Abstract
BACKGROUND
Subfertility affects approximately 15% of all couples, and a severe male factor is identified in 17% of these couples. While the etiology of a severe male factor remains largely unknown, prior gonadotoxic treatment and genomic aberrations have been associated with this type of subfertility. Couples with a severe male factor can resort to ICSI, with either ejaculated spermatozoa (in case of oligozoospermia) or surgically retrieved testicular spermatozoa (in case of azoospermia) to generate their own biological children. Currently there is no direct treatment for azoospermia or oligozoospermia. Spermatogonial stem cell (SSC) autotransplantation (SSCT) is a promising novel clinical application currently under development to restore fertility in sterile childhood cancer survivors. Meanwhile, recent advances in genomic editing, especially the clustered regulatory interspaced short palindromic repeats-associated protein 9 (CRISPR-Cas9) system, are likely to enable genomic rectification of human SSCs in the near future.
OBJECTIVE AND RATIONALE
The objective of this review is to provide insights into the prospects of the potential clinical application of SSCT with or without genomic editing to cure spermatogenic failure and to prevent transmission of genetic diseases.
SEARCH METHODS
We performed a narrative review using the literature available on PubMed not restricted to any publishing year on topics of subfertility, fertility treatments, (molecular regulation of) spermatogenesis and SSCT, inherited (genetic) disorders, prenatal screening methods, genomic editing and germline editing. For germline editing, we focussed on the novel CRISPR-Cas9 system. We included papers written in English only.
OUTCOMES
Current techniques allow propagation of human SSCs in vitro, which is indispensable to successful transplantation. This technique is currently being developed in a preclinical setting for childhood cancer survivors who have stored a testis biopsy prior to cancer treatment. Similarly, SSCT could be used to restore fertility in sterile adult cancer survivors. In vitro propagation of SSCs might also be employed to enhance spermatogenesis in oligozoospermic men and in azoospermic men who still have functional SSCs albeit in insufficient numbers. The combination of SSCT with genomic editing techniques could potentially rectify defects in spermatogenesis caused by genomic mutations or, more broadly, prevent transmission of genomic diseases to the offspring. In spite of the promising prospects, SSCT and germline genomic editing are not yet clinically applicable and both techniques require optimization at various levels.
WIDER IMPLICATIONS
SSCT with or without genomic editing could potentially be used to restore fertility in cancer survivors to treat couples with a severe male factor and to prevent the paternal transmission of diseases. This will potentially allow these couples to have their own biological children. Technical development is progressing rapidly, and ethical reflection and societal debate on the use of SSCT with or without genomic editing is pressing.
Keywords: spermatogonial stem cell autotransplantation, male infertility, male reproductive disorders, germline editing, CRISPR-Cas9
Introduction
Subfertility, defined as a failure to achieve a clinical pregnancy after at least 12 months of regular unprotected coitus (Zegers-Hochschild et al., 2009), affects ~15% of all couples. In ~17% of these couples, a severe male factor, defined as a total motile sperm count below 3 × 106, is present (van der Steeg et al., 2007). A severe male factor may present as azoospermia (complete absence of spermatozoa in the ejaculate) or oligozoospermia (low number of spermatozoa in the ejaculate). In case of a severe male factor, a patient’s own biological children can be generated by ICSI with either ejaculated spermatozoa (in case of oligozoospermia) or surgically retrieved testicular spermatozoa by means of testicular sperm extraction (TESE) (in case of azoospermia). Currently, no direct clinical treatment exists for a severe male factor.
A severe male factor is typically caused by a disturbance during spermatogenesis. Spermatogenesis occurs in the seminiferous tubules inside the testis. Essential to this process are spermatogonial stem cells (SSCs) that maintain a perfect balance between self-renewal and differentiation into mature sperm, thereby sustaining fertility throughout a man’s life. Both oligozoospermia and azoospermia can be due to a reduction in SSC numbers throughout the testis as seen in testicular biopsies taken from these men (Takagi et al., 2001; Yakirevich et al., 2003; Hentrich et al., 2011). In some of these men, the testicular biopsies display focal spermatogenesis, where sperm is only produced in a subset of seminiferous tubules. Although the production of spermatozoa is limited due to the low number of SSCs, the existing SSCs are still functional and capable of continuous self-renewal and differentiation (Silber, 2000).
While the etiology of a severe male factor remains largely unknown, a few genetic defects (Visser and Repping, 2010; Tuttelmann et al., 2011; Krausz and Chianese, 2014), including structural and numerical chromosomal abnormalities (de Kretser, 1997; Oates, 2008) and Y-chromosome deletions (Kuroda-Kawaguchi et al., 2001; Noordam and Repping, 2006), have been identified to associate with a severe male factor-induced subfertility. In addition, male subfertility can be attributed to previous gonadotoxic treatment (Meistrich, 2013). Both chemotherapy and radiotherapy are known to destroy the SSC pool, resulting in oligozoospermia or azoospermia in a large proportion of cancer survivors (Howell and Shalet, 2005).
A possible future application that could directly treat a severe male factor might be autotransplantation of SSCs. Transplantation of SSCs that are stored prior to cancer treatment is proposed as a means to restore fertility in childhood cancer survivors. This application involves transplantation of SSCs into the seminiferous tubules via the efferent duct or rete testis (Brinster, 2007; Dores et al., 2012). Upon SSC transplantation (SSCT), SSCs migrate to the basement membrane of recipient seminiferous tubules, colonize the epithelium and undergo self-renewal and differentiation so that permanent spermatogenesis is established. Therefore, SSCT should allow natural conception without further fertility treatment, making SSCT a direct treatment for a severe male factor.
In case the severe male factor is due to a genomic mutation, SSCT can only be successful if it is combined with correction of the mutation. Recent advances in genomic editing, especially those with the clustered regulatory interspaced short palindromic repeats-associated protein 9 (CRIPSR-Cas9) system, can allow for rapid, easy and highly efficient genetic alterations of a wide array of cell types and organisms including human cells (Sander and Joung, 2014). Thus, if genomic editing is combined with SSCT, it would in principle allow those suffering from spermatogenic failure to have their own biological children. Furthermore, SSCT with genomic editing may be used to prevent paternal transmission of genomic diseases.
In this review, the clinical prospects of SSCT with or without genomic editing for male adult cancer survivors, for men with oligozoospermia and azoospermia and for carriers of genomic diseases are discussed (Fig. 1).
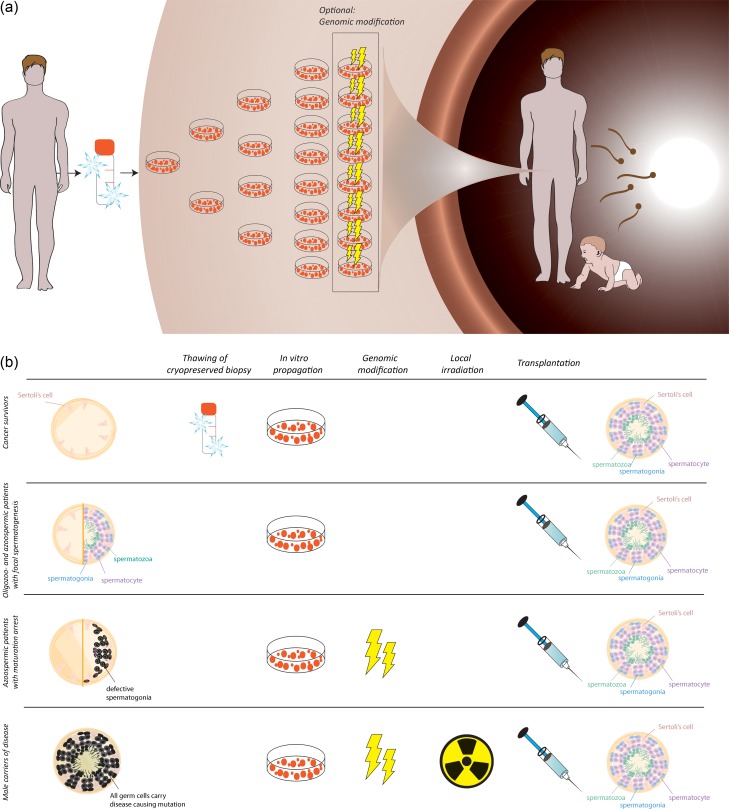
Figure 1.
A schematic depiction of the proposed SSCT therapy. (a) A testicular biopsy is taken from the patient and cryopreserved. From the biopsy, SSCs are propagated in vitro, during which endogenous genomic defects may be repaired. Propagated SSCs are subsequently autotransplanted to the testis and then colonize the testis and restore spermatogenesis, enabling the patient to father a child without additional therapy. (b) The testicular histology of men with a severe male factor in different patient groups. The histology may show various phenotypes throughout the testis. For male (childhood) cancer survivors, a biopsy is cryopreserved prior to cancer therapy. Hence, thawing of the cryopreserved biopsy is indispensable to the treatment. In vitro propagation is needed for all patient groups, while genomic modification is only needed for those with a maturation arrest or carriers of diseases. In male carriers of diseases with full spermatogenesis, all germ cells including spermatids express the mutated genes, and local irradiation of the testis is required prior to transplantation to remove the mutated endogenous spermatids. After (genomically modified) SSCT, testis histology should, in theory, restore to full spermatogenesis.
Methods
We performed a narrative review using the literature available on PubMed not restricted to any publishing year on topics of subfertility, fertility treatments, (molecular regulation of) spermatogenesis and SSCT, inherited (genetic) disorders, prenatal screening methods, genomic editing and germline editing. For germline editing, we focussed on the novel clustered regulatory interspaced short palindromic repeats-associated protein 9 (CRISPR-Cas9) system. We included papers written in English only.
Clinical prospects of SSCT to restore spermatogenesis
The first successful SSCT was reported in 1994 and resulted in fertility restoration in sterile recipient mice upon transplantation of donor-derived SSCs (Brinster and Zimmermann, 1994). Subsequent studies substantiated the findings by showing restoration of spermatogenesis after SSCT in mice (Kanatsu-Shinohara et al., 2003; Goossens et al., 2009; Kubota et al., 2009; Yuan et al., 2009; Wu et al., 2012) and other animal models (Honaramooz et al., 2002; Izadyar et al., 2003; Ryu et al., 2007; Nobrega et al., 2010; Kawasaki et al., 2012), including non-human primates (Schlatt et al., 1999; Hermann et al., 2012). In mice, SSCs can be transplanted into the recipient seminiferous tubules by injecting the efferent ducts, the rete testis or directly injecting seminiferous tubules (Ogawa et al., 1997). Injection via the efferent ducts is mostly employed in rodents (Gonzalez and Dobrinski, 2015). Yet, due to the anatomic and size difference in testes, ultrasound-guided transplantation via the intra-rete testis has been proved to be the least invasive and most efficient and successful protocol for non-rodent species such as pigs (Honaramooz et al., 2002), bulls (Izadyar et al., 2003), primates (Schlatt et al., 1999; Schlatt, 2002; Hermann et al., 2012) and ex vivo human testes (Schlatt et al., 1999; Ning et al., 2012). Crucial to this procedure is the injection of cells via the rete testis, and optimization is needed in this respect to augment success rates (Jahnukainen et al., 2011). In addition, the quality and quantity of SSC niches in the recipient testis makes a difference to the success of autologous transplantation (Jahnukainen et al., 2006). To date, multiple papers describe the generation of SSCT-induced healthy offspring in rodents (Goossens et al., 2009; Yuan et al., 2009; Lee et al., 2009; Kubota et al., 2009; Goossens et al., 2010; Wu et al., 2012) and non-rodent large animal models such as goats and sheep (Honaramooz et al., 2003; Herrid et al., 2009; Zheng et al., 2014). A breakthrough was recently achieved by Hermann et al. (2012), who successfully performed autologous and allogeneic transplantation of SSCs from rhesus monkey, leading to the generation of donor-derived sperm in both cases. Subsequently, ICSI was conducted to fertilize oocytes, and embryos with donor paternal origin were finally produced. This demonstration in primates provides prospects for future clinical translation of SSCT.
For humans, SSCT has been proposed as a future clinical application for those men with the risk of complete germ cell depletion that have no option to cryopreserve sperm, in particular, prepubertal cancer patients (Brinster, 2007; Goossens et al., 2013; Struijk et al., 2013; Sadri-Ardekani and Atala, 2014) and, theoretically, even in case of focal spermatogenesis (Nickkholgh et al., 2015). Prepubertal cancer patients especially rely on SSCT because spermatogenesis is not initiated yet, which means that semen cryopreservation prior to treatment is not an option. By opting for storage of a testicular biopsy before cancer therapy, SSCT can be applied later in life when the patient is cured and expresses the wish to have children (Struijk et al., 2013; Picton et al., 2015). In this case, cryopreservation of biopsied testicular tissues constitutes an important part of fertility preservation (Schlatt et al., 2009). Currently the most popular avenue to preserve pieces of testicular tissues is through controlled slow freezing (Keros et al., 2005, 2007; Wyns et al., 2007, 2011; Gassei and Orwig, 2016; Ginsberg et al., 2010), with the addition of cryoprotective agents such as dimethyl sulphoxide or ethylene glycol with or without sucrose (Keros et al., 2005, 2007; Kvist et al., 2006; Wyns et al., 2007, 2008; Poels et al., 2014; Picton et al., 2015). Vitrification instead of controlled slow freezing has also been tested with positive outcomes (Curaba et al., 2011; Baert et al., 2013; Poels et al., 2013). Nevertheless, protocols for tissue preservation require further optimization to maximize the viability of post-thawed human testicular tissues (Picton et al., 2015).
Following successful cryopreservation, conventional SSCT can be performed at a later stage, which involves the transplantation of the patient’s SSCs into one’s own testis (i.e. autotransplantation). After autotransplantation, SSCs migrate to the basement membrane of the recipient seminiferous epithelium, from where they reinitiate spermatogenesis (Fig. 1). In this way, the patient can (re)gain his fertility and generate their own biological children by natural conception, as shown in animal experiments (Honaramooz et al., 2003; Goossens et al., 2009).
The efficiency of SSCT is highly dependent on the number of transplanted SSCs (Dobrinski et al., 1999; Nagano, 2003). Previous reports suggest that SSCT is unlikely to become clinically applicable without an approach to successfully expanding human SSCs in vitro (Jahnukainen et al., 2006, 2011). In vitro expansion of SSCs has been successfully demonstrated in studies using rodent SSCs (Kanatsu-Shinohara et al., 2003, 2005, 2008; Kubota et al., 2004; Ryu et al., 2005). Even after 2 years of culture, the SSCs retained the capacity to colonize the basal membrane of seminiferous tubules and could further develop into healthy and functional sperm (Kanatsu-Shinohara et al., 2005). In human trials, SSCs can first be isolated from biopsies and then subjected to primary culture to increase their number for the sake of SSCT. Several culture systems have been established for human adult SSCs (Sadri-Ardekani et al., 2009; Lim et al., 2010; Kossack et al., 2013; Akhondi et al., 2013; Guo et al., 2015), as well as for human prepubertal SSCs (Sadri-Ardekani et al., 2011). Presently (xeno)transplantation is the well-acknowledged and only available assay for functionality of human SSCs. It is shown that in vitro propagated human SSCs are capable of migrating to the niche at the basal membrane of the seminiferous tubules upon xenotransplantation into the testis of immunodeficient mice (Sadri-Ardekani et al., 2009, 2011), indicating that these cultured testicular cells still have SSC capabilities. Nevertheless, human spermatogenesis is not initiated in the mouse model. This is not unexpected given the large phylogenetic distance between mice and humans, and because of this, the murine niche cannot support full development of human SSCs into sperm (Nagano et al., 2002). Despite that, by comparing the numbers of migrated SSCs in the recipient testis after xenotransplantation of early and late passages from primary human testicular cultures, it has been established that adult as well as prepubertal SSCs can indeed proliferate in culture (Sadri-Ardekani et al., 2009, 2011). Thus, the successful expansion of human SSCs in culture paves the way for clinical applications of SSCT.
Once SSCT is clinically implemented for childhood cancer survivors, other patient groups that have the risk of becoming subfertile or that suffer from subfertility might also benefit from this treatment. In a recent study in azoospermic men, it was shown that these men hold a positive attitude toward SSCT, which was persistent even after acknowledging that a new experimental technique might have some risks for themselves or their offspring (Hendriks et al., 2014).
SSCT to restore fertility in adult cancer survivors
Due to the high sensitivity of spermatogonia to DNA damage (van der Meer et al., 1992; Meistrich, 2013), spermatogonial apoptosis and subsequent subfertility is a major side-effect of most cancer treatments. The chance of becoming azoospermic temporarily or permanently after cancer therapy is highly dependent on the type and dosage of the treatment (Howell and Shalet, 2005). A recent study has shown that 25% of patients who underwent chemotherapy were azoospermic after 7–218 months (median: 40 months), with the highest chance of azoospermia in Hodgkin disease survivors (63%) (Tomlinson et al., 2015).
Cryopreservation of semen prior to treatment is historically an effective and inexpensive way to preserve fertility in adult male cancer patients. The cryopreserved semen can be used to achieve a pregnancy by cervical or intrauterine insemination (IUI) or, in case the quality of the semen is too low, by IVF or ICSI treatment. However, cancer patients often show decreased fertility at the time of cancer diagnosis. In fact, Ragni et al. (2003) report that 12% of patients who wish to store their semen are azoospermic at the time of cancer diagnosis. Moreover, semen cryopreservation severely reduces sperm motility (Keel and Webster, 1989; Boitrelle et al., 2012), sperm count (Keel and Webster, 1989) and DNA integrity (Valcarce et al., 2013). Hampered sperm motility significantly decreases the chance of live birth after IUI (Hendin et al., 2000), and therefore the majority of patients who make use of cryopreserved spermatozoa have to resort to IVF/ICSI. However, IVF/ICSI is costly and burdensome, and requires ovarian hyperstimulation of the healthy female to retrieve oocytes, which is not risk-free and leads to onset of the ovarian hyperstimulation syndrome in 1–8% of stimulated women (Mathur et al., 2000; Gelbaya, 2010). In addition, it is well known that IVF/ICSI is associated with adverse short-term outcomes including preterm birth, lower birthweight and a higher prevalence of birth defects (Helmerhorst et al., 2004; Hansen et al., 2013).
Although SSCT does require a testis biopsy and treatment of the affected male, it would avoid IVF/ICSI treatment of the healthy female partner since natural conception might be feasible after SSCT. In the future, SSCT may therefore become an appealing alternative for fertility preservation in adult cancer patients in a similar way as described for prepubertal cancer patients by cryopreserving a testis biopsy before onset of cancer treatment.
SSCT to enhance fertility in oligozoospermic and azoospermic men
Oligozoospermic and azoospermic men are capable of fertilization if they produce morphologically normal sperm in the testis. In couples where the male is oligozoospermic or azoospermic, ICSI is used to achieve fertilization with ejaculated or surgically retrieved spermatozoa, respectively. However, since oligozoospermia and azoospermia may be caused by a reduction in functional SSCs (Takagi et al., 2001; Yakirevich et al., 2003; Hentrich et al., 2011), a simple increase in SSCs may restore spermatogenesis and fertility in these men. This is especially relevant to the patients who display focal spermatogenesis at the histological level, in which some tubules show normal spermatogenesis, while others display Sertoli cell-only syndrome. The tubules with normal spermatogenesis harbor functional SSCs, and the hypothesis is that if these spermatogonia are propagated in vitro and transplanted back into the testis, they will repopulate the empty tubules and initiate spermatogenesis in these tubules. A recent article describes the characteristics of cultured SSCs deriving from patients who suffer from focal spermatogenesis due to a deletion of the azoospermia factor c (AZFc) region on the Y chromosome (Nickkholgh et al., 2015). In vitro propagated SSCs from these men with focal spermatogenesis behaved similarly during culture and showed comparable gene expression of key spermatogonial markers when compared to SSCs originating from healthy counterparts with normal spermatogenesis. These results suggest that patients with oligozoospermia or azoospermia as a result of focal spermatogenesis might also benefit from propagation and transplantation of their own SSCs. Yet, this hypothesis needs to be demonstrated in clinical trials. However, one drawback that should be accounted for is that if a mutation is present on the Y chromosome, as in the case of men with AZFc deletions, male offspring of these men will harbor the same mutation and are likely to be oligozoospermic or azoospermic too (Page et al., 1999). It makes sense that this also holds true for mutations on other chromosomes. However, this problem also arises with other contemporary fertility treatment, such as IVF/ICSI with or without TESE. In order to prevent transmission of these genetic aberrations, additional measures have to be taken.
Clinical prospects of SSCT and germline genomic editing
While the use of SSCT for adult cancer patients or oligozoospermic and azoospermic patients that display focal spermatogenesis seems rather straightforward, azoospermic patients suffering from a maturation arrest in spermatogenesis cannot directly benefit from SSCT because transplantation of the patient’s SSCs would result in the same arrested phenotype and not cure their spermatogenic failure (Fig. 1). However, in some cases, the maturation arrest may be attributed to genetic mutations or arises from epigenetic disturbances. Repair of these disorders in SSCs before SSCT would theoretically restore spermatogenesis and subsequent fertility in these patients and in addition prevent the transmission of the mutation to the offspring. The fact that human SSCs can propagate in culture for extended periods of time enables (epi)genetic editing prior to transplantation.
With recent advances in the field of genomic editing, in particular the use of novel techniques such as the CRISPR-Cas9 system, SSCT with genetically modified SSCs has become feasible (Fanslow et al., 2014; Wu et al., 2015; Chapman et al., 2015; Sato et al., 2015). In addition, recent work has shown that epigenetic editing is also possible with CRIPSR-Cas9 (Hilton et al., 2015).
CRISPR-Cas9 to genomically modify SSCs
Traditional genome editing mainly relies on homologous recombination in embryonic stem cells (ESCs). Over the last decade, novel genome editing platforms such as zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and most recently the CRISPR-Cas9 system have been developed. These techniques are based on engineered nucleases that can cause a double-strand break of DNA, and are much less laborious and time-consuming compared to traditional strategies. With the emergence of these novel techniques (Table 1), the majority of cell types, including SSCs, can now be targeted.
Table I.
An overview of different genome editing techniques.
| Genome editing systems | Homologous recombination without engineered nucleases | Conventional engineered nucleases (ZFNs/TALENs) | Novel engineered nucleases (CRISPR-Cas9) | Novel engineered nucleases (GeCKOa) |
|---|---|---|---|---|
| Target cells | Mostly ESCs | Most cell types | Most cell types | Most cell types |
| Approaches to delivering targeting vectors | Non-viral transfection/viral transduction/microinjection | Non-viral transfection/viral transduction/microinjection | Non-viral transfection/viral transduction/microinjection | Lentiviral transduction |
| Technical difficulty | High | High | Low | Intermediate |
| Targeting efficiency | Low | Variable | Generally high | High |
| Off-target effects | Low | Variable | Generally low | Variable |
| Possible to target a large scale of genes in parallel? | No | No | No | Yes |
| Suitable for the clinic? | No, due to low efficiency and the typical requirement of ESCs | Not optimal | Yes | Currently not due to lentiviral transduction |
aGeCKO, genome-scale CRISPR knockout; ESC, embryonic stem cell; ZFN, zinc-finger nuclease; TALEN, transcription activator-like effector nuclease; CRISPR-Cas9, clustered regulatory interspaced short palindromic repeats-associated protein 9.
Prior to clinical application of genomically modified SSCs, the safety of the patient needs to be guaranteed. In a clinical setting, off-target effects are not acceptable, and technical simplicity would be desirable. ZFNs and TALENs were utilized to successfully manipulate mouse SSCs (Fanslow et al., 2014). However, the required design and engineering of nucleases necessary for both ZFNs and TALENs is strenuous and of high technical difficulty. A better alternative to genetically modify SSCs seems to be the unprecedentedly simple CRISPR-Cas9 system (Fig. 2), and articles describing successful manipulation of rodent SSCs by way of CRISPR are now available (Wu et al., 2015; Chapman et al., 2015; Sato et al., 2015). The CRISPR-Cas9 system not only bypasses the engineering of nucleases but also generates far less off-target effects compared with ZFNs (Ul Ain et al., 2015). According to a recent report, with genome-wide screens, no obvious off-target genetic or epigenetic changes could be detected in a large SSCT experiment involving CRISPR-Cas9-mediated gene targeting and transplantation of modified mouse SSCs (Wu et al., 2015).
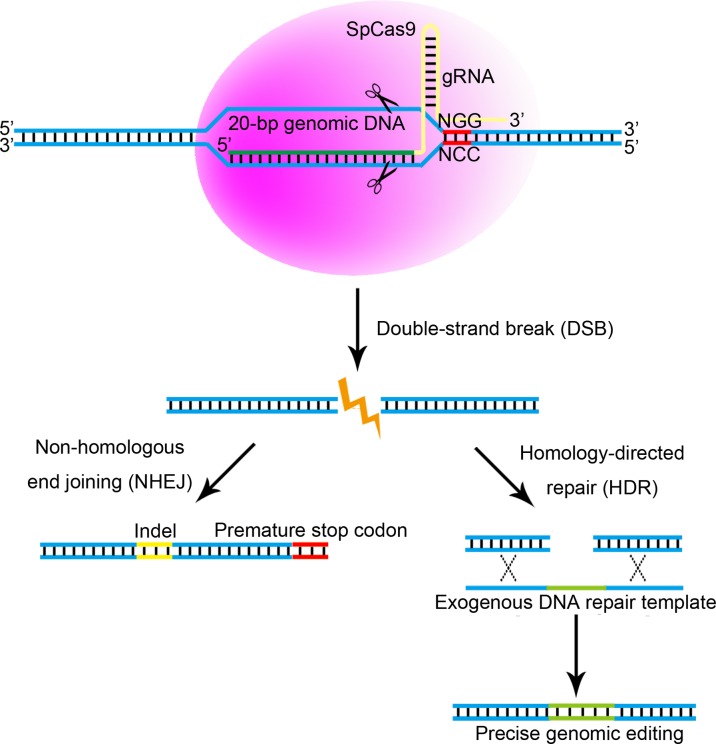
Figure 2.
The CRISPR-Cas9 system. The Type II Streptococcus pyogenes clustered regulatory interspaced short palindromic repeats-associated protein 9 (CRISPR-Cas9) (SpCas9) system, which is the simplest and most extensively used CRISPR-Cas9 technology, is based on a guide-RNA (gRNA) containing a specific 20 bp sequence to guide the DNA endonuclease Cas9 to a complementary target DNA sequence in the genome where it induces a DNA double-strand break (DSB). The 20-bp target genomic DNA must be upstream of a specific sequence (5′-NGG, where N represents a random nucleotide). The Cas9-induced DSB occurs ~3-bp upstream of the 5′-NGG, and can in theory be induced in any 20-bp genomic DNA sequence flanking 5′-NGG. The Cas9-induced DSB will then be repaired by either homology-directed repair (HDR), which can occur with the presence of DNA repair templates, or by non-homologous end joining (NHEJ). The error-prone NHEJ creates insertions/deletions (indels) around the DSB point. Indels, especially when occurring in early coding exons, can cause loss of gene function (gene knockout) by causing a frame shift that can lead to formation of a pre-mature stop codon. In contrast, HDR uses a template sequence for very precise repair of the DSB. Exogenous DNA repair templates (with the required sequences placed between homology arms) can be provided to the cells together with other components of the CRISPR-Cas9 system to create specific indels or modifications at target genomic loci. Thus, the CRISPR-Cas9 system can be used to insert sequences or correct disease-causing mutations in a very accurate way.
Curing spermatogenic failure by transplantation of genetically modified SSCs
A proportion of infertile men have non-obstructive azoospermia as a result of spermatogenic arrest. However, in 41% of these patients a few germ cells escape the arrest and form elongated spermatids, which can be extracted from testicular tissue by means of TESE and used for fertilization in an ICSI procedure (Vloeberghs et al., 2015). When no sperm is found during this procedure, no treatment options are currently available. Fortunately, Lim et al. (2010) have shown that spermatogonia from patients suffering from non-obstructive azoospermia due to a maturation arrest are able to proliferate in their long-term culture system in a similar manner as men with obstructive azoospermia. Therefore, SSCs might be used in future SSCT as a valid option to treat these men.
Very recently, Yuan et al. (2015) employed TALENs to successfully rectify a point mutation in the mouse c-kit gene that blocks spermatogonial differentiation, and after correction spermatogenesis could be rescued, for the first time demonstrating that spermatogenic failure-related genetic defects can be corrected by genome editing platforms. Thus, it makes sense that after CRISPR-Cas9-mediated correction of the genetic defects responsible for spermatogenic failure, SSCs from azoospermic men can subsequently be used for SSCT, offering men with spermatogenic failure a patient-tailored treatment option. Besides, in small cohorts of azoospermic men, various single nucleotide polymorphisms (SNPs) associated with arrests during spermatogenesis have been identified (Aston et al., 2010; Teng et al., 2012; Parada-Bustamante et al., 2012; He et al., 2012; Hu et al., 2012), forming additional candidate targets for genetic modification of SSCs. A recent article reports the use of CRISPR-Cas9 to interrogate male infertility-related SNPs in mice (Singh and Schimenti, 2015). In addition, the recently developed genome-scale CRISPR knockout (GeCKO) enables the targeting of a variety of genes in parallel (Shalem et al., 2014; Sanjana et al., 2014). Because infertility is often not believed to be a monogenic disorder but is rather thought to be caused by a spectrum of genes (Jan et al., 2012), the GeCKO system could serve as a prospective platform for gene therapy of such patients. Hence, when the infertility-causing genetic mutations are known, SSCs could first be isolated from testicular biopsies and propagated in vitro to increase their number. Subsequently, the propagated SSCs could be co-transfected with CRISPR-related vectors (to cut DNA) and exogenous DNA repair templates specific for the mutations (to induce homology-directed repair, Fig. 2) in a patient-specific manner. Finally, after selection and whole genome (epi)genetic off-target analysis, the modified SSCs can be autotransplanted into the testis to initiate spermatogenesis and produce corrected sperm.
Preventing diseases in offspring by transplantation of genetically modified SSCs
Carriers of inherited genetic diseases, albeit often fertile, have to make important decisions when it comes to reproduction. Couples can opt to remain childless to prevent diseases in their children, opt for adoption or resort to a germ cell donor. Alternatively, these couples can attempt to have their own biological children and detect whether their prospective children are carriers of the disease via prenatal testing during pregnancy. In case the fetus is a carrier of the disease, the parents have to make the emotionally laden decision whether to terminate their pregnancy. Couples can also opt for PGD as a preventive measure for the birth of a child with a genetic defect. PGD is well established for monogenic diseases such as cystic fibrosis (Handyside et al., 1992), beta thalassemia (Kuliev et al., 1999) and Huntington’s disease (Sermon et al., 1998). In PGD, couples undergo IVF treatment in which a single cell is aspirated from each embryo at the 6–8 cell stage to perform subsequent genetic testing for high-risk disease alleles. Only unaffected embryos are transferred to prevent the birth of children with these severe genetic diseases.
Even though PGD provides a solution for couples at risk for transmitting a genetic disease, IVF treatment of the women, including ovarian hyperstimulation, is indispensable in the process of PGD. Moreover, many embryos are created, while only a few will be used to induce pregnancy.
Currently there is no way to prevent genetic diseases in the offspring without creating affected embryos. Nevertheless, if the prospective father is the carrier of a disease allele, the disease-causing mutation can theoretically be corrected in isolated SSCs during in vitro culture. Subsequently, transplantation of the modified SSCs would result in genetically normal sperm and therefore prevent transfer of the disease allele to the next generations. Additionally, it would enable male carriers at risk for transmitting genetic diseases to naturally conceive a healthy child without IVF or prenatal genetic testing.
Inspiringly, CRISPR-Cas9 has been shown to successfully repair mutations in disease-causing genes in different species and cell types. In mice, mutations in the Crygc gene (which causes cataracts) (Wu et al., 2013), dystrophin gene (which causes Duchenne muscular dystrophy, DMD) (Long et al., 2014) and a Fah mutation in hepatocytes (Yin et al., 2014) have been repaired by the CRISPR-Cas9 system. In human trials, the CRISPR-Cas9 system has been used to precisely correct the hemoglobin beta and dystrophin gene, in β-thalassemia (Xie et al., 2014) and DMD patient-induced pluripotent stem cells (Li et al., 2015), respectively. Another report describes successful repair of the cystic fibrosis transmembrane conductor receptor locus in cultured intestinal stem cells from patients with cystic fibrosis (Schwank et al., 2013). These reports raise the possibility that CRISPR-Cas9 could be used to repair inheritable mutations through the germline.
Interestingly, a recent article reports successful genome editing of mouse SSCs with the CRISPR-Cas9 system (Wu et al., 2015). Transplantation of the genetically modified SSCs led to fertile offspring, in which a Crygc mutation causing cataracts was corrected. To our knowledge, this is the first report that describes CRISPR-Cas9-mediated genome editing in SSCs in combination with SSCT, thereby preventing diseases in the offspring. Furthermore, as the transplanted SSCs were the cell lines derived from isolated and corrected single cells, this method can generate healthy descendants at 100% efficiency, thereby averting the problem of mosaicism. In addition to this pioneering work, two other recent articles also provide the proof of concept by showing CRISPR-Cas9 and SSCT-induced germline transmission in rodents (Chapman et al., 2015; Sato et al., 2015). Hence, SSCT and the CRISPR-Cas9 system can be well combined in the future to prevent the transmission of inheritable diseases to the offspring.
Epigenetic editing of SSCs
Mechanisms that underlie cellular functioning are orchestrated by different layers of transcriptional regulation. Apart from genetic factors, epigenetic regulation is key for the proper functioning of a cell. Epigenetic traits are defined (Berger et al., 2009) as ‘heritable phenotypes resulting from changes in a chromosome without alterations in the DNA sequence’, mediated by several factors such as DNA methylation, histone modifications and higher order chromatin structuring. Disruption of epigenetic regulation has been shown in an array of complex diseases, including cancer, diabetes and cardiovascular diseases (reviewed in MacFarlane et al., 2009; Ordovas and Smith, 2010; Dawson and Kouzarides, 2012). Thus, in theory, epigenetic editing may be a valid alternative for genetic repair to modify aberrant gene expression in SSCs from those at risk of transmitting diseases to their own biological children.
The core of currently described epigenetic editing approaches is the fusion of an epigenetic modulatory enzyme to a protein with a DNA binding domain in order to affect gene expression and modulate local parameters, such as cytosine methylation and demethylation or histone modification once the enzymatic construct is in place. This powerful approach has been used to silence and/or activate specific DNA sequences by altering epigenetic parameters of target genes (reviewed in de Groote et al., 2012; Falahi et al., 2015). For example, a recent study describes the guidance of the Ten-Eleven Translocation 2 DNA demethylation enzyme to the promoter region of the intercellular adhesion molecule 1 gene, causing local demethylation and reactivation of the gene where it was normally silenced (Chen et al., 2014). Another group has reported the development of a programmable molecular construct consisting of a nuclease-deactivated Cas9 (dCas9) protein fused to the catalytic core of the acetyltransferase p300, which can be used to modulate histone acetylation of any Cas9-targetable genomic location (Hilton et al., 2015). One might imagine that epigenetic editing techniques might allow correction of the transcriptional regulation of pivotal genes in various biological processes, including germ cell development. Epigenetics have been shown to play a key role in normal germ cell development (Gan et al., 2013; An et al., 2014; Hammoud et al., 2014), and allele-specific DNA methylation was altered in semen from men that suffer from spermatogenic failure (Poplinski et al., 2010; Kläver et al., 2013; Ferfouri et al., 2013; Richardson et al., 2014; Urdinguio et al., 2015). Local correction of abnormal DNA methylation or histone modification of target genes in infertile men might improve the spermatogenic potential.
In theory, SSCT of epigenetically modified SSCs may also be applicable for inherited epigenetic diseases. Epidemiological data point to human transgenerational epigenetic inheritance, including the Dutch Famine Birth Cohort Study (Lumey, 1992; Heijmans et al., 2008; Painter et al., 2008; Veenendaal et al., 2013) and the Swedish Overkalix population (Pembrey et al., 2006). Additionally, a few (case) studies describe inheritance of a disease-associated epimutation of a specific locus, such as the SNURF-SNRPN locus in Prader–Willi and Angelman syndrome (Buiting et al., 2003) and the cancer predisposing gene MLH1 (Suter et al., 2004). However, one must realize that the epigenome is reset in an extensive way during early embryonic development (Daxinger and Whitelaw, 2012; Wei et al., 2015). Even though some inherited epigenetic marks seem to escape epigenetic reprogramming, it remains unclear whether epigenetic germline editing combined with SSCT may benefit patients in the future. Therefore, more research is needed before SSCT can be applied to cure heritable epigenetic diseases.
Clinical and technical hurdles and drawbacks
The field of SSCT is uprising, and a variety of patient groups may benefit from this therapy in the future. However, some hurdles still need to be overcome prior to its clinical implementation. For one thing, safety of the patient and his offspring is of major concern (Struijk et al., 2013). Human SSCs may change (epi)genetically when exposed to an in vitro environment. Yet, there is evidence of genetic stability of cultured human SSCs (Nickkholgh et al., 2014). DNA methylation of maternal and paternal imprinted genes in uncultured murine SSCs did not alter after transplantation (Goossens et al., 2009; Wu et al., 2012), whereas cultured human SSCs showed changes in DNA methylation in some selected regions of maternal and paternal imprinted genes (Nickkholgh et al., 2014). Conclusively, although the published data suggest that SSCT may be safe for the clinic, more (pre)clinical studies are needed in this field to ensure safety for patients, as well as for their offspring.
Another challenge in cancer patients is that primary testicular cultures may be contaminated with lingering cancer cells from leukemic or metastasized patients. While some researchers were unable to successfully sort out cancer cells (Geens et al., 2007), others succeeded in removing leukemic cells from testicular cultures (Sadri-Ardekani et al., 2014) or from cell suspensions (Hermann et al., 2011).
In comparison, the clinical implementation of genomically modified SSCT is even more challenging. Aside from the potential risks accompanying conventional SSCT, we currently do not know whether genomic manipulation of SSCs harbors any extra risks for the patient or his offspring. In addition, a major clinical drawback of germline therapy in fertile carriers of diseases remains that the patients have to undergo local irradiation to deplete the testis of endogenous mutated spermatogonia before SSCT. Otherwise, the testis of the recipient father would produce two populations of sperm cells: those that arise from endogenous SSCs carrying the disease-causing mutation as well as those from the corrected SSCs introduced by transplantation. As a consequence, the semen of the father would contain a mixed population of spermatozoa and children conceived by natural conception could be derived from either a corrected or endogenous sperm cell. Although local irradiation has been demonstrated to be an effective measure to deplete the testis of germ cells in animal studies (Zhang et al., 2006; Herrid et al., 2009), it can have a deleterious influence on outgrowth of seminiferous tubules, especially in prepubertal testes (Jahnukainen et al., 2011). Besides, it may cause damages to surrounding organs and cells. Moreover, some endogenous spermatogonia might survive the irradiation and are still capable of developing into spermatids, thereby risking the transfer of the genomic aberrations. In this sense, the development of alternatives to exclude endogenous SSCs is needed to better strike the balance between the benefits of SSCT and the potential risks of the required total depletion of endogenous spermatogenesis.
In terms of genomic manipulation, some technical problems remain to be addressed. First, the CRISPR-Cas9 components need to be delivered into cells. The way in which the Cas9 nuclease is introduced (viral or non-viral) has significant clinical implications (Table 1). In case of viral transduction, CRISPR-sequences, and possibly even residual viral sequences, integrate into the genome of the patient. This raises serious safety concerns and is not suitable for clinical application. However, non-viral delivery methods often fall short, as they can be inefficient in gene delivery (e.g. liposome-mediated transfection), are laborious (e.g. microinjection) or lead to significant cell mortality (e.g. electroporation). While SSCs have been demonstrated to be refractory to most non-viral transfection approaches (Kanatsu-Shinohara et al., 2005), novel electroporation devices that are currently being used in some laboratories may be the option to transfect SSCs with adequate efficiency (Zeng et al., 2012; Fanslow et al., 2014; Wu et al., 2015; Chapman et al., 2015; Sato et al., 2015). Alternatively, transfection of the mRNA instead of the corresponding DNA vectors has been shown to be more efficient for genome editing (Fanslow et al., 2014). Also the recently developed novel method regarding direct intracellular delivery of proteins might serve as another option for gene targeting (D’Astolfo et al., 2015).
In spite of the developments, one needs to realize that SSCT in combination with genomic editing to cure or prevent diseases is only feasible when the genetic mutation is known. Unfortunately, the genetic etiology remains elusive in many cases. More research on the identification of genomic mutations that cause subfertility is necessary in this respect (Visser and Repping, 2010; Tuttelmann et al., 2011; Krausz and Chianese, 2014). A recent article describes that CRISPR-Cas9 was exploited to successfully eradicate porcine endogenous retroviruses with the copy number as high as 62 in a porcine cell line (Yang et al., 2015), showing the robustness and versatility of CRISPR. However, various genetic diseases, including subfertility, are believed to be caused by an array of genes (Jan et al., 2012), which renders genetic correction substantially more difficult as it would require simultaneous targeting of various loci. A previous report shows that a maximum of five genes could be simultaneously disrupted by microinjection of CRISPR components into mouse ESCs (Wang et al., 2013). At present, the novel GeCKO system seems to be the only possible approach to targeting a wide array of genes in parallel. Nevertheless, GeCKO, which requires lentiviral transduction and subsequent integration of CRISPR components into the genome, is considered unsafe for clinical application at the moment. We still need to await further development in this regard before we can target a variety of genes in parallel safely for clinical purposes.
Another important issue is the potential off-target alterations induced by the CRISPR-Cas9 system. Recent studies have revealed that the 20-bp gRNA-DNA hybrid (Fig. 2) has the potential to tolerate 1–3 or even more sequence mismatches (Fu et al., 2013; Mali et al., 2013; Sander and Joung, 2014). As a consequence, normal genes containing high homology to the target sequence might also be targeted. Reassuringly, whole genome sequencing of the CRISPR-Cas9-modified SSCs showed no apparent off-target mutations (Wu et al., 2015). Moreover, novel versions of CRISPR with enhanced specificity but without the sacrifice of on-target activity have been developed recently (Slaymaker et al., 2015; Kleinstiver et al., 2016), which will further facilitate its broad applications in the clinic.
Also, the targeting spectrum of the CRISPR-Cas9 system needs to be expanded. The requirement of a specific sequence (e.g. 5′-NGG for type II SpCas9, Fig. 2) following the target is a principal constraint. While 5′-NGG occurs quite frequently in the human genome, further extensions of the targeting range by development of other types of CRISPR that recognize distinct sequences downstream of the targeting site (Zetsche et al., 2015; Nishimasu et al., 2015) would give broader options for genome editing.
Ethical issues
As genomic modification of SSCs leads to germline transmission of the modified trait to the next generation, elaborate ethical reflection and an intensive societal debate on the acceptability of a clinical application of germline gene editing should precede the actual clinical application of modified SSCs. Recently two groups employed the CRISPR-Cas9 system to achieve genome editing in human tripronuclear zygotes (Liang et al., 2015; Kang et al., 2016). Although the zygotes used were unable to develop into viable embryos, these reports still initiated major debates. Some people propose a complete ban on germline genomic editing (Lanphier et al., 2015), while others request a moratorium on the clinical application of germline editing but suggest permission for research in this field (Baltimore et al., 2015). To date, multiple articles have been published to broaden the discussion (Ishii, 2015; Miller, 2015; Pollack, 2015; Porteus and Dann, 2015; Vassena et al., 2016). We strongly support the establishment of a societal platform including molecular and (stem) cell biologists, medical professionals, ethicists, politicians, citizens and most importantly patients, to discuss under which circumstances and to what extent germline modification should be allowed.
Concluding remarks
In this review, we explore different patient groups that may benefit from SSCT with or without genomic editing. We conclude that the clinical implementation of SSCT can potentially reach far beyond fertility preservation in childhood cancer patients. Conventional SSCT could help adult cancer patients preserve their fertility, while fertility may also be enhanced in oligozoospermic or azoospermic patients using this technology. Successful genomic modification of SSCs by the novel CRISPR-Cas9 system in culture could repair detrimental mutations, thereby treating patients with non-obstructive azoospermia and carriers of diseases in the future.
The safety of the patient and the following generations is of paramount importance. From our perspective, by no means should these techniques be employed in the clinic until safety and efficiency has been demonstrated empirically. Therefore, more fundamental research remains to be conducted. In addition, a societal and ethical debate should precede the use of modified SSCs in a future clinical application of SSCT.
Nonetheless, SSCT, with or without genome editing, is a potential powerful platform that potentially can be employed to cure infertility or even inheritable mutations in various patient groups. Research in this field is thriving, and a revolution might be visible at the horizon.
Authors’ roles
C.L.M., Y.Z., G.H. and A.M.M.P. designed the outline of the review. C.L.M. and Y.Z. drafted the original manuscript. S.Z.J. and R.B.S. gave substantial contribution to the manuscript. C.L.M., Y.Z. and S.Z.J. designed and created the figures and table in this manuscript. S.R., A.M.M.P. and G.H. critically reviewed and revised the manuscript and approved the final version.
Funding
ZonMW (TAS116003002), the China Scholarship Counsel (CSC) (201306300081), an AMC Fellowship and the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme (CIG 293765).
Conflict of interest
The author(s) report no financial or other conflict of interest relevant to the subject of this article.
References
- Akhondi M, Mohazzab A, Jeddi-Tehrani M, Sadeghi M, Eidi A, Khodadadi A, Piravar Z. Propagation of human germ stem cells in long-term culture. Iran J Reprod Med 2013;11:551–558. [PMC free article] [PubMed] [Google Scholar]
- An J, Zhang X, Qin J, Wan Y, Hu Y, Liu T, Li J, Dong W, Du E, Pan C et al.. The histone methyltransferase ESET is required for the survival of spermatogonial stem/progenitor cells in mice. Cell Death Dis 2014;5:e1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston K, Krausz C, Laface I, Ruiz-Castané E, Carrell D. Evaluation of 172 candidate polymorphisms for association with oligozoospermia or azoospermia in a large cohort of men of European descent. Hum Reprod 2010;25:1383–1397. [DOI] [PubMed] [Google Scholar]
- Baert Y, Van Saen D, Haentjens P, In’t Veld P, Tournaye H, Goossens E. What is the best cryopreservation protocol for human testicular tissue banking? Hum Reprod 2013;28:1816–1826. [DOI] [PubMed] [Google Scholar]
- Baltimore D, Berg P, Botchan M, Carroll D, Charo R, Church G, Corn J, Daley G, Doudna J, Fenner M et al.. A prudent path forward for genomic engineering and germline gene modification. Science 2015;348:36–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev 2009;23:781–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boitrelle F, Albert M, Theillac C, Ferfouri F, Bergere M, Vialard F, Wainer R, Bailly M, Selva J. Cryopreservation of human spermatozoa decreases the number of motile normal spermatozoa, induces nuclear vacuolization and chromatin decondensation. J Androl 2012;33:1371–1378. [DOI] [PubMed] [Google Scholar]
- Brinster RL. Male germline stem cells: from mice to men. Science 2007;316:404–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Zimmermann JW. Spermatogenesis following male germ-cell transplantation. Proc Natl Acad Sci USA. 1994;91:11298–11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buiting K, Gross S, Lich C, Gillessen-Kaesbach G, el-Maarri O, Horsthemke B. Epimutations in Prader-Willi and Angelman syndromes: a molecular study of 136 patients with an imprinting defect. Am J Hum Genet 2003;72:571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KM, Medrano GA, Jaichander P, Chaudhary J, Waits AE, Nobrega MA, Hotaling JM, Ober C, Hamra FK. Targeted germline modifications in rats using CRISPR/Cas9 and spermatogonial stem cells. Cell Rep 2015;10:1828–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Kazemier H, de Groote M, Ruiters M, Xu G, Rots M. Induced DNA demethylation by targeting Ten-Eleven Translocation 2 to the human ICAM-1 promoter. Nucleic Acids Res 2014;42:1563–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curaba M, Poels J, van Langendonckt A, Donnez J, Wyns C. Can prepubertal human testicular tissue be cryopreserved by vitrification? Fertil Steril 2011;95:e2129–2112. [DOI] [PubMed] [Google Scholar]
- D’Astolfo DS, Pagliero RJ, Pras A, Karthaus WR, Clevers H, Prasad V, Lebbink RJ, Rehmann H, Geijsen N. Efficient intracellular delivery of native proteins. Cell 2015;161:674–690. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell 2012;150:12–27. [DOI] [PubMed] [Google Scholar]
- Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat Rev Genet 2012;13:153–162. [DOI] [PubMed] [Google Scholar]
- de Groote M, Verschure P, Rots M. Epigenetic editing: targeted rewriting of epigenetic marks to modulate expression of selected target genes. Nucleic Acids Res 2012;40:10596–10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kretser DM. Male infertility. Lancet 1997;349:787–790. [DOI] [PubMed] [Google Scholar]
- Dobrinski I, Ogawa T, Avarbock M, Brinster RL. Computer assisted image analysis to assess colonization of recipient seminiferous tubules by spermatogonial stem cells from transgenic donor mice. Mol Reprod Dev 1999;53:142–148. [DOI] [PubMed] [Google Scholar]
- Dores C, Alpaugh W, Dobrinski I. From in vitro culture to in vivo models to study testis development and spermatogenesis. Cell Tissue Res 2012;349:691–702. [DOI] [PubMed] [Google Scholar]
- Falahi F, Sgro A, Blancafort P. Epigenome engineering in cancer: fairytale or a realistic path to the clinic? Front Oncol 2015;5:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanslow DA, Wirt SE, Barker JC, Connelly JP, Porteus MH, Dann CT. Genome editing in mouse spermatogonial stem/progenitor cells using engineered nucleases. PLoS One 2014;9:e112652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferfouri F, Boitrelle F, Ghout I, Albert M, Molina Gomes D, Wainer R, Bailly M, Selva J, Vialard F. A genome-wide DNA methylation study in azoospermia. Andrology 2013;1:815–821. [DOI] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 2013;31:822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan H, Wen L, Liao S, Lin X, Ma T, Liu J, Song C, Wang M, He C, Han C et al.. Dynamics of 5-hydroxymethylcytosine during mouse spermatogenesis. Nat Commun 2013;4:1995 1-11. [DOI] [PubMed] [Google Scholar]
- Gassei K, Orwig KE. Experimental methods to preserve male fertility and treat male factor infertility. Fertil Steril 2016;105:256–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geens M, Van de Velde H, De Block G, Goossens E, Van Steirteghem A, Tournaye H. The efficiency of magnetic-activated cell sorting and fluorescence-activated cell sorting in the decontamination of testicular cell suspensions in cancer patients. Hum Reprod 2007;22:733–742. [DOI] [PubMed] [Google Scholar]
- Gelbaya TA. Short and long-term risks to women who conceive through in vitro fertilization. Hum Fertil 2010;13:19–27. [DOI] [PubMed] [Google Scholar]
- Ginsberg JP, Carlson CA, Lin K, Hobbie WL, Wigo E, Wu X, Brinster RL, Kolon TF. An experimental protocol for fertility preservation in prepubertal boys recently diagnosed with cancer: a report of acceptability and safety. Hum Reprod 2010;25:37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R, Dobrinski I. Beyond the mouse monopoly: studying the male germ line in domestic animal models. ILAR J 2015;56:83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens E, De Rycke M, Haentjens P, Tournaye H. DNA methylation patterns of spermatozoa and two generations of offspring obtained after murine spermatogonial stem cell transplantation. Hum Reprod 2009;24:2255–2263. [DOI] [PubMed] [Google Scholar]
- Goossens E, de Vos P, Tournaye H. Array comparative genomic hybridization analysis does not show genetic alterations in spermatozoa and offspring generated after spermatogonial stem cell transplantation in the mouse. Hum Reprod 2010;25:1836–1842. [DOI] [PubMed] [Google Scholar]
- Goossens E, Van Saen D, Tournaye H. Spermatogonial stem cell preservation and transplantation: from research to clinic. Hum Reprod 2013;28:897–907. [DOI] [PubMed] [Google Scholar]
- Guo Y, Liu L, Sun M, Hai Y, Li Z, He Z. Expansion and long-term culture of human spermatogonial stem cells via the activation of SMAD3 and AKT pathways. Exp Biol Med (Maywood) 2015;240:1112–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud S, Low D, Yi C, Carrell D, Guccione E, Cairns B. Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell 2014;15:239–53. [DOI] [PubMed] [Google Scholar]
- Handyside A, Lesko J, Tarín J, Winston R, Hughes MR. Birth of a normal girl after in vitro fertilization and preimplantation diagnostic testing for cystic fibrosis. N Engl J Med 1992;327:905–909. [DOI] [PubMed] [Google Scholar]
- Hansen M, Kurinczuk JJ, Milne E, de Klerk N, Bower C. Assisted reproductive technology and birth defects: a systematic review and meta-analysis. Hum Reprod Update 2013;19:330–353. [DOI] [PubMed] [Google Scholar]
- He X, Ruan J, Du W, Chen G, Zhou Y, Xu S, Zuo X, Cao Y, Zhang X. PRM1 variant rs35576928 (Arg>Ser) is associated with defective spermatogenesis in the Chinese Han population. Reprod Biomed Online 2012;25:627–34. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA 2008;105:17046–17049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmerhorst FM, Perquin DA, Donker D, Keirse MJ. Perinatal outcome of singletons and twins after assisted conception: a systematic review of controlled studies. Br Med J 2004;328:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendin B, Falcone T, Hallak J, Nelson D, Vemullapalli S, Goldberg J, Thomas AJ, Agarwal A. The effect of patient and semen characteristics on live birth rates following intrauterine insemination: a retrospective study. J Assist Reprod Genet 2000;17:245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks S, Dancet EA, Meissner A, Van der Veen F, Mochtar M, Repping S. Perspectives of infertile men on future stem cell treatments for nonobstructive azoospermia. Reprod Biomed Online 2014;28:650–657. [DOI] [PubMed] [Google Scholar]
- Hentrich A, Wolter M, Szardening-Kirchner C, Lüers G, Bergmann M, Kliesch S, Konrad L. Reduced numbers of sertoli, germ, and spermatogonial stem cells in impaired spermatogenesis. Mod Pathol 2011;24:1380–1389. [DOI] [PubMed] [Google Scholar]
- Hermann BP, Sukhwani M, Salati J, Sheng Y, Chu T, Orwig KE. Separating spermatogonia from cancer cells in contaminated prepubertal primate testis cell suspensions. Hum Reprod 2011;26:3222–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann BP, Sukhwani M, Winkler F, Pascarella JN, Peters KA, Sheng Y, Valli H, Rodriguez M, Ezzelarab M, Dargo G et al.. Spermatogonial stem cell transplantation into rhesus testes regenerates spermatogenesis producing functional sperm. Cell Stem Cell 2012;11:715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrid M, Olejnik J, Jackson M, Suchowerska N, Stockwell S, Davey R, Hutton K, Hope S, Hill JR. Irradiation enhances the efficiency of testicular germ cell transplantation in sheep. Biol Reprod 2009;81:898–905. [DOI] [PubMed] [Google Scholar]
- Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 2015;33:510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honaramooz A, Behboodi E, Megee SO, Overton SA, Galantino-Homer H, Echelard Y, Dobrinski I. Fertility and germline transmission of donor haplotype following germ cell transplantation in immunocompetent goats. Biol Reprod 2003;69:1260–1264. [DOI] [PubMed] [Google Scholar]
- Honaramooz A, Megee SO, Dobrinski I. Germ cell transplantation in pigs. Biol Reprod 2002;66:21–28. [DOI] [PubMed] [Google Scholar]
- Howell S, Shalet S. Spermatogenesis after cancer treatment: damage and recovery. J Natl Cancer Inst Monogr 2005;34:12–17. [DOI] [PubMed] [Google Scholar]
- Hu Z, Xia Y, Guo X, Dai J, Li H, Hu H, Jiang Y, Lu F, Wu Y, Yang X et al.. A genome-wide association study in Chinese men identifies three risk loci for non-obstructive azoospermia. Nat Genet 2012;44:183–186. [DOI] [PubMed] [Google Scholar]
- Ishii T. Germline genome-editing research and its socioethical implications. Trends Mol Med 2015;21:473–481. [DOI] [PubMed] [Google Scholar]
- Izadyar F, Den Ouden K, Stout TA, Stout J, Coret J, Lankveld DP, Spoormakers TJ, Colenbrander B, Oldenbroek JK, Van der Ploeg KD et al.. Autologous and homologous transplantation of bovine spermatogonial stem cells. Reproduction 2003;126:765–774. [PubMed] [Google Scholar]
- Jahnukainen K, Ehmcke J, Hou M, Schlatt S. Testicular function and fertility preservation in male cancer patients. Best Pract Res Clin Endocrinol Metab 2011;25:287–302. [DOI] [PubMed] [Google Scholar]
- Jahnukainen K, Ehmcke J, Quader MA, Saiful Huq M, Epperly MW, Hergenrother S, Nurmio M, Schlatt S. Testicular recovery after irradiation differs in prepubertal and pubertal non-human primates, and can be enhanced by autologous germ cell transplantation. Hum Reprod 2011;26:1945–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahnukainen K, Ehmcke J, Soder O, Schlatt S. Clinical potential and putative risks of fertility preservation in children utilizing gonadal tissue or germline stem cells. Pediatr Res 2006;59:40R–47R. [DOI] [PubMed] [Google Scholar]
- Jan SZ, Hamer G, Repping S, de Rooij DG, van Pelt AM, Vormer TL. Molecular control of rodent spermatogenesis. Biochim Biophys Acta 2012;1822:1838–1850. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Miki H, Inoue K, Ogonuki N, Toyokuni S, Ogura A, Shinohara T. Long-term culture of mouse male germline stem cells under serum-or feeder-free conditions. Biol Reprod 2005;72:985–991. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Muneto T, Lee J, Takenaka M, Chuma S, Nakatsuji N, Horiuchi T, Shinohara T. Long-term culture of male germline stem cells from hamster testes. Biol Reprod 2008;78:611–617. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Ogonuki N, Inoue K, Miki H, Ogura A, Toyokuni S, Shinohara T. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod 2003;69:612–616. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Toyokuni S, Shinohara T. Genetic selection of mouse male germline stem cells in vitro: offspring from single stem cells. Biol Reprod 2005;72:236–240. [DOI] [PubMed] [Google Scholar]
- Kang X, He W, Huang Y, Yu Q, Chen Y, Gao X, Sun X, Fan Y. Introducing precise genetic modifications into human 3PN embryos by CRISPR/Cas-mediated genome editing. J Assist Reprod Genet 2016;33:581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T, Saito K, Sakai C, Shinya M, Sakai N. Production of zebrafish offspring from cultured spermatogonial stem cells. Genes Cells 2012;17:316–325. [DOI] [PubMed] [Google Scholar]
- Keel B, Webster B. Semen analysis data from fresh and cryopreserved donor ejaculates: comparison of cryoprotectants and pregnancy rates. Fertil Steril 1989;52:100–105. [DOI] [PubMed] [Google Scholar]
- Keros V, Hultenby K, Borgstrom B, Fridstrom M, Jahnukainen K, Hovatta O. Methods of cryopreservation of testicular tissue with viable spermatogonia in pre-pubertal boys undergoing gonadotoxic cancer treatment. Hum Reprod 2007;22:1384–1395. [DOI] [PubMed] [Google Scholar]
- Keros V, Rosenlund B, Hultenby K, Aghajanova L, Levkov L, Hovatta O. Optimizing cryopreservation of human testicular tissue: comparison of protocols with glycerol, propanediol and dimethylsulphoxide as cryoprotectants. Hum Reprod 2005;20:1676–1687. [DOI] [PubMed] [Google Scholar]
- Kläver R, Tüttelmann F, Bleiziffer A, Haaf T, Kliesch S, Gromoll J. DNA methylation in spermatozoa as a prospective marker in andrology. Andrology 2013;1:731–740. [DOI] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016;529:490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kossack N, Terwort N, Wistuba J, Ehmcke J, Schlatt S, Scholer H, Kliesch S, Gromoll J. A combined approach facilitates the reliable detection of human spermatogonia in vitro. Hum Reprod 2013;28:3012–3025. [DOI] [PubMed] [Google Scholar]
- Krausz C, Chianese C. Genetic testing and counselling for male infertility. Curr Opin Endocrinol Diabetes Obes 2014;21:244–250. [DOI] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Brinster RL. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc Natl Acad Sci USA 2004;101:16489–16494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Schmidt JA, Brinster RL. Spermatogonial stem cells derived from infertile Wv/Wv mice self-renew in vitro and generate progeny following transplantation. Biol Reprod 2009;81:293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuliev A, Rechitsky S, Verlinsky O, Ivakhnenko V, Cieslak J, Evsikov S, Wolf G, Angastiniotis M, Kalakoutis G, Strom C et al.. Birth of healthy children after preimplantation diagnosis of thalassemias. J Assist Reprod Genet 1999;16:207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Silber S, Oates R, Rozen S et al.. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet 2001;29:279–286. [DOI] [PubMed] [Google Scholar]
- Kvist K, Thorup J, Byskov AG, Hoyer PE, Mollgard K, Yding Andersen C. Cryopreservation of intact testicular tissue from boys with cryptorchidism. Hum Reprod 2006;21:484–491. [DOI] [PubMed] [Google Scholar]
- Lanphier E, Urnov F, Haecker S, Werner M, Smolenski J. Don’t edit the human germ line. Nature 2015;519:410–411. [DOI] [PubMed] [Google Scholar]
- Lee J, Kanatsu-Shinohara M, Ogonuki N, Miki H, Inoue K, Morimoto T, Morimoto H, Ogura A, Shinohara T. Heritable imprinting defect caused by epigenetic abnormalities in mouse spermatogonial stem cells. Biol Reprod 2009;80:518–527. [DOI] [PubMed] [Google Scholar]
- Li HL, Fujimoto N, Sasakawa N, Shirai S, Ohkame T, Sakuma T, Tanaka M, Amano N, Watanabe A, Sakurai H et al.. Precise correction of the dystrophin gene in Duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports 2015;4:143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, Xu Y, Zhang X, Ding C, Huang R, Zhang Z, Lv J, Xie X, Chen Y, Li Y et al.. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell 2015;6:363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Sung S, Kim H, Song S, Hong J, Yoon T, Kim J, Kim K, Lee D. Long-term proliferation and characterization of human spermatogonial stem cells obtained from obstructive and non-obstructive azoospermia under exogenous feeder-free culture conditions. Cell Prolif 2010;43:405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science 2014;345:1184–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumey LH. Decreased birthweights in infants after maternal in utero exposure to the Dutch famine of 1944-1945. Paediatr Perinat Epidemiol 1992;6:240–253. [DOI] [PubMed] [Google Scholar]
- MacFarlane AJ, Strom A, Scott FW. Epigenetics: deciphering how environmental factors may modify autoimmune type 1 diabetes. Mamm Genome 2009;20:624–632. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science 2013;339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur R, Akande A, Keay S, Hunt L, Jenkins J. Distinction between early and late ovarian hyperstimulation syndrome. Fertil Steril 2000;73:901–907. [DOI] [PubMed] [Google Scholar]
- Meistrich M. Effects of chemotherapy and radiotherapy on spermatogenesis in humans. Fertil Steril 2013;100:1180–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller HI. Germline gene therapy: we’re ready. Science 2015;348:1325. [DOI] [PubMed] [Google Scholar]
- Nagano M. Homing efficiency and proliferation kinetics of male germ line stem cells following transplantation in mice. Biol Reprod 2003;69:701–707. [DOI] [PubMed] [Google Scholar]
- Nagano M, Patrizio P, Brinster RL. Long-term survival of human spermatogonial stem cells in mouse testes. Fertil Steril 2002;78:1225–1233. [DOI] [PubMed] [Google Scholar]
- Nickkholgh B, Korver C, van Daalen S, van Pelt A, Repping S. AZFc deletions do not affect the function of human spermatogonia in vitro Mol Hum Reprod 2015;21:553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickkholgh B, Mizrak SC, van Daalen SK, Korver CM, Sadri-Ardekani H, Repping S, van Pelt AM. Genetic and epigenetic stability of human spermatogonial stem cells during long-term culture. Fertil Steril 2014;102:1700–1707 e1701. [DOI] [PubMed] [Google Scholar]
- Ning L, Meng J, Goossens E, Lahoutte T, Marichal M, Tournaye H. In search of an efficient injection technique for future clinical application of spermatogonial stem cell transplantation: infusion of contrast dyes in isolated cadaveric human testes. Fertil Steril 2012;98:1443–1448. [DOI] [PubMed] [Google Scholar]
- Nishimasu H, Cong L, Yan WX, Ran FA, Zetsche B, Li Y, Kurabayashi A, Ishitani R, Zhang F, Nureki O. Crystal structure of Staphylococcus aureus Cas9. Cell 2015;162:1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega RH, Greebe CD, van de Kant H, Bogerd J, de Franca LR, Schulz RW. Spermatogonial stem cell niche and spermatogonial stem cell transplantation in zebrafish. PLoS One 2010;5:e12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordam MJ, Repping S. The human Y chromosome: a masculine chromosome. Curr Opin Genet Dev 2006;16:225–232. [DOI] [PubMed] [Google Scholar]
- Oates RD. The genetic basis of male reproductive failure. Urol Clin North Am 2008;35:257–270, ix. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Arechaga JM, Avarbock MR, Brinster RL. Transplantation of testis germinal cells into mouse seminiferous tubules. Int J Dev Biol 1997;41:111–122. [PubMed] [Google Scholar]
- Ordovas JM, Smith CE. Epigenetics and cardiovascular disease. Nat Rev Cardiol 2010;7:510–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page D, Silber S, Brown L. Men with infertility caused by AZFc deletion can produce sons by intracytoplasmic sperm injection, but are likely to transmit the deletion and infertility. Hum Reprod 1999;14:1722–1726. [DOI] [PubMed] [Google Scholar]
- Painter RC, Osmond C, Gluckman P, Hanson M, Phillips DI, Roseboom TJ. Transgenerational effects of prenatal exposure to the Dutch famine on neonatal adiposity and health in later life. Br J Obstet Gynaecol 2008;115:1243–1249. [DOI] [PubMed] [Google Scholar]
- Parada-Bustamante A, Lardone M, Valdevenito R, Ebensperger M, López P, Madariaga M, Piottante A, Castro A. Analysis of 6 single-nucleotide polymorphisms in the androgen receptor gene in Chilean patients with primary spermatogenic failure. J Androl 2012;33:88–95. [DOI] [PubMed] [Google Scholar]
- Pembrey ME, Bygren LO, Kaati G, Edvinsson S, Northstone K, Sjostrom M, Golding J. Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet 2006;14:159–166. [DOI] [PubMed] [Google Scholar]
- Picton HM, Wyns C, Anderson RA, Goossens E, Jahnukainen K, Kliesch S, Mitchell RT, Pennings G, Rives N, Tournaye H et al.. A European perspective on testicular tissue cryopreservation for fertility preservation in prepubertal and adolescent boys. Hum Reprod 2015;30:2463–2475. [DOI] [PubMed] [Google Scholar]
- Poels J, Abou-Ghannam G, Herman S, Van Langendonckt A, Wese FX, Wyns C. In search of better spermatogonial preservation by supplementation of cryopreserved human immature testicular tissue xenografts with N-acetylcysteine and testosterone. Front Surg 2014;1:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poels J, Van Langendonckt A, Many MC, Wese FX, Wyns C. Vitrification preserves proliferation capacity in human spermatogonia. Hum Reprod 2013;28:578–589. [DOI] [PubMed] [Google Scholar]
- Pollack R. Eugenics lurk in the shadow of CRISPR. Science 2015;348:871. [DOI] [PubMed] [Google Scholar]
- Poplinski A, Tüttelmann F, Kanber D, Horsthemke B, Gromoll J. Idiopathic male infertility is strongly associated with aberrant methylation of MEST and IGF2/H19 ICR1. Int J Androl 2010;33:642–649. [DOI] [PubMed] [Google Scholar]
- Porteus MH, Dann CT. Genome editing of the germline: broadening the discussion. Mol Ther 2015;23:980–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragni G, Somigliana E, Restelli L, Salvi R, Arnoldi M, Paffoni A. Sperm banking and rate of assisted reproduction treatment: insights from a 15-year cryopreservation program for male cancer patients. Cancer 2003;97:1624–1629. [DOI] [PubMed] [Google Scholar]
- Richardson M, Bleiziffer A, Tüttelmann F, Gromoll J, Wilkinson M. Epigenetic regulation of the RHOX homeobox gene cluster and its association with human male infertility. Hum Mol Genet 2014;23:12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu BY, Kubota H, Avarbock MR, Brinster RL. Conservation of spermatogonial stem cell self-renewal signaling between mouse and rat. Proc Natl Acad Sci USA 2005;102:14302–14307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu BY, Orwig KE, Oatley JM, Lin CC, Chang LJ, Avarbock MR, Brinster RL. Efficient generation of transgenic rats through the male germline using lentiviral transduction and transplantation of spermatogonial stem cells. J Androl 2007;28:353–360. [DOI] [PubMed] [Google Scholar]
- Sadri-Ardekani H, Akhondi MA, van der Veen F, Repping S, van Pelt AM. In vitro propagation of human prepubertal spermatogonial stem cells. J Am Med Assoc 2011;305:2416–2418. [DOI] [PubMed] [Google Scholar]
- Sadri-Ardekani H, Atala A. Testicular tissue cryopreservation and spermatogonial stem cell transplantation to restore fertility: from bench to bedside. Stem Cell Res Ther 2014;5:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadri-Ardekani H, Homburg CH, van Capel TM, van den Berg H, van der Veen F, van der Schoot CE, van Pelt AM, Repping S. Eliminating acute lymphoblastic leukemia cells from human testicular cell cultures: a pilot study. Fertil Steril 2014;101:1072–1078 e1071. [DOI] [PubMed] [Google Scholar]
- Sadri-Ardekani H, Mizrak SC, van Daalen SK, Korver CM, Roepers-Gajadien HL, Koruji M, Hovingh S, de Reijke TM, de la Rosette JJ, van der Veen F et al.. Propagation of human spermatogonial stem cells in vitro. J Am Med Assoc 2009;302:2127–2134. [DOI] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 2014;32:347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 2014;11:783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Sakuma T, Yokonishi T, Katagiri K, Kamimura S, Ogonuki N, Ogura A, Yamamoto T, Ogawa T. Genome editing in mouse spermatogonial stem cell lines using TALEN and double-nicking CRISPR/Cas9. Stem Cell Reports 2015;5:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlatt S. Spermatogonial stem cell preservation and transplantation. Mol Cell Endocrinol 2002;187:107–111. [DOI] [PubMed] [Google Scholar]
- Schlatt S, Ehmcke J, Jahnukainen K. Testicular stem cells for fertility preservation: preclinical studies on male germ cell transplantation and testicular grafting. Pediatr Blood Cancer 2009;53:274–280. [DOI] [PubMed] [Google Scholar]
- Schlatt S, Rosiepen G, Weinbauer GF, Rolf C, Brook PF, Nieschlag E. Germ cell transfer into rat, bovine, monkey and human testes. Hum Reprod 1999;14:144–150. [DOI] [PubMed] [Google Scholar]
- Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent CK et al.. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 2013;13:653–658. [DOI] [PubMed] [Google Scholar]
- Sermon K, Goossens V, Seneca S, Lissens W, De Vos A, Vandervorst M, Van Steirteghem A, Liebaers I. Preimplantation diagnosis for Huntington’s disease (HD): clinical application and analysis of the HD expansion in affected embryos. Prenat Diagn 1998;18:1427–36. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG et al.. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014;343:84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber SJ. Evaluation and treatment of male infertility. Clin Obstet Gynecol 2000;43:854–888. [DOI] [PubMed] [Google Scholar]
- Singh P, Schimenti JC. The genetics of human infertility by functional interrogation of SNPs in mice. Proc Natl Acad Sci U S A 2015;112:10431–10436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science 2015;351:84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struijk R, Mulder C, Van der Veen F, van Pelt A, Repping S. Restoring fertility in sterile childhood cancer survivors by autotransplanting spermatogonial stem cells: are we there yet? Biomed Res Int 2013;2013:903142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter CM, Martin DI, Ward RL. Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet 2004;36:497–501. [DOI] [PubMed] [Google Scholar]
- Takagi S, Itoh N, Kimura M, Sasao T, Tsukamoto T. Spermatogonial proliferation and apoptosis in hypospermatogenesis associated with nonobstructive azoospermia. Fertil Steril 2001;76:901–907. [DOI] [PubMed] [Google Scholar]
- Teng Y, Chang Y, Tseng J, Kuo P, Lee I, Lee M, Kuo P. A single-nucleotide polymorphism of the DAZL gene promoter confers susceptibility to spermatogenic failure in the Taiwanese Han. Hum Reprod 2012;27:2857–2865. [DOI] [PubMed] [Google Scholar]
- Tomlinson M, Meadows J, Kohut T, Haoula Z, Naeem A, Pooley K, Deb S. Review and follow-up of patients using a regional sperm cryopreservation service: ensuring that resources are targeted to those patients most in need. Andrology 2015;3:709–716. [DOI] [PubMed] [Google Scholar]
- Tuttelmann F, Simoni M, Kliesch S, Ledig S, Dworniczak B, Wieacker P, Ropke A. Copy number variants in patients with severe oligozoospermia and Sertoli-cell-only syndrome. PLoS One 2011;6:e19426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ul Ain Q, Chung JY, Kim YH. Current and future delivery systems for engineered nucleases: ZFN, TALEN and RGEN. J Control Release 2015;205:120–127. [DOI] [PubMed] [Google Scholar]
- Urdinguio RG, Bayon GF, Dmitrijeva M, Torano EG, Bravo C, Fraga MF, Bassas L, Larriba S, Fernandez AF. Aberrant DNA methylation patterns of spermatozoa in men with unexplained infertility. Hum Reprod 2015;30:1014–1028. [DOI] [PubMed] [Google Scholar]
- Valcarce D, Cartón-García F, Riesco M, Herráez M, Robles V. Analysis of DNA damage after human sperm cryopreservation in genes crucial for fertilization and early embryo development. Andrology 2013;1:723–730. [DOI] [PubMed] [Google Scholar]
- van der Meer Y, Huiskamp R, Davids J, Van der Tweel I, De Rooij D. The sensitivity to X rays of mouse spermatogonia that are committed to differentiate and of differentiating spermatogonia. Radiat Res 1992;130:296–302. [PubMed] [Google Scholar]
- van der Steeg JW, Steures P, Eijkemans MJ, Habbema JD, Hompes PG, Broekmans FJ, van Dessel HJ, Bossuyt PM, van der Veen F, Mol BW et al.. Pregnancy is predictable: a large-scale prospective external validation of the prediction of spontaneous pregnancy in subfertile couples. Hum Reprod 2007;22:536–542. [DOI] [PubMed] [Google Scholar]
- Vassena R, Heindryckx B, Peco R, Pennings G, Raya A, Sermon K, Veiga A. Genome engineering through CRISPR/Cas9 technology in the human germline and pluripotent stem cells. Hum Reprod Update 2016. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Veenendaal MV, Painter RC, de Rooij SR, Bossuyt PM, van der Post JA, Gluckman PD, Hanson MA, Roseboom TJ. Transgenerational effects of prenatal exposure to the 1944-45 Dutch famine. Br J Obstet Gynaecol 2013;120:548–553. [DOI] [PubMed] [Google Scholar]
- Visser L, Repping S. Unravelling the genetics of spermatogenic failure. Reproduction 2010;139:303–307. [DOI] [PubMed] [Google Scholar]
- Vloeberghs V, Verheyen G, Haentjens P, Goossens A, Polyzos NP, Tournaye H. How successful is TESE-ICSI in couples with non-obstructive azoospermia? Hum Reprod 2015;30:1790–1796. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013;153:910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Schatten H, Sun QY. Environmental epigenetic inheritance through gametes and implications for human reproduction. Hum Reprod Update 2015;21:194–208. [DOI] [PubMed] [Google Scholar]
- Wu X, Goodyear SM, Abramowitz LK, Bartolomei MS, Tobias JW, Avarbock MR, Brinster RL. Fertile offspring derived from mouse spermatogonial stem cells cryopreserved for more than 14 years. Hum Reprod 2012;27:1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Liang D, Wang Y, Bai M, Tang W, Bao S, Yan Z, Li D, Li J. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell 2013;13:659–662. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhou H, Fan X, Zhang Y, Zhang M, Wang Y, Xie Z, Bai M, Yin Q, Liang D et al.. Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res 2015;25:67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyns C, Curaba M, Martinez-Madrid B, Van Langendonckt A, Francois-Xavier W, Donnez J. Spermatogonial survival after cryopreservation and short-term orthotopic immature human cryptorchid testicular tissue grafting to immunodeficient mice. Hum Reprod 2007;22:1603–1611. [DOI] [PubMed] [Google Scholar]
- Wyns C, Curaba M, Petit S, Vanabelle B, Laurent P, Wese JF, Donnez J. Management of fertility preservation in prepubertal patients: 5 years’ experience at the Catholic University of Louvain. Hum Reprod 2011;26:737–747. [DOI] [PubMed] [Google Scholar]
- Wyns C, Van Langendonckt A, Wese FX, Donnez J, Curaba M. Long-term spermatogonial survival in cryopreserved and xenografted immature human testicular tissue. Hum Reprod 2008;23:2402–2414. [DOI] [PubMed] [Google Scholar]
- Xie F, Ye L, Chang JC, Beyer AI, Wang J, Muench MO, Kan YW. Seamless gene correction of beta-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res 2014;24:1526–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakirevich E, Sabo E, Dirnfeld M, Sova Y, Spagnoli G, Resnick M. Morphometrical quantification of spermatogonial germ cells with the 57B anti-MAGE-A4 antibody in the evaluation of testicular biopsies for azoospermia. Appl Immunohistochem Mol Morphol 2003;11:37–44. [DOI] [PubMed] [Google Scholar]
- Yang L, Guell M, Niu D, George H, Lesha E, Grishin D, Aach J, Shrock E, Xu W, Poci J et al.. Genome-wide inactivation of porcine endogenous retroviruses (PERVs). Science 2015;350:1101–1104. [DOI] [PubMed] [Google Scholar]
- Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol 2014;32:551–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Zhou Q, Wan H, Shen B, Wang X, Wang M, Feng C, Xie M, Gu T, Zhou T et al.. Generation of fertile offspring from Kit(w)/Kit(wv) mice through differentiation of gene corrected nuclear transfer embryonic stem cells. Cell Res 2015;25:851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Hou R, Wu J. Generation of mice by transplantation of an adult spermatogonial cell line after cryopreservation. Cell Prolif 2009;42:123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegers-Hochschild F, Adamson GD, de Mouzon J, Ishihara O, Mansour R, Nygren K, Sullivan E, Vanderpoel S, International Committee for Monitoring Assisted Reproductive Technology, and World Health Organisation . International Committee for Monitoring Assisted Reproductive Technology (ICMART) and the World Health Organization (WHO) revised glossary of ART terminology, 2009. Fertil Steril 2009;92:1520–1524. [DOI] [PubMed] [Google Scholar]
- Zeng W, Tang L, Bondareva A, Luo J, Megee SO, Modelski M, Blash S, Melican DT, Destrempes MM, Overton SA et al.. Non-viral transfection of goat germline stem cells by nucleofection results in production of transgenic sperm after germ cell transplantation. Mol Reprod Dev 2012;79:255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A et al.. Cpf1 Is a Single RNA-guided endonuclease of a Class 2 CRISPR-Cas system. Cell 2015;163:759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Shao S, Meistrich ML. Irradiated mouse testes efficiently support spermatogenesis derived from donor germ cells of mice and rats. J Androl 2006;27:365–375. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Zhang Y, Qu R, He Y, Tian X, Zeng W. Spermatogonial stem cells from domestic animals: progress and prospects. Reproduction 2014;147:65–74. [DOI] [PubMed] [Google Scholar]