SUMMARY
The shelterin protein protects telomeres against activation of the DNA damage checkpoint and recombinational repair. We show here that a dimer of the shelterin subunit TRF2 wraps ~90 bp of DNA through several lysine and arginine residues localized around its homodimerization domain. The expression of a wrapping-deficient TRF2 mutant, named Top-less, alters telomeric DNA topology, decreases the number of terminal loops (t-loops), and triggers the ATM checkpoint, while still protecting telomeres against non-homologous end joining (NHEJ). In Top-less cells, the protection against NHEJ is alleviated if the expression of the TRF2-interacting protein RAP1 is reduced. We conclude that a distinctive topological state of telomeric DNA, controlled by the TRF2-dependent DNA wrapping and linked to t-loop formation, inhibits both ATM activation and NHEJ. The presence of RAP1 at telomeres appears as a backup mechanism to prevent NHEJ when topology-mediated telomere protection is impaired.
In Brief
Benarroch-Popivker et al. show that TRF2 wraps DNA around its TRFH domain, thereby controlling telomeric DNA topology, t-loop formation, and ATM inhibition. In TRF2 wrapping-deficient cells, protection of telomeres against fusion relies on the recruitment of RAP1.
INTRODUCTION
Telomeres have evolved in eukaryotes from the need to protect chromosome ends and provide genome stability. Their maintenance requires protection against the DNA damage response (DDR) that would otherwise stop cell division by checkpoint activation and lead to end-to-end fusion by non-homologous end joining (NHEJ). In humans, telomeres consist of a repetitive DNA ending with a single-stranded 3′ overhang and organized in a peculiar chromatin structure involving the shelterin protein complex and the noncoding RNA TERRA (Giraud-Panis et al., 2013). Their main function is to protect chromosome ends against DNA damage checkpoint and recombinational repair as well as to assist terminal DNA replication and processing (de Lange, 2005; Gilson and Géli, 2007).
TRF2, one of the shelterin subunits, inhibits NHEJ and the ataxia telangiectasia mutated (ATM)-dependent DDR pathway (Celli and de Lange, 2005; Denchi and de Lange, 2007; Okamoto et al., 2013; van Steensel et al., 1998). TRF2 also protects telomeric sequences against replicative DNA damage, particularly those due to topological stress (Muraki et al., 2011; Saint-Léger et al., 2014; Ye et al., 2010). In order to achieve these functions, TRF2 exhibits numerous activities (Feuerhahn et al., 2015). At its N terminus, a basic domain (B domain) interacts with branched DNA structures and protects them against resolution (Fouché et al., 2006; Poulet et al., 2009). The homodimerization domain that forms a horseshoe structure in its dimeric form (TRFH for TRF homology domain) (Chen et al., 2008; Fairall et al., 2001) has been shown to suppress ATM activation (Okamoto et al., 2013) and to control TERRA transcription (Porro et al., 2014a, 2014b). This domain also acts as a binding hub for various repair proteins, such as Apollo, SLX4, or RTEL1 (Chen et al., 2008; Kim et al., 2009; Sarek et al., 2015; Wan et al., 2013; Wilson et al., 2013). The hinge domain harbors sites for other protein interactions such as the shelterin subunits RAP1 and TIN2 and also inhibits ATM signaling (Okamoto et al., 2013). Finally, at the C terminus a Myb/SANT domain (Telobox) is responsible for sequence-specific telomeric DNA binding (Bilaud et al., 1996, 1997; Court et al., 2005). TRF2 is also capable of folding telomeric DNA into a lasso-like structure called the t-loop (Griffith et al., 1999; Stansel et al., 2001). This higher-order telomeric DNA structure is believed to play a key role in telomere protection (Doksani et al., 2013) and has been proposed to be linked to the ability of TRF2 to stimulate invasion of duplex telomeric DNA by a homologous single strand (Amiard et al., 2007; Baker et al., 2009, 2011; Poulet et al., 2012; Verdun and Karlseder, 2006).
In this report, we show that ~90 base pairs (bp) of DNA is wrapped around a TRFH homodimer. This wrapping involves lysines and arginines located on a DNA path, whose mutation compromises TRF2 capacity to induce DNA wrapping in vitro. In human cells, expression of this mutant, named Top-less, causes changes in telomeric DNA topology, a decrease in the amount of t-loops, and defects in telomere protection against DDR. However, chromosome ends are still protected against NHEJ. A reduced expression of RAP1 alleviates this protection. These findings reveal that a distinctive topological state of telomeric DNA, controlled by TRF2-mediated DNA wrapping and linked to t-loop formation, inhibits both ATM activation and NHEJ. The presence of RAP1 at telomeres appears as a backup mechanism to prevent NHEJ when topology-mediated telomere protection is impaired.
RESULTS
TRF2 Condenses ~90 Bp of DNA through the TRFH Domain
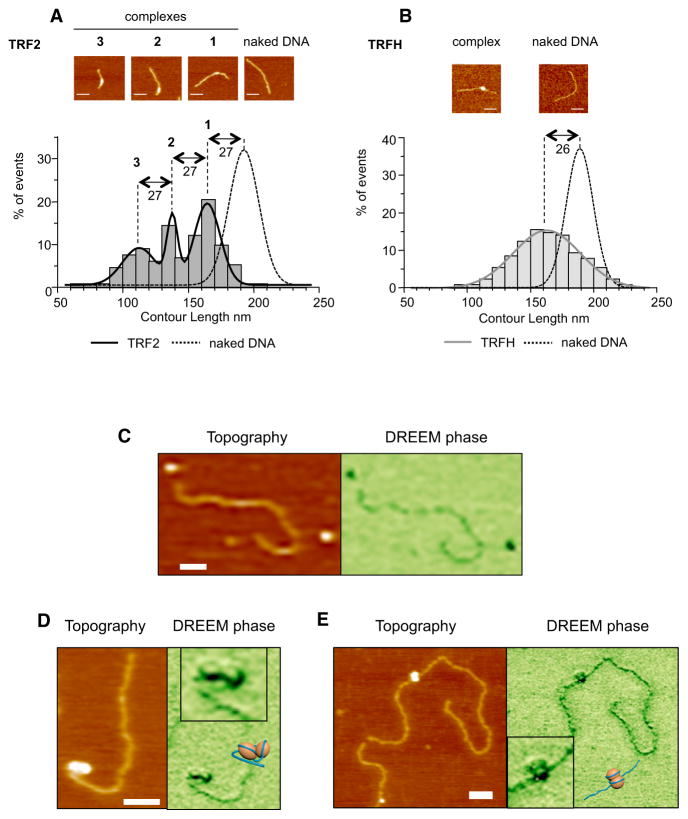
TRF2-mediated DNA condensation can be observed by measuring the length of DNA molecules (DNA contour length, CL) in TRF2-DNA complexes using atomic force microscopy (AFM). As seen in Figure 1A, TRF2 causes a large decrease in CL. Fitting the CL distribution with a multi-Gaussian curve reveals the presence of three types of complexes (CLs of 165 ± 10, 138 ± 4, and 111 ± 13 nm). Notably, these CL values and that of the naked DNA (192 ± 11 nm) all differ by multiples of 27 nm. Deconvoluted volumes of TRF2-DNA complexes (Figure S1A, available online) also showed a three-peaks distribution. Since the sum of the volumes of one TRFH dimer and two Telobox domains corresponds to 66 nm3, the mean deconvoluted volume of complexes in peak 1 (90 ± 34 nm3) is compatible with that of a dimer of the protein (Figure S1A). By inference, the two other types of complexes should correspond to two and three dimers bound to DNA. These analyses revealed that TRF2 dimers can form complexes with DNA, each condensing DNA by ~27 nm (~90 bp).
Figure 1. TRFH Domain of TRF2 Condenses ~90 Bp of DNA.
(A) AFM experiments show a decrease in the contour length (CL) of a 650 bp telomeric DNA fragment due to TRF2 binding. (Top) Representative AFM images; scale bars, 50 nm; (bottom) graph representing CL distribution for free and bound DNA (n = 133 for TRF2, n = 304 for DNA). Histograms correspond to raw data and curves to the sum of a Gaussian multipeak fitting.
(B) Same experiment as in (A) using the TRFH domain (n = 130 for TRFH, n = 154 for DNA).
(C) Topographic AFM (left panel) and DREEM phase (right panel) images of free TRFH protein molecules and DNA.
(D and E) Representative topographic AFM (left panels) and DREEM phase (right panels) images of TRFH-DNA complexes with telomeric sequences (D, 135 TTAGGG repeats) or a nontelomeric fragment (E, 3.8 kb).
The XY scale bars, 50 nm. Boxed regions in (D) and (E) are zoomed DREEM images from main figures. The TRFH-DNA models are as follows: orange spheres for TRFH dimers and dark blue lines for DNA.
Since TRF2 ability to condense DNA depends on the TRFH domain (Amiard et al., 2007; Poulet et al., 2012), we explored whether this domain is sufficient. Purified TRFH binds DNA, albeit with low affinity (Figures S1B and S1C), and leads to a DNA condensation similar to that of full-length TRF2 (Figure 1B). In agreement, the preferred length of DNA bound by this domain is ~92 bp (Figures S1D and S1E). We obtained a multipeak distribution for the deconvoluted volumes compatible with dimers and multimers (Figure S1F). As for the full-length protein, larger TRFH-DNA complexes show smaller contour lengths, and vice versa (Figure S1G). Hence, the Gaussian aspect of the TRFH CL distribution (Figure 1B) is probably a consequence of variations in condensation for the different TRFH complexes, likely due to the weak affinity of TRFH for DNA. Alternatively, other domains such as the N-terminal B domain or the C-terminal Myb/SANT domain of TRF2 may stabilize the wrapped structure and be accessory to this TRFH-driven reaction.
We found a good correspondence between circumference and DNA shortening of TRF2-DNA complexes (Figure S1H). Furthermore, the value of nearly 1 in the slope of the linear fit curve suggests that circumference and DNA shortening increase at the same rate. Thus, dimensions of TRF2-DNA complexes can be described by multiples of ~27 nm that correspond to both the length of condensed DNA and the circumference of the complexes.
This number is similar to the circumference of ~25 nm calculated from the 3D structure of the TRFH domain (PDB 1H6O and 3BUA) (Chen et al., 2008; Fairall et al., 2001). This suggested that the circumference of the TRFH/DNA complexes should be similar to that of the full-length protein, and, indeed, we obtained 26 ± 9 nm for the smallest TRFH/DNA complex and multiples of ~27 nm for multimeric complexes (Figure S1I).
Overall, these results strongly suggest that the TRFH domain is encircled by ~90 bp of DNA. In order to confirm this wrapping, we used a recently developed AFM imaging technique called dual resonance frequency enhanced electrostatic force microscopy (DREEM). In recent studies, DREEM was successfully used to observe DNA wrapping around histone proteins in chromatin, DNA passing through the hMutSα repair protein, and higher-order DNA looping at the edge of multiprotein full-length TRF2-DNA complexes (K.P., D. Wu, J. Lin, P. Countryman, R. Riehn, P.L. Opresdo, and H. Wang, unpublished data; Wu et al., 2016). We chose to analyze TRFH-DNA complexes rather than those formed with the full-length protein since the other domains of TRF2 may impede the visualization of the wrapping around TRFH. In DREEM imaging, both free proteins and DNA show a decrease in phase, but proteins show a greater contrast than DNA, thus allowing distinction of both molecules in a complex (Figure 1C). TRFH-telomeric DNA complexes in DREEM phase images show dark regions consistent with protein, and regions with decreased signal consistent with DNA (Figure 1D). The regions with decreased intensities show DNA paths on the TRFH consistent with the wrapping of DNA around this domain. We could also observe wrapping when using a nontelomeric linear DNA fragment (Figure 1E), showing that the DNA wrapping around TRFH is not telomeric DNA-sequence specific.
TRFH Contacts DNA through a Set of Lysine Residues
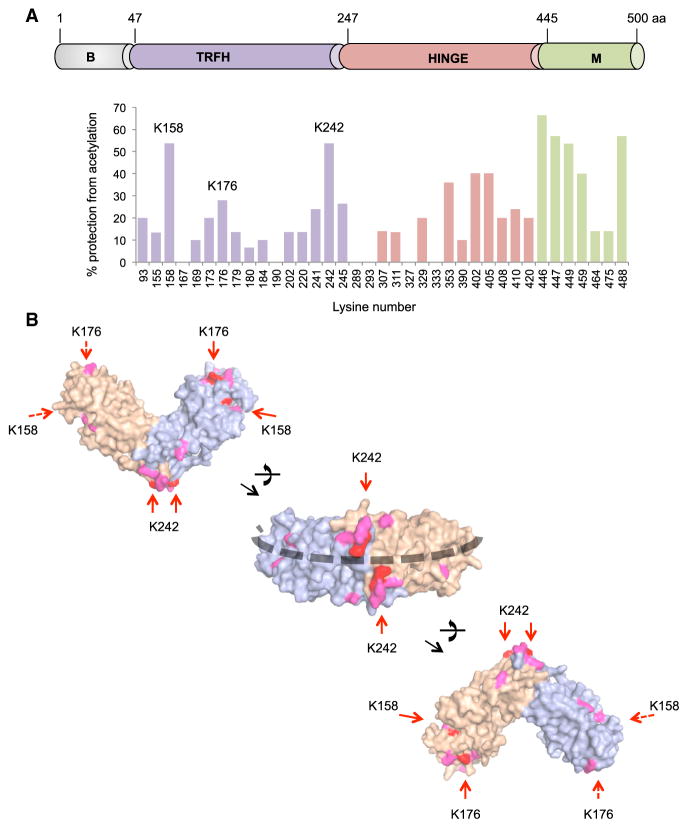
To identify the TRFH residues in contact with DNA, we performed protein footprinting using in vitro acetylation by sulfosuccinimidyl acetate (Figure S2A). This compound specifically acetylates lysines exposed to the solvent, which can be mapped using mass spectrometry (Mendoza and Vachet, 2009). We used lysine acetylation profiles to calculate probabilities of their acetylation (Figure S2B; Experimental Procedures). Physical contact of the protein with another molecule modifies lysine acetylation. Comparing acetylation profiles for unbound and bound TRF2 on a 650 bp of telomeric DNA, we determined the percentage of DNA-dependent protection for each acetylable lysine (Figure 2A). Lysines not present in the unbound protein profile due to lack of acetylation or partial coverage in mass spectrometry were not analyzed (K140, K495, and K180). TRF2 contains 44 lysines distributed along the sequence, with the exception of the N-terminal basic domain. Binding of the DNA causes variations in acetylation to different degrees. Lysines closer to the DNA in the Telobox structure (Court et al., 2005) are more protected from acetylation, validating this approach (Figure S2C). The acetylation of some lysines in the hinge domain is also modulated upon DNA binding, perhaps due to conformational changes in this domain or to DNA binding. Importantly, marked changes in acetylation were observed in three regions of the TRFH centered on K158, K176, and K242. When positioned on the 3D structure, these lysines could be aligned along a DNA path encircling this domain (Figure 2B). Interestingly, K173, K176, and K179 are located in front for one monomer and in the back for the other monomer, thus introducing chirality in the path around the dimer and forcing DNA strands to cross (Figure S2D).
Figure 2. Lysines Involved in DNA Binding Define a ‘‘DNA Path’’ around the TRFH Domain.
(A) (Top) Schematic view of TRF2 domains. (Bottom) Footprinting graph showing the percentage of DNA-dependent protection from acetylation for acetylable lysines (Figure S2).
(B) Positions of protected lysines on the 3D structure of the TRFH domain (PDB: 3BUA). Lysines in red show protection above 20%, and those in pink show protection between 10% and 20%. Lysines on the back of the structures are indicated by dashed arrows. The black dashed line marks the identified DNA path.
TRF2 Wraps DNA around Its TRFH Domain
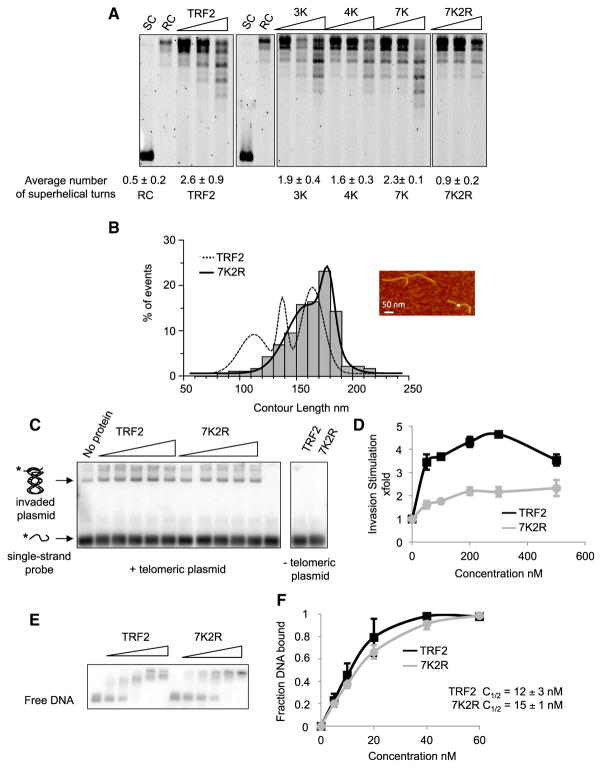
To go further, we constructed a set of TRF2 mutants containing lysine-to-alanine replacement. We focused on the lysines exhibiting highest signals in footprinting (K158, K176, and K242) and their surrounding lysines. Mutants with different numbers of mutated lysines were constructed (Figures S3A and S3B): K241, K242, and K245 in mutant 3K; K158, K173, K176, and K179 in mutant 4K; and all seven of them in mutant 7K. We analyzed the capacity of these mutants to bind and wrap DNA by EMSA and by monitoring their topological activity on a plasmid using the Topoisomerase I relaxation assay (Amiard et al., 2007; Poulet et al., 2012; Figures S3C and 3A; numbers below gels). All mutants were active to different degrees. We concluded that, if these lysines contributed to wrapping, other residues must be involved.
The TRF1 TRFH is also capable of condensing DNA, but in TRF1, this capacity is inhibited by the presence of an acidic N-terminal domain. This suggests that the residues involved in DNA wrapping might be conserved between TRF1 and TRF2. Indeed, lysines giving a strong signal in the footprinting assay are either conserved, replaced by an arginine, or only slightly shifted (Figure S3D). Two conserved arginines are located on the putative DNA path (R69 and R99 for TRF2; R91 and R121 in TRF1), and their symmetrical location strongly resembles that of the conserved lysines K245. We mutated these two arginines to alanines in combination with the seven lysines, giving the 7K2R mutant (Figure S3A). This mutant showed reduced topological activity (Figure 3A) and wrapping efficiency (Figure 3B). Similarly, the capacity of 7K2R to stimulate single-strand invasion into a telomeric double helix was strongly impaired (Figures 3C and 3D). These reduced activities did not originate from changes in affinities for telomeric DNA (Figures 3E and 3F) and were not due to the sole mutations of the two arginines since the 2K2R mutant (mutations of K158, K242, and the two arginines) was topologically active (Figure S3E). Overall, we conclude that a set of lysine and arginine residues located on the outer surface of the TRFH domain is required to wrap DNA around it and to confer the topological properties of TRF2. Thus, the 7K2R mutant was dubbed Top-less.
Figure 3. Biochemical Characterization of a Topology-Deficient TRF2 Mutant.
(A) Topoisomerase I assay showing the topological activity of TRF2 and of lysine/arginine to alanine mutants. Protein concentrations used were 100, 250, and 500 nM. Average number of helical turns was calculated at 500 nM for at least 3 experiments. SC stands for supercoiled, and RC stands for relaxed circular.
(B) AFM experiments showing the decreased wrapping activity of 7K2R. The graph represents CL distribution for the TRF2- and 7K2R-bound DNA (n = 133 for TRF2, n = 190 for 7K2R). Histograms correspond to raw data and curves to the sum of Gaussian curves fitting the raw data.
(C) Invasion assay showing the decrease in invasion caused by 7K2R mutations. Concentrations used were 50, 100, 200, 300, and 500 nM for both proteins.
(D) Quantitative analysis of (C). Error bars correspond to standard deviation from three experiments.
(E) EMSA using ds106Telo and either TRF2 or 7K2R. Concentrations used were 5, 10, 20, 40, and 60 nM of proteins.
(F) Quantitative analysis of (E). Error bars represent SD from three experiments.
In order to characterize Top-less, we compared its biochemical properties to those of the wild-type protein (Figures S3F–S3K). Circular dichroism experiments showed that mutations in Topless did not modify the overall folding of the protein (Figure S3G). We also showed that Top-less could bind RAP1 in vitro (Figure S3H). As expected, Top-less mutations caused a marked decrease in the affinity of the TRFH for DNA (Figures S3I and S3J). The capacity of TRF2 to promote formation of Holliday junctions and to inhibit their migration, a property a priori unrelated to DNA topology, was unaffected (Figure S3K). We also explored whether Top-less could bind telomeric DNA in vivo. For this purpose, we used a HeLa cell line where TRF2 expression could be severely decreased by expression of a doxycycline (DOX)-inducible shRNA directed against TERF2 (Grolimund et al., 2013). Cells treated with DOX were transduced with lentiviral vectors expressing either wild-type or Top-less Myc-tagged forms of TRF2 (resistant to the inducible shRNA). Ectopic expression of both wild-type TRF2 and Top-less restored a level of protein that exceeded the endogenous amount observed in cells not treated with DOX (Figure S4A). Binding to telomeres was examined using chromatin immuno-precipitation (ChIP) using either an anti-TRF2 or an anti-Myc antibody (Figures S4B and S4C, respectively). No obvious difference was observed between wild-type and Top-less. Finally, we checked that Top-less modified neither the expression of the other shelterin subunits nor the association of RAP1 and TIN2 at telomeres (Figures S4D–S4G).
Overall, these data show that Top-less is a valuable separation-of-function mutant of TRF2 and is deficient for DNA wrapping activity, but it still exhibits several of the known properties of this protein.
TRF2 Controls Telomeric DNA Topology in Human Cells
Next, we investigated whether DNA wrapping plays a role in the control of telomere DNA topology in human cells. To monitor changes in the DNA topological state, we used the capacity of Trioxsalen (4,5′,8-trimethylpsoralen) to bind preferentially to unwound genomic regions and to crosslink DNA strands when exposed to UV. To validate this approach, we performed experiments on cells treated with ICRF-193, a catalytic inhibitor of Topoisomerases 2 (Chen et al., 2015; d’Alcontres et al., 2014; Hsieh et al., 2015; Ye et al., 2010). HeLa cells were incubated with Trioxsalen for 5 min and immediately exposed to UV before recovery of the cells. Hence, the binding profile of Trioxsalen provides a snapshot of the topological state of DNA. As controls, cells were treated with Trioxsalen but not exposed to UV, or vice versa. Trioxsalen DNA crosslinking was quantified on sonicated genomic DNA after denaturation of DNA fragments by glyoxal and separation of crosslinked species (double stranded) and noncrosslinked species (single stranded) by electrophoresis (Kouzine et al., 2013). We verified that fragments were of equivalent length (between 210 and 230 bp) using a Bioanalyzer (an example is given in Figure S5A). After migration, gels were stained with SYBR green II following a denaturing step to remove Trioxsalen. The SYBR green II image obtained thus reflected genome-wide binding of Trioxsalen. To quantify the crosslinked (double stranded) material, we used a 0.6 kb threshold because it corresponded to an inflection point in the telomeric DNA profiles (Figure S5B). We analyzed telomeric DNA by hybridization of the membrane obtained by Southern blot of the SYBR gel with a telomeric probe (Figure S5D). Under our conditions, ~20% of genomic DNA was crosslinked (~1 Trioxsalen every kilobase). Interestingly, ICRF-193 treatment causes a detectable increase in Trioxsalen crosslinking of telomeric DNA but not of bulk DNA, indicative of a telomere-specific effect on DNA topology (Figure S5E). It may appear counterintuitive to observe an increase in Trioxsalen binding when inhibiting an enzyme that removes DNA-positive supercoils, but this could be due to topology-driven regression of replication forks (Yeeles et al., 2013) or replication/transcription forks stalling, resulting in the accumulation of unwound regions.
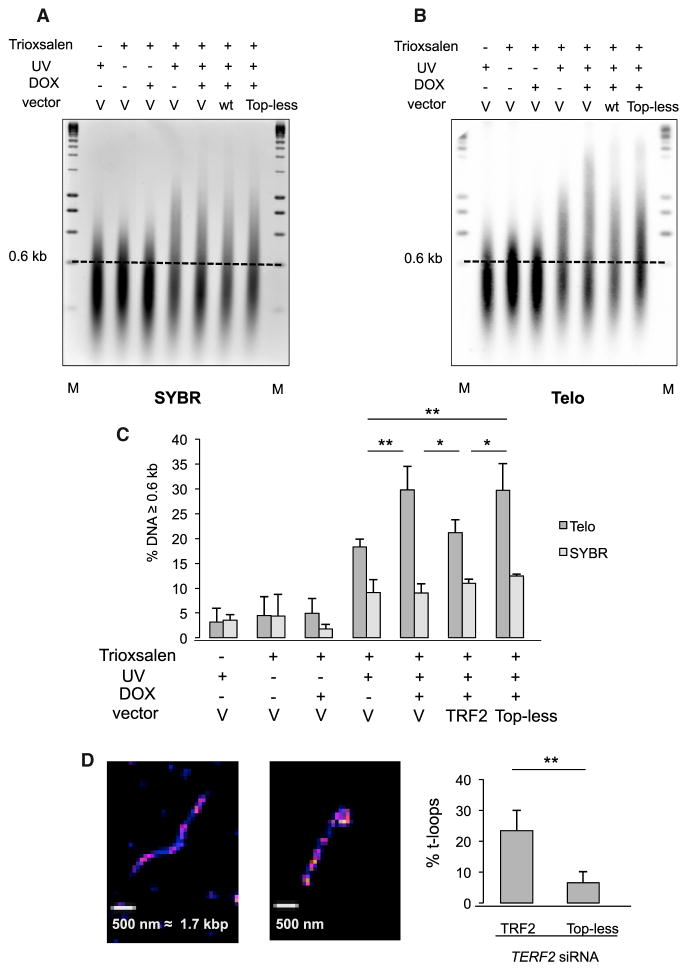
Next, HeLa cells were treated with DOX and transduced with either the empty, TRF2, or Top-less lentiviral vectors as above. The binding of Trioxsalen to global genomic DNA does not depend on TRF2 (Figures 4A and 4C), as expected. However, a nearly 2-fold increase in crosslinked telomeric species is observed when treating HeLa cells with DOX. This topological change is rescued by the expression of wild-type TRF2. In contrast, the expression of Top-less fails to rescue topological changes triggered by TRF2 downregulation (Figures 4B and 4C).
Figure 4. The TRFH-Wrapping Domain of TRF2 Controls Telomeric DNA Topology and t-Loops.
(A) SYBRII-stained glyoxal gel. M stands for molecular weight markers, V stands for empty vector, WT stands for wild type TRF2, and the dotted line marks the 0.6 kb threshold used for analysis. Of note, a nonrelevant lane was removed from the image, and glyoxal in the samples slows migration compared to the markers.
(B) Southern blot of the glyoxal gel hybridized using a telomeric probe (Telo). As above, a nonrelevant lane was removed from the image.
(C) Quantitative analysis of (B). The relative amount of DNA material above the 0.6 kb mark was measured for each condition. SYBR indicates the values obtained for the SYBRII stained gels, and Telo for the Southern blots. Error bars correspond to standard errors between three replicates. p values were calculated using the Mann-Whitney test (**p < 0.01, *p < 0.05; absence of mark indicates no significance).
(D) Representative images of linear (left) DNA and t-loop (right) obtained on spread chromatin of HT1080 super Telomerase cells by STORM and quantification of the percentage of t-loops in TRF2-(437 objects counted) or Top-less (634 objects counted)-expressing cells. Quantification of TERF2 transcripts was performed by RT-qPCR and corresponded to a 77% knockdown of the endogenous TERF2 transcript, while in TRF2 and Top-less conditions the ectopic mRNA was 9.5-fold and 6.5-fold more expressed, respectively, than in the endogenous TERF2 mRNA in the si-Control condition. Data represent the means ± SE. p values were calculated using the Mann-Whitney test (**p < 0.01).
It is unlikely that the effect of TRF2 knockdown on telomere DNA topology is related to a decrease in nucleosome occupancy, since we rather observe more H3 binding in this condition than when TRF2 is ectopically expressed (Figure S4C), in agreement with previous reports (Benetti et al., 2008; Galati et al., 2012), showing that Top-less is not impaired in at least some of the chromatin-remodeling properties of TRF2.
The topological change due to TRF2 dysfunction could be due to the increase in telomere transcription that was previously observed upon TRF2 depletion (Porro et al., 2014a, 2014b). However, Top-less fully rescues the increased TERRA expression observed in TRF2-compromised cells (Figures S5F and S5G).
These results demonstrate a functional link between the intrinsic ability of TRF2 to wrap DNA and the in vivo control of telomere DNA topology.
TRF2-Mediated DNA Wrapping Controls t-Loops
Two facts suggested that Top-less could lead to variations in the t-loop content in cells: (1) the reduced capacity of this mutant to stimulate single-strand invasion in vitro (Figure 3C), a property thought to be involved in t-loop formation; (2) the telomere topological change caused by this mutant that could be linked to a loss of constraining structures such as t-loops. In order to investigate this, we performed direct stochastic optical reconstruction microscopy (STORM) imaging as described by Doksani et al. (2013). In order to increase our chances to observe t-loops, we used HT1080 cells overexpressing telomerase which can harbor telomeres of more than 20 kb (Cristofari and Lingner, 2006). Endogenous TRF2 expression was reduced by transfection of a siRNA directed against TRF2, and wild-type TRF2 or Top-less was ectopically expressed. As seen in Figure 4D, the amount of t-loops is markedly decreased in Top-less cells as compared to wild-type TRF2 cells.
TRF2-Mediated DNA Wrapping Inhibits ATM Signaling
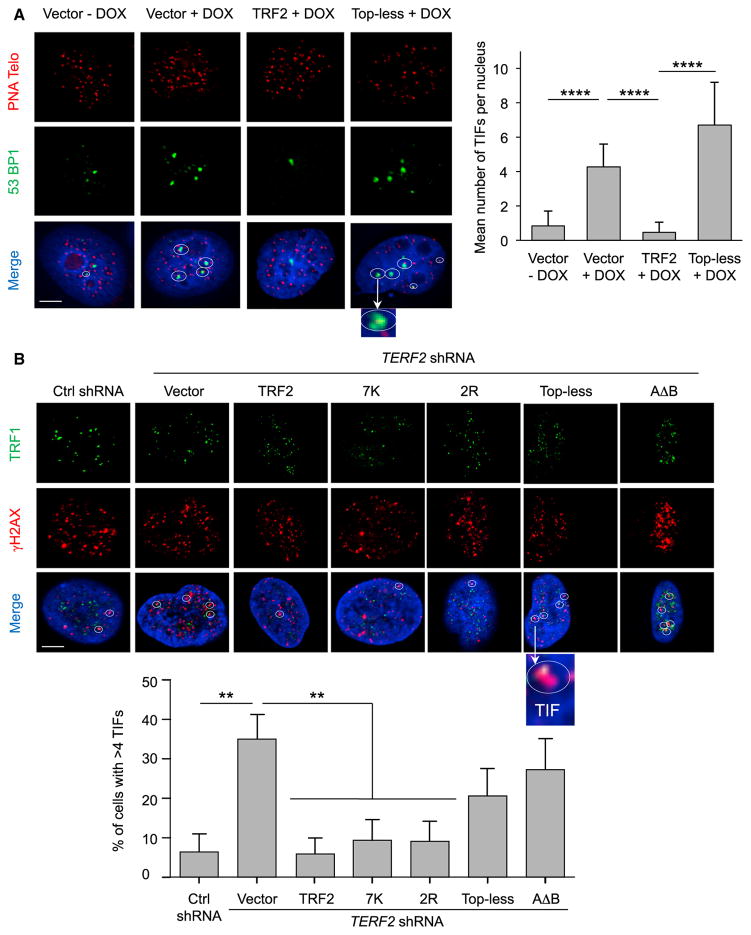
Next, we investigated DDR activation in the HeLa cell-line system used for Trioxsalen experiments (DOX-inducible expression of shTERF2, lentiviral expression of TRF2, or Top-less). We scored telomere dysfunction-induced foci (TIFs) observed through the recruitment of 53BP1 on telomeres. As expected, knockdown of TRF2 significantly increased TIFs (Takai et al., 2003; Figure 5A). This telomere deprotection is rescued by exogenous expression of TRF2, but not of Top-less. Monitoring phosphorylated ATM (pATM) gave similar results, showing that Top-less is impaired in ATM inhibition (Figure S6A). In agreement, the CHK2 phosphorylation triggered by TRF2 downregulation is not fully rescued by Top-less expression (Figure S6B). Of note, in the time frame of our experiment, we could not detect modifications of the cell cycle (Figure S6C) ruling out an indirect effect of Top-less on cell proliferation. DDR activation was also observed in other Top-less-expressing cells (HT1080 supertelomerase cells used for t-loops measurements, BJ-HELT, cells and HT1080 cells; Figures S6D, S6E, and S6F, respectively). We also observed an increased level of TIFs in cells expressing AΔB, a TRF2 mutant also compromised for DNA wrapping but through addition of the TRF1 acidic domain and not through TRFH mutations (as in Top-less) (Poulet et al., 2012).
Figure 5. Top-less Does Not Protect against DDR Activation.
(A) (Left) Representative section images of detection of 53BP1 by IF (green), telomeric DNA (red), and the merge with DAPI (blue) under the indicated conditions. TIFs are marked with a circle. Scale bar, 5 μm.
(Right) TIFs per nucleus were quantified. Data represent the means ± SE. p values were calculated using the Mann-Whitney test (****p < 0.0001).
(B) (Top) Representative section images of detection of TRF1 by IF (green), γ-H2AX by IF (red), and the merge with DAPI (blue) under the indicated conditions using HT1080 cells. TIFs are marked circles. Scale bar, 5 μm.
(Bottom) The percentage of cells showing more than four TIFs was quantified. Data represent the means ± SE. p values were calculated using the Mann-Whitney test (**p < 0.01; absence of mark indicates no significance). The quantification of TERF2 transcript level for the different conditions (control scramble shRNA with expression of empty vector, TERF2 shRNA with expression of either empty vector or TRF2, 7K, 2R, Top-less, or AΔB) was done by RT-qPCR and is, respectively, 1, 0.65, 20, 42, 76, 41.
We also analyzed this response in HT1080 cells by monitoring the colocalization of TRF1 and phosphorylated histone H2AX (γH2AX). Again, we obtained a similar response for the Topless mutant (Figure 5B). Of note, the expression of the 7K and 2R mutants in this setting rescued the telomere uncapping triggered by TRF2 inhibition. We concluded that the strong DDR activation at telomeres triggered by Top-less stems from the combination of both the 7K and 2R mutation sets.
We also explored whether Top-less could alter telomere length and cause formation of t-circles by 2D gel analysis. We did not observe overt production of t-circles and found no difference in mean telomere length upon TRF2 or Top-less expression (Figures S6G and S6H), suggesting that the decrease in t-loop number that we observed does not originate from t-loop excision. Finally, we measured the amount of the 3′ overhang using an in-gel assay. As expected, TRF2 knockdown decreases the amount of 3′ overhang, an effect rescued by both TRF2 and Top-less expression (Figure S6I), indicating that the decrease in t-loop formation is not caused by a decreased length of the 3′ overhang.
In summary, the DNA-wrapping activity of TRF2 is required for telomere protection against ATM activation but is involved neither in telomere length regulation nor in 3′ overhang formation.
TRF2-Mediated DNA Wrapping Inhibits NHEJ in RAP1-Compromised Cells
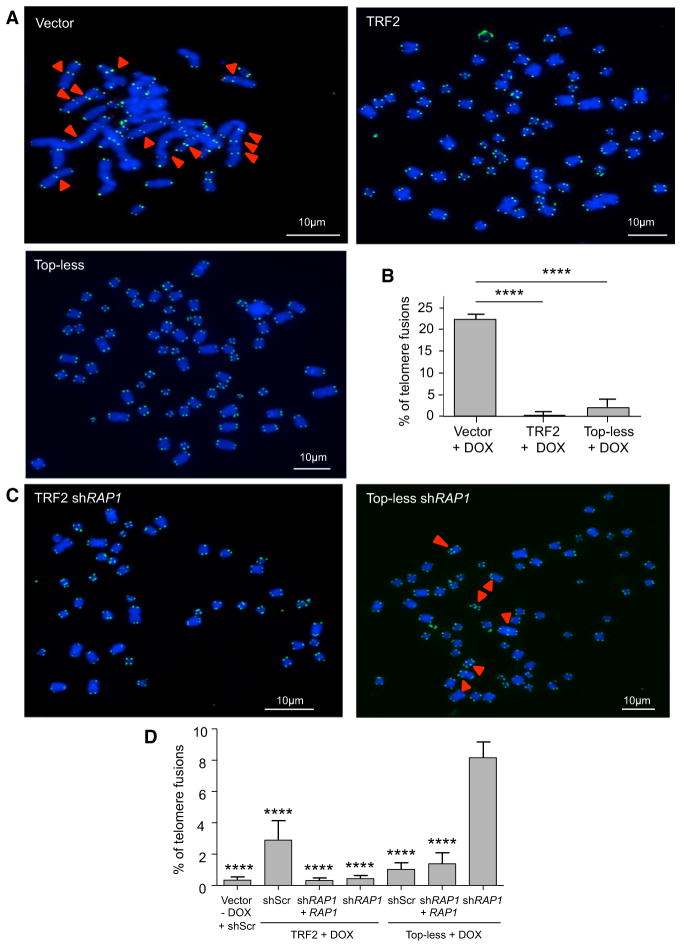
Then, we tested the ability of Top-less to prevent NHEJ by scoring telomere fusions in metaphase chromosomes. Upon TRF2 knockdown in HeLa cells, more than 20% of telomeres were fused (Figures 6A and 6B). This effect was rescued by both TRF2 and Top-less expression. Since RAP1 was previously shown to inhibit NHEJ independently of TRF2 (Bae and Baumann, 2007; Sarthy et al., 2009), we analyzed the effect of Top-less in RAP1-compromised cells. In agreement with previous reports showing that RAP1 is dispensable for NHEJ protection in mammalian cells (Kabir et al., 2014), reducing its expression did not increase fusions in wild-type TRF2-expressing cells (Figures 6C and 6D). However, a 10-fold increase in the percentage of chromosome fusions was observed in Topless cells upon RAP1 inhibition. This effect was rescued by an ectopic expression of RAP1, excluding an off-target effect of the RAP1 shRNA. These results indicate that TRF2-mediated DNA wrapping is involved in NHEJ inhibition independently of RAP1. Moreover, they reveal the anti-NHEJ activity of RAP1 as a backup mechanism for telomere protection in Top-less cells.
Figure 6. TRF2-Mediated DNA Wrapping Inhibits NHEJ in RAP1-Compromised Cells.
(A) Metaphase chromosome spreads of HeLa cells transduced with either empty vector, TRF2, or Top-less viruses upon TRF2 knockdown using doxycycline (DOX). Chromosomes were stained for telomeric DNA (green) and with DAPI (blue). The red arrows show examples of telomere fusions.
(B) Graph showing the percentage of fusions counted on 2,000 chromosomes. Data represent the means ± SE, and p values were calculated using Student’s t test (****p < 0.0001; absence of a mark indicates no significance).
(C) Metaphase chromosome spreads of HeLa cells transduced with TRF2 or Top-less viruses upon TRF2 knockdown using doxycycline (DOX) and knockdown of RAP1 by shRNA.
(D) Graph showing the percentage of fusions counted on 2,000 chromosomes for each condition. p value was calculated using a one-way ANOVA (****p < 0.0001). Downregulation of RAP1 was quantified by RT-qPCR and corresponded to a knockdown of 83%.
DISCUSSION
Although control of DNA topology is crucial for chromosomal integrity (Vos et al., 2011), our understanding of its role at telomeres is limited. Theoretically, the free DNA ends of telomeres should allow dissipation of torsional strain. The fact that we (Biroccio et al., 2011; Chen et al., 2015; Leonetti et al., 2008; Temime-Smaali et al., 2008; Ye et al., 2010) and others (d’Alcontres et al., 2014; Germe et al., 2009; Hsieh et al., 2015) have found that telomere integrity is particularly sensitive to topological stress suggests that telomeres may form topologically constrained chromatin entities. In agreement with this idea, telomeres harbor t-loop structures that may constitute topological barriers. In this report, we unveil that telomeres are topological objects that rely on a particular DNA-wrapping activity of TRF2 to be protected against ATM activation and NHEJ.
By combining AFM, DREEM, protein footprinting, and topology assays, we demonstrate that 90 bp of DNA wrap around the TRFH domain of TRF2 through an interaction with a set of lysines and arginines located on the surface of this domain. Interestingly, the localization of these residues on the TRFH domain imposes a chirality in the DNA-TRF2 complex (Figure S2D).
The identification of TRFH residues contacting DNA allowed us to design a mutant largely deficient in wrapping activity and therefore named Top-less. Top-less behaves as a valuable separation-of-function mutant to study the role of DNA topology at telomeres since, on one hand, it alters the topological state of telomeric DNA in vitro and in vivo, while on another hand, it conserves many TRF2 properties, including (1) proper folding according to CD analysis, (2) specific binding to telomeric DNA both in vitro and in vivo, (3) TIN2 and RAP1 recruitment at telomeres, (4) facilitation of Holliday junction formation and inhibition of their migration, and (5) unaltered expression of the other shelterin subunits.
Top-less causes a marked ATM activation at telomeres showing a loss of function for ATM inhibition. Of note, the parental mutants 7K and 2K, which bear separately the seven mutated lysines (7K) or the two arginines (2R) mutated in Topless, fully protect against ATM activation. Moreover, the wrapping-deficient AΔB mutant, bearing a wild-type TRFH domain, behaves similarly to Top-less in vivo. Overall, the behavior of these mutants indicates that Top-less-mediated telomere de-protection is not due to alterations in unidentified TRFH binding sites for cellular factors. Of note, Top-less cells not only recruit phosphorylated ATM and γ-H2AX at telomeres but also recruit 53BP1. Together with an increased amount of phosphorylated CHK2, these results show that Top-less telomeres are impaired in the inhibition of both the initiation and the propagation of ATM signaling. This might appear at odds with the preservation in Top-less of a small region of the hinge domain (iDDR domain, aa 407–431), which has been shown to inhibit the recruitment of 53BP1 (Okamoto et al., 2013). One explanation to reconcile these results could be that the iDDR domain fuction is somehow altered by the Top-less mutations. In agreement, the iDDR domain lies in a region where the lysine acetylation profile changes upon DNA binding (Figure 2A).
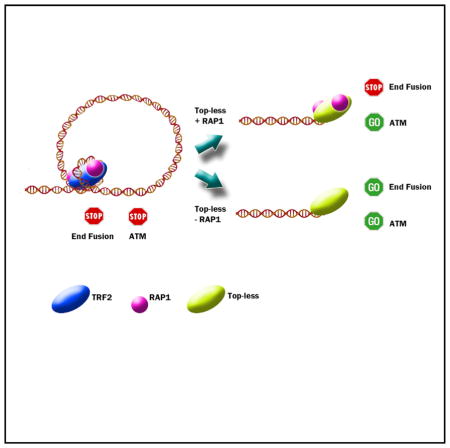
An important result of this study is that Top-less cells exhibit a decreased number of t-loops, indicating that TRF2-wrapping activity is required for t-loop folding. This is in agreement with the fact that Top-less is unable to facilitate strand invasion, a key mechanism in t-loop formation (Griffith et al., 1999). As an explanation, DNA wrapping around the TRFH domain could be involved in strand invasion and t-loop folding through the unwinding of DNA outside TRF2 binding sites as we suggested earlier (Amiard et al., 2007). The efficient protection against telomere fusion in Top-less cells seems contradictory to the previously proposed protective role of t-loops against NHEJ (Doksani et al., 2013). Since mammalian RAP1 was shown to protect against NHEJ in a TRF2-independent manner (Bae and Baumann, 2007; Sarthy et al., 2009) and Top-less can still recruit RAP1 at telomeres, RAP1 could provide a backup anti-NHEJ mechanism in Topless cells (Figure 6E). Indeed, a reduced expression of RAP1 triggers a marked increase in telomere fusions in Top-less. These results show that TRF2 can protect against NHEJ through different mechanisms, including the recruitment at telomeres of RAP1 and its capacity to wrap DNA around its TRFH domain.
Our results show that one of the mechanisms by which telomeres control their DNA topology and protect against ATM activation and NHEJ stems from the right-handed wrapping of telomeric DNA around the TRFH domain of TRF2. Three independent findings support this conclusion: (1) TRF2 wraps DNA in a right-handed manner, (2) TRF2 controls telomere DNA topology in human cells, and (3) the expression of TRF2 mutants specifically impaired in this wrapping activity fails to control telomere DNA topology and uncaps telomeres. Several nonexclusive mechanisms can be envisaged to link the topological properties of TRF2 to ATM signaling and NHEJ. One is suggested by the decreased amount of t-loops in Top-less cells. This is in agreement with the view that t-loops prevent ATM activation and constitute a poor substrate for NHEJ. Another, nonexclusive possibility is that TRF2 acts as a torsional strain sensor to orchestrate various activities required to resolve topological problems that may arise during DNA processing (replication, transcription, and repair).
In RAP1-proficient cells, Top-less uncouples ATM inhibition from the anti-NHEJ activity of TRF2. Interestingly, this partially uncapped telomere phenotype of Top-less cells is reminiscent of the phenotype of cells either exhibiting spontaneous DDR activation at telomeres (Cesare et al., 2009; Kaul et al., 2012; Thanasoula et al., 2010), either with a reduced expression of TRF2 (Cesare et al., 2013), either upon prolonged mitotic arrest (Hayashi et al., 2012) or upon deletion of the TIN2 gene (Takai et al., 2011). This phenotype is described as an ‘‘intermediate state’’ of telomere protection and was proposed to occur when telomeres of primary human cells become too short to efficiently protect against DDR activation and to lead to cell senescence (Cesare and Karlseder, 2012). A topology switch at telomeres may thus constitute a common mechanism leading to the appearance of such intermediate state telomeres. In this hypothesis, our results predict that RAP1 may be critical to protect telomeres of senescent cells from NHEJ.
This study reveals that telomeres directly use positively superhelical strain to escape from inappropriate activation of DDR. Such a functional link between telomere DNA topology and DDR control is reminiscent of the transcription of nuclear pore-associated genes in yeast (Bermejo et al., 2011). The involvement of mechanisms that control DNA topology in telomeric functions appears conserved during evolution since bacteria and yeast telomeres also rely on topoisomerase to maintain their integrity (Bankhead et al., 2006; Bao and Cohen, 2004; Chaconas and Kobryn, 2010; Germe et al., 2009; Mirabella and Gartenberg, 1997; Tsai et al., 2011). Thus, we propose that the folding of telomeres into topologically constrained superstructures is a universal feature of telomeres that may have been used as a mechanism for end protection during chromosome evolution.
EXPERIMENTAL PROCEDURES
Only specific techniques used in this study are presented in this section. Published protocols have been used for several experiments and are detailed in the Supplemental Information.
Proteins
All proteins were obtained using the plasmid pTrcHisB (Invitrogen), bearing an N-terminal His-tag fusion, and were produced from DH5α bacteria, as described (Poulet et al., 2012). The TRF2 protein used corresponds to a 500 aa peptide.
Cell Lines and Reagents
HT1080 cells were grown in DMEM supplemented with 10% fetal calf serum, penicillin (100 IU/ml), and streptomycin (100 μg/ml) at 37°C. shTERF2-inducible HeLa cells were a gift from Joachim Lingner and were used as described previously (Grolimund et al., 2013).
The sequence of TERF2 shRNA used in HT1080 cells was 5′-CCGGCAT TGGAATGATGACTCTGAACTCGAGTTCAGAGTCATCATTCCAATGTTTTT-3′. Lentivirus production was performed by transient cotransfection of 293T cells with the specified lentiviral-expression vector and two packaging plasmids, p8.91 and pVSVg, by calcium-phosphate precipitation. Viral supernatants were collected 24 hr after transfection. The transduction efficiency was determined for the pWPIR-GFP vectors (pWPIR-GFP, pWPIR-GFP-TRF2, pWPIR-GFP-7K, pWPIR-GFP-2R, pWPIR-GFP-Top-less, and pWPIR-GFP-AΔB) by flow-cytometry analysis of GFP-positive cells 3 days after infection and for the pLKO-shRNA plasmids (pLKO-shScramble and pLKO-shTERF2) by counting the number of clones after 1 week of selection with puromycin (1 μg/ml).
DREEM Imaging
Topographic signals are collected through mechanically driving cantilevers near its resonance frequency. Simultaneously, electrostatic signals are collected through applying AC and DC biases to a highly doped silicon cantilever with the frequency of the AC bias centered on cantilever’s first overtone. Importantly, there are no significant cross-talks between topographic and DREEM channels. The DNA substrates were a mixture of DNA (T135 DNA) fragments from digestion of the pSXneo 135 (T2AG3) plasmid DNA (a gift from Dr. Peter Lansdorp at the University of British Columbia) using XbaI and BglII restriction enzymes (NEB). The two fragments resulted from digestion and have distinct DNA contour lengths, which enable us to differentiate telomeric (263 nm) and plasmidic (1,150 nm) DNA fragments. The TRFH domain was diluted to a final concentration of 445 nM in 5 mM HEPES, 150 mM KCl (pH 7.5) and incubated with the T135 DNA fragments (2 nM) for 20 min at room temperature. The incubated samples were diluted 20-fold in 5 mM HEPES, 150 mM KCl, 10 mM Mg(OAc)2 (pH 7.5) and deposited onto freshly cleaved mica surface (SPI Supply). DREEM images were collected using a MFP-3D-Bio AFM (Asylum Research) and highly doped Pointprobe PPP-FMR probes (Nanosensors; results for force constant were as follows: ~2.8 N/m; results for resonant frequency were as follows: f1 = ~80 kHz; and results for first overtone were as follows: f2 ~500 kHz). Detailed description of DREEM imaging technique is described in two studies (K.P., D. Wu, J. Lin, P. Countryman, R. Riehn, P.L. Opresdo, and H. Wang, unpublished data; Wu et al., 2016). Briefly, AFM cantilevers were scraped with tweezers to remove the oxidized layer, and the top surface was coated with a thin layer of colloidal liquid silver (Ted Pella Inc.). A function generator (Sanford Research System, model DS335) and lock-in amplifier (Sanford Research System, model SR844 RF) were used to generate the AC and DC biases and monitor changes in vibration amplitude and phase signals near the first overtone frequency as a function of sample positions. While the AC and DC biases are applied to AFM tips, the mica substrate is grounded. To optimize DREEM signals, AC and DC biases were adjusted from 0 to 20 V and −1.5 to 1.5 V, respectively.
Protein Footprinting
In total, 8 pmol of TRF2 protein were incubated for 20 min at 25°C with or without 16 pmol of a linearized DNA plasmid containing 650 bp of telomeric sequences in 10 mM Tris-HCl (pH 8), 150 mM NaCl, 0.5 mM DTT, and 5% glycerol. Acetylation of lysines was performed by adding 0.5 mM of sulfosuc-cinimidyl acetate (Thermo scientific) for 30 min at 30°C. The reaction was stopped by adding 1% trifluoroacetic acid (Sigma). The samples were resuspended in Laemmli loading buffer and boiled for 5 min. Proteins were resolved by SDS-PAGE and submitted to trypsin proteolysis, and profiles of lysine acetylation were analyzed using mass spectrometry. We determined the probability of lysine acetylation and the probability of disappearance of lysine acetylation upon DNA interaction. The percentage of protection from acetylation presented in Figure 2 was calculated as follows: probability of disappearance of lysine acetylation upon DNA interaction × probability of lysine acetylation of the TRF2 protein. Data shown are the results of five independent experiments.
Trioxsalen Experiments
In total, two million HeLa cells were treated with or without doxycycline (1 μg/ml for 5 days) and ICRF-193 (3 μg/ml final concentration for 24 hr) and transduced by the Empty, TRF2, or Top-less expressing vectors. Treatment was performed in a 10 cm Petri dish in PBS with 280 μl of a saturated 0.9 mg/ml solution of 4,5′,8-trimethylpsoralen (Trioxsalen) for 4 min at 37°C in aluminum foil. Crosslinking was performed on a BioSun (Vilber Lourmat) at 350 nm at 0.36 J/cm2. Then, trioxsalen was removed and cells were washed, trypsinized, and pelleted. After classical extraction, DNA was resuspended in 75 μl of TE and sonicated using a Bioruptor (Diagenode) until fragments were around 200 bp in length. This length was checked using a Bioanalyzer (Agilent). A total of 8 μg of DNA was dried using a speed vac, resuspended in 10 μl of Glyoxal buffer (1 M Glyoxal, 50% DMSO), and incubated at 55°C for 90 min. Orange dye loading buffer was added, and samples were loaded on a 3% agarose 10 mM Na phosphate buffer (pH 7) gel. Migration was performed for 14 hr in 10 mM Na phosphate buffer (pH 7) at 2.5 V/cm. After migration, the gel was incubated for 3 hr at 65°C in 0.5 N NaOH and 1.5 M NaCl. After several washes in water, the gel was incubated 3 times for 20 min in 1× TBE, and 40 μl of SYBR Green II (life Technologies) was added to 200 ml of 1× TBE for staining. After rinsing with water, the gel was scanned using a Typhoon FLA 9500 (GE Healthcare). DNA in the gel was then transferred to a N+ Hybond membrane (Southern blotting), telomeric DNA was revealed using a telomeric radiolabeled probe, and the membrane was analyzed as for EMSA gels.
Supplementary Material
Highlights.
TRF2 modifies DNA topology by wrapping 90 base pairs of DNA around its TRFH domain
A mutant deficient in DNA wrapping, Top-less, causes relaxation of telomeric DNA
Top-less telomeres are deprotected and harbor fewer t-loops but are not fused by NHEJ
RAP1 protects Top-less telomeres against fusions
Acknowledgments
We greatly thank Joachim Lingner for the HeLa cell line used in this study. This work was performed using the genomic, PICMI (supported by ARC and the Conseil General 06 de la Région Provence Alpes-Côte d’Azur), and CYTOMED (supported by FEDER, le Minitère de l’Enseignement Supérieur, la région Provence Alpes-Côte d’Azur, and l’INSERM) facilities of IR-CAN; we are very grateful for the help they provided. We are also indebted to Isabelle Zanella-Cléon for mass spectrometry analysis (Centre Commun de Micro-analyse des Protéines, IBCP Lyon). We acknowledge the PICT-IBiSA platform of the Institut Curie, member of the French National Research Infrastructure France-BioImaging (ANR-10-INSB-04). We are grateful to Matteo de Chiara and Silvia Bottini for the script of 2D probability density map on R. The AFM facility of PICMI was supported by the ARC and Conseil General 06 de la Région Provence Alpes-Côte d’Azur. This work was supported by grants from the Fondation ARC, Ligue Contre le Cancer (EG Equipe labellisé), ANR (‘‘Teloloop’’ ANR-1582-30020690), Institut Nationale du Cancer (INCa) (TELOCHROM), ‘‘Investments for the Future’’ LABEX SIGNALIFE (reference ANR-11-LABX-0028-01), and an NIH grant for DREEM experiments (NIH R01GM107559).
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and six figures and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2015.12.009.
AUTHOR CONTRIBUTIONS
D.B.-P. performed cell biology and microscopy experiments with the help of E.J., K.F., S.B., N.D., C.M.L., S.P., A.M.-B., and J.Y.; S.P. performed AFM and STORM with the assistance of V.F., A.L.-V., and S.B.; D.B.-P., A.M.-B., L.L., and M.-J.G.-P. did the biochemical analysis of biological samples; D.B.-P., B.P., A.G., and M.-J.G.-P. performed in vitro biochemical experiments; S.M. and M.-H.L. conducted CD. E.A. constructed the HeLa cell line. P.K. and H.W. performed DREEM experiments. J.C.-V. assisted for data analysis. D.B.-P. and S.P. were involved in the writing of the manuscript. E.G. and M.-J.G.-P. coordinated the study and wrote the manuscript.
References
- Amiard S, Doudeau M, Pinte S, Poulet A, Lenain C, Faivre-Moskalenko C, Angelov D, Hug N, Vindigni A, Bouvet P, et al. A topological mechanism for TRF2-enhanced strand invasion. Nat Struct Mol Biol. 2007;14:147–154. doi: 10.1038/nsmb1192. [DOI] [PubMed] [Google Scholar]
- Bae NS, Baumann P. A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol Cell. 2007;26:323–334. doi: 10.1016/j.molcel.2007.03.023. [DOI] [PubMed] [Google Scholar]
- Baker AM, Fu Q, Hayward W, Lindsay SM, Fletcher TM. The Myb/SANT domain of the telomere-binding protein TRF2 alters chromatin structure. Nucleic Acids Res. 2009;37:5019–5031. doi: 10.1093/nar/gkp515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker AM, Fu Q, Hayward W, Victoria S, Pedroso IM, Lindsay SM, Fletcher TM. The telomere binding protein TRF2 induces chromatin compaction. PLoS ONE. 2011;6:e19124. doi: 10.1371/journal.pone.0019124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankhead T, Kobryn K, Chaconas G. Unexpected twist: harnessing the energy in positive supercoils to control telomere resolution. Mol Microbiol. 2006;62:895–905. doi: 10.1111/j.1365-2958.2006.05423.x. [DOI] [PubMed] [Google Scholar]
- Bao K, Cohen SN. Reverse transcriptase activity innate to DNA polymerase I and DNA topoisomerase I proteins of Streptomyces telomere complex. Proc Natl Acad Sci USA. 2004;101:14361–14366. doi: 10.1073/pnas.0404386101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetti R, Schoeftner S, Muñoz P, Blasco MA. Role of TRF2 in the assembly of telomeric chromatin. Cell Cycle. 2008;7:3461–3468. doi: 10.4161/cc.7.21.7013. [DOI] [PubMed] [Google Scholar]
- Bermejo R, Capra T, Jossen R, Colosio A, Frattini C, Carotenuto W, Cocito A, Doksani Y, Klein H, Gómez-González B, et al. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell. 2011;146:233–246. doi: 10.1016/j.cell.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud T, Koering CE, Binet-Brasselet E, Ancelin K, Pollice A, Gasser SM, Gilson E. The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res. 1996;24:1294–1303. doi: 10.1093/nar/24.7.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud T, Brun C, Ancelin K, Koering CE, Laroche T, Gilson E. Telomeric localization of TRF2, a novel human telobox protein. Nat Genet. 1997;17:236–239. doi: 10.1038/ng1097-236. [DOI] [PubMed] [Google Scholar]
- Biroccio A, Porru M, Rizzo A, Salvati E, D’Angelo C, Orlandi A, Passeri D, Franceschin M, Stevens MF, Gilson E, et al. DNA damage persistence as determinant of tumor sensitivity to the combination of Topo I inhibitors and telomere-targeting agents. Clin Cancer Res. 2011;17:2227–2236. doi: 10.1158/1078-0432.CCR-10-3033. [DOI] [PubMed] [Google Scholar]
- Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- Cesare AJ, Karlseder J. A three-state model of telomere control over human proliferative boundaries. Curr Opin Cell Biol. 2012;24:731–738. doi: 10.1016/j.ceb.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ, Kaul Z, Cohen SB, Napier CE, Pickett HA, Neumann AA, Reddel RR. Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat Struct Mol Biol. 2009;16:1244–1251. doi: 10.1038/nsmb.1725. [DOI] [PubMed] [Google Scholar]
- Cesare AJ, Hayashi MT, Crabbe L, Karlseder J. The telomere deprotection response is functionally distinct from the genomic DNA damage response. Mol Cell. 2013;51:141–155. doi: 10.1016/j.molcel.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaconas G, Kobryn K. Structure, function, and evolution of linear replicons in Borrelia. Annu Rev Microbiol. 2010;64:185–202. doi: 10.1146/annurev.micro.112408.134037. [DOI] [PubMed] [Google Scholar]
- Chen Y, Yang Y, van Overbeek M, Donigian JR, Baciu P, de Lange T, Lei M. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science. 2008;319:1092–1096. doi: 10.1126/science.1151804. [DOI] [PubMed] [Google Scholar]
- Chen L, Zhu X, Zou Y, Xing J, Gilson E, Lu Y, Ye J. The topoisomerase II catalytic inhibitor ICRF-193 preferentially targets telomeres that are capped by TRF2. Am J Physiol Cell Physiol. 2015;308:C372–C377. doi: 10.1152/ajpcell.00321.2014. [DOI] [PubMed] [Google Scholar]
- Court R, Chapman L, Fairall L, Rhodes D. How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: a view from high-resolution crystal structures. EMBO Rep. 2005;6:39–45. doi: 10.1038/sj.embor.7400314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Alcontres MS, Palacios JA, Mejias D, Blasco MA. TopoIIα prevents telomere fragility and formation of ultra thin DNA bridges during mitosis through TRF1-dependent binding to telomeres. Cell Cycle. 2014;13:1463–1481. doi: 10.4161/cc.28419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- Doksani Y, Wu JY, de Lange T, Zhuang X. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013;155:345–356. doi: 10.1016/j.cell.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairall L, Chapman L, Moss H, de Lange T, Rhodes D. Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol Cell. 2001;8:351–361. doi: 10.1016/s1097-2765(01)00321-5. [DOI] [PubMed] [Google Scholar]
- Feuerhahn S, Chen LY, Luke B, Porro A. No DDRama at chromosome ends: TRF2 takes centre stage. Trends Biochem Sci. 2015;40:275–285. doi: 10.1016/j.tibs.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Fouché N, Cesare AJ, Willcox S, Ozgür S, Compton SA, Griffith JD. The basic domain of TRF2 directs binding to DNA junctions irrespective of the presence of TTAGGG repeats. J Biol Chem. 2006;281:37486–37495. doi: 10.1074/jbc.M608778200. [DOI] [PubMed] [Google Scholar]
- Galati A, Magdinier F, Colasanti V, Bauwens S, Pinte S, Ricordy R, Giraud-Panis MJ, Pusch MC, Savino M, Cacchione S, Gilson E. TRF2 controls telomeric nucleosome organization in a cell cycle phase-dependent manner. PLoS ONE. 2012;7:e34386. doi: 10.1371/journal.pone.0034386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germe T, Miller K, Cooper JP. A non-canonical function of topoisomerase II in disentangling dysfunctional telomeres. EMBO J. 2009;28:2803–2811. doi: 10.1038/emboj.2009.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E, Géli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- Giraud-Panis MJ, Pisano S, Benarroch-Popivker D, Pei B, Le Du MH, Gilson E. One identity or more for telomeres? Front Oncol. 2013;3:48. doi: 10.3389/fonc.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- Grolimund L, Aeby E, Hamelin R, Armand F, Chiappe D, Moniatte M, Lingner J. A quantitative telomeric chromatin isolation protocol identifies different telomeric states. Nat Commun. 2013;4:2848–2859. doi: 10.1038/ncomms3848. [DOI] [PubMed] [Google Scholar]
- Hayashi MT, Cesare AJ, Fitzpatrick JA, Lazzerini-Denchi E, Karlseder J. A telomere-dependent DNA damage checkpoint induced by prolonged mitotic arrest. Nat Struct Mol Biol. 2012;19:387–394. doi: 10.1038/nsmb.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh MH, Tsai CH, Lin CC, Li TK, Hung TW, Chang LT, Hsin LW, Teng SC. Topoisomerase II inhibition suppresses the proliferation of telomerase-negative cancers. Cell Mol Life Sci. 2015;72:1825–1837. doi: 10.1007/s00018-014-1783-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabir S, Hockemeyer D, de Lange T. TALEN gene knockouts reveal no requirement for the conserved human shelterin protein Rap1 in telomere protection and length regulation. Cell Rep. 2014;9:1273–1280. doi: 10.1016/j.celrep.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul Z, Cesare AJ, Huschtscha LI, Neumann AA, Reddel RR. Five dysfunctional telomeres predict onset of senescence in human cells. EMBO Rep. 2012;13:52–59. doi: 10.1038/embor.2011.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Lee OH, Xin H, Chen LY, Qin J, Chae HK, Lin SY, Safari A, Liu D, Songyang Z. TRF2 functions as a protein hub and regulates telomere maintenance by recognizing specific peptide motifs. Nat Struct Mol Biol. 2009;16:372–379. doi: 10.1038/nsmb.1575. [DOI] [PubMed] [Google Scholar]
- Kouzine F, Gupta A, Baranello L, Wojtowicz D, Ben-Aissa K, Liu J, Przytycka TM, Levens D. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat Struct Mol Biol. 2013;20:396–403. doi: 10.1038/nsmb.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonetti C, Scarsella M, Riggio G, Rizzo A, Salvati E, D’Incalci M, Staszewsky L, Frapolli R, Stevens MF, Stoppacciaro A, et al. G-quadruplex ligand RHPS4 potentiates the antitumor activity of camptothecins in preclinical models of solid tumors. Clin Cancer Res. 2008;14:7284–7291. doi: 10.1158/1078-0432.CCR-08-0941. [DOI] [PubMed] [Google Scholar]
- Mendoza VL, Vachet RW. Probing protein structure by amino acid-specific covalent labeling and mass spectrometry. Mass Spectrom Rev. 2009;28:785–815. doi: 10.1002/mas.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabella A, Gartenberg MR. Yeast telomeric sequences function as chromosomal anchorage points in vivo. EMBO J. 1997;16:523–533. doi: 10.1093/emboj/16.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraki K, Nabetani A, Nishiyama A, Ishikawa F. Essential roles of Xenopus TRF2 in telomere end protection and replication. Genes Cells. 2011;16:728–739. doi: 10.1111/j.1365-2443.2011.01520.x. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Bartocci C, Ouzounov I, Diedrich JK, Yates JR, 3rd, Denchi EL. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature. 2013;494:502–505. doi: 10.1038/nature11873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porro A, Feuerhahn S, Delafontaine J, Riethman H, Rougemont J, Lingner J. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat Commun. 2014a;5:5379–5391. doi: 10.1038/ncomms6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porro A, Feuerhahn S, Lingner J. TERRA-reinforced association of LSD1 with MRE11 promotes processing of uncapped telomeres. Cell Rep. 2014b;6:765–776. doi: 10.1016/j.celrep.2014.01.022. [DOI] [PubMed] [Google Scholar]
- Poulet A, Buisson R, Faivre-Moskalenko C, Koelblen M, Amiard S, Montel F, Cuesta-Lopez S, Bornet O, Guerlesquin F, Godet T, et al. TRF2 promotes, remodels and protects telomeric Holliday junctions. EMBO J. 2009;28:641–651. doi: 10.1038/emboj.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulet A, Pisano S, Faivre-Moskalenko C, Pei B, Tauran Y, Haftek-Terreau Z, Brunet F, Le Bihan YV, Ledu MH, Montel F, et al. The N-terminal domains of TRF1 and TRF2 regulate their ability to condense telomeric DNA. Nucleic Acids Res. 2012;40:2566–2576. doi: 10.1093/nar/gkr1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Léger A, Koelblen M, Civitelli L, Bah A, Djerbi N, Giraud-Panis MJ, Londoño-Vallejo A, Ascenzioni F, Gilson E. The basic N-terminal domain of TRF2 limits recombination endonuclease action at human telomeres. Cell Cycle. 2014;13:2469–2474. doi: 10.4161/cc.29422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarek G, Vannier JB, Panier S, Petrini JH, Boulton SJ. TRF2 recruits RTEL1 to telomeres in S phase to promote t-loop unwinding. Mol Cell. 2015;57:622–635. doi: 10.1016/j.molcel.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarthy J, Bae NS, Scrafford J, Baumann P. Human RAP1 inhibits non-homologous end joining at telomeres. EMBO J. 2009;28:3390–3399. doi: 10.1038/emboj.2009.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansel RM, de Lange T, Griffith JD. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 2001;20:5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- Takai KK, Kibe T, Donigian JR, Frescas D, de Lange T. Telomere protection by TPP1/POT1 requires tethering to TIN2. Mol Cell. 2011;44:647–659. doi: 10.1016/j.molcel.2011.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temime-Smaali N, Guittat L, Wenner T, Bayart E, Douarre C, Gomez D, Giraud-Panis MJ, Londono-Vallejo A, Gilson E, Amor-Guéret M, Riou JF. Topoisomerase IIIalpha is required for normal proliferation and telomere stability in alternative lengthening of telomeres. EMBO J. 2008;27:1513–1524. doi: 10.1038/emboj.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanasoula M, Escandell JM, Martinez P, Badie S, Muñoz P, Blasco MA, Tarsounas M. p53 prevents entry into mitosis with uncapped telomeres. Curr Biol. 2010;20:521–526. doi: 10.1016/j.cub.2010.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HH, Huang CH, Tessmer I, Erie DA, Chen CW. Linear Streptomyces plasmids form superhelical circles through interactions between their terminal proteins. Nucleic Acids Res. 2011;39:2165–2174. doi: 10.1093/nar/gkq1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- Vos SM, Tretter EM, Schmidt BH, Berger JM. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat Rev Mol Cell Biol. 2011;12:827–841. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan B, Yin J, Horvath K, Sarkar J, Chen Y, Wu J, Wan K, Lu J, Gu P, Yu EY, et al. SLX4 assembles a telomere maintenance toolkit by bridging multiple endonucleases with telomeres. Cell Rep. 2013;4:861–869. doi: 10.1016/j.celrep.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JS, Tejera AM, Castor D, Toth R, Blasco MA, Rouse J. Localization-dependent and -independent roles of SLX4 in regulating telomeres. Cell Rep. 2013;4:853–860. doi: 10.1016/j.celrep.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Kaur P, Wang H, Erie DA. Visualizing the path of DNA through proteins using DREEM imaging. Mol Cell. 2016;61 doi: 10.1016/j.molcel.2015.12.012. this issue, ■■■–■■■. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Lenain C, Bauwens S, Rizzo A, Saint-Léger A, Poulet A, Benarroch D, Magdinier F, Morere J, Amiard S, et al. TRF2 and apollo cooperate with topoisomerase 2alpha to protect human telomeres from replicative damage. Cell. 2010;142:230–242. doi: 10.1016/j.cell.2010.05.032. [DOI] [PubMed] [Google Scholar]
- Yeeles JT, Poli J, Marians KJ, Pasero P. Rescuing stalled or damaged replication forks. Cold Spring Harb Perspect Biol. 2013;5:a012815. doi: 10.1101/cshperspect.a012815. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.