Abstract
New insights into the intrarenal renin-angiotensin system (RAS) have modified our traditional view of the system. However, many finer details of this network of peptides and associated peptidases remain unclear. We hypothesized that a computational systems biology approach, applied to peptidomic data, could help to unravel the network of enzymatic conversions. We built and refined a Bayesian network model and a dynamic systems model starting from a skeleton created with established elements of the RAS and further developed it with archived MALDI-TOF mass spectra from experiments conducted in mouse podocytes exposed to exogenous angiotensin (Ang) substrates. The model-building process suggested previously unrecognized steps, three of which were confirmed in vitro, including the conversion of Ang(2-10) to Ang(2-7) by neprilysin (NEP), and Ang(1-9) to Ang(2-9) and Ang(1-7) to Ang(2-7) by aminopeptidase A (APA). These data suggest a wider role of NEP and APA in glomerular formation of bioactive Ang peptides and/or shunting their formation. Other steps were also suggested by the model and supporting evidence for those steps was evaluated using model-comparison methods. Our results demonstrate that systems biology methods applied to peptidomic data are effective in identifying novel steps in the Ang peptide processing network, and these findings improve our understanding of the glomerular RAS.
Keywords: Proteomics, Renin Angiotensin System, Systems Biology, Mass Spectrometry
Introduction
The renin-angiotensin system (RAS) is one of the most important and well-studied regulatory hormonal systems in medicine and plays a role in blood pressure regulation, fluid and electrolyte balance, and development.1 The decapeptide angiotensin (Ang) I [Ang(1-10)], is cleaved from the N-terminus of angiotensinogen by renin, and further cleavage by Ang-converting enzyme (ACE), chymase, neprilysin (NEP), aminopeptidase A (APA), aminopeptidase N (APN), ACE2, and other RAS-associated peptidases produce a cocktail of bioactive fragments which act through various cell surface receptors (AT1, AT2, AT4 and AT7).2-6 Distinct receptor activation can lead to opposing biological effects (e.g., vasoconstriction vs. vasodilatation) and our understanding of how these effects associate with the signaling flux across this cocktail of peptides is limited. Pharmacological blockade of the RAS is a therapeutic modality extensively utilized in clinical practice and specifically aimed to antagonize the generation or actions of Ang II [Ang(1-8)] to treat essential hypertension, heart failure, and chronic kidney disease.7-11 Although the introduction of RAS antagonists, i.e., ACE inhibitors (ACEIs), Ang AT1 receptor blockers (ARBs) and direct renin inhibitors (DRIs) into clinical practice have significantly improved treatment options, there are certain scenarios where the lack of a complete understanding of the intricacies of the local RAS may be limiting the potential for designing therapeutically valuable manipulations of the system.12-15 To date, study of the RAS has focused primarily on Ang II. Nonetheless, pharmacological alterations of the RAS result in simultaneous changes in abundance of various Ang peptides, as well as in compensatory changes in expression and activity of the participating RAS enzymes. The final stimulatory signal at a given cell reflects the combined concentrations of a cocktail of Ang peptides and their corresponding receptors present locally. In addition to Ang II, other peptide fragments {Ang(1-7), Ang(1-9), Ang(2-10), Ang III [Ang(2-8)], Ang IV [Ang(3-8)], Ang(2-7), Ang(3-7), and Ang(3-4)} have been identified as potentially bioactive, with actions in concert or in opposition.16-22 Local RAS systems have been identified in the kidney, heart, lung, liver, vasculature, and nervous system, and are, presumably, part of nearly every organ system.23 Of note, the RAS localized in the kidney is characterized by the highest tissue concentration of Ang II.24,25 We are only now beginning to understand the complexity of the network of peptidases that produce this profile and how that network differs across localities.
Methods from the field of proteomics have significantly improved our ability to identify, classify, characterize, and quantify proteins and peptides in complex mixtures.26 Mass spectrometry (MS), coupled with stable, isotopically-labeled peptide references, and applied using the Absolute Quantification (AQUA) strategy27 provides an effective tool for targeted quantification of peptides and proteins. In our applications, a cocktail of labeled Ang peptides of known concentrations is mixed with cell media and spotted on a MALDI (Matrix Assisted Laser Desorption Ionization) plate from which a spectrum is collected. Peak areas from isotopic clusters associated with native and AQUA peptides are calculated and native peptide quantities estimated from the cluster area ratios and known AQUA peptide concentrations. This method has proven effective in our laboratory and has resulted in an extensive archive of MALDI spectra from cell culture experiments.28,29 A recent reexamination of the archive indicated that many of the Ang peptides with masses above 600 Da were clearly detectable and suggested that the normalized peak areas associated with these peptides might be informative as to the underlying peptide processing network. We therefore, sought to determine if a data mining and network modeling effort, applied to these data, might lead to the identification of novel steps in this important pathway.
The field of computational systems biology provides methods for integrating large data sets in support of efforts to identify, model, and understand the networks central to biological systems. One modeling approach, the Bayesian Network, has proven useful in identifying novel signaling paths in cell signaling networks.30-33 The underlying inference methods attempt to build statistical models of conditional dependencies among the constituents of the network (e.g., molecules of the signal transduction system) from observations of the network under various conditions of stimulation and inhibition.34 The resulting conditional dependencies give insight into potential interactions among the signaling system components and have led to novel discoveries in cell signaling systems. Another method, dynamic systems modeling, goes further, attempting to develop a mathematical model of the dynamic behavior of the system. Often, dynamic systems models are constructed using sets of ordinary differential or stochastic differential equations and the associated kinetic parameters.35 These models capture temporal relationships not easily represented in Bayesian networks but present a more challenging network identification and parameter estimation task.36
Here we report on efforts to identify and confirm novel steps in the Ang peptide processing network using a database of spectra from previously conducted experiments. The study was executed in two phases. During the first phase, Ang peptide peak areas were extracted from an archive of spectra and used to build a Bayesian network representation of angiotensin peptide processing. Predictions from that network focused further experimental efforts that confirmed new steps suggested by the model. In the second phase, spectra from these additional experiments were added to the archive and the network model rebuilt. Steps suggested in the second model were then assessed in the context of a dynamic systems model. The study considered all Ang peptides identifiable in our archive without consideration for bioactivity, as we attempted to identify paths that bypass bioactive molecules as well as those that lead to bioactive molecules. We employed computational methods from systems biology to build and refine the Bayesian networks and then to build a dynamic systems model from which the strength of evidence for novel edges can be assessed. Finally, we present confirmatory experimental evidence for three of the edges predicted through these methods thus expanding our understanding of the Ang network and demonstrating the potential for mining archives of proteomic data using methods from systems biology.
Materials and Methods
Mouse Podocyte Cell Culture
Immortalized mouse podocytes [generously provided by Dr. Peter Mundel (Massachusetts General Hospital, Boston, MA) and Dr. Jeffrey Kopp (National Institutes of Health, Bethesda, MD)] were grown under standard conditions as previously published.37 Cells were exposed to Ang substrates as previously described.38 See details in Online Supplement.
MALDI-TOF MS
Samples obtained at various time points were assayed by MALDI time of flight (TOF) MS, as previously described, with minor modifications.38,39 See details in Online Supplement.
Recombinant RAS enzyme activity
For an in vitro APA activity assay: 100 ng of human recombinant APA (rAPA; R&D Systems, Minneapolis, MN) in assay buffer (25 mmol/L Tris, 50 mmol/L CaCl2, 0.2 mol/L NaCl, pH 8.0) were pre-incubated in the presence or absence of 100 μmol/L amastatin (APA inhibitor, Sigma-Aldrich, St. Louis, MO) for 20 min at room temperature and subsequently exposed to 1 μmol/L Ang 1-7 for 30 – 120 min. Reactions were quenched with 1% trifluoroacetic acid (TFA) acidification. Cleavage of Ang peptide was analyzed by MALDI-TOF MS and isotope-labeled peptide quantification as described above. For an in vitro NEP activity assay: 100 ng of human recombinant NEP (rNEP; R&D Systems) in assay buffer (50 mmol/L Tris, pH 7.4) were pre-incubated in the presence or absence of 1 μmol/L SCH39370 (NEP inhibitor) or 50 - 100 nmol/L thiorphan (Sigma-Aldrich) for 20 min at room temperature and subsequently exposed to 1 μmol/L Ang 2-10 for 15 – 60 min. Reactions were quenched with 1% TFA acidification. Cleavage of Ang peptide was analyzed by MALDI-TOF MS and isotope-labeled peptide quantification as described above. All experiments were run in triplicate.
Mass Spectra Data Processing
Quantification in AQUA experiments and other peak area calculations were performed using a custom-developed software package developed in the R statistical computing language, version 2.11.40 Briefly, the elemental composition of each peptide (and its AQUA reference) was determined from the amino acid sequence and isotopic spectra were convolved to compute the relative abundance of each isotopic peak. A search algorithm was used to find the peak width (resolution), mass error, baseline, and isotopic cluster area. The algorithm searches the space of mass errors and peak widths in a −1 Da to +7 Da region about the monoisotopic mass and a linear fit to the observed intensity is computed giving the cluster area. The isotopic cluster area for the mass error and peak width giving the lowest R2 value is taken as the area estimate. To account for overlap in isotopic clusters from nearby native peptides [e.g. Ang(1-9), m/z 1183.6 and Ang(2-10), m/z 1181.7] and their associated AQUA peptides, groups of overlapping peptides were fit simultaneously, sharing a common baseline and peak width. Doing so facilitated decomposition of the overlapping isotopic clusters.
Estimated Peak Area Quality Assessment
As we are not able to confirm the identity of each peak from the archived MALDI spectra, we must consider the possibility that some non-Ang peptide or source of noise might yield fits comparable to that of the peptides of interest. To assess the quality of the fitted isotopic clusters, we compared the distribution of R2 for the Ang peptide fits with that obtained by fitting random peptide sequences of similar length. We also found that the R2 distribution for fits to angiotensin peptides had a “bathtub” shape indicative of a mixture of fits to noise (similar to the null case) and fits to peptides of interest. The minima of the bathtub occurred at R2 = 0.65. This value was used as a threshold for accepting a peak area estimate in our modeling effort. The dataset was then filtered, replacing peak areas having R2 values below this threshold with a small value.
Bayesian Network Inference
Bayesian network inference was performed using the bnlearn package, version 2.441, executed within the R statistical computing environment, version 2.1140. Network nodes included all Ang peptides with length greater than 5 amino acids (see Online Supplement, Table S1), all added substrates, all inhibitors, and time since substrate addition. We treat the data as continuous and employ Gaussian Bayesian Networks where the global distribution is taken to be multivariate normal and local distributions are normal and linked by linear constraints associated with edges.42 Peptide areas from spectra were normalized to total angiotensin ion current and log transformed. Times were also log transformed. All edges are assumed excluded unless either (1) the edge extends from a peptide to a shorter peptide that can be obtained through a single site cleavage, or (2) the edge extends from a stimulus to its associated peptide, or (3) the edge extends from an inhibitor to any peptide, or (4) the edge extends from the time node to any peptide. An edge inclusion list was constructed and included edges associated with processing steps that are part of the processing pathway established in the literature. The network was constructed using the bnlearn tabu function using the Bayesian Information Criterion as score. Network edge strength assessment was performed using the bnlearn arc.strength function.
Dynamic System Model Inference and Comparison
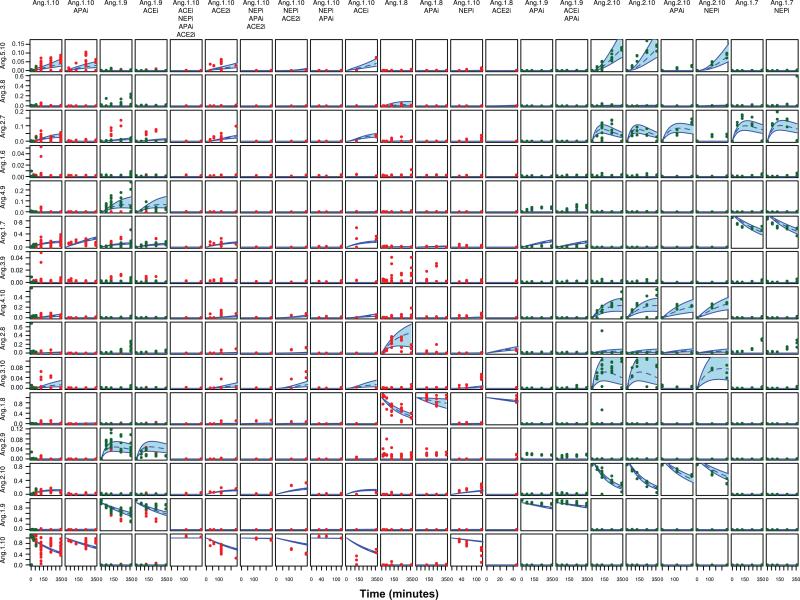
While the Bayesian network model employed here proved useful in identifying novel steps in the angiotensin pathway, its ability to capture the transient response is limited by the available data. Dynamic model parameter inference and time course calculations were performed using a custom-developed software package implemented in the R statistical computing language, version 2.1140. The model is expressed as a system of linear differential equations. In each reaction, the rate is assumed proportional to the available substrate. In experimental cases where inhibition is applied, the rate constant is set to 0 for all reactions associated with the inhibited peptidase. The time course for this model can be solved in closed form, eliminating the need for numerical integration. We assume the observed area is related to the underlying concentration through a scale factor. The scale factor is assumed constant across experiments and specific to a peptide. Rate constants and flyability scale factors for a given network structure were inferred using Markov Chain Monte Carlo (MCMC) sampling using the Metropolis Hastings algorithm.43 Prior distributions for kinetic parameters (rate constants) were based on the estimated linear response at low substrate concentrations (kcat/Km) using kinetic parameters found in the literature for peptidases of this system.44-48 Observed data for a given experimental condition (stimulus and inhibition) is assumed normally distributed with mean given by the associated model prediction (μ) and variance given by (δ + sμ)2. Both δ and s are inferred. For each Markov chain, 800,000 samples were collected following a burn-in of 200,000 samples which were then thinned by a factor of 800 to give 1000 samples used in the analysis. For each structural alternative, 5 chains starting from different random seeds were collected. Deviance Information Criterion (DIC) was computed for each chain and DIC mean and variance were computed from the set for use in model comparisons. Structural alternatives were considered to differ if their mean DIC values differed significantly. To visualize uncertainty in the predicted trajectories due to uncertainty in the inferred model parameters, time courses were computed for each experimental condition for the samples collected from the five chains and the credible intervals for each time point were then computed for plotting (see Figure 6).
Figure 6.
Compilation of normalized data for Ang peptides vs. time, organized by experimental condition shown with predicted time course uncertainty for the proposed network model. Experimental conditions across top indicate the added peptide and the inhibitors (e.g. ACEi indicates ACE inhibition.) Times are given in minutes and amounts in μmol/L, normalized to a 1 μmol/L substrate addition. Time course uncertainty bounds are taken from the 95% credible interval on trajectories computed from 5,000 kinetic and flyability parameter samples from the MCMC process (1,000 from each of 5 independent chains following burn-in and thinning). The dashed line gives the time course median. Red dots indicate data from the original spectrum archive, green dots from experiments performed as part of this study. Ang(2-10) data is shown in 2 columns, each containing data from separate groups of experiments.
Statistical Analysis
Results of confirmatory experiments are expressed as means ± standard deviation. Peptide abundances were compared using Student's t test. Significance was set at p < 0.05.
Results
Peptide Peak Area Estimation and Identification
Peak areas for 21 Ang peptides (length greater than 4 amino acids) and associated AQUA peptides were computed for all spectra from the archive using an isotopic cluster model. Of the 384 spectra, 228 were acceptable as they had no significant noise or elevated baselines in the low mass range, were collected from cultured mouse podocyte experiments, and were collected from samples taken less than 12 hours post stimulation. Peak areas and quality measures for 20 Ang peptides [Ang(3-7), Ang(5-9)/(6-10), Ang(1-5), Ang(4-8), Ang(2-6), Ang(5-10), Ang(3-8), Ang(2-7), Ang(1-6), Ang(4-9), Ang(1-7), Ang(3-9), Ang(4-10), Ang(2-8), Ang(3-10), Ang(1-8), Ang(2-9), Ang(2-10), Ang(1-9), Ang(1-10] were estimated from each of these spectra yielding a total of 4,560 data points. Ang(5-9) and Ang(6-10) have the same elemental composition and thus appear at the same mass in our MALDI spectra. We found evidence for all Ang fragments of length 5 and above except Ang(1-6) and Ang(4-9) among the isotopic clusters having R2 above the selected threshold for quality. Follow-up experiments (see below) found evidence of Ang(4-9). Surprisingly, the isotopic cluster associated with Ang(5-9)/(6-10) appeared in more spectra than any other peptide. The identity of this peak was confirmed by tandem mass spectrometry in subsequent experiments and found to contain a mixture of Ang(5-9) and Ang(6-10).
Gaussian Bayesian Network Inference
The set of edges exceeding the strength threshold suggested several processing steps not found in the baseline network. Extending from Ang(1-9), the set included edges from Ang(1-9) to Ang(2-9), Ang(2-9) to Ang(3-9), and Ang(3-9) to Ang(4-9). Also included was a step from Ang(1-7) to Ang(2-7) as well as steps from Ang(1-10) to Ang(5-10), Ang(2-10) to Ang(2-9), and Ang(1-10) to Ang(3-10) (see Online Supplement, Table S2).
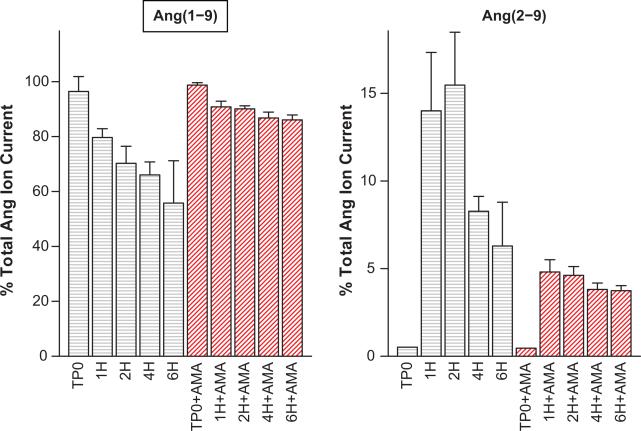
Confirmation of Ang(1-9) to Ang(2-9) via APA
Using Ang(1-9) as the substrate, we found evidence of Ang(2-9) production and significant inhibition of Ang(2-9) production in the presence of 100 μmol/L amastatin (APA inhibition) (Figure 1). These data suggest that Ang(1-9) is cleaved to Ang(2-9) by APA in mouse podocytes and therefore support the existence of the Ang(1-9) to Ang(2-9) step indicated in our network model. This finding is not surprising given the established Ang(1-10) to Ang(2-10) and Ang(1-8) to Ang(2-8) conversions catalyzed by APA. This step might be important as an alternative path for degradation of Ang(1-9), away from Ang(1-7).
Figure 1.
MALDI spectra peak areas from mouse podocytes, incubated with Ang(1-9) indicate production of Ang(2-9). Ang(1-9) label is inside a text box to indicate that it was the added substrate. Control, and amastatin-treated (AMA) cases were sampled just following substrate addition (TP0) and 1, 2, 4, and 6 hours post-addition (TP0, 1H, 2H, 4H, 6H). In the absence of cells, the peptide levels were stable over the 6 hour incubation period. Peak areas were normalized to total angiotensin peptide current and bars show mean of three experiments and one standard deviation about the mean. Ang(2-9) production is decreased and Ang(1-9) degradation rate is decreased at 1, 2, and 4 hours post addition of substrate following pretreatment with 100 μmol/L amastatin, an APA inhibitor.
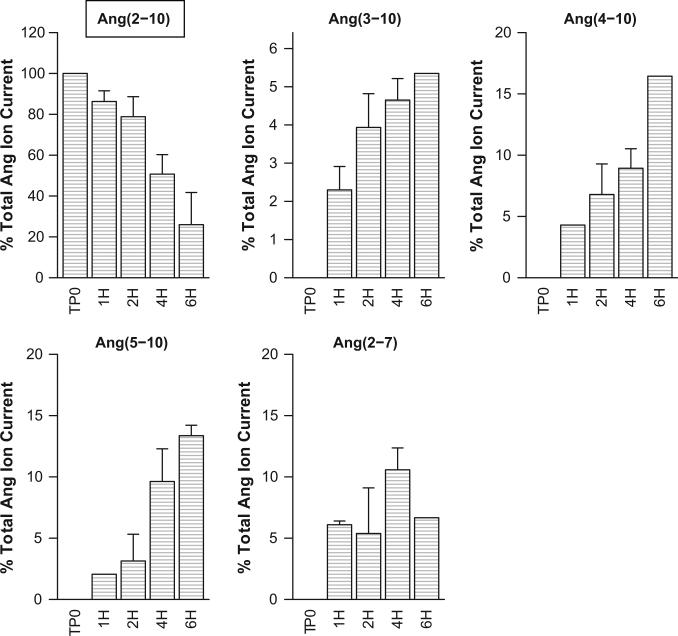
Path from Ang(2-10) to Ang(5-10) Established
Experiments involving addition of Ang(2-10) produced evidence of Ang(3-10), Ang(4-10), and Ang(5-10) indicating that Ang(5-10) is reachable from Ang(2-10) (Figure 2). This is consistent with the Bayesian network's indication of a weak Ang(5-10) dependency on Ang(4-10) (strength of 59%) but an associated processing step cannot be confirmed from this experiment as possible indirect paths might exist. More interesting in these data was evidence of Ang(2-7) production. While a step from Ang(2-10) to Ang(2-7) was not suggested by the Bayesian network, an initial bottom-up modeling effort performed by our group (unpublished) had identified the need for an Ang(2-10) to Ang(2-7) step, catalyzed by NEP, to explain a large increase in Ang(2-10) under NEP inhibition following Ang I addition. We also note that in the Bayesian network, Ang(2-10) was found to depend on NEP inhibition (appeared in 89% of the bootstrapped networks) and had a positive coefficient [Ang(2-10) increases in the presence of NEP inhibition] which would be consistent with our earlier observation.
Figure 2.
MALDI spectra peak areas from mouse podocytes, incubated with Ang(2-10) indicate production of Ang(3-10), Ang(4-10), Ang(5-10) and Ang(2-7). Ang(2-10) label is inside a text box to indicate that it was the added substrate. Control cases were sampled just following substrate addition (TP0) and 1, 2, 4, and 6 hours post-addition (TP0, 1H, 2H, 4H, 6H). In the absence of cells, the peptide levels were stable over the 6 hour incubation period. Peak areas were normalized to total angiotensin peptide current and bars show mean of three experiments and one standard deviation about the mean. No evidence of time dependent decay of Ang(2-10), production of other fragments, or post-source decay is evident in the cell-free cases.
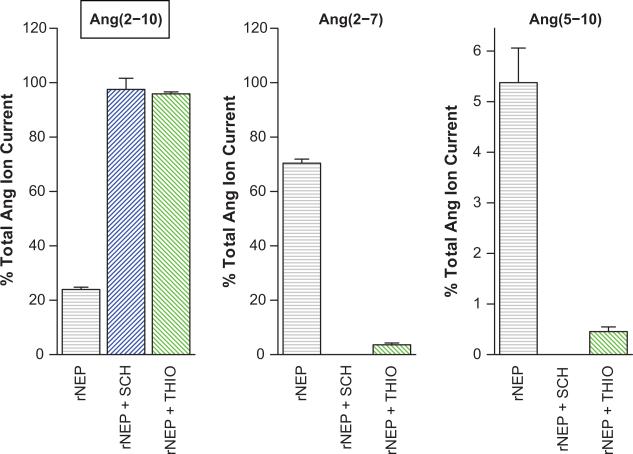
Confirmation of Ang(2-10) to Ang(2-7) Conversion by NEP
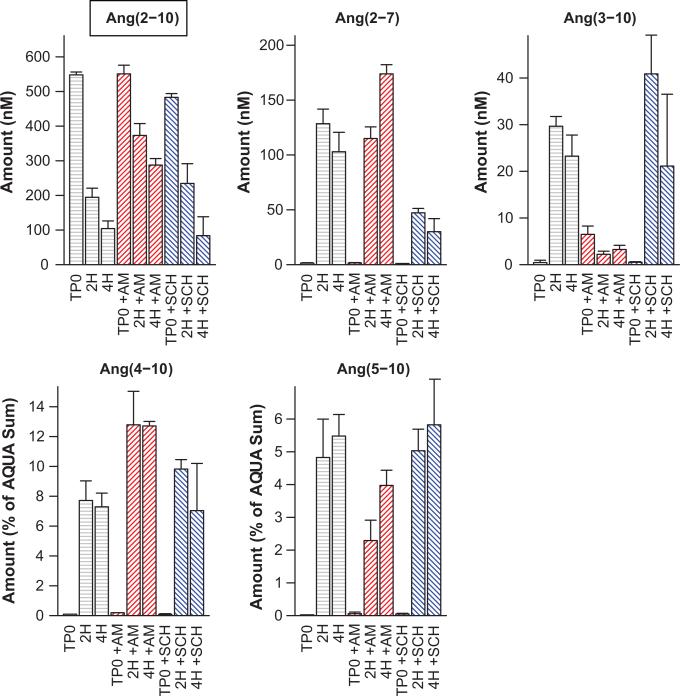
To confirm the proposed conversion of Ang(2-10) to Ang(2-7), additional experiments were performed. Ang(2-10) was exposed to human rNEP untreated or following pretreatment with 1 μmol/L SCH39370 (NEP inhibitor) or 70 nmol/L thiorphan (NEP inhibitor) for 15 minutes and subjected to MALDI mass spectrometry (Figure 3). For the conditions where inhibitor was omitted, the Ang(2-7) peak was 2.9 ± 0.16 fold that of the Ang(2-10) area, at 15 minutes. Under SCH39370 treatment, none of the spectra contained Ang(2 -7) peaks with R2 above our quality threshold indicating nearly complete inhibition (0.009 ± 0.002 fold, relative to Ang(2-10) peak area). Treatment with thiorphan yielded a similar result with a relative Ang(2-7) peak area of 0.037 ± 0.007. Together these findings support NEP as the enzyme responsible for Ang(2-10) conversion to Ang(2-7). Of note, the conversion of Ang(2-10) to Ang(5-10) via NEP was also evident when inhibitor was omitted, but the magnitude of the Ang(5-10) peak area was significantly smaller. These data support the possibility of a direct Ang(2-10) to Ang(5-10) conversion by NEP, in addition to the Ang(2-10) to Ang(2-7) conversion. We performed additional experiments adding the substrate Ang(2-10) with or without amastatin (100 μmol/L) or SCH39370 (10 μmol/L) pretreatment. Samples were collected immediately following the addition of substrate, 2, and hours post addition. The peptides Ang(2-10), Ang(2-7), and Ang(3-10) were quantified using AQUA peptides, and Ang(4-10) and Ang(5-10) were normalized to the sum of the included AQUA peptide signals (Figure 4). These experiments indicated a significant decrease in Ang(2-7) production following pretreatment with SCH39370, thus providing additional evidence of the NEP-catalyzed Ang(2-10) to Ang(2-7) conversion. We additionally note that a significant decrease in net Ang(3-10) conversion following treatment with amastatin is accompanied by a significant increase (at 4 hours) in Ang(2-7). Also, Ang(2-10) levels at 2 and 4 hours in the presence of amastatin were significantly higher than control. Because amastatin can inhibit both APA and APN 49-51, this observation suggests that APA/APN inhibition blocks the conversion of Ang(2-10) to Ang(3-10) providing additional substrate for conversion to Ang(2-7). Collectively, these data confirm NEP conversion of Ang(2-10) to Ang(2-7) in mouse podocytes.
Figure 3.
Recombinant human NEP produces Ang(2-7) and Ang(5-10) from Ang(2-10). Ang(2-10) label is inside a text box to indicate that it was the added substrate. Peak areas were normalized to total angiotensin peptide current and bars show mean of three experiments and one standard deviation about the mean. Ang(2-10) with recombinant human NEP alone, or pretreated with 1 μmol/L SCH39370 (NEP inhibitor) or 70 nmol/L thiorphan (NEP inhibitor) was sampled at 15 minutes. Production of both Ang(2-7) and Ang(5-10) was decreased in the presence of the NEP inhibitors.
Figure 4.
Ang peptide quantitation from mouse podocytes, incubated with Ang(2-10) in confirmatory experiments support production of Ang(3-10), Ang(4-10), Ang(5-10) and Ang(2-7), decreased Ang(2-7) production under NEP inhibition, and decreased production of Ang(3-10) under APA inhibition. Ang(2-10) label is inside a text box to indicate that it was the added substrate. Cells alone or following pretreatment with SCH39370 (SCH, NEP inhibitor) or amastatin (AM, APA inhibitor) were sampled immediately following substrate addition (TP0) and 2 and 4 hours post addition (2H, 4H). Ang(2-10), Ang(3-10), and Ang(2-7) were quantified using AQUA peptides, Ang(4-10) and Ang(5-10) were normalized to the sum of the AQUA peptide areas. Bars show the mean of three experiments and one standard deviation about the mean.
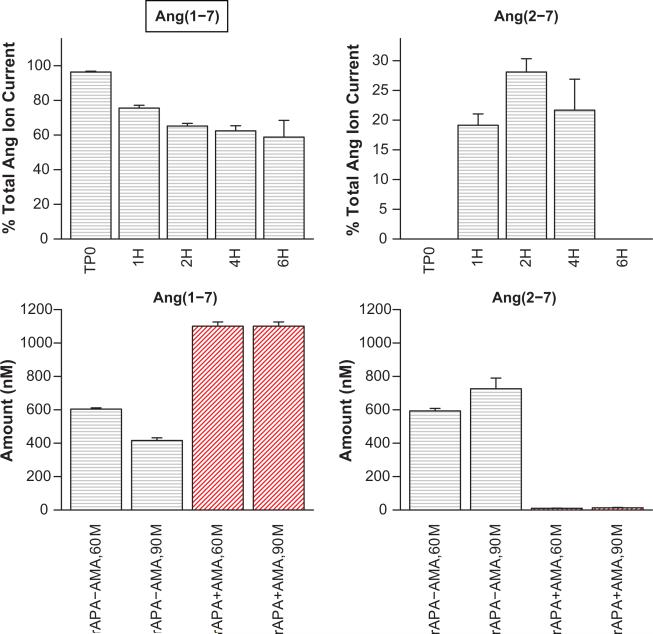
Confirmation of Ang(1-7) to Ang(2-7) Conversion via APA
From the cell culture experiments using Ang(1-7) as the substrate, we found evidence of Ang(2-7) production (Figure 5). The Ang(2-7) signal reached 28% of the total Ang ion current at 2 hours and was well above baseline (19-28% of Ang ion current) at 1, 2, and 4 hours. Inhibition with thiorphan (NEP inhibitor) had no significant effect on the Ang(2-7) response. To confirm the proposed conversion of Ang(1-7) to Ang(2-7), additional experiments were performed. Ang(1-7) was exposed to human rAPA untreated or following pretreatment with 10 μmol/L amastatin for 60 or 90 minutes, subjected to MALDI mass spectrometry (Figure 5, Panel B), and quantified using Ang(2-7) and Ang(1-7) AQUA peptides. Most of Ang(1-7) was converted to Ang(2-7) after 90 minutes (725 ± 64 nmol/L, 66%) and Ang(2-7) production was blocked following treatment with amastatin (12.8 ± 2.5 nmol/L, 1.2%). These data support Ang(1-7) to Ang(2-7) conversion by APA in mouse podocytes as indicated by our network model. Together with our findings regarding Ang(1-9), it appears that APA may play a role in converting Ang I, Ang II, Ang(1-9), and Ang(1-7) to Ang(2-10), Ang III, Ang(2-9), and Ang(2-7), respectively.
Figure 5.
Normalized angiotensin peak areas from mouse podocytes, incubated with Ang(1-7) indicate production of Ang(2-7) at 1, 2, and 4 hours post addition of substrate (Panel A) and Ang(1-7), incubated with recombinant APA shows production of Ang(2-7) at 60 and 90 minutes that is lost when pretreated with APA inhibitor amastatin (AMA) (Panel B). Ang(1-7) label is inside a text box to indicate that it was the added substrate. Culture media were sampled just following substrate addition (TP0) and 1, 2, 4, and 6 hours post-addition (TP0, 1H, 2H, 4H, 6H). In the absence of cells, the peptide levels were stable over the 6 hour incubation period. Areas were normalized to total angiotensin peptide current and bars show mean of three experiments and one standard deviation about the mean. No evidence of time dependent decay of Ang(1-7), production of other fragments, or post-source decay was evident in the cell-free cases (IC). In-vitro experiments were quantified relative to AQUA Ang(1-7) (500 nmol/L) and AQUA Ang(2-7) (200 nmol/L).
Network Revision and New Discoveries
Following confirmation of predictions from our initial Bayesian network model, we revised the model using data collected subsequent to the initial analysis. Repeating the procedure described above, we incorporated experimental data from the Ang(1-9), Ang(1-7), and Ang(2-10) substrate addition experiments. Additionally, we performed a short time series experiment in mouse podocyte cultures. In these experiments, cells were incubated with 1 μmol/L Ang I and sampled immediately after substrate addition and at 5, 10, 15, 30, and 60 minutes post addition. Samples were analyzed by MALDI-TOF MS and normalized areas extracted as before. The complete dataset included 665 spectra of which 465 were used in this analysis. The Bayesian network inference process was repeated using this subset. In addition to the steps suggested in the initial version of the Bayesian network, the revised version included Ang(2-10) to Ang(3-10), Ang(4-10) to Ang(5-10), Ang(2-10) to Ang(2-7), and Ang(2-9) to Ang(4-9) (see Online Supplement Table S2).
System Model Inference and Reaction Set
Of the combinations of additional steps analyzed using the dynamic systems model, we found that inclusion of Ang(4-10) to Ang(5-10), Ang(1-10) to Ang(5-10), Ang(2-10) to Ang(4-10), and Ang(2-9) to Ang(4-9) as a group provided the lowest DIC of the networks tested. Surprisingly, the edge from Ang(2-10) to Ang(2-9), which had a strength of 95% in the revised version of the Bayesian network, did not appear to improve the fit in the dynamic system model. Inclusion of steps from either Ang(1-10) to Ang(3-10) or Ang(2-9) to Ang(3-9) also failed to significantly improve the fit. A final model, built from the baseline and our confirmed steps, augmented with steps from Ang(4-10) to Ang(5-10), Ang(1-10) to Ang(5-10), Ang(2-10) to Ang(4-10) and Ang(2-9) to Ang(4-9) was examined further to better understand the remaining uncertainty and the 95% credible intervals for each of those trajectories were computed and are plotted, along with the observed data, in Figure 6.
Discussion
Our findings demonstrate that computational systems biology methods can be successfully utilized to model the multilayered Ang peptide processing network contained in mammalian cells. In addition to designing a model system that performed satisfactorily in reproducing biological experiments, using this approach, we identified enzymatic cleavages not previously described in the literature and with potential biological significance. Conversion of Ang(2-10) to Ang(2-7) by NEP, Ang(1-9) to Ang(2-9) by APA and Ang(1-7) to Ang(2-7) by APA, constitute paths within the network that expand on the well-recognized robust degrading capacity of aminopeptidases and endopeptidases localized in glomerular podocytes. Two of these unveiled steps involve generation of Ang(2-7). Importantly, there is evidence that indicates that Ang(2-7) may be a bioactive fragment that potentiates the hypotensive effect of bradykinin in a similar fashion to Ang(1-7).21 Another study reported that Ang(2-7) binds to the AT4 receptor, the receptor for Ang IV.22 Alternatively, it can be postulated that the conversion of Ang(2-10) into Ang(2-7) by NEP suggests the possibility of a bypass around the bioactive fragments Ang III and Ang IV. Notably, Ang III is known to exert vasoconstrictive effects and promote aldosterone stimulation 52,53 , whereas Ang IV has been linked to arterial thrombosis and biphasic pressor responses 54.
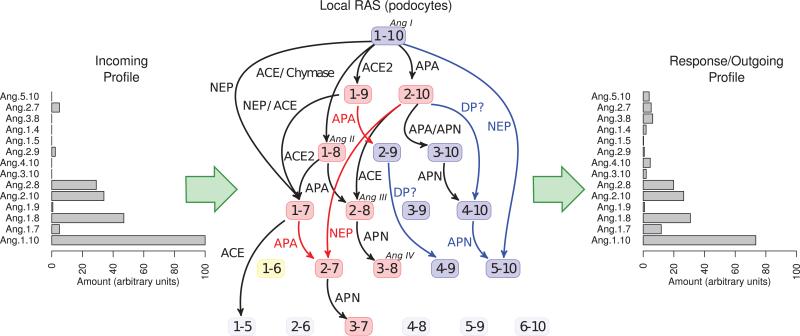
An illustration of the proposed network, based on the analysis described here, is given in Figure 7. We depart from the more standard representation of the RAS network and adopt the layout shown in the figure to emphasize (1) that placing the bioactive peptides in context with all other Ang peptides makes routes to and around those bioactive peptides more evident, and (2) that the RAS acts not only on incoming Ang I (or angiotensinogen), but against a profile of peptides that enter at intermediate points in this network. Viewed as a system of reactions within a local compartment, this network would likely act to filter an incoming profile, creating and responding to the filtered version of that profile based on the local pattern of expressed peptidases. To illustrate this notion, Figure 7 also shows a theoretical effect of podocyte-based Ang peptide processing on circulating Ang peptides as they pass through the site where podocytes reside. The histogram of the incoming peptide profile (left) is based on measured values55, whereas the outgoing profile (right) was computed from X̄ ≈ (I–V/F NK)− X0; where X0 is the incoming peptide profile, I is the identity matrix, V and F are the volume of flow rate through the local RAS compartment, N is the stoichiometric matrix, K is a matrix of estimated rate constants, and X̄ is the steady state of the peptide profile. Although based on a number of assumptions, these calculations exemplify a potential application our computational tool.
Figure 7.
Proposed network model showing all angiotensin peptides down to length 5 including bioactive peptides (red), undetected peptides (yellow), and all other peptides (blue). Edges in black were supported in the literature and summarized in Velez, et al 2. Edges in red were predicted in the first phase of our effort and confirmed by experiments reported here. Edges in blue were predicted in the revised model and found to significantly improve model fit (based on DIC) in the dynamic systems model. The histogram of the incoming peptide profile (left) is based on measured reported plasma values of Ang peptides. The response profile is included for illustration purposes only. DP? indicates an unidentified dipeptidyl aminopeptidase.
At present, our understanding of the bioactivity of Ang peptides suggests that only a small subset directly interact with cell surface receptors with high affinity. However, the paths both to and around this subset are extensive and most of these steps have the potential for influencing flux to the bioactive components. Additionally, possible regulatory feedback paths, such as inhibition of APA by Ang IV49, suggest that downstream peptides might act indirectly to modulate network peptidases and thus flux to bioactive peptides, influencing cellular responses without receptor interaction. While our efforts did indicate the presence of nearly all peptide fragments, we failed to find sufficient evidence in our existing dataset to include paths from Ang(2-9) to Ang(3-9) or Ang(2-8), or from Ang(2-7) to Ang(3-7). It is likely that with additional effort these steps will emerge and may result in paths back to the subset of bioactive components [back to Ang III, Ang IV, or Ang(3-7)]. At present, therapeutic modulation of this pathway has focused primarily on the steps from angiotensinogen to Ang I (DRIs), conversion of Ang I to Ang II (ACEIs), and Ang II action through the Ang AT1 receptor (ARBs). New approaches are emerging, such as enhancement of ACE2 activity56 and modulation of the prorenin receptor.57 Our increasing realization of the complexity of this pathway and the activity of its components calls into question a strictly Ang II-centric treatment approach and systems-based studies, as used here, will likely be necessary to design more directed targeting of therapeutics.
We find that in the application of ’omics’ techniques, we often collect much more data than are analyzed due in part to the act that experiments were designed to address a specific hypothesis. In this effort, we found that the totality of that data, when considered using a systems-based approach has value in network discovery efforts. While the current model is insufficient to capture the regulatory feedback mechanisms that must eventually be considered, we see it as an essential first step toward that goal. Key to the discovery process is this unifying model, that links observations across time and experimental conditions (stimulus and inhibition), and inference methods that provide a disciplined approach to assess the weight of evidence for structural variations within the model.
In this study, cell culture experiments, inhibitor-based perturbations of the system response, and use of data collected over 6 years of evolving experimental procedures presents challenges, limitations, and benefits. A key challenge was the development of complete and consistent characterizations of each of the spectra from the archive. While the spectra were readily recovered from data files recorded by the instrument, identifying the covariates from the associated experiment was somewhat more difficult. Generally, spectra used in earlier publications were well documented but many of the spectra from our archive came from experiments that might have been performed as a precursor to the published work. In our model-based approach, a critical underlying assumption is that the selected perturbations to the system are peptidase-specific and essentially complete. We recognize that some level of non-specific or incomplete inhibition is likely to occur thus contributing additional unexplained variation in our observations. We find that, even with these additional sources of error and variation, enzymatic steps suggested by the approach were readily confirmed in follow-up experiments thus suggesting that the approach has utility.
Conversions to tetrapeptides or smaller fragments were not included because matrix ions would often coexist in the same m/z window as the peptide. Because our archived samples mainly reflect ectoenzymatic activity, our analysis does not account for intracellular cleavage of Ang peptides58,59, although we cannot completely exclude a minor contribution of intracellular peptide processing to the detected spectra. Although other RAS enzymes, such as chymase, prolylendopeptidase, prolylcarboxypeptidase, thimet oligopeptidase, dipeptidyl aminopeptidase, etc., were not included in our model because of their low activity in podocytes28,29, they may be important in other kidney cells or disease states.29,60-63 It is conceivable that the podocyte RAS likely includes peptidases not considered in our analysis. In addition, we did not incorporate the potential enzymatic inhibitory actions of some Ang peptides. Notably, Ang IV has been reported to inhibit APA49, whereas ACE can be inhibited by Ang 1-7.64 Moreover, biologically active Ang peptides generated through enzymatic decarboxylation have also been reported to be detectable in human plasma.65 Altogether, these unaccounted aspects of the RAS further emphasize its complexity.
Perspectives
In summary, the Ang peptide processing network can be successfully modeled using computational tools and data mining. Although our effort was based on an artificial system, we were able to elucidate intermediate pathways within the network that were not previously recognized. Because in vivo maneuvers to alter the system involve inhibition or inactivation of RAS enzymes, we speculate that our methods may help predicting and/or better understanding systemic or local effects of those manipulations and guide future research. Further exploration of the Ang peptide processing network by refined computational systems biology methods could improve our understanding of the intricacies of the RAS with potential impact in medical therapies.
Supplementary Material
Novelty and Significance.
What is New?
Our study is the first to apply computational tools to model a cell-based network of Ang peptides and peptidases and, through the process, reveal novel steps within that network.
What is Relevant?
We demonstrated that conversion of Ang(2-10) to Ang(2-7) by NEP, Ang(1-9) to Ang(2-9) by APA, and Ang(1-7) to Ang(2-7) by APA, constitute elements of the cascade of fragmentation of Ang substrates in mouse podocytes.
Summary.
The Ang peptide processing network can be successfully modeled using Bayesian network inference and dynamic systems modeling coupled with peptidomic data mining. Our tools proved effective in revealing steps of the network that lead to either transient generation of bioactive peptides or shunting of their formation.
Acknowledgements
Jennifer Bethard of the MUSC Mass Spectrometry Core was instrumental in helping us acquire spectra for MALDI-TOF-TOF analysis. We thank Alison Bland for her assistance in collecting MALDI-TOF mass spectra.
Source of Funding
J.C.G.S was supported under a training grant from the National Institute of General Medicine (5T32GM74934-8). M.G.J. was supported by Career Development Awards through the Department of Veterans’ Affairs (CDA-2) and the Nephcure Foundation. J.C.Q.V is supported by a grant from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) of the National Institute of Health (NIH) (DK080944-04) and Dialysis Clinics Incorporated. J.H.S. was supported, in part, by a grant from NIDDK of the NIH (1R01DK080234).
Footnotes
Author Contributions
J.H.S., M. G. J. and J.C.Q.V.: conception and design of research; J.H.S., J.C.G.S., J.L.I., J.M.C. and J.C.Q.V.: performed experiments; J.H.S., J.C.G.S., M. G. J. and J.C.Q.V analyzed data; J.H.S., M. G. J., J.M.A. and J.C.Q.V. interpreted results of experiments; J.H.S. prepared figures and drafted manuscript; M. G. J. and J.C.Q.V edited and revised manuscript.
Disclosures
None.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Skrbic R, Igic R. Seven decades of angiotensin (1939-2009). Peptides. 2009;30:1945–1950. doi: 10.1016/j.peptides.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Velez JC. The importance of the intrarenal renin-angiotensin system. Nat Clin Pract Nephrol. 2009;5:89–100. doi: 10.1038/ncpneph1015. [DOI] [PubMed] [Google Scholar]
- 3.Ardaillou R. Angiotensin II receptors. J Am Soc Nephrol. 1999;10(Suppl 11):S30–39. [PubMed] [Google Scholar]
- 4.Handa RK, Handa SE, Elgemark MK. Autoradiographic analysis and regulation of angiotensin receptor subtypes AT(4), AT(1), and AT((1-7)) in the kidney. Am J Physiol Renal Physiol. 2001;281:F936–947. doi: 10.1152/ajprenal.2001.281.5.F936. [DOI] [PubMed] [Google Scholar]
- 5.Santos RA, Simoes e Silva AC, Maric C, Silva DM, Machado RP, de Buhr I, Heringer-Walther S, Pinheiro SV, Lopes MT, Bader M, Mendes EP, Lemos VS, Campagnole-Santos MJ, Schultheiss HP, Speth R, Walther T. Angiotensin-(1-7) is an endogenous ligand for the G protein-coupled receptor Mas. Proc Natl Acad Sci U S A. 2003;100:8258–8263. doi: 10.1073/pnas.1432869100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gwathmey TM, Alzayadneh EM, Pendergrass KD, Chappell MC. Novel roles of nuclear angiotensin receptors and signaling mechanisms. Am J Physiol Regul Integr Comp Physiol. 2012;302:R518–530. doi: 10.1152/ajpregu.00525.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis EJ, Hunsicker LG, Clarke WR, Berl T, Pohl MA, Lewis JB, Ritz E, Atkins RC, Rohde R, Raz I, Collaborative Study Group Renoprotective effect of the angiotensin-receptor antagonist irbesartan in patients with nephropathy due to type 2 diabetes. N Engl J Med. 2001;345:851–860. doi: 10.1056/NEJMoa011303. [DOI] [PubMed] [Google Scholar]
- 8.Lewis EJ, Hunsicker LG, Bain RP, Rohde RD. The effect of angiotensin-converting-enzyme inhibition on diabetic nephropathy. The Collaborative Study Group. N Engl J Med. 1993;329:1456–1462. doi: 10.1056/NEJM199311113292004. [DOI] [PubMed] [Google Scholar]
- 9.Parving HH, Persson F, Lewis JB, Lewis EJ, Hollenberg NK, Investigators AS Aliskiren combined with losartan in type 2 diabetes and nephropathy. N Engl J Med. 2008;358:2433–2446. doi: 10.1056/NEJMoa0708379. [DOI] [PubMed] [Google Scholar]
- 10.Dahlof B, Devereux RB, Kjeldsen SE, Julius S, Beevers G, de Faire U, Fyhrquist F, Ibsen H, Kristiansson K, Lederballe-Pederesen O, Lindholm LH, Nieminen MS, Omvik P, Oparil S, Wedel H, LIFE Study Group Cardiovascular morbidity and mortality in the Losartan Intervention For Endpoint reduction in hypertension study (LIFE): a randomised trial against atenolol. Lancet. 2002;359:995–1003. doi: 10.1016/S0140-6736(02)08089-3. [DOI] [PubMed] [Google Scholar]
- 11.The SOLVD Investigators Effect of enalapril on survival in patients with reduced left ventricular ejection fractions and congestive heart failure. N Engl J Med. 1991;325:293–302. doi: 10.1056/NEJM199108013250501. [DOI] [PubMed] [Google Scholar]
- 12.van den Meiracker AH, Man in 't Veld AJ, Admiraal PJ, Ritsema van Eck HJ, Boomsma F, Derkx FH, Schalekamp MA. Partial escape of angiotensin converting enzyme (ACE) inhibition during prolonged ACE inhibitor treatment: does it exist and does it affect the antihypertensive response? J Hypertens. 1992;10:803–812. [PubMed] [Google Scholar]
- 13.Mann JF, Schmieder RE, McQueen M, Dyal L, Schumacher H, Pogue J, Wang X, Maggioni A, Budaj A, Chaithiraphan S, Dickstein K, Keltai M, Metsarinne K, Oto A, Parkhomenko A, Piegas LS, Svendsen TL, Teo KK, Yusuf S, ONTARGET Investigators Renal outcomes with telmisartan, ramipril, or both, in people at high vascular risk (the ONTARGET study): a multicentre, randomised, double-blind, controlled trial. Lancet. 2008;372:547–553. doi: 10.1016/S0140-6736(08)61236-2. [DOI] [PubMed] [Google Scholar]
- 14.Borghi C, Boschi S, Ambrosioni E, Melandri G, Branzi A, Magnani B. Evidence of a partial escape of renin-angiotensin-aldosterone blockade in patients with acute myocardial infarction treated with ACE inhibitors. J Clin Pharmacol. 1993;33:40–45. doi: 10.1002/j.1552-4604.1993.tb03901.x. [DOI] [PubMed] [Google Scholar]
- 15.Ahmed AK, Kamath NS, El Kossi M, El Nahas AM. The impact of stopping inhibitors of the renin-angiotensin system in patients with advanced chronic kidney disease. Nephrol Dial Transplant. 2010;25:3977–3982. doi: 10.1093/ndt/gfp511. [DOI] [PubMed] [Google Scholar]
- 16.Haulica I, Bild W, Mihaila CN, Ionita T, Boisteanu CP, Neagu B. Biphasic effects of angiotensin (1-7) and its interactions with angiotensin II in rat aorta. J Renin Angiotensin Aldosterone Syst. 2003;4:124–128. doi: 10.3317/jraas.2003.013. [DOI] [PubMed] [Google Scholar]
- 17.Dharmani M, Mustafa MR, Achike FI, Sim MK. Effect of des-aspartateangiotensin I on the actions of angiotensin II in the isolated renal and mesenteric vasculature of hypertensive and STZ-induced diabetic rats. Regul Pept. 2005;129:213–219. doi: 10.1016/j.regpep.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 18.Tallant EA, Ferrario CM, Gallagher PE. Angiotensin-(1-7) inhibits growth of cardiac myocytes through activation of the mas receptor. Am J Physiol Heart Circ Physiol. 2005;289:H1560–1566. doi: 10.1152/ajpheart.00941.2004. [DOI] [PubMed] [Google Scholar]
- 19.Kemp BA, Bell JF, Rottkamp DM, Howell NL, Shao W, Navar LG, Padia SH, Carey RM. Intrarenal angiotensin III is the predominant agonist for proximal tubule angiotensin type 2 receptors. Hypertension. 2012;60:387–395. doi: 10.1161/HYPERTENSIONAHA.112.191403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Axelband F, Dias J, Miranda F, Ferrao FM, Reis RI, Costa-Neto CM, Lara LS, Vieyra A. Angiotensin-(3-4) counteracts the Angiotensin II inhibitory action on renal Ca(2+)-ATPase through a cAMP/PKA pathway. Regul Pept. 2012;177:27–34. doi: 10.1016/j.regpep.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Paula RD, Lima CV, Britto RR, Campagnole-Santos MJ, Khosla MC, Santos RA. Potentiation of the hypotensive effect of bradykinin by angiotensin-(1-7)-related peptides. Peptides. 1999;20:493–500. doi: 10.1016/s0196-9781(99)00031-5. [DOI] [PubMed] [Google Scholar]
- 22.Handa RK. Angiotensin-(1-7) can interact with the rat proximal tubule AT(4) receptor system. Am J Physiol. 1999;277:F75–83. doi: 10.1152/ajprenal.1999.277.1.F75. [DOI] [PubMed] [Google Scholar]
- 23.Paul M, Poyan Mehr A, Kreutz R. Physiology of local renin-angiotensin systems. Physiol Rev. 2006;86:747–803. doi: 10.1152/physrev.00036.2005. [DOI] [PubMed] [Google Scholar]
- 24.Navar LG, Lewis L, Hymel A, Braam B, Mitchell KD. Tubular fluid concentrations and kidney contents of angiotensins I and II in anesthetized rats. J Am Soc Nephrol. 1994;5:1153–1158. doi: 10.1681/ASN.V541153. [DOI] [PubMed] [Google Scholar]
- 25.Seikaly MG, Arant BS, Jr., Seney FD., Jr. Endogenous angiotensin concentrations in specific intrarenal fluid compartments of the rat. J Clin Invest. 1990;86:1352–1357. doi: 10.1172/JCI114846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gstaiger M, Aebersold R. Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat Rev Genet. 2009;10:617–627. doi: 10.1038/nrg2633. [DOI] [PubMed] [Google Scholar]
- 27.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci U S A. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Velez JC, Bland AM, Arthur JM, Raymond JR, Janech MG. Characterization of renin-angiotensin system enzyme activities in cultured mouse podocytes. Am J Physiol Renal Physiol. 2007;293:F398–407. doi: 10.1152/ajprenal.00050.2007. [DOI] [PubMed] [Google Scholar]
- 29.Velez JC, Ierardi JL, Bland AM, Morinelli TA, Arthur JM, Raymond JR, Janech MG. Enzymatic processing of angiotensin peptides by human glomerular endothelial cells. Am J Physiol Renal Physiol. 2012;302:F1583–1594. doi: 10.1152/ajprenal.00087.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawhon SD, Khare S, Rossetti CA, Everts RE, Galindo CL, Luciano SA, Figuereido JF, Nunes JE, Gull T, Davidson GS, Drake KL, Garner HR, Lewin HA, Baumler AJ, Adams LG. Role of SPI-1 secreted effectors in acute bovine response to Salmonella enterica Serovar Typhimurium: a systems biology analysis approach. PLoS One. 2011;6:e26869. doi: 10.1371/journal.pone.0026869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parikh A, Huang E, Dinh C, Zupan B, Kuspa A, Subramanian D, Shaulsky G. New components of the Dictyostelium PKA pathway revealed by Bayesian analysis of expression data. BMC Bioinformatics. 2010;11:163. doi: 10.1186/1471-2105-11-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sachs K, Perez O, Pe'er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- 33.Woolf PJ, Prudhomme W, Daheron L, Daley GQ, Lauffenburger DA. Bayesian analysis of signaling networks governing embryonic stem cell fate decisions. Bioinformatics. 2005;21:741–753. doi: 10.1093/bioinformatics/bti056. [DOI] [PubMed] [Google Scholar]
- 34.Pe'er D. Bayesian network analysis of signaling networks: a primer. Sci STKE. 2005;2005:pl4. doi: 10.1126/stke.2812005pl4. [DOI] [PubMed] [Google Scholar]
- 35.Voit EO. Computational analysis of biochemical systems : a practical guide for biochemists and molecular biologists. Cambridge University Press; New York: 2000. [Google Scholar]
- 36.Voit EO. Optimization and Systems Biology. Vol. 9. World Publishing Corp.; 2008. Model Identification: A Key Challenge is Computational Systems Biology. pp. 1–12. [Google Scholar]
- 37.Mundel P, Reiser J, Zuniga Mejia Borja A, Pavenstadt H, Davidson GR, Kriz W, Zeller R. Rearrangements of the cytoskeleton and cell contacts induce process formation during differentiation of conditionally immortalized mouse podocyte cell lines. Exp Cell Res. 1997;236:248–258. doi: 10.1006/excr.1997.3739. [DOI] [PubMed] [Google Scholar]
- 38.Velez JC, Bland AM, Arthur JM, Raymond JR, Janech MG. Characterization of renin-angiotensin system enzyme activities in cultured mouse podocytes. Am J Physiol Renal Physiol. 2007;293:F398–407. doi: 10.1152/ajprenal.00050.2007. [DOI] [PubMed] [Google Scholar]
- 39.Velez JC, Ryan KJ, Harbeson CE, Bland AM, Budisavljevic MN, Arthur JM, Fitzgibbon WR, Raymond JR, Janech MG. Angiotensin I is largely converted to angiotensin (1-7) and angiotensin (2-10) by isolated rat glomeruli. Hypertension. 2009;53:790–797. doi: 10.1161/HYPERTENSIONAHA.109.128819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.R: A Language and Environment for Statistical Computing [computer program] R Foundation for Statistical Computing; Viena, Austria: 2009. [Google Scholar]
- 41.Scutari M. Learning Bayesian Networks with the bnlearn R Package. Journal of Statistical Software. 2010;35:1–22. [Google Scholar]
- 42.Grzegorczyk M. An introduction to Gaussian Bayesian networks. Methods Mol Biol. 2010;662:121–147. doi: 10.1007/978-1-60761-800-3_6. [DOI] [PubMed] [Google Scholar]
- 43.Hastings WK. Monte Carlo sampling methods using Markov chains and their applications. Biometrika. 1970;57:97–109. [Google Scholar]
- 44.Ryan JW, Chung AY, Nearing JA, Valido FA, Shun-Cun C, Berryer P. A radiochemical assay for aminopeptidase N. Anal Biochem. 1993;210:27–33. doi: 10.1006/abio.1993.1145. [DOI] [PubMed] [Google Scholar]
- 45.Rice GI, Thomas DA, Grant PJ, Turner AJ, Hooper NM. Evaluation of angiotensin-converting enzyme (ACE), its homologue ACE2 and neprilysin in angiotensin peptide metabolism. Biochem J. 2004;383:45–51. doi: 10.1042/BJ20040634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iturrioz X, Rozenfeld R, Michaud A, Corvol P, Llorens-Cortes C. Study of asparagine 353 in aminopeptidase A: characterization of a novel motif (GXMEN) implicated in exopeptidase specificity of monozinc aminopeptidases. Biochemistry. 2001;40:14440–14448. doi: 10.1021/bi011409j. [DOI] [PubMed] [Google Scholar]
- 47.Chiu AT, Ryan JW, Stewart JM, Dorer FE. Formation of angiotensin III by angiotensin-converting enzyme. Biochem J. 1976;155:189–192. doi: 10.1042/bj1550189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bausback HH, Churchill L, Ward PE. Angiotensin metabolism by cerebral microvascular aminopeptidase A. Biochem Pharmacol. 1988;37:155–160. doi: 10.1016/0006-2952(88)90712-5. [DOI] [PubMed] [Google Scholar]
- 49.Goto Y, Hattori A, Ishii Y, Mizutani S, Tsujimoto M. Enzymatic properties of human aminopeptidase A. Regulation of its enzymatic activity by calcium and angiotensin IV. J Biol Chem. 2006;281:23503–23513. doi: 10.1074/jbc.M603191200. [DOI] [PubMed] [Google Scholar]
- 50.Bauvois B, Dauzonne D. Aminopeptidase-N/CD13 (EC 3.4.11.2) inhibitors: chemistry, biological evaluations, and therapeutic prospects. Med Res Rev. 2006;26:88–130. doi: 10.1002/med.20044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahmad S, Ward PE. Role of aminopeptidase activity in the regulation of the pressor activity of circulating angiotensins. J Pharmacol Exp Ther. 1990;252:643–650. [PubMed] [Google Scholar]
- 52.Yatabe J, Yoneda M, Yatabe MS, Watanabe T, Felder RA, Jose PA, Sanada H. Angiotensin III stimulates aldosterone secretion from adrenal gland partially via angiotensin II type 2 receptor but not angiotensin II type 1 receptor. Endocrinology. 2011;152:1582–1588. doi: 10.1210/en.2010-1070. [DOI] [PubMed] [Google Scholar]
- 53.Plovsing RR, Wamberg C, Sandgaard NC, Simonsen JA, Holstein-Rathlou NH, Hoilund-Carlsen PF, Bie P. Effects of truncated angiotensins in humans after double blockade of the renin system. Am J Physiol Regul Integr Comp Physiol. 2003;285:R981–991. doi: 10.1152/ajpregu.00263.2003. [DOI] [PubMed] [Google Scholar]
- 54.Handa RK. Biphasic actions of angiotensin IV on renal blood flow in the rat. Regul Pept. 2006;136:23–29. doi: 10.1016/j.regpep.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 55.Campbell DJ, Lawrence AC, Towrie A, Kladis A, Valentijn AJ. Differential regulation of angiotensin peptide levels in plasma and kidney of the rat. Hypertension. 1991;18:763–773. doi: 10.1161/01.hyp.18.6.763. [DOI] [PubMed] [Google Scholar]
- 56.Nadarajah R, Milagres R, Dilauro M, Gutsol A, Xiao F, Zimpelmann J, Kennedy C, Wysocki J, Batlle D, Burns KD. Podocyte-specific overexpression of human angiotensin-converting enzyme 2 attenuates diabetic nephropathy in mice. Kidney Int. 2012;82:292–303. doi: 10.1038/ki.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaneshiro Y, Ichihara A, Sakoda M, Takemitsu T, Nabi AH, Uddin MN, Nakagawa T, Nishiyama A, Suzuki F, Inagami T, Itoh H. Slowly progressive, angiotensin II-independent glomerulosclerosis in human (pro)renin receptor-transgenic rats. J Am Soc Nephrol. 2007;18:1789–1795. doi: 10.1681/ASN.2006091062. [DOI] [PubMed] [Google Scholar]
- 58.Singh VP, Le B, Khode R, Baker KM, Kumar R. Intracellular angiotensin II production in diabetic rats is correlated with cardiomyocyte apoptosis, oxidative stress, and cardiac fibrosis. Diabetes. 2008;57:3297–3306. doi: 10.2337/db08-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cristovam PC, Arnoni CP, de Andrade MC, Casarini DE, Pereira LG, Schor N, Boim MA. ACE-dependent and chymase-dependent angiotensin II generation in normal and glucose-stimulated human mesangial cells. Exp Biol Med (Maywood) 2008;233:1035–1043. doi: 10.3181/0708-RM-229. [DOI] [PubMed] [Google Scholar]
- 60.Ramirez M, Prieto I, Martinez JM, Vargas F, Alba F. Renal aminopeptidase activities in animal models of hypertension. Regul Pept. 1997;72:155–159. doi: 10.1016/s0167-0115(97)01054-9. [DOI] [PubMed] [Google Scholar]
- 61.Oliveira V, Ferro ES, Gomes MD, Oshiro ME, Almeida PC, Juliano MA, Juliano L. Characterization of thiol-, aspartyl-, and thiol-metallo-peptidase activities in Madin-Darby canine kidney cells. J Cell Biochem. 2000;76:478–488. [PubMed] [Google Scholar]
- 62.Chappell MC, Gomez MN, Pirro NT, Ferrario CM. Release of angiotensin-(1-7) from the rat hindlimb: influence of angiotensin-converting enzyme inhibition. Hypertension. 2000;35:348–352. doi: 10.1161/01.hyp.35.1.348. [DOI] [PubMed] [Google Scholar]
- 63.Park S, Bivona BJ, Kobori H, Seth DM, Chappell MC, Lazartigues E, Harrison-Bernard LM. Major role for ACE-independent intrarenal ANG II formation in type II diabetes. Am J Physiol Renal Physiol. 2010;298:F37–48. doi: 10.1152/ajprenal.00519.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deddish PA, Marcic B, Jackman HL, Wang HZ, Skidgel RA, Erdos EG. N-domain-specific substrate and C-domain inhibitors of angiotensin-converting enzyme: angiotensin-(1-7) and keto-ACE. Hypertension. 1998;31:912–917. doi: 10.1161/01.hyp.31.4.912. [DOI] [PubMed] [Google Scholar]
- 65.Jankowski V, Vanholder R, van der Giet M, Tolle M, Karadogan S, Gobom J, Furkert J, Oksche A, Krause E, Tran TN, Tepel M, Schuchardt M, Schluter H, Wiedon A, Beyermann M, Bader M, Todiras M, Zidek W, Jankowski J. Mass-spectrometric identification of a novel angiotensin peptide in human plasma. Arterioscler Thromb Vasc Biol. 2007;27:297–302. doi: 10.1161/01.ATV.0000253889.09765.5f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.