Publisher's Note: There is an Inside Blood Commentary on this article in this issue.
Key Points
PTNFL is a biologically distinct indolent lymphoma characterized by common MEK/ERK pathway mutations.
The biology of PTNFL is not defined by age, as the mutational profile is similar in pediatric and adult cases.
Abstract
Pediatric-type nodal follicular lymphoma (PTNFL) is a variant of follicular lymphoma (FL) characterized by limited-stage presentation and invariably benign behavior despite often high-grade histological appearance. It is important to distinguish PTNFL from typical FL in order to avoid unnecessary treatment; however, this distinction relies solely on clinical and pathological criteria, which may be variably applied. To define the genetic landscape of PTNFL, we performed copy number analysis and exome and/or targeted sequencing of 26 PTNFLs (16 pediatric and 10 adult). The most commonly mutated gene in PTNFL was MAP2K1, encoding MEK1, with a mutation frequency of 43%. All MAP2K1 mutations were activating missense mutations localized to exons 2 and 3, which encode negative regulatory and catalytic domains, respectively. Missense mutations in MAPK1 (2/22) and RRAS (1/22) were identified in cases that lacked MAP2K1 mutations. The second most commonly mutated gene in PTNFL was TNFRSF14, with a mutation frequency of 29%, similar to that seen in limited-stage typical FL (P = .35). PTNFL was otherwise genomically bland and specifically lacked recurrent mutations in epigenetic modifiers (eg, CREBBP, KMT2D). Copy number aberrations affected a mean of only 0.5% of PTNFL genomes, compared with 10% of limited-stage typical FL genomes (P < .02). Importantly, the mutational profiles of PTNFLs in children and adults were highly similar. Together, these findings define PTNFL as a biologically and clinically distinct indolent lymphoma of children and adults characterized by a high prevalence of MAPK pathway mutations and a near absence of mutations in epigenetic modifiers.
Introduction
Pediatric-type nodal follicular lymphoma (PTNFL) is currently considered a variant of follicular lymphoma (FL) that is characterized by localized presentation and invariably benign behavior despite its often high-grade (ie, grade 3) histological appearance.1 Until recently, specific and objective criteria for defining this indolent variant have not been available. We recently identified high proliferation index and the absence of BCL2, BCL6, and IRF4 gene rearrangements as characteristic features of PTNFL in both children and adults.2 PTNFL has a follicular architecture with no component of diffuse large B-cell lymphoma, and follicles are often composed of medium-sized blastoid cells, rather than classical centrocytes or centroblasts.1 Cases defined by these criteria present with localized lymphadenopathy and are associated with an invariably excellent prognosis.1,2
Recent studies have shown that PTNFL is not restricted to the pediatric age group: PTNFLs frequently present in young adults between 18 and 30 years of age and occasionally in older adults with the same indolent behavior. Patients with PTNFL consistently have indefinite remissions if treated with surgical excision alone.3 However, most adults with PTNFL continue to be treated with standard chemoimmunotherapy and/or radiation therapy, irrespective of BCL2 translocation status, because of its histological mimicry of high-grade typical FL. Therefore, objective biological markers are needed to distinguish PTNFL from typical FL in both children and adults in order to avoid potentially unnecessary treatment.
Considering the unique clinical behavior of PTNFL, we hypothesized that its mutational landscape would differ from that of typical FL. In contrast to the well-characterized landscape of typical FL,3-16 the molecular genetic features of PTNFL are essentially undefined. Therefore, we performed comprehensive mutation analysis and copy number variant analysis on PTNFLs from children and adults to address this problem.
Methods
Patients
Formalin-fixed, paraffin-embedded (FFPE) blocks and stained slides from 44 cases of stage I or II FL were collected from the pathology archives of the following academic medical centers: Massachusetts General Hospital, Ann & Robert H. Lurie Children's Hospital of Chicago, Weill Cornell Medical College/New York–Presbyterian Hospital, Brigham & Women's Hospital/Dana-Farber Cancer Institute, Boston Children's Hospital, Stanford University School of Medicine, State University of New York Downstate Medical Center, and University of Pittsburgh Medical Center. Initial exclusion criteria included (1) evidence of advanced stage (ie, stage III or IV) disease at the time of diagnosis, (2) histological evidence of a diffuse large B-cell component, and (3) insufficient material for comprehensive mutation analysis.
Whole exome capture and next-generation sequencing (NGS)
Six tumor/germ-line pairs of PTNFL exome libraries were generated, sequenced, and analyzed at the Broad Institute, as previously outlined.17 Sixteen additional tumor exomes were sequenced and analyzed by the Dana-Farber Cancer Institute Center for Cancer Genome Discovery. Whole exome capture was performed using the Agilent SureSelect Human All Exon 44 Mb v2.0 bait set (Agilent Technologies). In summary, genomic DNA was sheared, end repaired, ligated with barcoded Illumina sequencing adapters, amplified, size selected, and subjected to in solution hybrid capture using the Agilent SureSelect Human All Exon v2.0 bait set.18 Resulting exome Illumina sequencing libraries were then quantitative polymerase chain reaction quantified, pooled, and sequenced with 76 base paired-end reads using Illumina GAII or HiSequation 2000 sequencers to a minimum of 80% of exome targets covered to 20× (after removal of duplicates and low-quality reads). Reads were aligned to the hg19/GRCh37 build of the reference human genome sequence and annotated using Oncotator. These and other tools used for exome sequence analysis are described on the Broad Institute Cancer Genome Analysis Web site.
Targeted NGS
Targeted exon capture and NGS of all coding exons of 113 genes (supplemental Table 1, available on the Blood Web site) previously described to be mutated in FL were performed as previously described19-21 on PTNFL cases. Additional sequencing of TNFRSF14 and CREBBP was performed on limited-stage typical follicular lymphoma (LSTFL) cases.
Mutation calling
Variants were rejected as germ line if they were present in the matched nontumor sample, if available. Otherwise, they were rejected if they were known germ-line polymorphisms from the Exome Sequencing Project and/or dbSNP (build 142) databases. We used the evolutionary conservation of the affected amino acid in protein homologs to predict the functional effect of detected variants. We only considered variants predicted to be nonsilent (ie, missense, nonsense, translation start site, or splice site alterations, or in-frame or frame-shift insertions/deletions) and with variant allele frequencies (VAFs) of 4% or more. For every alteration meeting these criteria, sequencing reads were individually visualized to evaluate mapping quality and the phase and spatial distribution of alterations within reads. Mutations were then called by cross-referencing candidate variants to ClinVar, COSMIC, and a review of published literature. To obtain high confidence mutation calls, we also applied the following filters as applied in Louissaint et al2: (1) Panel of normals: Variant calls from ExAC project (ExAC_nonTCGA.r0.3.1)3 were used to filter germ-line variants. Variants observed with frequency >0.01% in ExAC were excluded. (2) Blacklisted mutations: Variants blacklisted by TCGA pan-cancer analysis4 were also excluded. On the basis of a bimodal distribution of VAFs (supplemental Figure 1), in which there was a large peak centered at ∼50% with a minimal VAF of 35%, and a smaller peak centered at ∼8%, we excluded variants with a VAF of >35%. The final sets of curated mutations are shown in supplemental Table 1.
Copy number analysis
Analysis was performed on DNA isolated as described previously from FFPE tissue blocks. Tumor content was determined by pathologist (A.L.) review of hematoxylin and eosin (H&E)–stained sections. Approximately 80 ng of DNA was used for hybridization. Genomic microarray analysis was performed using the OncoScan FFPE Assay Kit (Affymetrix, Santa Clara, CA). This kit utilizes molecular inversion probe (MIP) technology to interrogate both copy number and genotype by single nucleotide polymorphism (SNP) analysis. Data analysis was performed using Nexus Express Software for OncoScan (Biodiscovery, El Segundo, CA). Files were processed using the TuScan or SNP-FASST2 algorithms and using a minimum of 20 probes per segment. All calls were manually reviewed. In cases where false or inaccurate calls were identified because of poor probe performance or misinterpretation of genomic context by the software, certain calls were manually omitted or edited in the software.
Statistical analysis
Associations between categorical variables were assessed by Fisher’s exact test. Estimates of failure-free survival were determined using the Kaplan-Meier method and compared with the log-rank test. All P values are 2 sided.
Results
Clinicopathological features of PTNFL
We collected 44 cases of limited-stage (stage I/II) FL from 8 academic medical centers. Cases were defined as PTNFL based on the following criteria2: (1) nodal involvement only, (2) architectural effacement by a clonal follicular proliferation (with clonality based on polymerase chain reaction for IgH gene rearrangement), (3) presence of high proliferation index (ie, >30% by Ki-67 staining), and (4) absence of MUM1/IRF4 expression by immunohistochemistry and BCL2 or BCL6 rearrangements by fluorescence in situ hybridization (FISH). Twenty-six of the 44 cases met these criteria for PTNFL. The remaining 18 cases were classified as LSTFL.
The clinicopathological features of the 2 groups are summarized in Table 1. Subjects with PTNFL included 16 children (4-18 years, median 15 years), and 10 adults (20-60 years; median 30 years). Patients with LSTFL were older (33-85 years, median 62.9 years) than those with PTNFL (P < .0001). Patients with PTNFL were almost all male (92%), and 92% presented with localization to head and neck lymph nodes. In contrast, patients with LSTFL were predominantly female (P < .0001 compared with PTNFLs) and less commonly had head and neck involvement (P < .0001 compared with PTNFLs).
Table 1.
Clinical, laboratory, and treatment characteristics of PTNFL and LSTFL cases included in this series
| PTNFL |
LSTFL |
P |
PTNFL ≤18 |
PTNFL >18 |
P |
|
|---|---|---|---|---|---|---|
| N (%) | N (%) | PTNFL vs typical FL | N (%) | N (%) | PTNFL <18 vs >18 | |
| Number of cases | 26 | 18 | 16 | 10 | ||
| Male:female | 24:2 | 5:13 | <.0001 | 16:0 | 8:2 | .14 |
| Median age, y (range) | 16.0 (4-60) | 62.9 (33-85) | <.0001 | 14.7 (4-18) | 30 (20-60) | <.0001 |
| Age >18 y | 10 (38) | 18 (100) | <.0001 | 0 | 10 (100) | |
| Stage | .008 | 1 | ||||
| I | 26 (100) | 13 (72) | 14 (100) | 9 (100) | ||
| II | 0 | 5 (28) | 0 | 0 | ||
| Nodal location | ||||||
| Head and neck | 24 (92) | 4 (22) | <.0001 | 15 (94) | 9 (90) | 1 |
| Supraclavicular | 1 (4) | 0 | 1 (6) | 0 | ||
| Inguinal | 1 (4) | 9 (50) | 0 | 1 (10) | ||
| Axillary | 0 | 1 (6) | 0 | 0 | ||
| Other* | 0 | 4 (22) | 0 | 0 | ||
| Grade | ||||||
| 1-2 | 2 (8) | 15 (83) | 0 | 2 (20) | ||
| 3 | 24/26 (92) | 3 (17) | 16/16 | 8 (80) | ||
| Immunohistochemistry | ||||||
| CD10+ | 26/26 (100) | 17/18 (94) | .41 | 16/16 (100) | 10/10 (100) | 1 |
| BCL6+ | 26/26 (100) | 16/16 (100) | 1 | 16/16 (100) | 10/10 (100) | 1 |
| BCL2+ | 1/26 (4) | 16/18 (89) | <.0001 | 1/16 (6) | 0/10 (0) | 1 |
| MUM1+ | 0/26 (0) | 0/18 (0) | 1 | 0/16 (0) | 0/10 (0) | 1 |
| PI >30% (Ki-67) | 26/26 (100) | 5/15 (33) | <.0001 | 16/16 (100) | 10/10 (100) | 1 |
| Gene rearrangements | ||||||
| BCL2 rearrangement | 0/26 (0) | 16/18 (89) | <.0001 | 0 | 0 | 1 |
| BCL6 rearrangement | 0/26 (0) | 2/16 (13) | .1617 | 0 | 0 | 1 |
| Therapeutic approach | <.001 | <.01 | ||||
| Excision alone | 17 (65) | 2 (11) | 14 (88) | 3 (30) | ||
| Excision + rituximab | 1 (4) | 1 (6) | 1 (6) | 0 | ||
| Local RT (25-36 Gy) | 3 (12) | 11 (61) | 0 | 3 (30) | ||
| R-CHOP/+/−IFRT | 3 (12) | 4 (22) | 0 | 3 (30) | ||
| Unknown | 2 (8) | 0 | 1 (6) | 1 (10) | ||
| Outcome | ||||||
| Follow-up (mo) | 39 (4-126) | 38 (10-144) | .94 | 26 (4-126) | 43 (6-101) | .31 |
| Remission/NED | 24 (92) | 7 (35) | <.0001 | 15 (94) | 9 (90) | 1 |
| Recurrence/progression | 0 | 11 (65) | 0 | 0 | ||
| Unknown | 2 (8) | 0 | 1 (6) | 1 (10) | ||
| Transformation | 0 | 3 (18) | <.05 | 0 | 0 | 1 |
P values designating statistical significance are highlighted bold and italics.
IFRT, involved field RT; NED, no evidence of disease; PI, proliferation index; R-CHOP, rituximab, cyclophosphamide, hydroxydaunorubicin, oncovin, prednisone; RT, radiation therapy.
Other includes saphenous, femoral, abdominal, and antecubital lymph nodes.
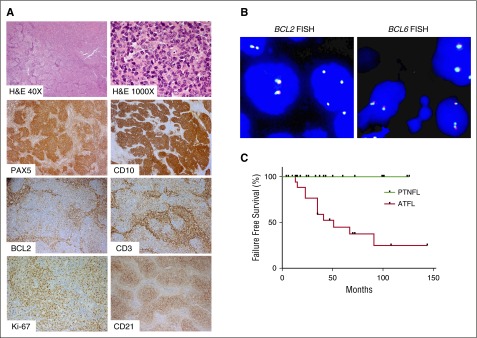
Histologically, PTNFLs had effacement of the lymph node by a follicular proliferation of CD20+CD10+BCL6+ B cells that lacked BCL2 expression and BCL2 or BCL6 rearrangement in all 26 cases (Figure 1A-B; Table 1). Follicles in 24 cases (92%) were composed of centroblasts and/or medium-sized cells with blastoid features, and the remaining 2 cases (8%) had grade 1 to 2 of 3 histology (Figure 1A; Table 1). There were no cases with pure centroblastic morphology (ie, grade 3B). Cases of LSTFL mostly had grade 1 to 2 histology (83%). As expected, LSTFLs had BCL2 expression (89% of cases), generally low proliferation index by Ki-67 staining (10 of 15 with data available), and the presence of BCL2 and/or BCL6 gene rearrangements (16 of 17 with data available).
Figure 1.
Morphology and immunophenotype of PTNFL. (A) A representative PTNFL showing architectural effacement by follicular proliferation (H&E, ×40). Follicles were composed of medium-sized cells with blastoid features (H&E, ×1000) that were PAX5+ CD10+ with dim or no BCL2 expression. There were scattered occasional CD3+ BCL2+ T cells within follicles. Follicles harbored a tight network of CD21+ follicular dendritic cell meshworks and had a relatively high proliferation index, on the basis of Ki-67 staining. Magnification: ×100 for PAX5, CD10, BCL2, CD3, CD2; ×200 for Ki-67. (B) FISH for BCL2 rearrangement, using LSI BCL2 Dual color break apart rearrangement probe (left), and FISH for BCL6 rearrangement, using LSI BCL6 Dual color break apart rearrangement probe (right). PTNFLs lacked both BCL2 and BCL6 rearrangements as evidenced by colocalization of probes. (C) LSTFL had significantly lower progression-free survival than PTNFL (P < .001).
Differences in treatment of PTNFL based on patient age
Overall, 88% of patients ≤18 years old with PTNFL were treated with excision alone compared with only 30% of patients >18 years old (Table 1; P < .01). Among the 24 patients with PTNFL and known outcome (median follow-up, 39 months; range, 4-126 months), all 24 (100%) remain in complete remission, including 17 treated with excision alone (median follow-up, 33 months; range, 4-126 months).
Most patients with LSTFL were treated with involved-field radiotherapy (61%) and/or R-CHOP (22%). Despite being treated more aggressively than patients with PTNFL, failure-free survival was significantly lower in patients with LSTFL (Figure 1C; P < .001). There was also a significantly greater likelihood of histological transformation to diffuse large B-cell lymphoma in patients with LSTFL compared with those with PTNFL (18% vs 0%, P < .01; Table 1).
PTNFL is characterized by frequent MAPK pathway mutations
We performed whole-exome sequencing (WES) on tumor/normal pairs from the 6 patients with PTNFL and matched controls available. WES produced an average of 132.5 million and 122.2 million reads per tumor and matched control sample, respectively. The mean coverage depth of the exome was 99 for tumor samples and 100 for matched control (median, 89 and 99, respectively), with an average of 85% and 87% of the target tumor and control exome, respectively, covered by >20 reads. WES on the 6 exomes revealed 267 putative coding or splice-site, nonsynonymous somatic mutations within 252 genes (supplemental Table 1A).
At the behest of an astute reviewer, we performed WES on 15 additional PTNFL cases that lacked paired germ-line material. The average mean coverage depth of the tumor exomes was 121.5× (range, 67-172.2); an average of 94% of all targets were covered by ≥30× depth. WES on the 16 additional exomes revealed 1945 variants. Of these variants, 1571 had a VAF >35%, likely representing germ-line variants (supplemental Figure 1; see “Methods”). The 374 remaining variants had a median VAF of 0.10 (supplemental Table 1B; supplemental Figure 1). These included 305 (82%) missense single nonsynonymous variants, 39 (10%) insertions or deletions, and 30 (8%) nonsense single nonsynonymous variants.
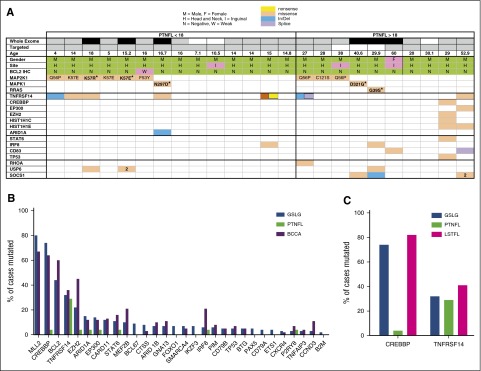
Taking into consideration all 21 cases that underwent exome sequencing, the only 2 genes mutated in ≥4 samples were TNFRSF14 and MAP2K1 (supplemental Table 1B; Figure 2). Overall, MAP2K1 (which encodes the MEK1 protein) was the most commonly mutated gene in PTNFL, with mutations in 9 (43%) of 21 cases, 2 of which occurred in samples with paired germ-line sequencing (supplemental Figure 2). MAP2K1 mutations were present at a median VAF of 0.12 by WES (range, 5% to 24%). All MAP2K1 mutations were known activating mutations in the negative regulatory (F53Y, Q56P [3], K57E [3], K57R) region and the catalytic core (C121S) domain, encoded by exons 2 and 3, respectively (supplemental Figure 2). PTNFLs (10%) had known activating mutations in MAPK1, which encodes extracellular signal-regulated kinase 2 (ERK2) and is immediately downstream of MEK1 in the MEK/ERK pathway, and 1 PTNFL had a known activating mutation in RRAS. Both MAPK1 mutations and the RRAS mutation were identified in samples with paired germ-line sequencing (supplemental Figure 2). The mutations in MAP2K1, MAPK1, and RRAS were mutually exclusive, such that in aggregate, somatic mutations in MAPK pathway genes were identified in 57% of samples (Figure 2A; supplemental Table 1B). Notably, mutations in BRAF, which are recurrent in other malignancies with frequent MAP2K1 mutations,22 were not identified in any case.
Figure 2.
Mutational profile of PTNFL. (A) Mutational profile of PTNFL. The table includes genes with somatic nonsynonymous variants identified in >2 cases by targeted and WES of PTNFL samples. A known gain-of-function mutation in RHOA is also included. Top rows delineate whether whole exome (black, initial 6 exomes; gray, subsequent exomes; white, indicates not sequenced) or targeted sequencing was performed. Amino acid substitutions are indicated for MAPK pathway alterations. MAPK variants identified in samples with paired sequencing are delineated by bold text. The number “2” indicates multiple missense variants. (B) Comparison of mutation frequencies among PTNFLs and 2 cohorts of advanced stage FL (GSLG and BCCA). The differences in mutational frequencies between PTNFL vs GSLG, and PTNFL vs BCCA were statistically significant for KMT2D (P < 10−6, P < 10−5), CREBBP (P < 10−5, P < 10−6), BCL2 (P < 10−5, P < 10−6), and EZH2 (P = .05, P < 10−4). (C) Mutational frequencies of CREBBP and TNFRSF14 in PTNFL vs LSTFL. CREBBP mutations were present in 83% of LSTFLs vs 4% of PTNFL (P < 10−6). TNFRSF14 mutations were present in 29% of PTNFLs vs 41% of LSTFLs (P = .35).
TNFRSF14 mutations were present in 7 (33%) of 21 cases (Figure 2). SOCS1, P2RY8, and USP6 were each mutated in 3 cases. Interestingly, 1 PTNFL harbored a RHOA P5W mutation previously described to have gain-of-function properties in gastric cancers. Consistent with the lack of BCL2 translocation, there was a complete absence of BCL2 mutations in PTNFL, which occur at high frequency in typical FL.
PTNFLs lack mutations in epigenetic modifier genes commonly mutated in typical FL
In order to more rigorously assess for mutations in genes recurrently mutated in typical FL, we performed targeted sequencing of 21 PTNFL cases, including 18 of the 21 analyzed by exome sequencing, for 113 genes previously found to be mutated in typical FL23 (supplemental Table 2). With the targeted strategy, we achieved an average target coverage of 250× (range, 97× to 557×) and an average of 98% of the targets were covered at ≥30× depth. Among the 21 samples, only 13 had nonsynonymous, coding, or splice site variants in any of the 113 genes. These 13 samples harbored 37 somatic nonsynonymous variants within 20 of the 113 genes. This translated into a median of 1 nonsilent mutation per patient (range, 0-7; Figure 2A; supplemental Table 3). In comparison, our previous analysis of ∼300 typical advanced-stage FLs biopsied within 12 months of receiving up-front treatment demonstrated a median of 4 (German Low Grade Lymphoma Study Group [GSLG]2000 cohort) or 5 (British Columbia Cancer Agency [BCCA] cohort) nonsynonymous mutations per FL using the same 113 gene panel (Figure 2; supplemental Figure 4).23 The median number of nonsynonymous mutations per PTNFL did not differ between patients ≤18 or >18 years old (1 vs 1 mutation per case; Figure 2A; supplemental Table 5).
Nonsilent mutations in any of the epigenetic modifier genes EP300, CREBBP, EZH2, KMT2D, or ARID1A were identified in only 3 of the 21 PTNFLs analyzed by targeted sequencing (Figure 2A). Two of the 3 patients (a 29-year-old and a 53-year-old) each harbored 3 different epigenetic mutations, and the third patient harbored a single ARID1A mutation. The 29-year-old also harbored STAT6, TP53, and EZH2 mutations, and the 53-year-old also harbored mutations in SOCS1 and CD83. There were no significant differences in the frequency of mutation for any epigenetic modifier genes between patients ≤18 or >18 years of age (supplemental Table 5).
CREBBP is mutated in a majority of patients with advanced stage typical FL,10,23 but in the PTNFL cohort, the only CREBBP mutation was from the aforementioned 29-year-old (Figure 2A). To determine whether the relative absence of CREBBP mutations was unique to PTNFL or could simply be a marker of limited-stage disease, we sequenced all exons of CREBBP and TNFRSF14 in 18 cases of LSTFL. In contrast to PTNFLs, 83% of LSTFLs harbored nonsynonymous CREBBP mutations (P < 10−6; Figure 2C; supplemental Tables 3 and 4) and 40% of LSTFLs had multiple nonsynonymous CREBBP mutations (P < 10−6). In contrast with CREBBP, TNFRSF14 was mutated in PTNFLs (29%) at a similar frequency to that of LSTFL cases (41%, P = .35; Figure 2C; supplemental Tables 3 and 4).
Copy number variants are uncommon in PTNFL
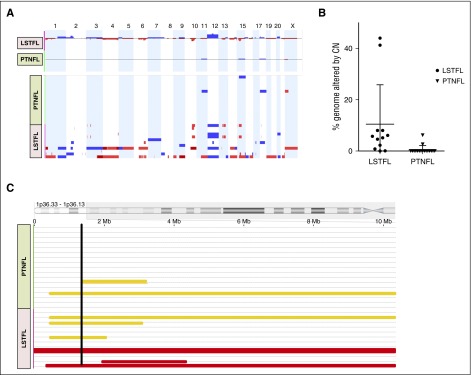
We used an MIP assay to quantify copy number alterations (CNAs) and copy number neutral loss of heterozygosity (CN-LOH) in 17 PTNFLs and 11 LSTFLs. CNAs were detected in 9 of 11 LSTFLs and were often complex (Figure 3A). In contrast, only 2 of 17 PTNFLs harbored detectable alterations (P < 10−4 vs LSTFLs), and these were limited to trisomies and a small X chromosome deletion in 1 case (Figure 3A). Overall, a mean of 10% of the genome of LSTFLs and 0.5% of the genome of PTNFLs was altered by copy number aberrations (P = .012; Figure 3B). Across all PTNFL and LSTFL samples, foci of CN-LOH were present at multiple loci, including chromosomes 1p, 1q, 3p, 3q, 4p, 5q, 6p, 11p, 12q, 14p, 15q, 16p, and 19p. CN-LOH involving the same region in >1 case occurred only at 6p (3 LSTFLs), 11p (3 LSTFLs), and 1p (3 LSTFLs, 2 PTNFLs). Foci of CN-LOH overlapped at chromosome 1p36, which includes the TNFRSF14 locus (Figure 3C). Both PTNFLs and all 3 LSTFLs with CN-LOH at chromosome 1p36 also harbored nonsynonymous TNFRSF14 mutations.
Figure 3.
CNAs in PTNFL. (A) CNAs in PTNFL and LSTFL. Copy number gains (blue) and copy number losses (red) for each of the PTNFLs (n = 17) and LSTFLs (n = 11) assessed with the MIP assay (bottom). Copy number gains and losses are superimposed to show aggregate CNAs for each cohort (top). There were fewer copy number changes in PTNFL vs LSTFL (P < .0001). (B) Percent genome altered by copy number in each case. A greater percentage of the genome was altered by CNAs for LSTFLs in comparison with PTNFLs (P < .05). (C) CN-LOH at chromosome 1p36 in PTNFL vs LSTFL. Foci of CN-LOH (yellow bars) overlapped the TNFRSF14 locus (black line). Each of the 3 LSTFLs and 2 PTNFLs with CN-LOH also harbored a mutation in TNFRSF14. Two additional LSTFLs (red bars) had copy number loss at this locus.
Discussion
PTNFL is characterized by limited-stage presentation and invariably excellent behavior despite its often high-grade histological appearance. In contrast to typical low-grade FL, which is considered indolent but incurable, and high-grade FL, which is clinically aggressive and requires chemoimmunotherapy, PTNFL is clinically indolent and cured with surgical excision alone. On this basis, we hypothesized that its mutational landscape would differ from that of typical FL. The goal of this study was to define the genomic differences that may distinguish it from typical FL. We discovered that PTNFL has a unique mutational profile that is fundamentally distinct from other non-Hodgkin lymphomas, including typical FL.
In this study, we find that PTNFL features a striking absence of mutations in many genes recurrently altered in typical FL, most notably in epigenetic modifier genes. In contrast to typical FL, in which CREBBP and KMT2D mutations occur in ∼80% of cases,6,7,9,23 recurrent mutations in these genes were not identified within our PTNFL cohort. We also show that CREBBP mutations occur at high frequency in typical FL with limited-stage presentation, just as observed in advanced stage FL. Thus, the relative absence of common FL mutations in PTNFL is not solely a reflection of an early stage of FL pathogenesis but suggests a distinctive PTNFL biology.
The only exceptions to the absence of typical FL mutations in PTNFL were mutations in TNSFRSF14 and to a lesser extent mutations in P2RY8. Both PTNFLs and LSTFLs harbored TNFRSF14 mutations and CN-LOH at chr.1p36 at similar frequencies. Mutations in TNFRSF14 may alter the response of immune effector cells to FL,24 suggesting that these mutations may confer a mechanism of immune escape rather than directly contributing to FL phenotypes like proliferation or metastasis.
Two of the cases included in the PTNFL group harbored 6 of the 7 epigenetic modifier mutations identified, as well as STAT6, HIST1H1C/E, SOCS1, and TP53 mutations. Based on this mutation profile, it is possible that these cases are in fact biologically more similar to LSTFL than PTNFL. One of the cases occurred in a 29-year-old man with localized cervical lymphadenopathy. Biopsy of the lesion showed mostly centroblasts, and the patient was treated with chemotherapy. Only 6 months of follow-up could be obtained for this case. The other case was that of a 53-year-old man with a periauricular mass. Biopsy of the lesion showed grade 1 to 2 of 3 histology. The patient was lost to follow-up, and it is not clear how he was treated.
A major finding in this study is the identification of recurrent somatic mutations in the MAPK pathway in PTNFL. MAP2K1 encodes MEK1, the immediate target of BRAF and regulator of ERK1/2. Although several tumor types harbor mutations in various genes within the MAPK pathway, RAS and BRAF mutations are usually the most frequent, with MAP2K1 mutations occurring less commonly in most tumor types.22,25-27 Therefore, the frequency of MAP2K1 mutations in PTNFL was striking, as MAP2K1 mutations occurred in 9/21 (43%) PTNFLs that underwent exome sequencing while BRAF mutations were not identified. MAP2K1 mutations have been recently identified in 50% of cases of hairy cell leukemia variant (HCLv) as well as cases of BRAF wild-type Langerhans cell histiocytosis (LCH) and other histiocytic disorders.22,25,27 Unlike PTNFL, BRAF mutations predominate over MAP2K1 mutations in HCLv and LCH. Of note, germ-line mutations in MAP2K1 are also associated with cardio-facio-cutaneous syndrome, which is characterized by developmental delay, cardiac defects, and craniofacial abnormalities.28
All MAP2K1 mutations identified in our cases of PTNFL are missense mutations that target known hotspots in the negative regulatory domain and the catalytic core of MEK1 (supplemental Figure 2).27 These particular mutations are recurrent in other malignancies, such as melanoma, HCLv, and LCH, as well as in cardio-facio-cutaneous syndrome. The same missense mutations in MAP2K1 increase basal MEK enzyme activity and promote cell proliferation in various in vitro assays.27
In addition to the 9 MAP2K1 mutations, 2 MAPK1 mutations (encoding ERK2) and 1 RRAS mutation were also identified in PTNFL. Both MAPK1 mutations identified (N297D and D321G) have been previously described in several other types of cancers and lymphomas, including mycosis fungoides.29 RRAS is a 23-kDa monomeric guanosine triphosphatase that transduces signals from activated cell surface receptors to the RAF/MEK/ERK pathway, and has a 55% to 60% homology to RAS. RRAS G39 is within the G1 motif that mediates guanosine triphosphate/guanosine diphosphate binding and guanosine triphosphatase activity and is a major hotspot in human cancers. Mutations at that RRAS G39 promote enhanced serum-dependent MEK, ERK, and AKT phosphorylation in vitro.30
We identified mutations in several genes (eg, PAK3, USP6, ABCF1, STK39) that were uncommon but may confer important effects within individual PTNFLs. Further study is needed to determine the contribution of these mutations within cases of PTNFL. In addition, important mutations may have been missed in our exome sequencing because of low coverage, their presence within subclones, and/or our VAF cutoff of ≥4%.
Taken together, the MAPK mutations were present in 57% of PTNFLs assessed by exome sequencing. The high prevalence of MAP2K1 mutation in PTNFL and the mutual exclusivity with mutations in other components of the same pathway strongly suggests the importance of MAPK pathway activation in the pathogenesis of PTNFL.
In 2008, the World Health Organization classification recognized “pediatric follicular lymphoma” as a provisional variant of FL, mainly defined by age, with some characteristic features including effacement of the lymph node architecture by expansile follicles with a predominance of mitotically active cells, absence of BCL2 protein expression, and frequent presentation in young boys with localized cervical lymphadenopathy.31 The features defining this entity have recently been refined in patients with nodal disease; these include high proliferative index, absence of BCL2, BCL6 and IRF4 rearrangements, and the presence of medium-sized to large blastoid cells rather than typical centroblasts.1,2 It was also recently recognized that “pediatric-type” FL may also occur in adults, most commonly among those <30 years of age.1,2 As a result of these recent observations, the term PTNFL was proposed to describe a single entity that occurs in both children and adults and is associated with benign clinical behavior.2
Children with PTNFL have an excellent response to surgical excision alone without subsequent progression or recurrence of disease.3 However, as seen in our PTNFL cohort, adults with PTNFL continue to be treated with chemoimmunotherapy and/or radiotherapy, because of its morphological overlap with high-grade FL, and the lack of definite evidence that these cases are biologically equivalent to PTNFL in children. In the current study, we provide strong evidence that PTNFL is frequently driven by aberrant activation of the MAPK pathway, rather than by genetic alteration of epigenetic modifiers as seen in typical FL. Moreover, we show for the first time that PTNFLs occurring in children and adults have essentially identical mutational profiles, including MEK/ERK pathway mutations, TNFRSF14 mutations, and an absence of recurrent mutations in epigenetic modifiers.
These findings confirm that PTNFL is a biologically distinct indolent lymphoma defined not by age of presentation, but by its unique biology that is associated with particularly indolent behavior. On this basis, the therapeutic approach and management of adults with PTNFL should be reevaluated. Activation of the MEK/ERK pathway in PTNFL by activating mutations may serve as an important objective parameter to confirm the diagnosis of PTNFL and guide a more conservative therapeutic approach. Whether therapeutic inhibitors of MEK will play a role in the treatment of this disease remains unclear.
Acknowledgments
The authors thank Paul van Hummelen, Aaron Thorner, Matthew Ducar, Jaegil Kim, Mara Rosenberg, and Maegan Harden for assistance with sequencing; Abha Aggarwal, Shumei Wang, Anita L. Hawkins, and Cynthia C. Morton for assistance with FISH; and Michelle F. Lee for assistance with images.
This work was supported by a grant from the National Institutes of Health, National Cancer Institute (K23CA184279). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. A.L. receives research support from American Cancer Society, the American Society of Hematology/Amos Medical Faculty Development Program, and Harvard Catalyst Program for Faculty Development and Diversity Inclusion. A.L. and E.P.H. gratefully acknowledge funding support from an anonymous foundation. D.M.W. is a Leukemia and Lymphoma Society Scholar.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.L. and D.M.W. designed the research and wrote manuscript; R.M. contributed to the design of research; K.T.S., J.T.G., A.E.K., D.G., C.G.R., E.A.M., R.P.H., N.L.H., E.P.H., and M.H.H. designed the research and contributed vital reagents; A.L., C.N.P., C.N., and S.K. performed research; and D.S.N., S.T.S., L.A.G., M.G., and A.L. analyzed the data.
Conflict-of-interest disclosure: L.A.G. is a consultant for Foundation Medicine, Novartis, Boehringer Ingelheim, and Third Rock; an equity holder in Foundation Medicine; and a member of the Scientific Advisory Board at Warp Drive. L.A.G. receives sponsored research support from Novartis, Astellas, BMS, and Merck. The remaining authors declare no competing financial interests.
Correspondence: Abner Louissaint Jr, Massachusetts General Hospital, Department of Pathology, 55 Fruit St, Warren 2, Boston, MA 02114; e-mail: alouissaint@partners.org.
References
- 1.Liu Q, Salaverria I, Pittaluga S, et al. Follicular lymphomas in children and young adults: a comparison of the pediatric variant with usual follicular lymphoma. Am J Surg Pathol. 2013;37(3):333–343. doi: 10.1097/PAS.0b013e31826b9b57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louissaint A, Jr, Ackerman AM, Dias-Santagata D, et al. Pediatric-type nodal follicular lymphoma: an indolent clonal proliferation in children and adults with high proliferation index and no BCL2 rearrangement. Blood. 2012;120(12):2395–2404. doi: 10.1182/blood-2012-05-429514. [DOI] [PubMed] [Google Scholar]
- 3.Attarbaschi A, Beishuizen A, Mann G, et al. European Intergroup for Childhood Non-Hodgkin Lymphoma (EICNHL) and the international Berlin-Frankfurt-Münster (i-BFM) Study Group. Children and adolescents with follicular lymphoma have an excellent prognosis with either limited chemotherapy or with a “Watch and wait” strategy after complete resection. Ann Hematol. 2013;92(11):1537–1541. doi: 10.1007/s00277-013-1785-2. [DOI] [PubMed] [Google Scholar]
- 4.Cheung KJ, Johnson NA, Affleck JG, et al. Acquired TNFRSF14 mutations in follicular lymphoma are associated with worse prognosis. Cancer Res. 2010;70(22):9166–9174. doi: 10.1158/0008-5472.CAN-10-2460. [DOI] [PubMed] [Google Scholar]
- 5.Morin RD, Johnson NA, Severson TM, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42(2):181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Annibali O, Romeo AA, Agostinelli C, et al. A case of primary MALT lymphoma of the endometrium presenting as an asymptomatic polyp. Ann Hematol. 2009;88(5):491–493. doi: 10.1007/s00277-008-0615-4. [DOI] [PubMed] [Google Scholar]
- 7.Wright TM, Rule S, Liu H, Du MQ, Smith ME. Extranodal marginal zone lymphoma of the uterine corpus. Leuk Lymphoma. 2012;53(9):1831–1834. doi: 10.3109/10428194.2012.660750. [DOI] [PubMed] [Google Scholar]
- 8.Ryan RJ, Nitta M, Borger D, et al. EZH2 codon 641 mutations are common in BCL2-rearranged germinal center B cell lymphomas. PLoS One. 2011;6(12):e28585. doi: 10.1371/journal.pone.0028585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iyengar P, Deodhare S. Primary extranodal marginal zone B-cell lymphoma of MALT type of the endometrium. Gynecol Oncol. 2004;93(1):238–241. doi: 10.1016/j.ygyno.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 10.Heeren JH, Croonen AM, Pijnenborg JM. Primary extranodal marginal zone B-cell lymphoma of the female genital tract: a case report and literature review. Int J Gynecol Pathol. 2008;27(2):243–246. doi: 10.1097/PGP.0b013e3181569a0a. [DOI] [PubMed] [Google Scholar]
- 11.Guo S, Chan JK, Iqbal J, et al. EZH2 mutations in follicular lymphoma from different ethnic groups and associated gene expression alterations. Clin Cancer Res. 2014;20(12):3078–3086. doi: 10.1158/1078-0432.CCR-13-1597. [DOI] [PubMed] [Google Scholar]
- 12.Karube K, Martínez D, Royo C, et al. Recurrent mutations of NOTCH genes in follicular lymphoma identify a distinctive subset of tumours. J Pathol. 2014;234(3):423–430. doi: 10.1002/path.4428. [DOI] [PubMed] [Google Scholar]
- 13.Frey NV, Svoboda J, Andreadis C, et al. Primary lymphomas of the cervix and uterus: the University of Pennsylvania’s experience and a review of the literature. Leuk Lymphoma. 2006;47(9):1894–1901. doi: 10.1080/10428190600687653. [DOI] [PubMed] [Google Scholar]
- 14.Hamadani M, Kharfan-Dabaja M, Kamble R, Kern W, Ozer H. Marginal zone B-cell lymphoma of the uterus: a case report and review of the literature. J Okla State Med Assoc. 2006;99(4):154–156. [PubMed] [Google Scholar]
- 15.Pasqualucci L, Khiabanian H, Fangazio M, et al. Genetics of follicular lymphoma transformation. Cell Rep. 2014;6(1):130–140. doi: 10.1016/j.celrep.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yildiz M, Li H, Bernard D, et al. Activating STAT6 mutations in follicular lymphoma. Blood. 2015;125(4):668–679. doi: 10.1182/blood-2014-06-582650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapman MA, Lawrence MS, Keats JJ, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gnirke A, Melnikov A, Maguire J, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27(2):182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagle N, Berger MF, Davis MJ, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2(1):82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Odejide O, Weigert O, Lane AA, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. 2014;123(9):1293–1296. doi: 10.1182/blood-2013-10-531509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lane AA, Odejide O, Kopp N, et al. Low frequency clonal mutations recoverable by deep sequencing in patients with aplastic anemia. Leukemia. 2013;27(4):968–971. doi: 10.1038/leu.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chakraborty R, Hampton OA, Shen X, et al. Mutually exclusive recurrent somatic mutations in MAP2K1 and BRAF support a central role for ERK activation in LCH pathogenesis. Blood. 2014;124(19):3007–3015. doi: 10.1182/blood-2014-05-577825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vang R, Medeiros LJ, Ha CS, Deavers M. Non-Hodgkin’s lymphomas involving the uterus: a clinicopathologic analysis of 26 cases. Mod Pathol. 2000;13(1):19–28. doi: 10.1038/modpathol.3880005. [DOI] [PubMed] [Google Scholar]
- 24.Collette Y, Gilles A, Pontarotti P, Olive D. A co-evolution perspective of the TNFSF and TNFRSF families in the immune system. Trends Immunol. 2003;24(7):387–394. doi: 10.1016/s1471-4906(03)00166-2. [DOI] [PubMed] [Google Scholar]
- 25.Brown NA, Furtado LV, Betz BL, et al. High prevalence of somatic MAP2K1 mutations in BRAF V600E-negative Langerhans cell histiocytosis. Blood. 2014;124(10):1655–1658. doi: 10.1182/blood-2014-05-577361. [DOI] [PubMed] [Google Scholar]
- 26.Alayed K, Medeiros LJ, Patel KP, et al. BRAF and MAP2K1 mutations in Langerhans cell histiocytosis: a study of 50 cases. Hum Pathol. 2016;52:61–67. doi: 10.1016/j.humpath.2015.12.029. [DOI] [PubMed] [Google Scholar]
- 27.Waterfall JJ, Arons E, Walker RL, et al. High prevalence of MAP2K1 mutations in variant and IGHV4-34-expressing hairy-cell leukemias. Nat Genet. 2014;46(1):8–10. doi: 10.1038/ng.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cave H. Gene symbol: MAP2K1. Disease: cardio-facio-cutaneous syndrome. Hum Genet. 2008;123(5):551. [PubMed] [Google Scholar]
- 29.da Silva Almeida AC, Abate F, Khiabanian H, et al. The mutational landscape of cutaneous T cell lymphoma and Sézary syndrome. Nat Genet. 2015;47(12):1465–1470. doi: 10.1038/ng.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flex E, Jaiswal M, Pantaleoni F, et al. Activating mutations in RRAS underlie a phenotype within the RASopathy spectrum and contribute to leukaemogenesis. Hum Mol Genet. 2014;23(16):4315–4327. doi: 10.1093/hmg/ddu148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris NL, Swerdlow SH, Jaffe ES, et al. Follicular lymphoma. In: Swerdlow SH, Campo E, Harris NL, et al., editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC; 2008. pp. 220–226. [Google Scholar]