. 2003;107:339–45. doi: 10.1161/01.cir.0000044942.94957.87. [DOI] [PubMed] [Google Scholar]
- 52.Fang DA, Wang Q, He L, Wang J, Wang Y. Characterization of heat shock protein 70 in the red claw crayfish (Cherax quadricarinatus): evidence for its role in regulating spermatogenesis. Gene. 2012;492:138–47. doi: 10.1016/j.gene.2011.10.036. [DOI] [PubMed] [Google Scholar]
- 53.Naaby-Hansen S, Herr JC. Heat shock proteins on the human sperm surface. J Reprod Immunol. 2010;84:32–40. doi: 10.1016/j.jri.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan CC, Sun GH, Shui HA, Wu GJ. Differential spermatozoal protein expression profiles in men with varicocele compared to control subjects: upregulation of heat shock proteins 70 and 90 in varicocele. Urology. 2013;81:1379.e1–8. doi: 10.1016/j.urology.2013.01.031. [DOI] [PubMed] [Google Scholar]
- 55.Eddy EM, Toshimori K, O’Brien DA. Fibrous sheath of mammalian spermatozoa. Microsc Res Tech. 2003;61:103–15. doi: 10.1002/jemt.10320. [DOI] [PubMed] [Google Scholar]
- 56.Miki K, Eddy EM. Single amino acids determine specificity of binding of protein kinase A regulatory subunits by protein kinase A anchoring proteins. J Biol Chem. 1999;274:29057–62. doi: 10.1074/jbc.274.41.29057. [DOI] [PubMed] [Google Scholar]
- 57.Carr DW, Fujita A, Stentz CL, Liberty GA, Olson GE, et al. Identification of sperm-specific proteins that interact with A-kinase anchoring proteins in a manner similar to the type II regulatory subunit of PKA. J Biol Chem. 2001;276:17332–8. doi: 10.1074/jbc.M011252200. [DOI] [PubMed] [Google Scholar]
- 58.Li YF, He W, Mandal A, Kim YH, Digilio L, et al. CABYR binds to AKAP3 and Ropporin in the human sperm fibrous sheath. Asian J Androl. 2011;13:266–74. doi: 10.1038/aja.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ben Khelifa M, Coutton C, Zouari R, Karaouzene T, Rendu J, et al. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am J Hum Genet. 2014;94:95–104. doi: 10.1016/j.ajhg.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Linck R, Fu X, Lin J, Ouch C, Schefter A, et al. Insights into the structure and function of ciliary and flagellar doublet microtubules: tektins, Ca2+-binding proteins, and stable protofilaments. J Biol Chem. 2014;289:17427–44. doi: 10.1074/jbc.M114.568949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roy A, Lin YN, Agno JE, Demayo FJ, Matzuk MM. Tektin 3 is required for progressive sperm motility in mice. Mol Reprod Dev. 2009;76:453–9. doi: 10.1002/mrd.20957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanaka H, Iguchi N, Toyama Y, Kitamura K, Takahashi T, et al. Mice deficient in the axonemal protein Tektin-t exhibit male infertility and immotile-cilium syndrome due to impaired inner arm dynein function. Mol Cell Biol. 2004;24:7958–64. doi: 10.1128/MCB.24.18.7958-7964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bhilawadikar R, Zaveri K, Mukadam L, Naik S, Kamble K, et al. Levels of tektin 2 and CatSper 2 in normozoospermic and oligoasthenozoospermic men and its association with motility, fertilization rate, embryo quality and pregnancy rate. J Assist Reprod Genet. 2013;30:513–23. doi: 10.1007/s10815-013-9972-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cao W, Ijiri TW, Huang AP, Gerton GL. Characterization of a novel tektin member, TEKT5, in mouse sperm. J Androl. 2011;32:55–69. doi: 10.2164/jandrol.109.009456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dacheux JL, Belleannee C, Jones R, Labas V, Belghazi M, et al. Mammalian epididymal proteome. Mol Cell Endocrinol. 2009;306:45–50. doi: 10.1016/j.mce.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 66.Dacheux JL, Belleannee C, Guyonnet B, Labas V, Teixeira-Gomes AP, et al. The contribution of proteomics to understanding epididymal maturation of mammalian spermatozoa. Syst Biol Reprod Med. 2012;58:197–210. doi: 10.3109/19396368.2012.663233. [DOI] [PubMed] [Google Scholar]
- 67.Belleannee C, Labas V, Teixeira-Gomes AP, Gatti JL, Dacheux JL, et al. Identification of luminal and secreted proteins in bull epididymis. J Proteomics. 2011;74:59–78. doi: 10.1016/j.jprot.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 68.Muciaccia B, Pensini S, Culasso F, Padula F, Paoli D, et al. Higher clusterin immunolabeling and sperm DNA damage levels in hypertensive men compared with controls. Hum Reprod. 2012;27:2267–76. doi: 10.1093/humrep/des173. [DOI] [PubMed] [Google Scholar]
- 69.Thacker S, Yadav SP, Sharma RK, Kashou A, Willard B, et al. Evaluation of sperm proteins in infertile men: a proteomic approach. Fertil Steril. 2011;95:2745–8. doi: 10.1016/j.fertnstert.2011.03.112. [DOI] [PubMed] [Google Scholar]
- 70.Han Z, Wang Z, Cheng G, Liu B, Li P, et al. Presence, localization, and origin of clusterin in normal human spermatozoa. J Assist Reprod Genet. 2012;29:751–7. doi: 10.1007/s10815-012-9779-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Bryan MK, Murphy BF, Liu DY, Clarke GN, Baker HW. The use of anticlusterin monoclonal antibodies for the combined assessment of human sperm morphology and acrosome integrity. Hum Reprod. 1994;9:1490–6. doi: 10.1093/oxfordjournals.humrep.a138736. [DOI] [PubMed] [Google Scholar]
- 72.Sylvester SR, Morales C, Oko R, Griswold MD. Localization of sulfated glycoprotein-2 (clusterin) on spermatozoa and in the reproductive tract of the male rat. Biol Reprod. 1991;45:195–207. doi: 10.1095/biolreprod45.1.195. [DOI] [PubMed] [Google Scholar]
- 73.Carlsson L, Ronquist G, Nilsson BO, Larsson A. Dominant prostasome immunogens for sperm-agglutinating autoantibodies of infertile men. J Androl. 2004;25:699–705. doi: 10.1002/j.1939-4640.2004.tb02844.x. [DOI] [PubMed] [Google Scholar]
- 74.Bailey R, Griswold MD. Clusterin in the male reproductive system: localization and possible function. Mol Cell Endocrinol. 1999;151:17–23. doi: 10.1016/s0303-7207(99)00016-7. [DOI] [PubMed] [Google Scholar]
- 75.Griffiths GS, Galileo DS, Aravindan RG, Martin-Deleon PA. Clusterin facilitates exchange of glycosyl phosphatidylinositol-linked SPAM1 between reproductive luminal fluids and mouse and human sperm membranes. Biol Reprod. 2009;81:562–70. doi: 10.1095/biolreprod.108.075739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lambard S, Galeraud-Denis I, Martin G, Levy R, Chocat A, et al. Analysis and significance of mRNA in human ejaculated sperm from normozoospermic donors: relationship to sperm motility and capacitation. Mol Hum Reprod. 2004;10:535–41. doi: 10.1093/molehr/gah064. [DOI] [PubMed] [Google Scholar]
- 77.Dorin JR, Barratt CL. Importance of b-defensins in sperm function. Mol Hum Reprod. 2014;20:821–6. doi: 10.1093/molehr/gau050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proteins detected in four fractions of fertile men ejaculated semen
The cellular development and cell motility functions associated network identified by IPA network generation algorithm (a). The proteins marked in “blue” are the ones from our dataset while the remaining ones in “black” are the proteins in the Ingenuity Knowledge Base (IKB) that interact with ours. The nine proteins in blue show increasing trends expression during spermatozoa maturation process. ACR: acrosin; AKAP3: a kinase anchor protein 3; AKAP4: a kinase anchor protein 3; ALAT: dihydrolipoamids S-acetyltransferase; PDHA2: pyruvate dehydrogenase (lipoamide) alpha 2; PRSS12: protease, serine, 12; SLC 25A3: solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3; APAG6: sperm associated antigen 6; SPAG 16: sperm associated antigen 16. (b) Molecular transport and cellular assembly functions the seven proteins in blue showed significant decreasing trends in spermatozoa maturation. CSE1L: CSE1 chromosome segregation 1-like; PRDX1: peroxiredoxin 1; PRDX2: peroxiredoxin 2; PRDX4: peroxiredoxin 4: RANBP1: RAN binding protein 1; XPO1: exportin 1; KPNB1: karyopherin (importin) beta 1.


