Publisher's Note: There is an Inside Blood Commentary on this article in this issue.
Key Points
PTFL is a monoclonal B-cell neoplasia with low genomic complexity and recurrent TNFRSF14 mutations/deletions.
The genetic profiles of conventional t(14;18)− and t(14;18)+ FL are similar but distinct from PTFL.
Abstract
Pediatric-type follicular lymphoma (PTFL) is a variant of follicular lymphoma (FL) with distinctive clinicopathological features. Patients are predominantly young males presenting with localized lymphadenopathy; the tumor shows high-grade cytology and lacks both BCL2 expression and t(14;18) translocation. The genetic alterations involved in the pathogenesis of PTFL are unknown. Therefore, 42 PTFL (40 males and 2 females; mean age, 16 years; range, 5-31) were genetically characterized. For comparison, 11 cases of conventional t(14:18)− FL in adults were investigated. Morphologically, PTFL cases had follicular growth pattern without diffuse areas and characteristic immunophenotype. All cases showed monoclonal immunoglobulin (IG) rearrangement. PTFL displays low genomic complexity when compared with t(14;18)− FL (mean, 0.77 vs 9 copy number alterations per case; P < .001). Both groups presented 1p36 alterations including TNFRSF14, but copy-number neutral loss of heterozygosity (CNN-LOH) of this locus was more frequently observed in PTFL (40% vs 9%; P = .075). TNFRSF14 was the most frequently affected gene in PTFL (21 mutations and 2 deletions), identified in 54% of cases, followed by KMT2D mutations in 16%. Other histone-modifying genes were rarely affected. In contrast, t(14;18)− FL displayed a mutational profile similar to t(14;18)+ FL. In 8 PTFL cases (19%), no genetic alterations were identified beyond IG monoclonal rearrangement. The genetic landscape of PTFL suggests that TNFRSF14 mutations accompanied by CNN-LOH of the 1p36 locus in over 70% of mutated cases, as additional selection mechanism, might play a key role in the pathogenesis of this disease. The genetic profiles of PTFL and t(14;18)− FL in adults indicate that these are two different disorders.
Introduction
Follicular lymphoma (FL) is among the most common non-Hodgkin lymphomas in adults (22%).1 However, it is very rare in the pediatric population, representing only 1% to 2% of B-cell NHL in children and young adults.2 Pediatric FL was recognized many years ago as an indolent disease affecting predominantly young males with a median age of 15 years, and presenting with isolated lymphadenopathy in the head and neck regions.3,4 Further studies demonstrated that the cells within the follicles showed mostly high-grade cytology (grade 3), IGH monoclonality, and lacked the characteristic t(14;18)(q32;q21) chromosomal alteration detectable in >80% of adult FL.5-7 BCL2 protein expression is usually absent, but ∼20% of the cases show weak positivity.5,7 Although this lymphoma is typically seen in children and adolescents, cases with similar morphology and phenotype have also been described in adults; therefore, the name pediatric-type FL (PTFL) has been proposed to highlight the lack of an age cutoff in the definition of the disease.6 In early studies, PTFL was treated with chemotherapy with excellent response.7 However, recent studies recommend a “watch-and-wait” strategy for cases in which a complete lymph node (LN) resection is achieved in stage I patients.8
The genetic alterations involved in the pathogenesis of nodal PTFL are currently largely unknown. Due to the indolent disease course, it has been questioned whether nodal PTFL represents a true lymphoma or rather an atypical “monoclonal” lymphoid hyperplasia. A recent study of FL patients under 18 years of age described TNFRSF14 gene mutations in a subset of cases.9 However, some of these cases were associated with a diffuse large B-cell component and advanced stage, not really representing the usual indolent nodal PTFL. Interestingly, the TNFRSF14 gene is localized on chromosome 1p36 and alterations of this locus and mutations of the gene are very common in adult FL (18%-46%).10,11
In past years, the genetic landscape of conventional FL, beyond the well-known BCL2 translocation,12 has been characterized comprehensively using next-generation sequencing (NGS).13-18 This and similar high-throughput approaches revealed that germinal center (GC)-derived lymphomas are characterized by frequent mutations of histone-modifying genes. In adult-type FL, histone methyltransferase KMT2D (formerly MLL2) and EZH2, and the histone acetylases CREBBP, EP300, and MEF2B, are the most commonly mutated genes. More recently, recurrent mammalian target of rapamycin complex 1–activating RRAGC mutations in FL have been described.19 This suggests that progression from t(14;18)+ premalignant precursor cells to FL is likely the result of epigenetic alterations.14,15 PTFL is also a lymphoma derived from GC cells, but without the characteristic t(14;18) translocation. However, mutational analyses of histone-modifying genes, important for the pathogenesis of FL, have not been performed so far in PTFL. Another important issue is the differential diagnosis between PTFL and conventional t(14;18)− FL in adults. It has been suggested that t(14;18)− FL in adults often presents with low clinical stage and excellent prognosis, and therefore might represent a form of PTFL.6,20 However, a recent study reported similar clinical features in FL with and without t(14;18) translocation.21 The aim of this study was to perform a comprehensive genetic analysis of a large series of nodal PTFL to better understand the pathogenesis of this disorder. Comparison with genetic alterations of conventional t(14;18)− FL should allow us to identify similarities and differences between these 2 related disorders.
Material and methods
Lymphoma samples and clinical data
Forty-two cases of nodal PTFL with material available for genetic studies were collected from several institutions, including University of Tübingen (Tübingen, Germany), National Cancer Institute, National Institutes of Health (NIH; Bethesda, MD), Hospital Clinic (Barcelona, Spain), and University College of London (London, United Kingdom). Eight additional cases were obtained from the workshop organized by the European Association for Haematopathology.20 The cases were reviewed by 4 of the authors (T.M., E.C., E.S.J., and L.Q.-M.) following the criteria of the 2008 World Health Organization Classification (WHO).22 All cases have >70% tumor cells. In addition, 10 LNs with florid reactive hyperplasia (RH) in children and young adults without demonstrable monoclonal IGH or IGK gene rearrangements and 11 conventional t(14;18)− FL23 were included as controls. The morphology, growth pattern, cytology, and immunohistochemical stainings for CD20, CD3, immunoglobulin D, Ki-67, BCL6, BCL2, MUM1, and CD10 protein expression were evaluated in formalin-fixed paraffin-embedded (FFPE) tissue sections performed as part of the diagnostic workup. The study was performed in accordance with the Declaration of Helsinki, and was approved by the local ethics review committee and the institutional review board panels of the contributing institutions.
DNA extraction and clonality analysis
DNA from FFPE tissue was extracted after dewaxing and protein K digestion applying standard phenol/chloroform purification procedures or either with the Qiagen FFPE DNA Tissue kit (Qiagen, Valencia, CA) or the Maxwell FFPE Tissue LEV DNA Purification kit (Promega, Madison, WI) according to the manufacturer’s protocol.
Polymerase chain reaction (PCR) amplifications for detecting monoclonal IGH chain gene rearrangements were performed according to the BIOMED-2 protocol24 using a D4 fluorescent dye modified JH consensus primer (Sigma-Aldrich, St. Louis, MO) and 2 different concentrations (30 or 60 ng) of genomic DNA. PCR products were analyzed on the GenomeLab GeXP Genetic Analysis System (Beckman Coulter, Pasadena, CA).
Cases with a polyclonal pattern in IGH clonality analysis were further analyzed for monoclonal rearrangements in the IGK gene using the BIOMED-2 protocol.24 In 2 multiplex PCRs, the family-specific Vκ primers were used in combination with either Jκ or Kde primers, which in addition amplify JΚ-CΚ intron-Kde rearrangements. Presence of IGH and/or IGK monoclonal rearrangements was part of the inclusion criteria.
Cytogenetics and fluorescence in situ hybridization
Interphase fluorescence in situ hybridization (FISH) analysis for the detection of BCL2 translocation was performed using a LSI BCL2 Dual Color Break Apart Rearrangement Probe (Vysis; Abbott Molecular, Wiesbaden, Germany). Screening for IRF4 rearrangements was performed by a self-labeled probe as described previously.25 In some cases, FISH for the t(14;18)/IGH/BCL2 and t(8;14)/IGH/MYC translocations was performed as part of the diagnostic workup. FISH analysis on metaphases using the D7S486/CEP 7 FISH Probe kit (Vysis) was performed in case PTFL24 (supplemental Figure 1, available on the Blood Web site).
Copy-number analysis
DNAs extracted from FFPE from 40 PTFL, 6 reactive LNs (RH), and 11 conventional t(14;18)− FLs were hybridized on the molecular inversion probe assay using Oncoscan FFPE according to standard protocols (Affymetrix, Santa Clara, CA). Gains and losses and copy-number neutral loss of heterozygosity (CNN-LOH) regions were evaluated and visually inspected using Nexus Biodiscovery version 7.5 software (Biodiscovery, Hawthorne, CA). Human reference genome was GRCh37/hg19. The copy number alterations (CNAs) with minimum size of 100 kb and CNN-LOH larger than 5 Mb were considered informative. Physiological deletions of the immunoglobulin loci were excluded from the analysis. T-cell receptor locus deletions were also excluded, most probably representing physiological deletions of accompanying reactive T cells.
NGS analysis
NGS analysis was done on the Ion Torrent PGM from Thermo Fisher Scientific (Schwerte, Germany). Thirty-seven PTFL cases, 11 conventional t(14;18)− FL, and 9 RH with good DNA quality (>200 bp) were subjected to NGS analysis. In the 5 remaining PTFL cases not analyzable by NGS due to low-quality DNA, TNFRSF14 exons 1-8 were analyzed by Sanger sequencing. NGS libraries were amplified using 2 primer pools of an Ion AmpliSeq custom panel covering 12 genes that have been shown to be frequently mutated in FL. The panel covered 98.24% of all exons of TNFRSF14, KMT2D (MLL2), FOXO1, EP300, MEF2B, HIST1H1B-E, and GNA13 as well as hotspot regions of EZH2 (exon 16) and CREBBP (exons 24-38 and 30) (for more information, see supplemental Table 1). The custom panel was designed using the Ion AmpliSeq Designer from Thermo Fisher Scientific (version 3.4). For description of library preparation, sequencing, and raw data analyses, see supplemental Methods.
Targeted resequencing and Sanger sequencing
For validation of the NGS results, variants with low allelic frequencies (<15%) were reanalyzed as single amplicons using a targeted resequencing approach on the Ion Torrent PGM. Amplicons were prepared using the Ion Amplicon Library Preparation Fusion Method protocol from Thermo Fisher Scientific. For description of primer design, see supplemental Methods and supplemental Table 2.
Variants with high allelic frequencies (≥15%) were validated with Sanger sequencing. M13-tailed primers were designed with Primer3 software (version 4.0.0; primer3.ut.ee/) obtained from Sigma-Aldrich (Steinheim, Germany) (see supplemental Table 3). PCRs were performed using 100 ng of DNA in a final volume of 25 µL with 200 µM dNTPs (Thermo Fisher Scientific), 0.2 µM per primer, 0.5 mM MgCl2 (Thermo Fisher Scientific), and 0.02 U/µL Phusion Hot Start High Fidelity Polymerase (Thermo Fisher Scientific). Purification of PCR products and analysis of sequencing reactions was performed as previously described.23
Statistical analysis
Comparison between clinical and biological parameters was performed using the Fisher exact or Mann-Whitney tests using IBM SPSS v.22.0. All statistical tests were 2-sided and statistical significance was concluded for values of P < .05.
Results
Clinical and morphological findings in PTFL
The clinical information is summarized in Table 1. A total of 42 patients were included in the study of which 40 were males and 2 females with a median age of 16 years (range, 5-31 years). LNs of the head and neck were most frequently involved (35 of 42 cases). All patients except for 2 had stage I disease. Ten patients were treated. Twelve patients have been followed with a watch-and-wait approach after the excision of the enlarged LN. The 23 patients with follow-up are alive with no evidence of disease with a median follow-up of 36 months (range, 2-96 months).
Table 1.
Clinical features of 42 PTFL patients
| Case no. | Sex | Age, y | Anatomic site | Stage | Treatment | Follow-up, mo |
|---|---|---|---|---|---|---|
| PTFL1 | M | 16 | Midline cervical LN | NA | NA | NA |
| PTFL2 | M | 19 | Submental LN | I | Excision | 38 NED |
| PTFL3 | M | 12 | Left cervical LN | I | Excision | 31 NED |
| PTFL4 | M | 13 | Left cervical LN | NA | NA | NA |
| PTFL5 | M | 18 | Left cervical LN | NA | NA | NA |
| PTFL6 | M | 17 | Submandibular LN | NA | NA | NA |
| PTFL7 | M | 14 | Cervical LN | NA | NA | NA |
| PTFL8 | M | 27 | Right cervical LN | NA | NA | NA |
| PTFL9 | M | 19 | Cervical LN | NA | NA | NA |
| PTFL10 | M | 17 | Left cervical LN | NA | NA | NA |
| PTFL11 | M | 23 | Cervical LN | NA | NA | NA |
| PTFL12 | M | 5 | Right femoral LN | NA | NA | NA |
| PTFL13* | M | 12 | Posterior auricular LN | II | CHOP | 36 NED |
| PTFL14 | M | 12 | Left submandibular LN | NA | NA | NA |
| PTFL15* | M | 14 | Cervical LN | I | R, C, doxo | 8 NED |
| PTFL16* | M | 15 | Axillary LN | NA | R-CHOP | 96 NED |
| PTFL17* | M | 17 | Left cervical LN | I | CHOP | 30 NED |
| PTFL18* | M | 20 | Right thigh LN | I | R-CHOP, Rx | 78 NED |
| PTFL19* | M | 20 | Right jugular LN | I | Excision | 2 NED |
| PTFL20* | M | 21 | Occipital LN | I | R-CHOP | 18 NED |
| PTFL21 | M | 14 | Left intraparotid LN | I | Excision | 19 NED |
| PTFL22 | M | 15 | Cervical LN | I | Excision | 51 NED |
| PTFL23 | M | 5 | Right cervical LN | I | NA | 36 NED |
| PTFL24 | M | 6 | Upper extremity mass | I | Excision | 44 NED |
| PTFL25 | M | 21 | Right inguinal LN | NA | NA | NA |
| PTFL26 | F | 17 | Left cervical LN | II | Rx | 48 NED |
| PTFL27 | M | 17 | Right prehyoidal LN | I | Excision | 48 NED |
| PTFL28 | M | 10 | Right cervical LN | I | Excision | 24 NED |
| PTFL29 | M | 15 | Right maxillary sinus LN | I | Excision | 24 NED |
| PTFL30 | M | 23 | Inguinal LN | I | R, Rx | 7 NED |
| PTFL31 | M | 17 | Left cervical LN | I | R, Rx | 51 NED |
| PTFL32 | M | 17 | Cervical LN | NA | NA | NA |
| PTFL33 | M | 15 | Parotid gland LN | I | R-CHOP | 48 NED |
| PTFL34 | M | 11 | Post auricular LN | NA | NA | NA |
| PTFL35 | M | 8 | Left LN axillary | NA | NA | NA |
| PTFL36 | M | 14 | Submandibular LN | I | NA | NA |
| PTFL37 | M | 16 | Left parotid LN | I | Excision | 36 NED |
| PTFL38* | M | 17 | Submandibular LN | I | Excision | 71 NED |
| PTFL39 | F | 20 | Parotid gland LN | I | NA | NA |
| PTFL40 | M | 22 | Cervical LN | NA | NA | NA |
| PTFL41 | M | 20 | Parotid gland LN | I | Excision | 12 NED |
| PTFL42 | M | 31 | Cervical LN | I | NA | NA |
C, cyclophosphamide; CHOP, cyclophosphamide, adriamycin, vincristine, prednisone; doxo, doxorubicin; F, female; M, male; NA, not available; NED, not evidence of disease; R, rituximab; Rx, local radiation.
These cases were previously reported by Liu et al.5
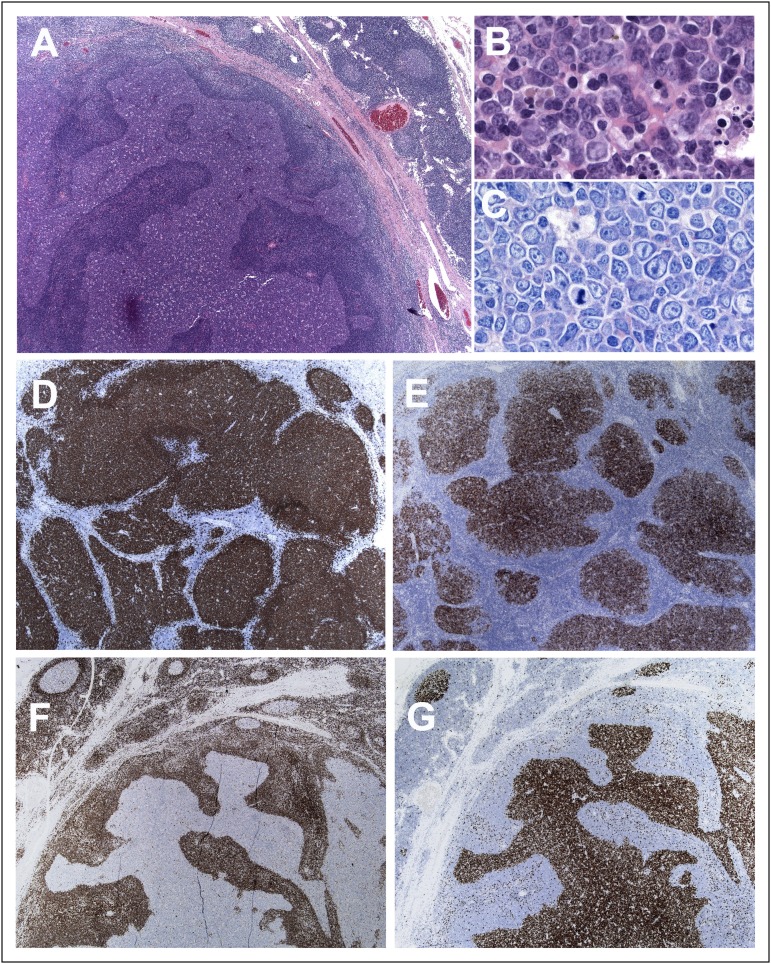
The morphology and immunophenotype in all cases were characteristic of PTFL (Figure 1). The LN architecture was partially or totally effaced by expansile, serpiginous, and sometimes confluent follicles with a starry sky pattern but without polarization (Figure 1A). The atypical follicles were composed of medium-sized to large blastoid cells but in some cases centroblasts predominated, characteristic of grade 3B cytology (Figure 1B-C). All cases had a follicular growth pattern without diffuse areas. Immunohistochemically, all cases were CD20 and/or CD79a+ (Figure 1D). The GC markers BCL6 and CD10 were strongly expressed in the majority of cases (Figure 1E). BCL2 was negative in 30 cases (Figure 1F) and weakly, heterogeneously positive in 12 cases without BCL2 translocation. IRF4/MUM1 was negative in 27 of 32 cases investigated. In 5 cases, a focal positivity was observed. Ki-67 demonstrated a high proliferation rate with some cases showing a rim of polarization in the periphery of the neoplastic follicles (Figure 1G).
Figure 1.
Histologic features of nodal PTFL. (A) The nodal architecture is effaced by ill-defined, coalescent follicles. A starry sky pattern is evident. The mantle zone is attenuated. Note a rim of residual normal nodal tissue with residual GCs (hematoxylin-and-eosin [H&E] stain; original magnification, ×25). (B) Cytologic features of the tumor cells within the abnormal follicles. The infiltrate is composed of medium-sized cells with blastic chromatin and inconspicuous nucleoli (H&E stain; original magnification, ×400). (C) Giemsa stain highlights the presence of medium-sized blastoid cells with few scattered centroblasts (original magnification, ×400). (D) CD20 shows the abnormal follicles, but CD20+ cells do not extend to the interfollicular region. (E) The follicles are strongly CD10+. (F) The follicular cells are BCL2−. (G) MIB1 stain demonstrates the high proliferation within the follicles without polarization (D-G, immunoperoxidase; original magnification, ×25).
IGH/IGK clonality, FISH, and cytogenetic analysis in PTFL
All 42 cases were monoclonal. In 39 cases, a monoclonal IGH gene rearrangement was demonstrated. Three cases were polyclonal using FR1-3 IGH primers but monoclonal when κ chain gene (IGK) primers were used. The 10 RH used as controls rendered polyclonal results with all IG primer sets used.
Conventional cytogenetics was available in 3 cases; 2 of them (PTFL25, PTFL36) presented a normal karyotype 46,XY in all cells studied whereas case PTFL24 displayed a 47,XY,add(7)(q36),+8[15]/46,XY[5] altered karyotype (supplemental Figure 1A).
FISH analysis demonstrated absence of BCL2 rearrangement in all 39 cases analyzed using BCL2 break apart probe (32 cases) and/or t(14;18) dual-color dual fusion probes (9 cases). IGH breaks were analyzed in 10 cases including the 3 cases in which BCL2 was not analyzed. The 10 cases gave negative results. Twenty-three cases analyzed for BCL6 rearrangements were negative for the translocation. Finally, 10 cases analyzed for IRF4 translocations, including 3 cases with focal expression of IRF4/MUM1, were negative.
Copy-number and CNN-LOH alterations in PTFL and RH
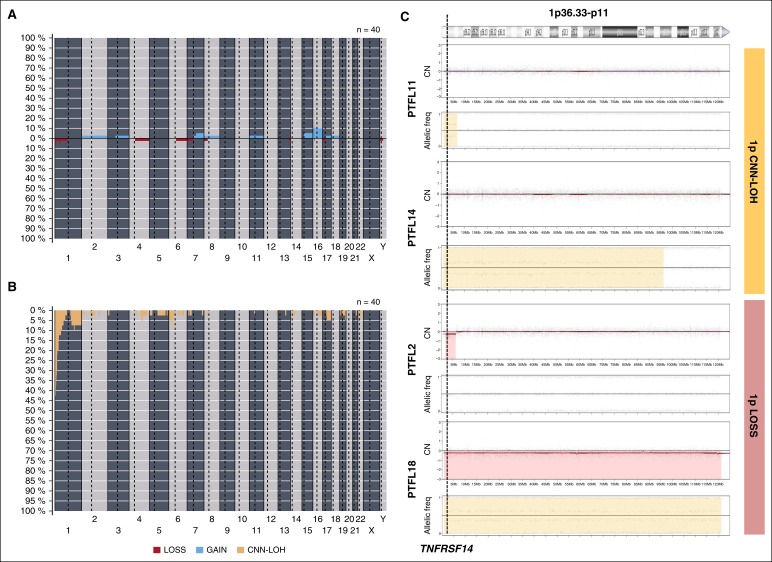
Copy-number analysis detected 31 alterations, 18 gains, and 13 losses in 16 of 40 cases analyzed (mean, 0.77 alterations per case; range, 0-6 alterations) (supplemental Table 4; Figure 2). No amplifications or homozygous deletions were observed. Recurrent regions of alterations were gains of 7q21-q36 (2 cases), 15q (2 cases), and whole chr16 (3 cases) and losses of 1p36 (3 cases). Two of the 3 cases with 1p36 deletion included the TNFRSF14 gene region. Sixty-five regions of CNN-LOH were detected. Recurrent regions of CNN-LOH were located at 1p36 (16 cases), 1q25.2-q25.3 (4 cases), and 6p22.1-p21.31 (4 cases). In total, 18 of 40 cases analyzed (45%) carried deletions (2 cases) or CNN-LOH (16 cases) of 1p36.32 including the TNFRSF14 gene. Of note, 10 cases (24%) carried 1p36 alterations (9 cases with CNN-LOH and 1 case deletion) as the only genetic abnormality. From the 6 RH successfully analyzed by copy-number array, 5 displayed no copy-number alterations or CNN-LOH regions whereas 1 case (RH3) displayed a region of CNN-LOH in 3q.
Figure 2.
Copy-number and CNN-LOH alterations in PTFL. Frequency of (A) copy number (CN) and (B) CNN-LOH alterations of 40 PTFL analyzed by Oncoscan Copy number molecular inversion probe assay. Each probe is aligned from chromosome 1 to Y and p to q. The vertical axis indicates frequency of the genomic aberration among the analyzed cases. Gains are depicted in blue, losses are depicted in red, and regions of CNN-LOH are represented in yellow. (C) Different patterns of 1p CNN-LOH and loss in 4 cases of PTFL. In the first panel for each case, CN calls are represented. In the second panel, allelic events are displayed along the x-axis. Germ-line homozygosity is given in calls at the 0 and 1 levels, respectively, whereas germ-line heterozygosity is given in calls around 0.5. CNN-LOH in the tumor leads to loss of calls around 0.5 and to the presence of allelic imbalance calls derived from a sum of heterozygous normal cell and homozygous tumor cell calls for a given locus, resulting in values between 0 and 0.5 or 0.5 and 1 depending on the percentage of cells carrying the alteration.
Identification of recurrent mutations by targeted NGS
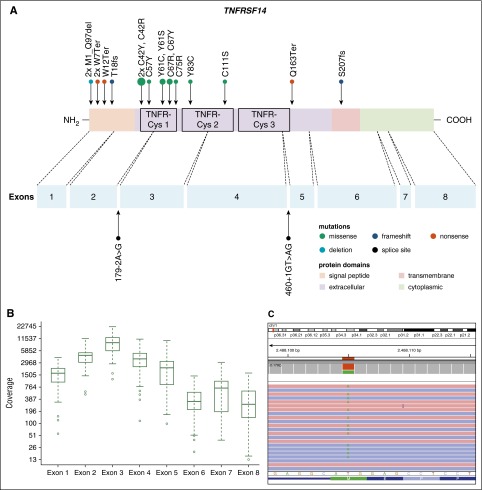
Thirty-seven PTFL cases were analyzed by NGS. The mean average read depth of the NGS sequence analysis was 3290 (range, 294-6989; for the mean coverage of TNFRSF14 exons, see Figure 4B) with over 96% of bases in the target region covered by at least 20 reads. Thirty-eight mutations were identified in 27 of 37 cases analyzed (73%). Alterations were most frequently found in TNFRSF14 with 20 mutated cases (54%) and allele frequencies ranging from 7% to 59%, followed by 7 KMT2D mutations in 6 cases (PTFL37 carrying 2 mutations, 16%), and GNA13 mutated in 4 cases (11%). Other mutated genes were EP300 (2 of 37, 5%), FOXO1 (2 of 37, 5%), CREBBP (1 of 37, 3%), HIST1H1B (1 of 37, 3%), and HIST1H1C (1 of 37, 3%) (Figure 3; supplemental Table 5). Four of the 5 cases that could not be analyzed by NGS were Sanger sequenced for all 8 TNFRSF14 exons. One case displayed a nonsense mutation in exon 5 of the TNFRSF14 gene, the remaining 3 cases were wild type. Taken together, TNFRSF14 was mutated in 21 of 41 cases analyzed (51%), including 13 missense mutations (2 affecting the start codon leading to a deletion of the first 97 amino acids), 4 nonsense mutations, 2 splice site mutations, and 2 frameshift mutations. Mutations were most frequently found in the first 3 exons (16 mutations) coding for the signal peptide and part of the extracellular domain containing the tumor necrosis factor (TNF) receptor (TNFR) cystein repeats (TNFR-Cys) 1 and 2 (Figure 4).
Figure 4.
Distribution of TNFRSF14 mutations on protein and exon level in PTFL. (A) TNFRSF14 protein with its different domains and the cysteine repeats TNFR-Cys 1-3 above. Below, localization of exons is indicated by dashed lines with position of splice site mutations. Domains of the protein are represented according to the Uniprot database (www.uniprot.org). Exact positions of each TNFRSF14 mutation found in 21 PTFL cases are given. (B) Coverage representation for TNFRSF14 exons 1-8 of all PTFL and RH samples included in the study. The spacing of the scale on the y-axis is proportional to the logarithm of the number. (C) Exemplary view of the Integrative Genomics Viewer (IGV) showing the mutation p.M1_97del of PTFL11.
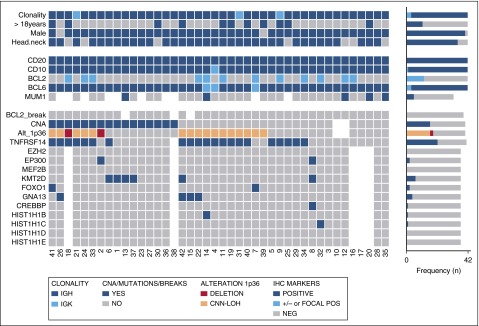
Figure 3.
Overview of clinical and histological findings, copy number (Oncoscan) and NGS results in 42 PTFL cases. Each column of the heat map represents 1 PTFL case and each line 1 specific analysis. On the right side of the figure, the frequency of the particular result of the analysis is shown.
Seven KMT2D mutations found in 6 cases displayed high allelic frequencies of 50% or more (range 24%-51%, 4 homozygous mutations). However, no alterations of chr12 were observed in these cases. Analysis by SIFT and Polyphen2 predicted a damaging effect for all of these mutations (supplemental Table 6). Mutations were scattered randomly across the whole gene without an apparent hotspot region. Nine cases (24%) displayed >1 mutation and 10 cases (27%) showed no genetic alteration in any of the 12 genes investigated. All 31 mutations further analyzed were validated either by Sanger sequencing or targeted resequencing (supplemental Table 5). In contrast, no mutations were found in any of the 9 LNs with RH used as controls.
Correlation between copy-number and TNFRSF14 mutation analyses in PTFL
Fifteen of 21 TNFRSF14 mutated cases had concomitant 1p36 CNN-LOH (14 cases) or deletion (1 cases). Of note, in 3 cases, 2 (PTFL7 and PTFL39) with a region of CNN-LOH including 1p36 and 1 (PTFL2) with 1p36 deletion including TNFRSF14, we could not demonstrate mutations in the TNFRSF14 gene (Figure 3; supplemental Figure 2), suggesting either the presence of an additional target in the 1p36 region or an alternative mechanism of TNFRSF14 silencing. In total, 22 cases (54%) showed either TNFRSF14 mutations with and without 1p36 CNN-LOH or deletion.
PTFL and conventional t(14;18)− FL are genetically different
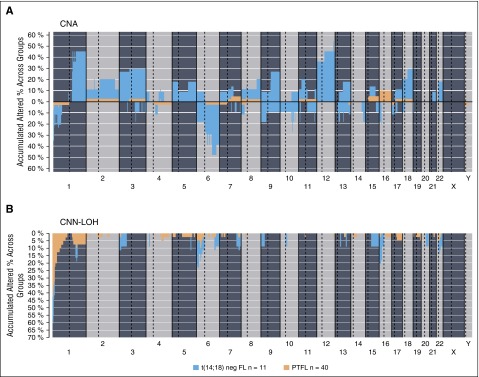
The clinicopathological information of the 11 t(14;18)− FL cases is summarized in supplemental Table 7. There were 8 women and 3 men, with a median age of 65 years (range, 48-83 years). These cases revealed the typical morphology of conventional FL grade 1-3A. In contrast to PTFL, women were more frequently affected and the tumor cells were more often CD10−. Comparative analysis of genetic features demonstrated significantly higher levels of genetic complexity in the 11 t(14;18)− FLs as compared with PTFL (mean, 9 vs 0.77 CNA; P < .001) (Figure 5). Both groups presented 1p36 alterations including TNFRSF14 but CNN-LOH alterations of this locus were more frequently observed in PTFL (40% vs 9%; P = .075), whereas FL without t(14;18) displayed more frequently losses of the locus (27% vs 5%; P = .061). In terms of mutated genes, t(14;18)− displayed a mutational profile similar to adult t(14;18)+ FL (Table 2; supplemental Table 8). FL without t(14;18) translocation more frequently carried mutations of CREBBP (45% vs 3%; P = .001), EZH2 (18% vs 0%; P = .049), and KMT2D (36 vs 16%; P= .206) when compared with PTFL.
Figure 5.
Comparative plot of copy number (CN) and CNN-LOH aberrations between PTFL and conventional t(14;18)− FL analyzed by CN array. (A) Copy number and (B) CNN-LOH. X-axis depicts chromosome positions with dotted lines pointing centromeres. Y-axis indicates frequency of the genomic aberration among the analyzed cases. PTFL are depicted in yellow whereas conventional t(14;18)− FL are depicted in dark blue.
Table 2.
Distribution of mutations in FL
Discussion
In this study, we performed a genome-wide analysis of nodal PTFL in pediatric and young adult patients. We found that nodal PTFL is a monoclonal B-cell disorder with extremely low genomic complexity and TNFRSF14 alterations as the major genetic feature present in 54% of the cases. Mutations of histone-modifying genes important for the pathogenesis of adult FL were rarely found in the pediatric counterpart. In contrast, conventional t(14;18)− FL displayed a significantly higher level of genetic complexity, and a mutational profile similar to BCL2+ FL with frequent mutations in CREBBP and EZH2 genes. In 8 PTFL cases (19%), no genetic alterations were identified, beyond IG monoclonal rearrangement. Further studies are needed to elucidate the genetic alterations involved in the pathogenesis of these lymphomas. The low genomic complexity found in PTFL correlates well with the indolent clinical course of the disease.5 Furthermore, the reactive LNs used as control samples showed no genetic alterations, indicating that PTFL is in fact a neoplasia with low malignant potential rather than just a benign clonal expansion in the course of an immune reaction/dysregulation.
FL with t(14;18) translocation is extremely rare in children and young adults. Most FLs in the pediatric age group are BCL2 translocation negative and can be divided into 2 different diseases with overlapping morphological features. One involves predominantly the Waldeyer ring; may be purely follicular, follicular/diffuse, or diffuse; has tumor cells expressing IRF4/MUM1 and often BCL2; and is associated with IRF4 translocations in the absence of BCL2 alterations.5,25 The marked male predominance characteristic of nodal PTFL is not observed.5,26 The term “large B-cell lymphoma with IRF4 rearrangement” has been proposed for this entity.20,27 In contrast, the nodal counterpart is mostly follicular; does not express IRF4/MUM1 and rarely BCL2; and the genetic alterations are largely unknown. In the present study, we analyzed a large cohort with characteristic clinical and morphological features and demonstrated that the most frequent genetic alterations in nodal PTFL are mutations/deletions in the TNFRSF14 gene.
TNFRSF14 mutations scattered across the whole gene but were more frequently found in the first 3 exons similar to the distribution seen in other studies.9,11,28 TNFRSF14 is a member of the TNFR superfamily, also known as herpes virus entry mediator, with important roles in the immune system, such as T-cell costimulation, regulation of dendritic cell homeostasis, autoimmune-mediated inflammatory responses, as well as host defense against pathogens.29 TNFRSF14 interacts with 2 ligands of the TNF family, lymphotoxin-α (LT-α) and LIGHT (also known as tumor necrosis factor superfamily member 14, TNFSF14), and the immunoglobulin domain-containing receptors B- and T-lymphocyte attenuator (BTLA) and CD160. These ligands bind to the extracellular domain of TNFRSF14 including the TNFR-Cys repeats 1-3.30-32 Interestingly, we found that these extracellular regions are frequently mutated, suggesting that these mutations may interfere with the ligand-binding activity. Mutations affecting the signal peptide all change the protein structure significantly, leading to an early translation stop or to a truncated protein missing a large part of the N-terminal amino acids, whereas mutations affecting the TNFR-Cys domains represent only single amino acid changes. This distribution of nonsense vs missense mutations indicates that the cysteine-rich domains TNFR-Cys 1 and TNFR-Cys 2 must be affected in order to promote tumor growth or to confer a tumor survival advantage.
The exact mode of action of TNFRSF14 signaling is unclear because it can deliver both costimulatory (via LIGHT and LT-α) and inhibitory signals (via BTLA and CD160) to T cells,29,33 and the protein is not expressed in normal GC B cells.34 In a study of TNFRSF14 mutations in de novo adult FL,10 the authors suggested that these mutations occur early in FL pathogenesis and might contribute to FL progression. Another study34 suggested that the disruption of the BTLA-TNFRSF14 pathway is involved in GC B-cell activation and that there might be some pressure in FL to inactivate TNFRSF14 to prevent interactions with BTLA-expressing T follicular helper cells. There are also conflicting data about TNFRSF14 mutations and its possible influence in FL prognosis with studies showing either worse prognosis or better overall survival in patients with TNFRSF14 mutations.10,11 Although the number of patients with follow-up in this study was relatively small, there were no clinical differences between cases with and without mutation, suggesting that the presence of the mutation in PTFL does not confer a bad prognosis. Nevertheless, the fact that TNFRSF14 mutations are found in the majority of PTFL indicates that they might be important for the pathogenesis of the disease; however, how these mutations contribute to PTFL pathogenesis remains to be elucidated.
Interestingly, TNFRSF14 mutations were associated with 1p36 CNN-LOH or deletions in over 70% of cases (15 of 21), pointing to a selection of the mutated allele and indicating a tumor suppressor role of TNFSRF14. In 2 cases, however, no TNFRSF14 mutation could be demonstrated despite the presence of CNN-LOH in the region where the gene is localized. This raises the possibility of another target gene in this region or another mechanism inhibiting TNFRSF14 protein function or gene expression. The 1p36 altered region contains 2 genes, CHD5 and UBE4B, which have been identified as candidate tumor suppressor genes in neuroblastoma,35,36 but no association so far has been shown with lymphoid neoplasias. Although PTFL and conventional t(14;18)− FL showed frequent alterations of TNFRSF14, CNN-LOH alterations of this locus were characteristically found in PTFL whereas t(14;18)− FL revealed more frequently losses of the locus similarly to BCL2+ FL. Furthermore, PTFL showed a lower genetic complexity when compared with conventional t(14;18)− FL, suggesting different mechanisms of lymphomagenesis.
Mutations in other histone-modifying genes frequently affected in adult FL were rarely encountered in nodal PTFL. In contrast, CREBBP mutations were identified in 45% of conventional t(14;18)− FL (P = .001), highlighting the different genetic landscape of these 2 disorders. The most frequently mutated histone-modifying gene in PTFL was KMT2D identified in 6 of 37 cases (16%). No hotspots were identified and mutations showed high allelic frequencies of ≥50% in 4 cases indicating homozygous mutations. The relatively high allelic frequency of KMT2D mutations was also observed in a recent study in adult FL.37 It has recently been shown that KMT2D sustains a gene expression program that represses B-cell lymphoma development.37,38 KMT2D catalyzes the methylation of lysine 4 on histone H3, a modification associated with transcriptionally active chromatin.39 Mutations of KMT2D in diffuse large B-cell lymphoma and FL impair the catalytic function of the enzyme and, as a result, drive GC expansion due to enhanced proliferation and impair B-cell terminal differentiation.38 Interestingly, KMT2D-deficient mice show an abnormal persistence of GCs, a defect in class switch recombination and reduced antibody production similar to Kabuki syndrome, linked to KMT2D mutations. Of special interest is the fact that one of the targets of KMT2D is TNFRSF14, and it has been recently suggested that mutations in KMT2D perturb TNFRSF14 expression and/or function.37 Four cases in our study had KMT2D mutations alone or in combination with gene mutations other than TNFRSF14. One can speculate that KMT2D disruptions may represent another way for the cells to overcome TNFRSF14 gene function. Of note, EZH2 mutations frequently found in adult FL16,40 and in conventional t(14;18)− FL in this study were not identified in our PTFL cohort. Our findings support previous studies suggesting that conventional FL with and without BCL2 rearrangement in adults are closely related but differ clinically and genetically from PTFL.21,41
In conclusion, this study demonstrates that nodal PTFL is a monoclonal B-cell neoplasia with low genetic complexity and a limited number of gene mutations that correlates well with the indolent nature of the disease. In contrast to adult FL with and without t(14;18) translocation, mutations in histone-modifying genes are rare, whereas there seems to be high selection pressure on mutations in the TNFRSF14 gene, whose exact role in the pathogenesis of the disease remains to be elucidated.
Acknowledgments
The authors are indebted to the Genomics Core Facility of Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS) and to Hospital Clínic de Barcelona–IDIBAPS Biobank-Tumor Bank and Hematopathology Collection for sample procurement. The authors are grateful to Noelia Garcia, Helena Suarez-Cisneros, Montse Sanchez, Laura Plà, Sieglinde Baisch, and Sema Colak for excellent technical assistance and to the Sociedad Española de Hematopatología y Oncología Pediátrica for excellent work in the coordination of Spanish specimens.
This work was supported by Fondo de Investigaciones Sanitarias, Instituto de Salud Carlos III (Miguel Servet contract CP13/00159 and PI15/00580), Generalitat de Catalunya Suport Grups de Recerca (2013-SGR-378), the European Regional Development Fund “Una manera de fer Europa” and the Wilhelm-Sander-Stiftung (2015.058.1; L.Q.-M. and F.F.). This work was partially developed at the Centro Esther Koplowitz, Barcelona, Spain.
Footnotes
The copy-number data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE78872).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: I.S. and L.Q.-M. conceived and designed the study, supervised the experimental work, and wrote the manuscript; L.Q.-M., E.S.J., and T.M. provided cases and reviewed all the cases; J.S., I.B., J.E.R.-Z., V.E., R.P., and I.S. performed genetic analysis and interpreted the data; B.M. performed the FISH analysis; G.C. performed statistical analysis; F.N. performed bioinformatics analysis; S.G., B.G.-F., O.B., A.M., C.B., V.P.-A., J.C., J.v.d.W., D.H., A.R., G.O., S.D., C.E., C.L.-M., F.F., and E.C. contributed with cases and performed pathological review; and J.S., E.S.J., F.F., and E.C. analyzed the data and helped writing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Leticia Quintanilla-Martinez, Institute of Pathology, University Hospital Tübingen, Eberhard Karls University of Tübingen and Comprehensive Cancer Center, Liebermeisterstr 8, 72076 Tübingen, Germany; e-mail: leticia.quintanilla-fend@med.uni-tuebingen.de; and Itziar Salaverria, Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), Rosselló 153, 08036 Barcelona, Spain; e-mail: isalaver@clinic.ub.es.
References
- 1.Harris NL, Swerdlow SH, Jaffe E. Follicular lymphoma. In: Jaffe E, Harris NL, Stein H, Vardiman JW, editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC; 2008. pp. 220–226. [Google Scholar]
- 2.Oschlies I, Salaverria I, Mahn F, et al. Pediatric follicular lymphoma--a clinico-pathological study of a population-based series of patients treated within the Non-Hodgkin’s Lymphoma--Berlin-Frankfurt-Munster (NHL-BFM) multicenter trials. Haematologica. 2010;95(2):253–259. doi: 10.3324/haematol.2009.013177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winberg CD, Nathwani BN, Bearman RM, Rappaport H. Follicular (nodular) lymphoma during the first two decades of life: a clinicopathologic study of 12 patients. Cancer. 1981;48(10):2223–2235. doi: 10.1002/1097-0142(19811115)48:10<2223::aid-cncr2820481018>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 4.Frizzera G, Murphy SB. Follicular (nodular) lymphoma in childhood: a rare clinical-pathological entity. Report of eight cases from four cancer centers. Cancer. 1979;44(6):2218–2235. doi: 10.1002/1097-0142(197912)44:6<2218::aid-cncr2820440634>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 5.Liu Q, Salaverria I, Pittaluga S, et al. Follicular lymphomas in children and young adults: a comparison of the pediatric variant with usual follicular lymphoma. Am J Surg Pathol. 2013;37(3):333–343. doi: 10.1097/PAS.0b013e31826b9b57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Louissaint A, Jr, Ackerman AM, Dias-Santagata D, et al. Pediatric-type nodal follicular lymphoma: an indolent clonal proliferation in children and adults with high proliferation index and no BCL2 rearrangement. Blood. 2012;120(12):2395–2404. doi: 10.1182/blood-2012-05-429514. [DOI] [PubMed] [Google Scholar]
- 7.Lorsbach RB, Shay-Seymore D, Moore J, et al. Clinicopathologic analysis of follicular lymphoma occurring in children. Blood. 2002;99(6):1959–1964. doi: 10.1182/blood.v99.6.1959. [DOI] [PubMed] [Google Scholar]
- 8.Attarbaschi A, Beishuizen A, Mann G, et al. European Intergroup for Childhood Non-Hodgkin Lymphoma (EICNHL) and the international Berlin-Frankfurt-Münster (i-BFM) Study Group. Children and adolescents with follicular lymphoma have an excellent prognosis with either limited chemotherapy or with a “Watch and wait” strategy after complete resection. Ann Hematol. 2013;92(11):1537–1541. doi: 10.1007/s00277-013-1785-2. [DOI] [PubMed] [Google Scholar]
- 9.Martin-Guerrero I, Salaverria I, Burkhardt B, et al. Recurrent loss of heterozygosity in 1p36 associated with TNFRSF14 mutations in IRF4 translocation negative pediatric follicular lymphomas. Haematologica. 2013;98(8):1237–1241. doi: 10.3324/haematol.2012.073916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Launay E, Pangault C, Bertrand P, et al. High rate of TNFRSF14 gene alterations related to 1p36 region in de novo follicular lymphoma and impact on prognosis. Leukemia. 2012;26(3):559–562. doi: 10.1038/leu.2011.266. [DOI] [PubMed] [Google Scholar]
- 11.Cheung KJ, Johnson NA, Affleck JG, et al. Acquired TNFRSF14 mutations in follicular lymphoma are associated with worse prognosis. Cancer Res. 2010;70(22):9166–9174. doi: 10.1158/0008-5472.CAN-10-2460. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt J, Salaverria I, Haake A, et al. Increasing genomic and epigenomic complexity in the clonal evolution from in situ to manifest t(14;18)-positive follicular lymphoma. Leukemia. 2014;28(5):1103–1112. doi: 10.1038/leu.2013.307. [DOI] [PubMed] [Google Scholar]
- 13.Kridel R, Sehn LH, Gascoyne RD. Pathogenesis of follicular lymphoma. J Clin Invest. 2012;122(10):3424–3431. doi: 10.1172/JCI63186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green MR, Kihira S, Liu CL, et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Natl Acad Sci USA. 2015;112(10):E1116–E1125. doi: 10.1073/pnas.1501199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loeffler M, Kreuz M, Haake A, et al. HaematoSys-Project. Genomic and epigenomic co-evolution in follicular lymphomas. Leukemia. 2015;29(2):456–463. doi: 10.1038/leu.2014.209. [DOI] [PubMed] [Google Scholar]
- 16.Okosun J, Bödör C, Wang J, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46(2):176–181. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pasqualucci L, Khiabanian H, Fangazio M, et al. Genetics of follicular lymphoma transformation. Cell Reports. 2014;6(1):130–140. doi: 10.1016/j.celrep.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Kaminski MS, Li Y, et al. Mutations in linker histone genes HIST1H1 B, C, D, and E; OCT2 (POU2F2); IRF8; and ARID1A underlying the pathogenesis of follicular lymphoma. Blood. 2014;123(10):1487–1498. doi: 10.1182/blood-2013-05-500264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okosun J, Wolfson RL, Wang J, et al. Recurrent mTORC1-activating RRAGC mutations in follicular lymphoma. Nat Genet. 2016;48(2):183–188. doi: 10.1038/ng.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quintanilla-Martinez L, Sander B, Chan JK, et al. Indolent lymphomas in the pediatric population: follicular lymphoma, IRF4/MUM1+ lymphoma, nodal marginal zone lymphoma and chronic lymphocytic leukemia. Virchows Arch. 2016;468(2):141–157. doi: 10.1007/s00428-015-1855-z. [DOI] [PubMed] [Google Scholar]
- 21.Leich E, Hoster E, Wartenberg M, et al. German Low Grade Lymphoma Study Group (GLSG) Similar clinical features in follicular lymphomas with and without breaks in the BCL2 locus. Leukemia. 2016;30(4):854–860. doi: 10.1038/leu.2015.330. [DOI] [PubMed] [Google Scholar]
- 22.Swerdlow SH, Campo E, Harris NL, et al. World Health Organization Classification of Tumours. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Vol. 4. Lyon, France: IARC Press; 2008. [Google Scholar]
- 23.Adam P, Baumann R, Schmidt J, et al. The BCL2 E17 and SP66 antibodies discriminate 2 immunophenotypically and genetically distinct subgroups of conventionally BCL2-“negative” grade 1/2 follicular lymphomas. Hum Pathol. 2013;44(9):1817–1826. doi: 10.1016/j.humpath.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 24.van Dongen JJ, Langerak AW, Brüggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17(12):2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 25.Salaverria I, Philipp C, Oschlies I, et al. Molecular Mechanisms in Malignant Lymphomas Network Project of the Deutsche Krebshilfe; German High-Grade Lymphoma Study Group; Berlin-Frankfurt-Münster-NHL Trial Group. Translocations activating IRF4 identify a subtype of germinal center-derived B-cell lymphoma affecting predominantly children and young adults. Blood. 2011;118(1):139–147. doi: 10.1182/blood-2011-01-330795. [DOI] [PubMed] [Google Scholar]
- 26.Karube K, Guo Y, Suzumiya J, et al. CD10-MUM1+ follicular lymphoma lacks BCL2 gene translocation and shows characteristic biologic and clinical features. Blood. 2007;109(7):3076–3079. doi: 10.1182/blood-2006-09-045989. [DOI] [PubMed] [Google Scholar]
- 27.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green MR, Gentles AJ, Nair RV, et al. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood. 2013;121(9):1604–1611. doi: 10.1182/blood-2012-09-457283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinberg MW, Cheung TC, Ware CF. The signaling networks of the herpesvirus entry mediator (TNFRSF14) in immune regulation. Immunol Rev. 2011;244(1):169–187. doi: 10.1111/j.1600-065X.2011.01064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Compaan DM, Gonzalez LC, Tom I, Loyet KM, Eaton D, Hymowitz SG. Attenuating lymphocyte activity: the crystal structure of the BTLA-HVEM complex. J Biol Chem. 2005;280(47):39553–39561. doi: 10.1074/jbc.M507629200. [DOI] [PubMed] [Google Scholar]
- 31.Kojima R, Kajikawa M, Shiroishi M, Kuroki K, Maenaka K. Molecular basis for herpesvirus entry mediator recognition by the human immune inhibitory receptor CD160 and its relationship to the cosignaling molecules BTLA and LIGHT. J Mol Biol. 2011;413(4):762–772. doi: 10.1016/j.jmb.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 32.Rooney IA, Butrovich KD, Glass AA, et al. The lymphotoxin-beta receptor is necessary and sufficient for LIGHT-mediated apoptosis of tumor cells. J Biol Chem. 2000;275(19):14307–14315. doi: 10.1074/jbc.275.19.14307. [DOI] [PubMed] [Google Scholar]
- 33.Cai G, Freeman GJ. The CD160, BTLA, LIGHT/HVEM pathway: a bidirectional switch regulating T-cell activation. Immunol Rev. 2009;229(1):244–258. doi: 10.1111/j.1600-065X.2009.00783.x. [DOI] [PubMed] [Google Scholar]
- 34.M’Hidi H, Thibult ML, Chetaille B, et al. High expression of the inhibitory receptor BTLA in T-follicular helper cells and in B-cell small lymphocytic lymphoma/chronic lymphocytic leukemia. Am J Clin Pathol. 2009;132(4):589–596. doi: 10.1309/AJCPPHKGYYGGL39C. [DOI] [PubMed] [Google Scholar]
- 35.Fujita T, Igarashi J, Okawa ER, et al. CHD5, a tumor suppressor gene deleted from 1p36.31 in neuroblastomas. J Natl Cancer Inst. 2008;100(13):940–949. doi: 10.1093/jnci/djn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krona C, Ejeskär K, Abel F, et al. Screening for gene mutations in a 500 kb neuroblastoma tumor suppressor candidate region in chromosome 1p; mutation and stage-specific expression in UBE4B/UFD2. Oncogene. 2003;22(15):2343–2351. doi: 10.1038/sj.onc.1206324. [DOI] [PubMed] [Google Scholar]
- 37.Ortega-Molina A, Boss IW, Canela A, et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med. 2015;21(10):1199–1208. doi: 10.1038/nm.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Dominguez-Sola D, Hussein S, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med. 2015;21(10):1190–1198. doi: 10.1038/nm.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bödör C, Grossmann V, Popov N, et al. EZH2 mutations are frequent and represent an early event in follicular lymphoma. Blood. 2013;122(18):3165–3168. doi: 10.1182/blood-2013-04-496893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leich E, Salaverria I, Bea S, et al. Follicular lymphomas with and without translocation t(14;18) differ in gene expression profiles and genetic alterations. Blood. 2009;114(4):826–834. doi: 10.1182/blood-2009-01-198580. [DOI] [PMC free article] [PubMed] [Google Scholar]