Abstract
Background
Streptomycetes are soil-dwelling Gram-positive bacteria that are best known as the major producers of antibiotics used in the pharmaceutical industry. The evolution of exceptionally powerful transporter systems in streptomycetes has enabled their adaptation to the complex soil environment.
Results
Our comparative genomic analyses revealed that each of the eleven Streptomyces species examined possesses a rich repertoire of from 761-1258 transport proteins, accounting for 10.2 to 13.7 % of each respective proteome. These transporters can be divided into seven functional classes and 171 transporter families. Among them, the ATP-binding Cassette (ABC) superfamily and the Major Facilitator Superfamily (MFS) represent more than 40 % of all the transport proteins in Streptomyces. They play important roles in both nutrient uptake and substrate secretion, especially in the efflux of drugs and toxicants. The evolutionary flexibility across eleven Streptomyces species is seen in the lineage-specific distribution of transport proteins in two major protein translocation pathways: the general secretory (Sec) pathway and the twin-arginine translocation (Tat) pathway.
Conclusions
Our results present a catalog of transport systems in eleven Streptomyces species. These expansive transport systems are important mediators of the complex processes including nutrient uptake, concentration balance of elements, efflux of drugs and toxins, and the timely and orderly secretion of proteins. A better understanding of transport systems will allow enhanced optimization of production processes for both pharmaceutical and industrial applications of Streptomyces, which are widely used in antibiotic production and heterologous expression of recombinant proteins.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-016-2899-4) contains supplementary material, which is available to authorized users.
Keywords: Streptomyces, Transport proteins, Comparative genomics, Drug efflux, Protein translocation
Background
Streptomyces is a group of soil-dwelling Gram-positive bacteria, which are well known for their ability to produce a broad array of secondary metabolites including antibiotics, antifungals, antiparasitic drugs, anticancer agents, immunosuppressants, and herbicides [1, 2]. They are also ideal systems in biotechnology for heterologous expression of recombinant proteins with simple downstream processing and high yields [3, 4]. In order to survive in the complex soil environment, streptomycetes have evolved exceptionally powerful transport systems [5, 6]. For example, in Streptomyces coelicolor, there are more than 600 predicted transport proteins with a large proportion being the ATP-binding Cassette (ABC) and Major Facilitator Superfamily (MFS) transporters, which have been implicated in the transport of secondary metabolites including antibiotics [7]. In addition to secondary metabolites, streptomycetes also secret to the environment a mass of proteins through the general secretory (Sec) pathway and the twin-arginine translocation (Tat) pathway [8–10]. These secretory systems are known to facilitate nutrient acquisition. For example, secreted cellulases and chitinases can degrade otherwise insoluble nutrient sources.
Transporters are of critical importance to all living organisms in facilitating metabolism, intercellular communication, biological synthesis and reproduction. They are involved in the uptake of nutrients from the environment, the secretion of metabolites, the efflux of drugs and toxins, the maintenance of ion concentration gradient across membranes, the secretion of macromolecules, such as sugars, lipids, proteins and nucleic acids, signaling molecules, the translocation of membrane proteins, and so on [11]. A Transporter Classification (TC) system has been developed by the Saier group [11, 12]. To date, more than 10,000 non-redundant transport proteins comprising about 750 families are collected in their Transporter Classification Database (TCDB) [13]. These families are divided among seven major classes: Channels/Pores (Class 1), Electrochemical Potential-driven Transporters (Class 2), Primary Active Transporters (Class 3), Group Translocators (Class 4), Transmembrane Electron Carriers (class 5), Accessory Factors Involved in Transport (Class 8), and Incompletely Characterized Transport Systems (Class 9). This classification system has been applied to in-depth studies of transporters in a number of microbial genomes [14–17], and is being adopted in this study for Streptomyces.
The availability of genomes from closely related Streptomyces species enables comprehensive analysis of the transport protein families in Streptomyces. In this study, we report a catalog and comparative genomic analysis of transporters in eleven Streptomyces species with complete genome sequences and annotations, including S. coelicolor (SCO), S. avermitilis (SAV), S. bingchenggensis (SBI), S. cattleya (SCAT), S. flavogriseus (SFLA), S. griseus (SGR), S. hygroscopicus (SHJG), S. scabiei (SCAB), S. sp. SirexAA-E (SACTE), S. venezuelae (SVEN) and S. violaceusniger (STRVI) [7, 18–24]. We identified and classified these Streptomyces transporters, using the nomenclature in the TCDB. The class, transmembrane topology and substrate specificity of these transporters are investigated in detail. An improved understanding of Streptomyces transporters will bring new insights into the mechanisms underlying the unique and powerful secretion systems of secondary metabolites and proteins in this group of bacteria of enormous economic and biomedical significance.
Results and discussion
Abundant transporters are present in eleven Streptomyces genomes
Strong material intake and secretion capacity powered by transport systems is an adaptive attribute of soil-dwelling bacteria [1]. We used the coding sequences from eleven Streptomyces genomes to query the TCDB [13, 25] using BLASTP and identified 761-1258 transporters in these eleven genomes, which accounted for 10.2 to 13.7 % of each respective proteome (Table 1 and Additional file 1). S. bingchenggensis, which has the largest genome, and the largest number of protein-coding genes, has the largest number of transporters, whereas S. cattleya contains only 761 transporters, the lowest number and proportion of transporters among the eleven Streptomyces species.
Table 1.
Distribution of transporters in eleven Streptomyces genomes
| Organisms | Accession ID | Genome size (Mbp) | # ORFs | # Transporters | % Transporters |
|---|---|---|---|---|---|
| S. avermitilis | NC_003155 (chr) | 9.1 | 7676 | 989 | 12.9 % |
| NC_004719 (pSAP1) | |||||
| S. bingchenggensis | NC_016582 (chr) | 11.9 | 10022 | 1258 | 12.6 % |
| S. cattleya | NC_016111(chr) | 8.1 | 7475 | 761 | 10.2 % |
| NC_016113(pSCAT) | |||||
| S. coelicolor | NC_003888(chr) | 9.1 | 8153 | 990 | 12.1 % |
| NC_003903 (pSCP1) | |||||
| NC_003904 (pSCP2) | |||||
| S. flavogriseus | NC_016114 (chr) | 7.7 | 6572 | 888 | 13.5 % |
| NC_016110 (pSFLA01) | |||||
| NC_016115 (pSFLA02) | |||||
| S. griseus | NC_010572 (chr) | 8.5 | 7136 | 975 | 13.7 % |
| S. hygroscopicus | NC_017765 (chr) | 10.4 | 9108 | 999 | 11.0 % |
| NC_017766 (pSHJG1) | |||||
| NC_016972 (pSHJG2) | |||||
| S. scabiei | NC_013929 (chr) | 10.1 | 8746 | 1021 | 11.7 % |
| S. sp. SirexAA-E | NC_015953 (chr) | 7.4 | 6357 | 869 | 13.7 % |
| S. venezuelae | NC_018750 (chr) | 8.2 | 7453 | 935 | 12.5 % |
| S. violaceusniger | NC_015957 (chr) | 11.0 | 8985 | 989 | 11.0 % |
| NC_015951(pSTRVI01) | |||||
| NC_015952(pSTRVI02) |
Streptomyces transporters show diverse transmembrane topology
The capacity of a transporter is often associated with the complexity and topology of its transmembrane region(s) where the major events of substrate uptake or output across the cell membranes take place. Using the TMHMM (TransMembrane prediction using Hidden Markov Models) algorithm [26], we performed the transmembrane topology analysis for Streptomyces transporters to identify the transmembrane segments (TMSs). The number of TMSs ranges from 0 to 24. The largest number of TMSs observed in a transporter in the eleven Streptomyces genomes varies from 16 to 24 (Table 2). Except for intra-/extra-cellular transporters which have no TMS, transporters with 6 and 12 TMSs are predominant. Most transporters with 6 TMSs are ABC transporters (TC 3.A.1), and transporters with 12 TMSs are mainly members of the Major Facilitator Superfamily (MFS) (TC 2.A.1), the Amino Acid-Polyamine-Organocation (APC) superfamily (TC 2.A.3), the Resistance-Nodulation-Cell Division (RND) superfamily (TC 2.A.6) and the ABC superfamily (TC 3.A.1). It is possible that these 12-TMS transporters have arisen from the primordial 6-TMS form via intragenic duplication [27]. Among the transporters with more than 6 TMSs, the transporters with an even number of TMSs are more abundant than those with an odd number of TMSs (Fig. 1). The distribution of TMSs in S. griseus transporters is unique: this bacterium has 53 transporters with 9 TMSs, mostly ABC transporters, accounting for 5.4 % of the total transporters. This proportion is significantly higher than that of the other ten sibling species. On the other hand, S. griseus has the lowest proportion of 12-TMS transporters (7.3 %), most of which are also ABC transporters. These topology patterns suggest that during the evolution of transporters in S. griseus, the “6 + 3” events may be more frequent than the typical “6 + 6” events observed in ten other Streptomyces species [27, 28].
Table 2.
Distribution of topological types of transporters in eleven Streptomyces genomes
| TMS | SACTE | SAV | SBI | SCAB | SCAT | SCO | SFLA | SGR | SHJG | STRVI | SVEN |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 322 | 382 | 482 | 424 | 280 | 344 | 332 | 372 | 392 | 371 | 350 |
| 1 | 41 | 41 | 55 | 33 | 51 | 47 | 45 | 33 | 28 | 37 | 44 |
| 2 | 14 | 17 | 18 | 21 | 19 | 19 | 21 | 15 | 16 | 15 | 16 |
| 3 | 26 | 28 | 32 | 20 | 21 | 26 | 28 | 30 | 22 | 26 | 13 |
| 4 | 27 | 29 | 31 | 36 | 23 | 32 | 26 | 29 | 36 | 28 | 30 |
| 5 | 49 | 55 | 62 | 52 | 40 | 58 | 42 | 58 | 56 | 58 | 55 |
| 6 | 119 | 130 | 201 | 141 | 72 | 135 | 124 | 122 | 116 | 143 | 113 |
| 7 | 22 | 29 | 23 | 15 | 12 | 24 | 23 | 22 | 20 | 19 | 24 |
| 8 | 26 | 32 | 35 | 30 | 26 | 34 | 27 | 25 | 35 | 31 | 22 |
| 9 | 33 | 25 | 36 | 28 | 16 | 35 | 34 | 53 | 30 | 21 | 30 |
| 10 | 41 | 55 | 62 | 46 | 44 | 54 | 41 | 43 | 48 | 41 | 55 |
| 11 | 20 | 28 | 42 | 32 | 27 | 30 | 24 | 26 | 40 | 35 | 29 |
| 12 | 66 | 72 | 109 | 89 | 68 | 89 | 70 | 71 | 97 | 106 | 85 |
| 13 | 18 | 26 | 24 | 19 | 15 | 20 | 14 | 21 | 25 | 23 | 21 |
| 14 | 40 | 37 | 43 | 31 | 45 | 38 | 32 | 46 | 35 | 32 | 44 |
| 15 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 2 | 0 | 0 | 1 |
| 16 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 2 | 2 | 1 |
| 17 | 0 | 0 | 1 | 2 | 0 | 2 | 2 | 1 | 1 | 1 | 2 |
| 18 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 24 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 |
| Total | 869 | 989 | 1258 | 1021 | 761 | 990 | 888 | 975 | 999 | 989 | 935 |
Note: SACTE (S. sp. SirexAA-E), SAV (S. avermitilis), SBI (S. bingchenggensis), SCAB (S. scabiei), SCAT (S. cattleya), SCO (S. coelicolor), SFLA (S. flavogriseus), SGR (S. griseus), SHJG (S. hygroscopicus), STRVI (S. violaceusniger), SVEN (S. venezuelae)
Fig. 1.
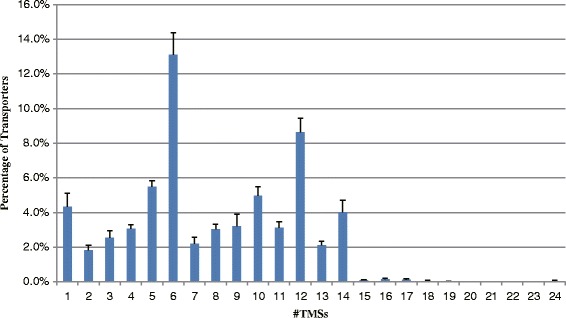
Distribution of transporter topologies in eleven Streptomyces genomes. The abbreviations for species are: S. sp. SirexAA-E (SACTE), S. avermitilis (SAV), S. bingchenggensis (SBI), S. scabiei (SCAB), S. cattleya (SCAT), S. coelicolor (SCO), S. flavogriseus (SFLA), S. griseus (SGR), S. hygroscopicus (SHJG), S. violaceusniger (STRVI), and S. venezuelae (SVEN)
Transporters in eleven Streptomyces genomes can be divided into seven classes and 171 families
The Streptomyces transporters fall into seven classes and 171 transporter families according to the TCDB system (Table 3 and Additional file 2). The distribution of transporters in each species is depicted in Fig. 2.
Table 3.
Distribution of Streptomyces transporters in each TC class and subclass
| Class | Subclass | SACTE | SAV | SBI | SCAB | SCAT | SCO | SFLA | SGR | SHJG | STRVI | SVEN |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1: Channels/Proes | 22 | 31 | 29 | 34 | 22 | 29 | 26 | 28 | 26 | 31 | 30 | |
| 1.A: α-Type Channels | 18 | 24 | 20 | 25 | 15 | 22 | 21 | 20 | 21 | 24 | 21 | |
| 1.B: β-Barrel Porins | 3 | 6 | 7 | 8 | 6 | 6 | 4 | 5 | 4 | 6 | 6 | |
| 1.C: Pore-Forming Toxins (Proteins and Peptides) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | 3 | |
| 1.I: Membrane-bounded channels | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 2: Electrochemical Potential-driven Transporters | 212 | 266 | 330 | 251 | 239 | 274 | 217 | 242 | 305 | 271 | 269 | |
| 2.A: Porters (uniporters, symporters, antiporters) | 212 | 266 | 328 | 251 | 239 | 274 | 217 | 242 | 305 | 271 | 269 | |
| 2.C: Ion-gradient-driven energizers | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 3: Primary Active Transporters | 500 | 544 | 705 | 553 | 365 | 552 | 498 | 555 | 489 | 528 | 494 | |
| 3.A: P-P-bond-hydrolysis-driven transporters | 455 | 492 | 656 | 505 | 304 | 497 | 451 | 508 | 433 | 476 | 449 | |
| 3.B: Decarboxylation-driven transporters | 6 | 6 | 5 | 6 | 10 | 7 | 6 | 6 | 6 | 4 | 6 | |
| 3.D: Oxidoreduction-driven transporters | 39 | 46 | 43 | 42 | 51 | 48 | 41 | 41 | 50 | 47 | 39 | |
| 3.E: Light absorption-driven transporters | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | |
| 4: Group Translocators | 27 | 46 | 62 | 54 | 35 | 30 | 36 | 40 | 46 | 37 | 43 | |
| 4.A: Phosphotransfer-driven group translocators | 5 | 7 | 4 | 5 | 2 | 8 | 8 | 6 | 5 | 7 | 6 | |
| 4.B: Nicotinamide ribonucleoside uptake transporters | 1 | 1 | 0 | 1 | 1 | 1 | 3 | 3 | 1 | 0 | 3 | |
| 4.C: Acyl CoA ligase-coupled transporters | 21 | 38 | 58 | 48 | 32 | 21 | 25 | 31 | 40 | 30 | 34 | |
| 5: Transmembrane Electron Carriers | 12 | 13 | 21 | 19 | 18 | 26 | 15 | 13 | 20 | 16 | 16 | |
| 5.A: Transmembrane 2-electron transfer carriers | 12 | 12 | 21 | 18 | 17 | 26 | 14 | 13 | 19 | 15 | 16 | |
| 5.B: Transmembrane 1-electron transfer carriers | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | |
| 8: Accessory Factors Involved in Transport | 4 | 4 | 5 | 6 | 5 | 5 | 5 | 6 | 6 | 4 | 4 | |
| 8.A: Auxiliary transport proteins | 4 | 4 | 5 | 6 | 5 | 5 | 5 | 6 | 6 | 4 | 4 | |
| 9: Incompletely Characterized Transport Systems | 60 | 63 | 74 | 67 | 75 | 67 | 68 | 66 | 63 | 69 | 55 | |
| 9.A: Recognized transporters of unknown biochemical mechanism | 27 | 25 | 44 | 27 | 33 | 31 | 32 | 35 | 27 | 33 | 25 | |
| 9.B: Putative transport proteins | 33 | 38 | 30 | 40 | 42 | 36 | 35 | 31 | 36 | 36 | 30 | |
| 9.C: Functionally characterized transporters lacking identified sequences | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| N/A | 32 | 22 | 32 | 37 | 2 | 7 | 23 | 25 | 44 | 33 | 24 | |
| Total | 869 | 989 | 1258 | 1021 | 761 | 990 | 888 | 975 | 999 | 989 | 935 | |
Fig. 2.
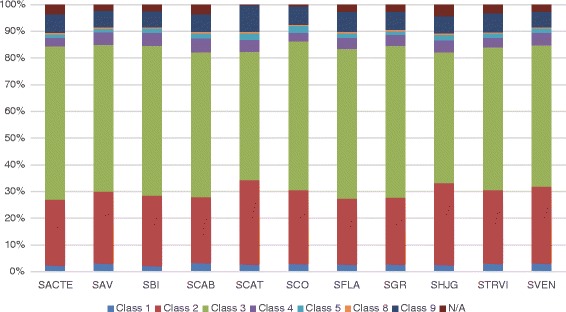
Distribution of transporter types according to the TC system in eleven Streptomyces genomes. Class 1: Channels/Proes; Class 2: Electrochemical Potential-driven Transporters; Class 3: Primary Active Transporters; Class 4: Group Translocators; Class 5: Transmembrane Electron Carriers; Class 8: Accessory Factors Involved in Transport; Class 9: Incompletely Characterized Transport Systems; N/A: Not assigned
The Primary Active Transporters (Class 3) is the most abundant class of transporters in Streptomyces, which includes 365-705 transporters (representing about 48.0-57.5 % of the total transport machinery). This class of transporters plays important roles in various aspects of bacterial life cycle, especially in the import and export of secondary metabolites, and cation transportation.
Class 2 transporters, the electrochemical potential-driven transporters, are also widely found in Streptomyces. 212-330 transporters in eleven Streptomyces genomes belong to this class, which account for 24.4 %-31.4 % of all the transporters. The porters in this class include uniporters, symporters and antiporters. The most abundant family, MFS, in Class 2 transporters has been implicated in drug efflux. Lineage-specificity is also observed in this class of transporters. For example, S. bingchenggensis possesses two Ion-gradient-driven Energizers (TC 2.C), while the other ten Streptomyces species only have Porters (uniporters, symporters, antiporters) (TC 2.A).
Class 1 transporters are not abundant, but are functionally important for Streptomyces. 22-34 channel/pore transporters are present in these eleven genomes, accounting for 2.3 %-3.2 % of all the transporters. The majority of these channel-type proteins are alpha-type channels (TC 1.A), which have been implicated in stress responses of Gram-positive bacteria, especially responses to osmotic pressure [27]. A small number of proteins belong to β-type porins and a fewer are putative Channel-Forming Toxins (TC 1.C). The membrane-bounded channel (TC 1.I) subclass is rare in Streptomyces; only S. bingchenggensis has a transport protein from this subclass.
Classes 4, 5, and 8 are relatively less abundant. About 3.0 %-5.3 % of all the transport proteins are Class 4 transporters. Two major subclasses observed in Class 4 are the PTS Glucose-Glucoside (Glc) family (4.A.1) and the Fatty Acid Transporter (FAT) family (4.C.1), which are responsible for the transport of glucoses-glucosides and fatty acids, respectively. Notably, S. cattleya, which has the smallest repertoire of transporters among the eleven Streptomyces, does not seem to contain any Glc transporters; it remains unknown if it uses an alternative system. Only 12-21 members of the Class 4 transporters, the Transmembrane Electron Carriers, are found in Streptomyces. Two subclasses are present, including the Prokaryotic Molybdopterin-containing Oxidoreductase (PMO) family (TC 5.A.3) and the Prokaryotic Succinate Dehydrogenase (SDH) family (TC 5.A.4), which transfer electrons mainly by redox reactions. Class 8, the Accessory Factors Involved in Transport, is the least abundant transporter class (0.4 %-0.7 %) in Streptomyces.
A significant number (60-75) of transporters in Streptomyces can be grouped into Class 9, an incompletely characterized class. While their exact physiological roles are yet to be elucidated, they might be involved in the transport of ions, implicated by their sequence similarities with the members of the HlyC/CorC (HCC) family (TC 9.A.40), and the Tripartite Zn2+ Transporter (TZT) family (TC 9.B.10).
Examples of important transporter families
Many of the 171 transporter families are involved in the transfer of ions, saccharides, amino acids, polypeptides, proteins, drugs, toxins and other compounds. The two most abundant and perhaps also the most important families are in the ABC (TC 3.A.1) and MFS (TC 2.A.1) superfamilies. They are responsible for the secretion of a wide array of antibiotics in Streptomyces [29, 30].
The ABC transporters
32.7 %-47.5 % (249-597) of all the transport proteins in the eleven Streptomyces genomes are members of ABC superfamily. ABC transporters are characterized by a conserved ATP hydrolyzing domain for energy provision, pore-forming membrane-integrated domain(s), and a substrate-binding domain [31, 32]. The ABC transport system is composed of the intake system and the efflux system.
The 30 intake families (TC 3.A.1-3.A.33) that we identified in the Streptomyces genomes are specialized in the uptake of diverse nutrient substances. This intake system includes families of Carbohydrate Uptake Transporters (TC 3.A.1.1, 3.A.1.2) that transport saccharides, Polar Amino Acid Uptake Transporters and Hydrophobic Amino Acid Uptake Transporters (TC 3.A.1.3, 3.A.1.4) that transfer amino acids, Polyamine/Opine/ Phosphonate Uptake Transporters and Quaternary Amine Uptake Transporters (TC 3.A.1.11, 3.A.1.12) that transfer amine substances, Iron Chelate Uptake Transporters and Manganese/Zinc/Iron Chelate Uptake Transporters (TC 3.A.1.14, 3.A.1.15) that transfer metal ions.
Unlike the intake system, the 35 Streptomyces efflux families are involved in the transport of macromolecular substances. These transporters are believed to be essential for Streptomyces due to their roles in drug efflux and protein secretion. The drug efflux system regulates various aspects of the response to drug compounds mediated by Drug Exporters (TC 3.A.1.105, 3.A.1.117, 3.A.1.119, 3.A.1.135), Drug Resistance ATPases (TC 3.A.1.120, 3.A.1.121), Macrolide Exporters (TC 3.A.1.122), β-Exotoxin I Exporters (TC 3.A.1.126), Multidrug Resistance Exporters (TC 3.A.201) and Pleiotropic Drug Resistance transporters (TC 3.A.1.205). Potent protein transport in Streptomyces is regulated by Protein/Peptide Exporters (TC 3.A.1.109, 110, 111, 112, 123, 124, 134), Lipoprotein Translocases (TC 3.A.1.125), AmfS Peptide Exporters (TC 3.A.1.127), and SkfA Peptide Exporters (TC 3.A.1.128).
The MFS transporters
Unlike the ABC transporters, the MFS transporters are driven by an electrochemical potential formed by ion concentration gradients across the cytomembrane [30]. There are 90-169 (10.1 %- 15.0 %) MFS transporters in eleven Streptomyces genomes. Streptomyces possesses 39 subfamilies of MFS transporters, including 20 intake systems, 13 efflux systems and 6 systems whose transport direction is unknown. The substances transported by the intake systems are mainly saccharides and organic acids.
One of the most important roles of the MFS transporters is drug efflux [30]. Diverse subfamilies of drug efflux MFS transporters are present in Streptomyces, with varying mechanisms of action, including Drug:H+ Antiporters (TC 2.A.1.2, 2.A.1.3, 2.A.1.21), Aromatic Compound/Drug Exporters (TC 2.A.1.32), Fosmidomycin Resistance transporters (TC 2.A.1.35), Acriflavin-sensitivity transporters (TC 2.A.1.36), and Microcin C51 Immunity Proteins (TC 2.A.1.61), to name a few.
The wide distribution of substrates for Streptomyces transporters
The capacity of the complex and powerful transporter system in Streptomyces is evidenced by the broad scope of the substrates being transported. Figure 3a shows the distribution of transporters that transport different type of substrates in Streptomyces, including carbon sources, drugs, toxicants, electrons, inorganic molecules, macromolecules, amino acids and derivatives, nucleotides and derivatives, vitamins, and accessory factors. The carbon source transporters are the most abundant, with their proportion of all the transport proteins ranging from 21.7 to 31.6 % in eleven genomes. Notably, the substrates of an average of 6.4 % of the transporters in Streptomyces genomes examined cannot be determined based on genomic analysis, and await advanced structural and biochemical characterization.
Fig. 3.
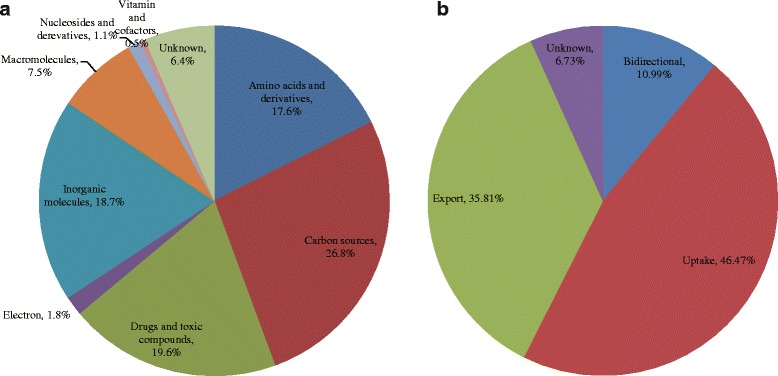
a Distribution of substrate types and (b) predicted polar characteristics: bidirectional transport, uptake or export in eleven Streptomyces genomes
Streptomyces transporters can be divided into three classes, uptake, efflux and bidirectional, according to the direction of the substrates transported (Fig. 3b). Among the transporters of the eleven Streptomyces genomes, on average 46.5 % are involved in the uptake of substrates, 35.8 % are involved in the efflux of substrates, and 11.0 % are in charge of the bidirectional transport of substrates. The direction of 6.7 % of these proteins remains undetermined.
Streptomyces have lineage-specific protein secretion systems
Streptomyces have two major lineage-specific protein transport systems, the Tat system (TC 2.A.64) and the Sec system (TC 3.A.5) [8, 9]. The Tat system was shown to be related to the pathogenicity of pathogenic bacteria [33]. In S. scabies, the transporters in the Tat pathway secrete several toxicity-associated proteins [34]. While the key component proteins of the Tat system, TatA, TatB and TatC, are present in all eleven Streptomyces genomes we looked at, lineage-specificity is clearly shown with respect to the copy number variation of these genes (Table 4). Only one copy of the tatB and tatC genes is present in nine Streptomyces genomes; S. flavogriseus has two copies of the tatB genes and S. hygroscopicus has two copies of the tatC genes. The copy number of the tatA gene ranges from one to three in eleven genomes (Table 4). Phylogenetic analysis shows that the multiple copies of the tatA genes may have different evolutionary origins and can be divided into three independent clades, namely tatA1, tatA2 and tatA3 (Fig. 4a). The tatA paralogous genes in the majority of the Streptomyces genomes belong to different clades. Notably, all the three tatA paralogous genes in S. cattleya are clustered into the tatA3 clade, indicative of recent gene duplication events.
Table 4.
The Tat translocation system in Streptomyces (TC 2.A.64)
| Species | tatA1 | tatA2 | tatA3 | tatB1 | tatB2 | tatC1 | tatC2 |
|---|---|---|---|---|---|---|---|
| SACTE | SACTE_1063 | SACTE_6092 | SACTE_3032 | SACTE_4381 | SACTE_1062 | ||
| SAV | SAV_6692 | SAV_3114 | SAV_6693 | ||||
| SBI | SBI_08493 | SBI_04079 | SBI_08494 | ||||
| SCAB | SCAB_73591 | SCAB_31121 | SCAB_73601 | ||||
| SCAT | SCAT_3206 | SCAT_2668 | SCAT_4914 | SCAT_4007 | SCAT_5184 | ||
| SCO | SCO1633 | SCO3768 | SCO5150 | SCO1632 | |||
| SFLA | Sfla_5203 | Sfla_0514 | Sfla_5510 | Sfla_5507 | Sfla_2146 | Sfla_5204 | |
| SGR | SGR_5870 | SGR_6484 | SGR_340 | SGR_2375 | SGR_5871 | ||
| SHJG | SHJG_2368 | SHJG_3070 | SHJG_0499 | SHJG_6250 | SHJG_2367 | SHJG_3069 | |
| STRVI | Strvi_6639 | Strvi_3352 | Strvi_1468 | Strvi_6638 | |||
| SVEN | SVEN_1225 | SVEN_4796 | SVEN_1224 |
Fig. 4.
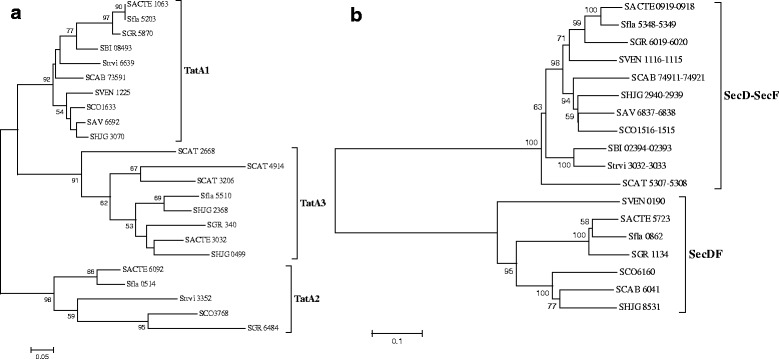
a Phylogenetic tree of the TatA system. b Phylogenetic tree of the SecD/SecF (b) system in eleven Streptomyces genomes. The trees were constructed using the neighbor-joining method by MEGA6 [43]. The Maximum Parsimony and Maximum Likelihood methods gave virtually the same topology (data not shown)
Similarly, the Sec system is also species-specific. This system includes SecA, SecY, SecE, SecG, SecD, SecF, YajC, FtsY, etc. [35], all of which are highly conserved in Streptomyces (Table 5). There is only one copy of the secE, secG, secD, secF, yajC and ftsY genes in each of the eleven Streptomyces genomes. Interestingly, there is a second set of secA2/secY2 genes in several species, which may be involved in the secretion of proteins with specific functions, for example, the secretion of toxic proteins [36]. In S. avermitilis, for instance, there are two copies of the secA genes, and S. venezuelae has two copies of the secY genes.
Table 5.
The Sec translocation system in Streptomyces (TC 3.A.5)
| Species | secA1 | secA2 | secY | secY2 | secE | secG |
|---|---|---|---|---|---|---|
| SACTE | SACTE_2472 | SACTE_3988 | SACTE_3949 | SACTE_1366 | ||
| SAV | SAV_5071 | SAV_2565 | SAV_4312 | SAV_4908 | SAV_6299 | |
| SBI | SBI_06502 | SBI_06209 | SBI_06158 | SBI_08032 | ||
| SCAB | SCAB_55371 | SCAB_36741 | SCAB_37261 | SCAB_69731 | ||
| SCAT | SCAT_2009 | SCAT_3612 | SCAT_3559 | SCAT_1102 | ||
| SCO | SCO3005 | SCO4722 | SCO4646 | SCO1944 | ||
| SFLA | Sfla_3902 | Sfla_2503 | Sfla_2541 | Sfla_4882 | ||
| SGR | SGR_4531 | SGR_2814 | SGR_2876 | SGR_5576 | ||
| SHJG | SHJG_4468 | SHJG_5817 | SHJG_5775 | SHJG_3400 | ||
| STRVI | Strvi_8396 | Strvi_0893 | Strvi_0854 | Strvi_7031 | ||
| SVEN | SVEN_2748 | SVEN_4399 | SVEN_0354 | SVEN_4338 | SVEN_1573 | |
| Species | secD | secF | secDF | yajC | ftsY | |
| SACTE | SACTE_0919 | SACTE_0918 | SACTE_5723 | SACTE_0920 | SACTE_4801 | |
| SAV | SAV_6837 | SAV_6838 | SAV_6836 | SAV_2654 | ||
| SBI | SBI_02394 | SBI_02393 | SBI_02395 | SBI_03477 | ||
| SCAB | SCAB_74911 | SCAB_74921 | SCAB_6041 | SCAB_74901 | SCAB_26291 | |
| SCAT | SCAT_5307 | SCAT_5308 | SCAT_5306 | SCAT_4417 | ||
| SCO | SCO1516 | SCO1515 | SCO6160 | SCO1517 | SCO5580 | |
| SFLA | Sfla_5348 | Sfla_5349 | Sfla_0862 | Sfla_5347 | Sfla_1718 | |
| SGR | SGR_6019 | SGR_6020 | SGR_1134 | SGR_6018 | SGR_1898 | |
| SHJG | SHJG_2940 | SHJG_2939 | SHJG_8531 | SHJG_2941 | SHJG_6701 | |
| STRVI | Strvi_3032 | Strvi_3033 | Strvi_3031 | Strvi_1937 | ||
| SVEN | SVEN_1116 | SVEN_1115 | SVEN_0190 | SVEN_1117 | SVEN_5276 | |
The evolutionary pattern in the secD and the secF genes is particularly interesting (Fig. 4b). In bacteria, these genes encode accessory factors in the Sec pathway that can accelerate the translocation of protein substrates. There are two forms of the secD and secF genes: in the first form, these two genes are adjacent but separate, while in the second form, the two genes are fused into a single secDF gene. The fused secDF is present in seven Streptomyces genomes. Unlike most bacteria that have one of the two forms, the majority of Streptomyces species have both the separated form and the fused form [37]. The acquisition of a second copy may confer a selective advantage to Streptomyces by enhancing the capacity and the effectiveness of protein transport.
Conclusions
Comparative genomic analyses of eleven Streptomyces genomes revealed an abundant repertoire of 761-1258 transporters, belonging to seven transporter classes and 171 transporter families. The powerful transport systems in Streptomyces play critical roles in drug efflux, protein secretion and stress response. A better understanding of transport systems will allow enhanced optimization of production processes for both pharmaceutical and industrial applications of Streptomyces.
Methods
Data
The completed whole genome data of the eleven Streptomyces species (Table 1), including amino acid sequences and functional annotations of all the proteins were downloaded from the NCBI database (http://www.ncbi.nlm.nih.gov/genome/browse/). The transporter classification and amino acid sequences of all classified transporters were downloaded from the TCDB database (http://www.tcdb.org/) [13]. We also collected data from the TransporterDB database [38] (http://www.membranetransport.org/) which included the transporter classification data of S. coelicolor and S. avermitilis, and from the Transporter Inference Parser database [39] (http://biocyc.org/), which identified transporter according to their function annotation and included the relevant data of S. coelicolor, S. avermitilis, S. griseus and S. scabies.
Identification and classification of transporters
The BLASTP search of all the proteins in eleven Streptomyces species versus all the transport proteins in TCDB database was conducted to identify transporters in Streptomyces that are homologs to known and predicted transporters in the TCDB [13, 25]. The threshold for homologous genes was set as follow: E-value ≤ 10-5, similarity ≥ 50 %, and the sequence coverage ≥ 30 %. We classified a Streptomyces transporter based on its homologous gene with known function in the TCDB that had the lowest expected value, the highest similarity score and the highest coverage. The classification of Streptomyces transporters in the TransporterDB and the Transporter Inference Parser, the annotations and the conserved domain information helped to filter false negative and false positive predictions. The Pfam search program based on the Hidden Markov Models (HMMs) (http://pfam.xfam.org/) [40] was used to identify conserved structure domains of Streptomyces transporters, with Pfam GA as the threshold. TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) [26] was used to analyze the transmembrane structures and the number of putative TMSs of Streptomyces transporters.
On the basis of the degree of similarities with known or predicted transporters in the TCDB, as well as the conserved domains and the number and location of TMSs, we further classified the Streptomyces transporters into families and subfamilies of homologous transporters according to the TC system [13]. The TC number generally has five components: V.W.X.Y.Z, representing the transporter class, subclass, family, subfamily and the substrate or range of substrates transported [11, 12]. Most Streptomyces transporters were classified at the transporter family level. The transporters in superfamilies such as ABC and MFS were classified at the subfamily level.
The substrate and transport direction of each Streptomyces transporter was predicted based on homology to functionally characterized transporters in the TCDB. Classification of a putative transporter into a family or subfamily according to the TC system allows for the prediction of substrate types and transport direction with confidence [13, 17, 41].
Phylogenetic analysis of transport protein families
Multiple sequence alignments were obtained using Clustal X 2.1 [42]. Phylogenetic trees were reconstructed using MEGA6 with neighbor-joining (NJ), maximum parsimony (MP) and maximum likelihood (ML) methods [43].
Acknowledgements
We thank the Computational Biology Initiative at UTSA for providing computational support. This work was supported by grants from the National Natural Science Foundation of China (31501021) and the Zhejiang Provincial Natural Sciences Foundation of China (LY15C060001) to ZZ, grants from the National Basic Research Program of China (973 Program, 2012CB721005) and the National Natural Science Foundation of China (30870033) to YQL, grants from the National Institutes of Health (GM100806, AI080579, and GM081068) to YW. ZZ was also supported by a government scholarship from the China Scholarship Council. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Declarations
Publication charges for this article have been funded by the National Natural Science Foundation of China (31501021) to ZZ.
This article has been published as part of BMC Genomics Volume 17 Supplement 7, 2016: Selected articles from the International Conference on Intelligent Biology and Medicine (ICIBM) 2015: genomics. The full contents of the supplement are available online at http://bmcgenomics.biomedcentral.com/articles/supplements/volume-17-supplement-7.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files.
Authors’ contributions
YW, YQL and ZZ conceived and designed the study. ZZ, NS, SW and YW performed data analysis. YW and ZZ drafted the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Additional files
A detailed description of transporters in eleven Streptomyces genomes. The file includs protein IDs, names, annotations, protein lengths, Pfam domains, number of TMSs, and their homologs in TCDB with the BLASTP E-value. (XLSX 918 kb)
The classification of Streptomyces transporters. (XLSX 76 kb)
Contributor Information
Zhan Zhou, Email: zhanzhou@zju.edu.cn.
Ning Sun, Email: 21007079@zju.edu.cn.
Shanshan Wu, Email: swodylm@zju.edu.cn.
Yong-Quan Li, Email: lyq@zju.edu.cn.
Yufeng Wang, Email: yufeng.wang@utsa.edu.
References
- 1.Hopwood DA. Streptomyces in Nature and Medicine: The Antibiotic Makers. New York: Oxford University Press; 2007. [Google Scholar]
- 2.Garrity GM, Lilburn TG, Cole JR, Harrison SH, Euzéby J., B.J. T. Part 10 - The Bacteria: Phylum “Actinobacteria”: Class Actinobacteria. In: Taxonomic Outline of the Bacteria and Archaea. 2007: Release 7.7.: 399-539.
- 3.Anne J, Maldonado B, Van Impe J, Van Mellaert L, Bernaerts K. Recombinant protein production and streptomycetes. J Biotechnol. 2012;158(4):159–167. doi: 10.1016/j.jbiotec.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 4.Li YD, Zhou Z, Lv LX, Hou XP, Li YQ. New approach to achieve high-level secretory expression of heterologous proteins by using Tat signal peptide. Protein Pept Lett. 2009;16(6):706–710. doi: 10.2174/092986609788490096. [DOI] [PubMed] [Google Scholar]
- 5.Zhou Z, Gu J, Li YQ, Wang Y. Genome plasticity and systems evolution in Streptomyces. BMC Bioinformatics. 2012;13(Suppl 10):S8. doi: 10.1186/1471-2105-13-S10-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Z, Gu J, Du YL, Li YQ, Wang Y. The -omics era- toward a systems-level understanding of streptomyces. Curr Genomics. 2011;12(6):404–416. doi: 10.2174/138920211797248556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417(6885):141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 8.Chater KF, Biro S, Lee KJ, Palmer T, Schrempf H. The complex extracellular biology of Streptomyces. FEMS Microbiol Rev. 2010;34(2):171–198. doi: 10.1111/j.1574-6976.2009.00206.x. [DOI] [PubMed] [Google Scholar]
- 9.Widdick DA, Dilks K, Chandra G, Bottrill A, Naldrett M, Pohlschroder M, Palmer T. The twin-arginine translocation pathway is a major route of protein export in Streptomyces coelicolor. Proc Natl Acad Sci U S A. 2006;103(47):17927–17932. doi: 10.1073/pnas.0607025103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morosoli R, Shareck F, Kluepfel D. Protein secretion in streptomycetes. FEMS Microbiol Lett. 1997;146(2):167–174. doi: 10.1111/j.1574-6968.1997.tb10188.x. [DOI] [PubMed] [Google Scholar]
- 11.Busch W, Saier MH., Jr The transporter classification (TC) system, 2002. Crit Rev Biochem Mol Biol. 2002;37(5):287–337. doi: 10.1080/10409230290771528. [DOI] [PubMed] [Google Scholar]
- 12.Busch W, Saier MH., Jr The IUBMB-endorsed transporter classification system. Mol Biotechnol. 2004;27(3):253–262. doi: 10.1385/MB:27:3:253. [DOI] [PubMed] [Google Scholar]
- 13.Saier MH, Jr, Reddy VS, Tamang DG, Vastermark A. The transporter classification database. Nucleic Acids Res. 2014;42(Database issue):D251–258. doi: 10.1093/nar/gkt1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paulsen IT, Nguyen L, Sliwinski MK, Rabus R, Saier MH., Jr Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J Mol Biol. 2000;301(1):75–100. doi: 10.1006/jmbi.2000.3961. [DOI] [PubMed] [Google Scholar]
- 15.Kumar U, Saier MH., Jr Comparative genomic analysis of integral membrane transport proteins in ciliates. J Eukaryot Microbiol. 2015;62(2):167–187. doi: 10.1111/jeu.12156. [DOI] [PubMed] [Google Scholar]
- 16.Paparoditis P, Vastermark A, Le AJ, Fuerst JA, Saier MH., Jr Bioinformatic analyses of integral membrane transport proteins encoded within the genome of the planctomycetes species, Rhodopirellula baltica. Biochim Biophys Acta. 2014;1838(1 Pt B):193–215. doi: 10.1016/j.bbamem.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Getsin I, Nalbandian GH, Yee DC, Vastermark A, Paparoditis PC, Reddy VS, Saier MH., Jr Comparative genomics of transport proteins in developmental bacteria: Myxococcus xanthus and Streptomyces coelicolor. BMC Microbiol. 2013;13:279. doi: 10.1186/1471-2180-13-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbe V, Bouzon M, Mangenot S, Badet B, Poulain J, Segurens B, Vallenet D, Marliere P, Weissenbach J. Complete genome sequence of Streptomyces cattleya NRRL 8057, a producer of antibiotics and fluorometabolites. J Bacteriol. 2011;193(18):5055–5056. doi: 10.1128/JB.05583-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang XJ, Yan YJ, Zhang B, An J, Wang JJ, Tian J, Jiang L, Chen YH, Huang SX, Yin M, et al. Genome sequence of the milbemycin-producing bacterium streptomyces bingchenggensis. J Bacteriol. 2010;192(17):4526–4527. doi: 10.1128/JB.00596-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohnishi Y, Ishikawa J, Hara H, Suzuki H, Ikenoya M, Ikeda H, Yamashita A, Hattori M, Horinouchi S. Genome sequence of the streptomycin-producing microorganism streptomyces griseus IFO 13350. J Bacteriol. 2008;190(11):4050–4060. doi: 10.1128/JB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Ōmura S. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol. 2003;21(5):526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 22.Bignell DRD, Seipke RF, Huguet-Tapia JC, Chambers AH, Parry RJ, Loria R. Streptomyces scabies 87-22 contains a coronafacic acid-like biosynthetic cluster that contributes to plant-microbe interactions. Mol Plant Microbe Interact. 2010;23(2):161–175. doi: 10.1094/MPMI-23-2-0161. [DOI] [PubMed] [Google Scholar]
- 23.Wu H, Qu S, Lu CY, Zheng HJ, Zhou XF, Bai LQ, Deng ZX. Genomic and transcriptomic insights into the thermo-regulated biosynthesis of validamycin in Streptomyces hygroscopicus 5008. BMC Genomics. 2012;13:337. doi: 10.1186/1471-2164-13-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yaxley AM. Study of the complete genome sequence of Streptomyces scabies (or scabiei) 87.22. University of Warwick, Coventry, UK; 2009.
- 25.Saier MH, Jr, Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34(Database issue):D181–186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 27.Saier MH., Jr Tracing pathways of transport protein evolution. Mol Microbiol. 2003;48(5):1145–1156. doi: 10.1046/j.1365-2958.2003.03499.x. [DOI] [PubMed] [Google Scholar]
- 28.Lam VH, Lee JH, Silverio A, Chan H, Gomolplitinant KM, Povolotsky TL, Orlova E, Sun EI, Welliver CH, Saier MH., Jr Pathways of transport protein evolution: recent advances. Biol Chem. 2011;392(1-2):5–12. doi: 10.1515/bc.2011.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendez C, Salas JA. The role of ABC transporters in antibiotic-producing organisms: drug secretion and resistance mechanisms. Res Microbiol. 2001;152(3-4):341–350. doi: 10.1016/S0923-2508(01)01205-0. [DOI] [PubMed] [Google Scholar]
- 30.Saidijam M, Benedetti G, Ren Q, Xu Z, Hoyle CJ, Palmer SL, Ward A, Bettaney KE, Szakonyi G, Meuller J, et al. Microbial drug efflux proteins of the major facilitator superfamily. Curr Drug Targets. 2006;7(7):793–811. doi: 10.2174/138945006777709575. [DOI] [PubMed] [Google Scholar]
- 31.Tomii K, Kanehisa M. A comparative analysis of ABC transporters in complete microbial genomes. Genome Res. 1998;8(10):1048–1059. doi: 10.1101/gr.8.10.1048. [DOI] [PubMed] [Google Scholar]
- 32.Wang B, Dukarevich M, Sun EI, Yen MR, Saier MH., Jr Membrane porters of ATP-binding cassette transport systems are polyphyletic. J Membr Biol. 2009;231(1):1–10. doi: 10.1007/s00232-009-9200-6. [DOI] [PubMed] [Google Scholar]
- 33.De Buck E, Lammertyn E, Anne J. The importance of the twin-arginine translocation pathway for bacterial virulence. Trends Microbiol. 2008;16(9):442–453. doi: 10.1016/j.tim.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 34.Joshi MV, Mann SG, Antelmann H, Widdick DA, Fyans JK, Chandra G, Hutchings MI, Toth I, Hecker M, Loria R, et al. The twin arginine protein transport pathway exports multiple virulence proteins in the plant pathogen Streptomyces scabies. Mol Microbiol. 2010;77(1):252–271. doi: 10.1111/j.1365-2958.2010.07206.x. [DOI] [PubMed] [Google Scholar]
- 35.Driessen AJ, Fekkes P, van der Wolk JP. The Sec system. Curr Opin Microbiol. 1998;1(2):216–222. doi: 10.1016/S1369-5274(98)80014-3. [DOI] [PubMed] [Google Scholar]
- 36.Rigel NW, Braunstein M. A new twist on an old pathway--accessory secretion systems. Mol Microbiol. 2008;69(2):291–302. doi: 10.1111/j.1365-2958.2008.06294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Z, Li Y, Sun N, Sun Z, Lv L, Wang Y, Shen L, Li YQ. Function and evolution of two forms of SecDF homologs in Streptomyces coelicolor. PLoS One. 2014;9(8):e105237. doi: 10.1371/journal.pone.0105237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren Q, Chen K, Paulsen IT. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res. 2007;35(Database issue):D274–279. doi: 10.1093/nar/gkl925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee TJ, Paulsen I, Karp P. Annotation-based inference of transporter function. Bioinformatics. 2008;24(13):i259–267. doi: 10.1093/bioinformatics/btn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saier MH., Jr A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol Mol Biol Rev. 2000;64(2):354–411. doi: 10.1128/MMBR.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 43.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files.


