Summary
Mitochondrial DNA is replicated by the nuclear encoded DNA polymerase γ (pol γ) which is composed of a single 140 kDa catalytic subunit and a dimeric 55 kDa accessory subunit. Mitochondrial DNA is vulnerable to various forms of damage, including several types of oxidative lesions, UV-induced photoproducts, chemical adducts from environmental sources, as well as alkylation and inter-strand crosslinks from chemotherapy agents. Although many of these lesions block DNA replication, Pol γ can bypass some lesions by nucleotide incorporation opposite a template lesion and further extension of the DNA primer past the lesion. This process of translesion synthesis (TLS) by Pol γ can occur in either an error-free or an error-prone manner. Assessment of TLS requires extensive analysis of oligonucleotide substrates and replication products by denaturing polyacrylamide sequencing gels. This chapter presents protocols for the analysis of translesion DNA synthesis.
Keywords: DNA polymerase γ, mitochondrial DNA polymerase, DNA replication, Translesion synthesis, DNA repair, enzyme assays, POLG
1. Introduction
The mitochondrial (mt) genome is a multicopy closed circular genome of 16,569 bp that encodes 13 proteins involved in the electron transport chain, 22 transfer RNA genes, and 2 ribosomal RNAs required for mitochondrial protein synthesis of the 13 polypeptides. Cells contain several thousand copies of mtDNA distributed within hundreds of mitochondria. The mtDNA is located in the mitochondrial matrix within discrete nucleoids, each containing 1–2 copies of mtDNA (1).
Mitochondrial DNA incurs chemical damage from both endogenous and exogenous sources, which can result in mutations (2). Leakage of electrons from the electron transport chain generates reactive oxygen species, which are the major source of oxidative damage to mtDNA. Mitochondria are able to repair oxidative lesions because they possess a robust base excision repair (BER) system, but the lack of an efficient mismatch repair system and the marked absence of nucleotide excision repair result in the persistence of miscoding lesions in mtDNA (3). Because unrepaired lesions can promote mtDNA mutations or even block DNA replication, the ability of pol γ to bypass such lesions by TLS is vital to maintaining the genetic integrity of mtDNA. In contrast to the nucleus, which possesses several specialized DNA polymerases to efficiently bypass different DNA lesions, mammalian mitochondria appears to contain a single DNA polymerase, pol γ, which bears the burden both to replicate the 16.5 kb circular mitochondrial genome and to participate in DNA repair (4–13). The human DNA pol γ holoenzyme is a heterotrimeric complex comprised of a catalytic subunit, encoded by POLG at chromosomal locus 15q25, and a homodimeric accessory subunit, encoded by POLG2 at chromosomal locus 17q24.1. The catalytic subunit, a member of the A family of DNA polymerases, is a 140 kDa enzyme (p140) that possesses DNA polymerase, 3'→5' exonuclease and 5'-dRP lyase activities (reviewed in (13)). The accessory subunit, a homodimer of two 55 kDa proteins (two p55 monomers) binds asymmetrically to the catalytic subunit, where the proximal p55 enables tight DNA binding and the distal monomer confers processive DNA synthesis to the holoenzyme (14, 15).
Translesion DNA synthesis by the human DNA pol γ has been studied for a number of DNA lesions, including 7,8-dihydro-8-oxo-2'-deoxyguanosine (16), benzopyrene adducts (17), UV photoadducts (18), acrolein adducts (19), and several others that are reviewed in (2). We evaluate translesion DNA synthesis by pol γ with synthetic oligonucleotide substrates that contain specific lesions of interest. DNA polymerization reactions are utilized to measure the efficiency of pol γ to incorporate opposite and to extend beyond specific DNA lesions in vitro. This paper describes the general procedures needed to measure translesion DNA synthesis and efficiency of lesion bypass with the purified DNA polymerase γ.
2. Materials
2.1 Enzymes
Recombinant catalytic subunit of human DNA pol γ (exonuclease-proficient and exonuclease-deficient forms) containing a His6 affinity-tag at its N-terminus was overproduced in baculovirus-infected Sf9 cells, and the protein was purified to homogeneity as described previously (20–22).
The p55 accessory subunit containing a His6 affinity-tag at its C-terminus was expressed in E. coli and purified to homogeneity as described (20–23).
2.2 Primer-template substrates
- Primer-template substrates with and without damage were designed and synthesized. A control oligonucleotide primer template pair with no damage is designed, constructed and purchased from Integrated DNA Technologies. An identical template with a site-specific lesion replacing a normal nucleotide is also synthesized. As an example, investigation of translesion synthesis opposite uv-photodimers utilized the following undamaged DNA template (18):
5’-AATTTCTGCAGGTCGACTCCAAAGGCT-3’ Primer 3’-TTAAAGACGTCCAGCTGAGGTTTCCGATTGGGCCATGGCTCGACC-5’ Template - Our investigation into translesion synthesis opposite an acrolein adduct (γ-hydroxy-1,N2-propano-2’-deoxyguanosine (γ-HOPdG)) utilized the following primer template pair (19):
5’-GGG GGC TCG TAA GGA TTC-3’ Primer 3’-CCC CCG AGC ATT CCT AAG (γ -HOPdG) CT GA-5’ Template - 3’-CCC CCG AGC ATT CCT AAG GCT GA-5’
2.3 Primer extension assays
Enzyme dilution buffer: 50 mM Tris-HCl, pH 7.5, 10% glycerol, 1 mM EDTA, 1 mM 2-mercaptoethanol, 50 µg/mL acetylated bovine serum albumin (BSA), 0.1 M NaCl.
T4 polynucleotide kinase.
Primer template extension buffer: 25 mM HEPES-KOH (pH 7.5), 2 mM 2-mercaptoethanol, 0.1 mM EDTA, 5 mM MgCl2.
Phosphor storage screen.
Typhoon 9400 phosphorimager.
NIH Image or ImageJ software (http://rsb.info.nih.gov/ij/).
Non-linear kinetic curves are generated using KaleidaGraph software, Version 4.1.3 (Synergy Software).
2.4 Sequencing Gel Electrophoresis
Model S2 Sequencing Gel Electrophoresis apparatus (Life Technologies) or equivalent
40% acrylamide/Bis solution, 19:1.
10x Tris-Borate EDTA (TBE) solution: 108g Tris base, 55g boric acid, 40 mL 0.5M EDTA (pH 8.0) in 1 liter deionized H2O.
Ultrapure Urea.
TEMED (N,N,N’,N’-tetra-methyl-ethylenediamine).
10% ammonium persulfate solution: 1 g ammonium persulfate dissolved in 10 mL dH2O.
0.4 mm×50 well flat-tooth comb (CBS Scientific).
3. Methods
3.1 5’-End labeling of the primer
In a chilled 1.5 mL polypropylene microfuge tube, add 10x T4 polynucleotide kinase buffer (to produce a final concentration of 1x), 20 pmol of the gel-purified primer, 25 pmol [γ-32P]ATP, 12 U T4 polynucleotide kinase and dH20 to 25 µL.
Incubate reactions at 37°C for 60 min.
Heat at 95°C for 5 min to inactivate the kinase.
Cool tube on ice.
3.2 Hybridization of primer-template
Mix 10 pmol 32P-labeled primer with 12 pmol template oligonucleotide. Add TE buffer to 100 µL.
Vortex gently. Heat tube in a 400 mL beaker containing 95°C H2O for 5 min.
Cool slowly to room temperature by leaving the beaker at room temperature.
Store primer-template at −20°C until needed.
3.3 Primer-Template Extension Reactions
-
1
Dilute and premix the polymerase subunits (gently mixing p140 and p55 in a 1:2 molar ratio) in enzyme dilution buffer to desired concentration. Premixed enzymes are added last to assembled reaction mixtures (10 µL) on ice (see Notes 1 and 2).
-
2
Combine 50 nM radiolabeled primer template substrate, 10 nM exonuclease-deficient p140 and 20 nM p55 in 10 µL of primer template extension buffer. Reactions are initiated by the addition of one of the four dNTPs (at different concentrations depending on the substrate and analysis). Reactions lacking p55 accessory subunit are performed with no added NaCl, whereas reactions containing the p55 accessory subunit are supplemented to a final concentration of 100 mM NaCl.
-
2
Reactions are incubated at 37°C for 2–10 min, depending on the desired extent of incorporation.
-
3
Reactions are stopped by the addition (10 µL) of 95% deionized formamide and 10 mM EDTA and stored at −20°C prior to further analysis.
3.4 Separation of reaction products on a sequencing gel
-
1
Prepare a polyacrylamide sequencing gel containing 8 M urea by dissolving 33.6 g urea with 28 mL of 40% acrylamide/Bis solution (19:1), 7 mL of 10x TBE and dH2O to a final volume of 70 mL. Warm slightly to dissolve urea.
-
2
Filter solution through Whatman paper in a Buchner funnel to remove impurities and degas for fifteen minutes while sitting in a chilled solution of water (see Note 3). Add 630 µL 10% APS and 15 µL of TEMED, mix gently and quickly pipet solution between sequencing gel plates with 0.4 mm spacers. Insert a 0.4 mm×50 well flat-tooth comb (not a sharks tooth comb) to form the wells. Allow gel to polymerize for 1 hour.
-
3
Remove the comb and clamp the gel in the Model S2 Sequencing Gel apparatus. Fill chambers with 1x TBE buffer and pre-run gel for 45 minutes at 70 W.
-
4
Heat samples at > 95°C for 5 min, load samples (3 uL per well), and resolve the products by electrophoresis through the denaturing 16% polyacrylamide gel.
-
6
Dry the gel and expose to a phosphor storage screen (see Note 4).
-
7
Image the radioactivity on a Typhoon 9400 phosphorimager (Fig 1).
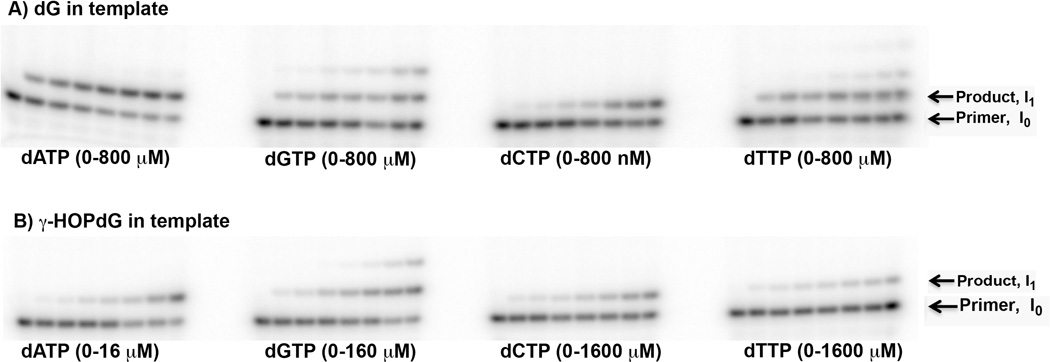
Fig. 1.
Representative single nucleotide incorporation reactions catalyzed by the exonuclease deficient form of DNA pol γ opposite a control primer template with a normal dG (Panel A) and the γ-HOPdG lesion in the DNA template (Panel B) (19). Reactions were performed with increasing concentrations of dATP, dGTP, dCTP and dTTP, as indicated.
3.5 Calculations
Quantify bands using NIH ImageJ software.
Kinetic constants are determined by integration of the band intensities (see Fig. 1). Velocity is determined from the ratio of the band intensities using the NIH Image J software as described (24, 25) where I0 is the intensity of the primer band and I1 is the intensity of the primer +1 incorporation product. Reaction velocity is v = I1/(I0 + I1) (24, 25). This is termed the standing start reaction (see Note 5).
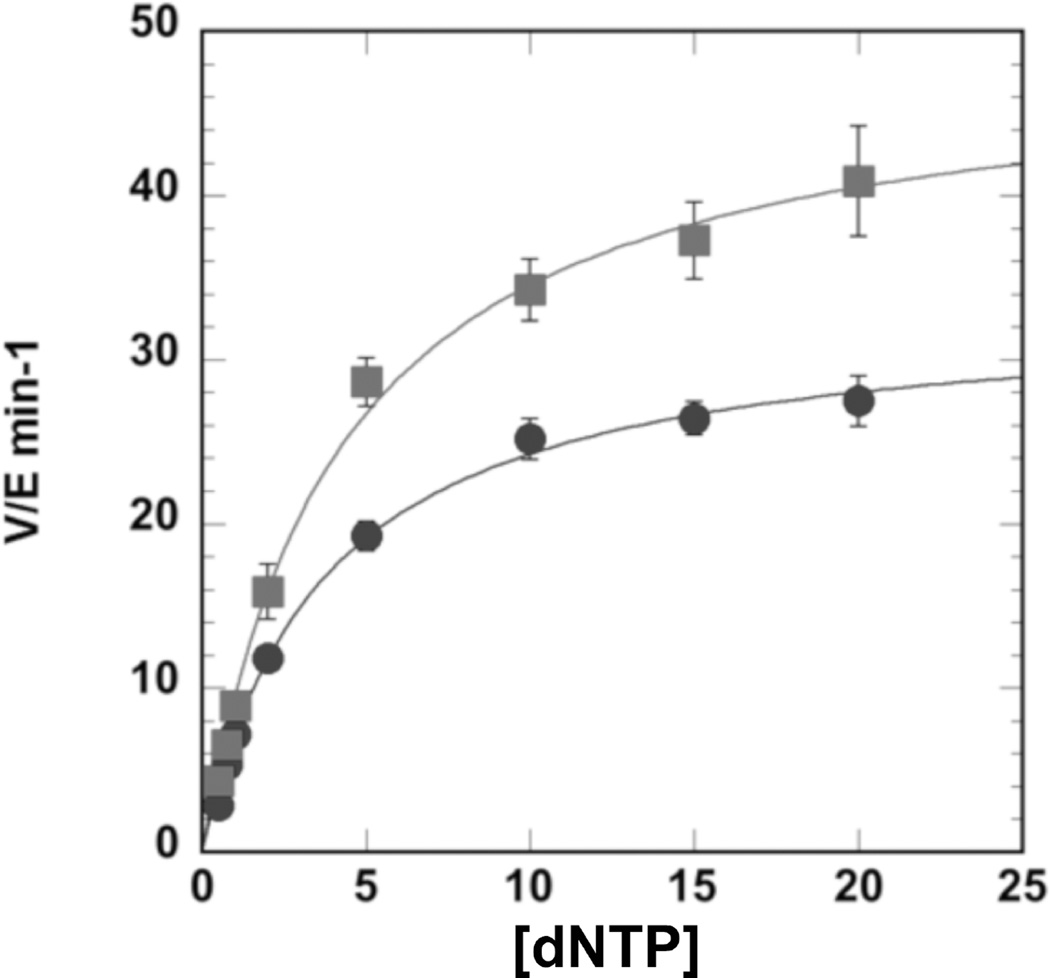
The steady-state kinetic parameters Km and Vmax were determined by fitting the data to the Michaelis-Menten model using KaleidaGraph (Version 4.1.3, Synergy) (see Fig. 2). The relative efficiency of incorporation opposite a specific DNA adduct compared to incorporation opposite a normal template nucleotide is estimated as the ratio of the kinetic constants for adducted DNA divided by the ratio for undamaged DNA (fin = (kcat/Km (adduct))/(kcat/Km (normal)).
Fig. 2.
Representative steady-state kinetics of pol γ. Steady-state incorporation of dATP (squares) and dGTP (circles) opposite (γ-HOPdG) (19), as fit to the Michaelis-Menten equation. The kcat values (V/E min−1) were plotted versus µM amounts of dATP or dGTP.
Acknowledgments
This work was supported by NIH, NIEHS intramural research funds (ES 065078 and ES 065080).
Footnotes
Assemble all reactions in an ice bucket. Keeping the enzymes cold is essential, as Pol γ has a functional half-life at 42°C of less than 2 minutes in the absence of the accessory subunit and DNA (26).
Refreeze enzyme stock tubes as soon as possible in liquid nitrogen and store at −80°C. Enzyme preparations begin to lose DNA polymerase activity after approximately 3 freeze-thaw cycles. The enzyme buffer contains glycerol, salt and 2-mercaptoethanol, which are very important for stability.
Degassing is needed to remove oxygen that will slow the polymerization reaction while chilling on ice is needed to slow down the polymerization once TEMED and APS are added. This provides sufficient time to pour the gel between the plates prior to polymerization.
Transferring the polyacrylamide gel to Whatman paper for drying is difficult and can result in tearing of the gel if done improperly. Ensure that the smaller glass plate is siliconized, which will easily allow the gel to detach while still sticking to the larger plate. The gel can then be transferred from the glass to plastic wrap, where any wrinkles can be flattened out, and finally sandwiched with a sheet of Whatman paper.
Running start reactions can also be performed with a primer terminus recessed three nucleotides proximal to the position of the lesion in the template. In this case, velocity is determined from the ratio of I3/I2 (24, 25). Similarly, extension past a lesion can also be assessed using a primer extended by one nucleotide where the primer’s 3’-end is positioned opposite the lesion.
References
- 1.Kukat C, Wurm CA, Spahr H, Falkenberg M, Larsson NG, Jakobs S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl. Acad. Sci. U S A. 2011;108:13534–13539. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cline SD. Mitochondrial DNA damage and its consequences for mitochondrial gene expression. Biochim Biophys Acta. 2012;1819:979–991. doi: 10.1016/j.bbagrm.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Copeland WC, Longley MJ. Mitochondrial genome maintenance in health and disease. DNA Repair (Amst) 2014;19:190–198. doi: 10.1016/j.dnarep.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanuri M, Minko IG, Nechev LV, Harris TM, Harris CM, Lloyd RS. Error prone translesion synthesis past gamma-hydroxypropano deoxyguanosine, the primary acrolein-derived adduct in mammalian cells. J. Biol. Chem. 2002;277:18257–18265. doi: 10.1074/jbc.M112419200. [DOI] [PubMed] [Google Scholar]
- 5.Minko IG, Washington MT, Kanuri M, Prakash L, Prakash S, Lloyd RS. Translesion synthesis past acrolein-derived DNA adduct, gamma - hydroxypropanodeoxyguanosine, by yeast and human DNA polymerase eta. J. Biol. Chem. 2003;278:784–790. doi: 10.1074/jbc.M207774200. [DOI] [PubMed] [Google Scholar]
- 6.Washington MT, Minko IG, Johnson RE, Wolfle WT, Harris TM, Lloyd RS, Prakash S, Prakash L. Efficient and error-free replication past a minorgroove DNA adduct by the sequential action of human DNA polymerases iota and kappa. Molecular and cellular biology. 2004;24:5687–5693. doi: 10.1128/MCB.24.13.5687-5693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Washington MT, Minko IG, Johnson RE, Haracska L, Harris TM, Lloyd RS, Prakash S, Prakash L. Efficient and error-free replication past a minorgroove N2-guanine adduct by the sequential action of yeast Rev1 and DNA polymerase zeta. Molecular and cellular biology. 2004;24:6900–6906. doi: 10.1128/MCB.24.16.6900-6906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolfle WT, Johnson RE, Minko IG, Lloyd RS, Prakash S, Prakash L. Human DNA polymerase iota promotes replication through a ring-closed minorgroove adduct that adopts a syn conformation in DNA. Molecular and cellular biology. 2005;25:8748–8754. doi: 10.1128/MCB.25.19.8748-8754.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCulloch SD, Kokoska RJ, Garg P, Burgers PM, Kunkel TA. The efficiency and fidelity of 8-oxo-guanine bypass by DNA polymerases delta and eta. Nucleic acids research. 2009;37:2830–2840. doi: 10.1093/nar/gkp103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCulloch SD, Kokoska RJ, Masutani C, Iwai S, Hanaoka F, Kunkel TA. Preferential cis-syn thymine dimer bypass by DNA polymerase eta occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 11.Takata K, Arana ME, Seki M, Kunkel TA, Wood RD. Evolutionary conservation of residues in vertebrate DNA polymerase N conferring low fidelity and bypass activity. Nucleic acids research. 2010;38:3233–3244. doi: 10.1093/nar/gkq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stone JE, Kumar D, Binz SK, Inase A, Iwai S, Chabes A, Burgers PM, Kunkel TA. Lesion bypass by S. cerevisiae Pol zeta alone. DNA repair. 2011;10:826–834. doi: 10.1016/j.dnarep.2011.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graziewicz MA, Longley MJ, Copeland WC. DNA polymerase gamma in Mitochondrial DNA Replication and Repair. Chemical Reviews. 2006;106:383–405. doi: 10.1021/cr040463d. [DOI] [PubMed] [Google Scholar]
- 14.Lee YS, Kennedy WD, Yin YW. Structural insight into processive human mitochondrial DNA synthesis and disease-related polymerase mutations. Cell. 2009;139:312–324. doi: 10.1016/j.cell.2009.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee YS, Lee S, Demeler B, Molineux IJ, Johnson KA, Yin YW. Each monomer of the dimeric accessory protein for human mitochondrial DNA polymerase has a distinct role in conferring processivity. J. Biol. Chem. 2010;285:1490–1499. doi: 10.1074/jbc.M109.062752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graziewicz MA, Bienstock RJ, Copeland WC. The DNA polymerase gamma Y955C disease variant associated with PEO and parkinsonism mediates the incorporation and translesion synthesis opposite 7,8-dihydro-8-oxo-2'-deoxyguanosine. Hum. Mol. Genet. 2007;16:2729–2739. doi: 10.1093/hmg/ddm227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graziewicz MA, Sayer JM, Jerina DM, Copeland WC. Nucleotide incorporation by human DNA polymerase gamma opposite benzo[a]pyrene and benzo[c]phenanthrene diol epoxide adducts of deoxyguanosine and deoxyadenosine. Nucleic Acids Res. 2004;32:397–405. doi: 10.1093/nar/gkh213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kasiviswanathan R, Gustafson MA, Copeland WC, Meyer JN. Human mitochondrial DNA polymerase gamma exhibits potential for bypass and mutagenesis at UV-induced cyclobutane thymine dimers. J. Biol. Chem. 2012;287:9222–9229. doi: 10.1074/jbc.M111.306852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasiviswanathan R, Minko IG, Lloyd RS, Copeland WC. Translesion Synthesis Past Acrolein-derived DNA Adducts by Human Mitochondrial DNA Polymerase gamma. J. Biol. Chem. 2013;288:14247–14255. doi: 10.1074/jbc.M113.458802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasiviswanathan R, Longley MJ, Young MJ, Copeland WC. Purification and functional characterization of human mitochondrial DNA polymerase gamma harboring disease mutations. Methods. 2010;51:379–384. doi: 10.1016/j.ymeth.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim SE, Ponamarev MV, Longley MJ, Copeland WC. Structural Determinants in Human DNA Polymerase gamma Account for Mitochondrial Toxicity from Nucleoside Analogs. J. Mol. Biol. 2003;329:45–57. doi: 10.1016/s0022-2836(03)00405-4. [DOI] [PubMed] [Google Scholar]
- 22.Longley MJ, Ropp PA, Lim SE, Copeland WC. Characterization of the native and recombinant catalytic subunit of human DNA polymerase gamma: identification of residues critical for exonuclease activity and dideoxynucleotide sensitivity. Biochemistry. 1998;37:10529–10539. doi: 10.1021/bi980772w. [DOI] [PubMed] [Google Scholar]
- 23.Lim SE, Longley MJ, Copeland WC. The mitochondrial p55 accessory subunit of human DNA polymerase gamma enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance. J. Biol. Chem. 1999;274:38197–38203. doi: 10.1074/jbc.274.53.38197. [DOI] [PubMed] [Google Scholar]
- 24.Boosalis MS, Petruska J, Goodman MF. DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J. Biol. Chem. 1987;262:14689–14696. [PubMed] [Google Scholar]
- 25.Mendelman LV, Petruska J, Goodman MF. Base mispair extension kinetics. Comparison of DNA polymerase alpha and reverse transcriptase. J. Biol. Chem. 1990;265:2338–2346. [PubMed] [Google Scholar]
- 26.Chan SSL, Longley MJ, Copeland WC. The common A467T mutation in the human mitochondrial DNA polymerase (POLG) compromises catalytic efficiency and interaction with the accessory subunit. J. Biol. Chem. 2005;280:31341–31346. doi: 10.1074/jbc.M506762200. [DOI] [PubMed] [Google Scholar]