Abstract
We previously found that in utero caffeine exposure causes down-regulation of DNA methyltransferases (DNMTs) in embryonic heart and results in impaired cardiac function in adulthood. To assess the role of DNMTs in these events, we investigated the effects of reduced DNMT expression on embryonic cardiomyocytes. siRNAs were used to knock down individual DNMT expression in primary cultures of mouse embryonic cardiomyocytes. Immunofluorescence staining was conducted to evaluate cell morphology. A video-based imaging assay and multielectrode array were used to assess cardiomyocyte contractility and electrophysiology, respectively. RNA-Seq and multiplex bisulfite sequencing were performed to examine gene expression and promoter methylation, respectively. At 72 h after transfection, reduced DNMT3a expression, but not DNMT1 or -3b, disrupted sarcomere assembly and decreased beating frequency, contractile movement, amplitude of field action potential, and cytosolic calcium signaling of cardiomyocytes. RNA-Seq analysis revealed that the DNMT3a-deficient cells had deactivated gene networks involved in calcium, endothelin-1, renin-angiotensin, and cardiac β-adrenergic receptor signaling, which were not inhibited by DNMT3b siRNA. Moreover, decreased methylation levels were found in the promoters of Myh7, Myh7b, Tnni3, and Tnnt2, consistent with the up-regulation of these genes by DNMT3a siRNA. These data show that DNMT3a plays an important role in regulating embryonic cardiomyocyte gene expression, morphology and function.—Fang, X., Poulsen, R. R., Wang-Hu, J., Shi, O., Calvo, N. S., Simmons, C. S., Rivkees, S. A., Wendler, C. C. Knockdown of DNA methyltransferase 3a alters gene expression and inhibits function of embryonic cardiomyocytes.
Keywords: DNA methylation, cellular morphology, contractility, transcriptome
Cardiomyocyte development is an orchestrated multistep process, with characteristic gene expression profiles at different stages (1). Expression of these genes is tightly controlled by multiple mechanisms, including DNA methylation (2, 3). DNA methylation is critical for cardiac development and plays a role in adult cardiac disease by regulating gene expression (2, 3). Increasing evidence indicates a link between aberrant DNA methylation and cardiac disease in humans, including dilated cardiomyopathy and heart failure (4, 5). However, the significance of DNA methylation in shaping cardiomyocyte morphology and function remains largely unknown.
DNA methylation is the process of adding a methyl group to a cytosine of the dinucleotide sequence CG (6). Methylation of normally unmethylated CpG sites located in the promoter region is associated with transcriptional inactivation of genes by suppressing transcription factor binding or by recruiting histone deacetylases that cause chromatin condensation (6). Methylation in intragenic regions also contributes to gene regulation and alternative splicing (7).
DNA methylation patterns are established and maintained by DNA methyltransferases (DNMTs) (6). DNMT1 is associated with maintaining methylation patterns, as it primarily methylates hemimethylated DNA (6). DNMT3a and -3b are de novo DNMTs that primarily methylate unmethylated DNA (6). DNMTs are essential for development and deletion of DNMT1 or 3b leads to embryonic lethality (8, 9). DNMT3a-knockout (KO) mice appear normal at birth, but become runted and die at 4 wk of age of an unknown cause (9).
Our understanding of the roles of de novo DNMTs in cardiac development and disease is in its early stages. DNMT3b KO mice die between E13.5 and 16.5 with abnormal heart structure (9). Tamoxifen-induced cardiomyocyte-specific loss of DNMT3b in adult heart for 3–4 wk results in compromised systolic function, increased interstitial fibrosis, and myosarcomeric disarray (10).
Mutations in the DNMT3a gene cause an overgrowth syndrome (Tatton-Brown-Rahman Syndrome) in humans, which causes intellectual disability, ventriculomegaly, and mitral and tricuspid regurgitation (11). In the heart, DNMT3a is down-regulated in patients with tetralogy of Fallot, a condition associated with DNA hypomethylation (12). In addition, DNMT3a is overexpressed in activated cardiac fibroblasts, leading to cardiac fibrosis (13). Phenotype analysis by the International Mouse Phenotyping Consortium (IMPC; http://www.mousephenotype.org/data/genes/MGI:1261827) revealed that Dnmt3a heterozygous KO mice have reduced heart rate variability, which is a predictor of adult heart failure (14).
DNMT3a is essential for the normal function of many cell types, but our understanding of the role of DNMT3a in cardiomyocytes is limited. DNMT3a activity is influenced by factors that include hypoxia (15), cadmium (16), vitamin A-deficiency (17), caffeine (18, 19), and cAMP (20). Our previous research demonstrated that in utero caffeine exposure during early embryogenesis decreases the expression of DNMTs in embryonic ventricles, alters DNA methylation patterns and cardiac gene expression in adult heart, and decreases adult cardiac function (18, 19). We hypothesize that these in utero caffeine effects on cardiac function are mediated by down-regulation of DNMTs.
To investigate the role of DNMTs during cardiac development and their influence on cardiac function, we used siRNAs to reduce individual DNMT expression in embryonic mouse cardiomyocytes. After transient DNMT knockdown, we examined morphology, contractility, electrophysiology, calcium signaling, whole-genome transcription, and target gene promoter methylation on these cardiomyocytes. We now show that disruption of DNMT3a activity results in direct adverse effects on cardiomyocyte gene expression, cellular structure, and function.
MATERIALS AND METHODS
Materials
Fibronectin, anti-α-actinin (sarcomeric) mAb (A7811), and anti-β-actin mAb (A2228) were purchased from Sigma-Aldrich (St. Louis, MO, USA). DNMT1 antibody (5119) was purchased from Cell Signaling Technology (Danvers, MA, USA). DNMT3a (ab13888), DNMT3b (ab16049), and anti-cardiac troponin T [1F11] (ab10214) antibodies were from Abcam (Cambridge, MA, USA). Goat anti-mouse IgG Alexa 488 secondary antibody, DAPI, lipofectamine RNAiMax, RNAqueous-Micro Total RNA Isolation kit, Fluo-4 Direct Calcium Assay kit, and Power SYBR Green PCR Master Mix were from Thermo Fisher Scientific Life Sciences (Waltham, MA, USA). Accutase and Accumax were from Innovative Cell Technologies (San Diego, CA, USA).
Isolation and culture of primary embryonic cardiomyocytes
Timed mating was carried out with CD-1 mice (Charles River Laboratories, Wilmington, MA, USA), and the day a vaginal plug was observed was designated embryonic day (E)0.5. All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Florida. Primary cardiomyocytes were isolated from mouse E13.5 embryonic ventricles, as described (18).
Small interfering RNA knockdown of DNMT1, DNMT3a, and DNMT3b
FlexiTube GeneSolution [with 4 small interfering (si)RNAs] DNMT1 siRNA (GS13433), DNMT3a siRNA (GS13435), DNMT3b siRNA (GS13436), AllStars negative control siRNA (1027281), and AllStars Mm/Rn Cell Death Control siRNA (SI04939025) were obtained from Qiagen (Valencia, CA, USA). Cardiomyocytes were seeded at a density of 6.0 × 105 cells per well in 12-well culture plates or at 6.0 × 106 per 12.5 mm2 flask. Cells were cultured for 48 h at 37°C to reach 70–80% confluence. DNMT1, -3a, -3b, negative control, or positive control siRNA at 12 nM were transfected into the cells using lipofectamine RNAiMax.
Cell viability, cytotoxicity, and apoptosis assay
Primary cardiomyocytes were seeded in 12-well plates and treated with various siRNAs. Cell viability, cytotoxicity, and apoptosis were measured with an ApoTox-Glo Triplex Assay (Promega, Madison, WI, USA) at 24 and 72 h after siRNA transfection. The fluorescent and luminescent signals were measured with a Synergy HT Multimode Microplate Reader (BioTek, Winooski, VT, USA).
Immunofluorescence staining and cell imaging
Cardiomyocytes were stained with an anti-α-actinin antibody and DAPI, as described (18). The cells were imaged with an Axio Vert.A1 inverted microscope (Zeiss, Jena, Germany). In addition, a Cytation 5 Cell Imaging Multimode Reader (BioTek) was used in montage mode, to scan the cardiomyocytes. The percent cardiomyocyte (α-actinin+ cells/DAPI+ cells) and total size of cardiomyocyte area (α-actinin+ area) were calculated with the Gen5 software (BioTek) and an in-house R script.
Video-based imaging assay for cardiomyocyte contractility
The beating rate of cardiomyocytes was counted under the Evos Cell Imaging Systems (Thermo Fisher Scientific Life Sciences). In addition, a video-based imaging assay was developed to quantitate cardiomyocyte contractility. First, high-speed video images [∼100 frames per second (fps)] were taken of the beating cell monolayers by phase-contrast microscope (Nikon, Tokyo, Japan), and the images were analyzed with a series of custom MatLab algorithms (MathWorks, Natick, MA, USA) (21). This method generated results of contraction frequency; velocity of cell contraction and relaxation; the force of contraction, as measured by the degree of movement; the percentage of regions that move at contraction; and cell contraction synchronicity.
Calcium signaling imaging and analysis
Calcium transient peak signaling in siRNA-treated cardiomyocytes was visualized with the Fluo-4 Direct Calcium Assay kit. High-speed video images (∼100 fps) were taken of the cardiomyocytes with a fluorescence microscope (Nikon), and images were analyzed with a series of custom MatLab algorithms for peak frequency and intensity amplitude.
Field potential recordings
Primary cardiomyocytes were seeded on multielectrode arrays (MEAs with 60 TiN electrodes; Multichannel Systems, Reutlingen, Germany) coated with 10 μg/ml fibronectin, allowed to grow to 70–80% confluence, and treated with DNMT3a siRNA or negative siRNA. At 72 h after transfection, electrical activity was measured with an MEA-2100 amplifier (Multichannel Systems), with the temperature kept at 37°C. Data were recorded and analyzed with MCRack software (Multichannel Systems).
Sorting of cardiomyocytes, RNA isolation, and reverse transcription
Cardiomyocytes were treated with various siRNAs and released with Accutase and dissociated with Accumax. To sort cardiomyocytes, we stained the dissociated cells with anti-VCAM1 antibody conjugated with allophycocyanin (APC; BioLegend, San Diego, CA, USA) and magnetically sorted using anti-APC microbeads and magnetic-assisted cell sorting (MACS) columns (Miltenyi Biotec, Bergisch Gladbach, Germany). For validation of cell purity, sorted cells were fixed, stained with anti-cardiac troponin T [1F11] antibody and goat anti-mouse IgG Alexa 488 secondary antibody, and analyzed on an FACS Calibur system (BD Biosciences, Franklin Lakes, NJ, USA). Data were analyzed with FCS Express 4 Flow Cytometry (De Novo Software, Glendale, CA, USA) software.
Total RNA was isolated from the sorted cardiomyocytes with the RNAqueous-Micro Total RNA Isolation kit. Total RNA was used for RNA-sequencing (RNA-Seq) library preparation or reverse transcribed to cDNA libraries by using iScript cDNA Synthesis kit (Bio-Rad, Hercules, CA, USA).
Illumina transcriptomic RNA sequencing
mRNA was isolated from total RNA using NextFlex Poly(A) Beads (Bio Scientific, Austin, TX, USA). Sequencing libraries were prepared with the NebNext mRNA Library Prep Master Mix Set for Illumina (New England Biolabs, Ipswich, MA, USA) and the NebNext Multiplex Oligos for Illumina (NEB). Nine Illumina-adapted libraries, including cardiomyocyte samples treated with negative, DNMT3a, or DNMT3b siRNA (n = 3/treatment), were pooled at an equal molar ratio and sequenced with a High Output 1 × 75 run on a NextSeq500 sequencer (Illumina, San Diego, CA, USA). All RNA-Seq data were uploaded to the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo), and the accession number is GSE77514.
RNA-Seq data analysis for differential gene expression
The fastq files generated from RNA-Seq were uploaded to the UF Research Computing Galaxy instance developed by the University of Florida (22). The data were cleaned with the FastQC program and mapped to the mouse genome (mm10) with the Tophat2 tool. Counting of RNA-seq reads was performed with HTSeq (23). Differential expression (DE) of genes between treatments was analyzed by 2 methods: R packages EdgeR (24) and DEseq2 (25), with Ensembl Mus_GRCm38.79.gtf as the reference annotation. A false-discovery rate (FDR) <0.05 and absolute fold change >1.5 was regarded as significant. Unique DE genes were identified by combining the results generated from the 2 analytical methods. Functional ontology was conducted with the unique DE genes by using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (26) and Ingenuity pathway analysis (IPA; Qiagen) (27). The enriched pathways for DNMT3a or DNMT3b siRNA treatment were compared by using IPA. The significance criterion for IPA analysis is P < 0.05.
Analysis of gene regulatory network
iRegulon (http://iregulon.aertslab.org/) was used to analyze the coexpressed DE genes (up- or down-regulated) for presence of enriched transcription factor binding sites/motifs and master transcription factors that coregulate these genes (28). Identification of transcription factor binding sites in target gene promoters was performed by using Promo software (29).
RNA-Seq data analysis for differential exon usage
Differential exon usage (DEU) analysis was performed with R package DEXSeq to identify changes in the relative usage of each exon after siRNA treatments (30). Significant changes were those with FDR <0.05 and absolute fold change >1.5. Functional ontology analysis on DEU genes was conducted with DAVID.
Overlap of DE and DEU genes
Distribution of DE and DEU genes across the chromosomes was analyzed. DE and DEU genes were compared, and the overlap of affected genes in both expression and alternative splicing by siRNA treatments was identified. DAVID analysis was performed on the overlap genes.
Quantitative real-time PCR analysis
Published qPCR primer pairs were used (18). β-actin primers were used as an internal control. Power SYBRGreen PCR Master Mix was used to perform quantitative PCR (qPCR) analysis in a GeneAmp 7300 Real Time PCR System (18, 31).
Western blot analysis
Protein was isolated, quantitated, gel separated, and transferred to a nitrocellulose membrane (Bio-Rad), as described (18). Blots were probed with antisera to target proteins and against β-actin, as a control for sample loading, and imaged with a ChemiDoc XRS+ Imaging System (Bio-Rad).
Genomic DNA extraction
Cardiomyocytes were treated with various siRNAs and sorted with the Miltenyi MACS technology, as described above. DNA was extracted with the Quick-gDNA MicroPrep kit (Zymo Research, Irvine, CA, USA), and DNA concentrations were quantitated with Nanodrop 2000 (Thermo Fisher Scientific Life Sciences).
Measurement of global cytosine methylation level
Genomic DNA was treated with RNAase to remove RNA contaminants. The percentage of cytosine methylation in genomic DNA samples was quantified with the MethylFlash Methylated DNA Quantification kit (Colorimetric; Epigentek, Farmingdale, NY, USA) and the Synergy HT Multimode Microplate Reader (BioTek) (20).
Sodium bisulfite PCR amplification of target gene regions
Genomic DNA was treated with sodium metabisulfite for bisulfite conversion (32). Bisulfite-specific PCR primers for 15 target genes (2 promoter regions per gene) were designed with Methyl Primer Express, v1.0 (Thermo Fisher Scientific Life Sciences; Table 1). Target gene regions were PCR amplified from the bisulfite-converted DNA with ZymoTaq PreMix (Hot Start DNA Taq Polmerase; Zymo Research). PCR products were cleaned with the ZR-96 DNA Clean & Concentrator-5 kit (Zymo Research).
TABLE 1.
Primers used for multiplex sodium bisulfite sequencing
| Gene and ID | Primer, 5′–3′ |
CG sites (n) | Location | |
|---|---|---|---|---|
| Forward | Reverse | |||
| Myh6 (17888) | AATAATTGAGGTAAGGGTTTGG | ACTTCCAAAAAAAACAATTTCC | 4 | Across TSS |
| Myh6 (17888) | TTTTTTATTAGGAGTGGAGGGT | ATCCTCCAAAAACCTTACTACC | 3 | Promoter |
| Myh7 (140781) | GTTTAGAGTTGTGTTTGGGAAG | CAACAAATAAAAAATTAATCCCAA | 9 | Promoter |
| Myh7 (140781) | AGGGTTAGGTTAGTATTGGGGG | CCCAATCCCTAACCAAATTTAA | 5 | Promoter |
| Myh7b (668940) | TTAGGGGTTGAGTTAGAGGAGA | CCTCCAAACATAAAAACATCCT | 3 | Promoter |
| Myh7b (668940) | TTATGTTGGATTTTTTTTTGGA | CAAAACACAAAAAACAAATTCC | 4 | Promoter |
| Nppa (230899) | GATTTTTGTAGTTGAGGGTTTG | TCCTAACCCTACCTTAAAATCC | 11 | Across TSS |
| Nppa (230899) | TTGTTTGGGGTTAGAGGTTTAT | CCTTAAACAACTCCACTCACC | 6 | Promoter |
| Nppb (18158) | TGGAAGGTGGGTATTAAAATTA | CCCTTCTTTCATACAAAACCTA | 10 | Promoter |
| Nppb (18158) | GTGTGTGTTTTTTTTGGGTTAG | CCCCACCATAAATAAAAAAAAA | 6 | Promoter |
| Tnnc1 (21924) | GAGTTTAAGGTGAGGAGGAGAT | AATCCTACCAAAAAACACTTCC | 6 | Promoter |
| Tnnc1 (21924) | GTTGGGTTTATAGTAGGGAAGG | TCCTCACCTTAAACTCTTACAAAA | 9 | Promoter |
| Tnni3 (21954) | TTTGGGTATGAATATGGGAAGT | TAAAATCTCCACCCCAAAACTA | 7 | Across TSS |
| Tnni3 (21954) | TTGAAGTAGGTTTAGGGATTGA | CTCCTATAAAATTCAACCAAACC | 7 | Promoter |
| Tnnt2 (21956) | GGGTTGGTGGATATAAATGTTTA | CAAAAATCAACCTCAAAAACAC | 6 | Promoter |
| Tnnt2 (21956) | GGGTTAGGGATTAAAGTTATTGG | CAACAAATCCCAACAAAATCTA | 9 | Promoter |
| Camta1 (100072) | GGGGTTGAGGTTAGGAAGG | AACCAATCAAAAAAAATTCC | 44 | Promoter |
| Camta1 (100072) | ATGTTGAGGAATTAAATAGTGTAAGTT | AACTCTCCTACCTATAAATCCACAC | 9 | Promoter |
| Camta2 (216874) | AGGATTTAGGATAATGAAGTTAGGTT | AACTAAAAACCCTAACTCTTACCTC | 34 | Across TSS |
| Camta2 (216874) | TAAGAAATTTGGAGAGAGTTTTG | AAAAAACACAAAACCAAATTCT | 8 | Promoter |
| Cdkn1A (12575) | TTAATGATGGAGTTGAGATTGG | CAAAACTAAACAAACCCAAACC | 5 | Promoter |
| Cdkn1A (12575) | TTTTTTTAGTGTTGGTTTGTGA | TACACACACCACACACATAAAC | 4 | Promoter |
| Cdkn1B (12576) | GAGATTTTTGGGGTTAGGTTAAG | ACCAACAAACCTACTCTAACTAACC | 28 | Promoter |
| Cdkn1B (12576) | AGTAATGGTTTTTATTGTGTTTAGG | CTAATTCAACCCACCTACATTC | 12 | Promoter |
| Cdkn1C (12577) | AGTTGGTGTAGTTTTAGGGTTAG | ATATAATATTTTCAATTTCAACAACAC | 55 | Promoter |
| Cdkn1C (12577) | TTAAGGTAGGTAGAATTGGGG | ACCATAAAACTAAACACAACCC | 14 | Promoter |
| Mef2c (17260) | AGAAATTATGATTGTTAAAGTGGA | AAAAAAATTAATCATACTCCCAA | 6 | Promoter |
| Mef2c (17260) | AGTTTGTGTGAAATGAGGAAATT | AAAACCCCCCAATTTAAACTAC | 7 | Across TSS |
| Mef2d (17261) | GGTTAAAGGTTTAGAAGTAATGGAG | AATAAACCCCAACTCCTACAC | 17 | Promoter |
| Mef2d (17261) | TGTTTTTTTTAGAAATAGTGGAAA | AAAACATCCAAAAACCTTATCT | 10 | Promoter |
TSS, transcription start site.
Illumina DNA library preparation and sequencing
PCR products from the same genomic DNA were pooled at an equal concentration ratio and made into an indexed sequencing library with the Nextera XT DNA Library Preparation kit and the Nextera XT Index kit (Illumina). Nine Illumina-adapted libraries, including cardiomyocyte samples treated with negative, DNMT3a, or DNMT3b siRNA (n = 3/treatment), were pooled at equal molar ratio, spiked with 20% PhiX control libraries (Illumina), and sequenced with one 1 × 150 run (v3) on an MiSeq sequencer (Illumina). All RNA-Seq data were uploaded to GEO (accession number GSE77621).
Multiplex bisulfite sequencing data analysis
The fastq files generated from MiSeq were uploaded to the UF Research Computing Galaxy instance (https://www.rc.ufl.edu/resources/software/interfaces/galaxy-web/). The data were cleaned with the FastQC program. A reference genome with the amplicon sequences was built and bisulfite converted in silico with Bismark Bisulfite Mapper (33). The high-quality sequence reads were aligned to the reference genome. Cytosine methylation (CpG) counting was performed with the Bismark methylation extractor. Differential methylation was analyzed with the Methylkit package (34). Significance was set at FDR <0.05 and absolute percentage methylation difference >5%.
Statistical analysis
All experiments were performed at least 3 times. Results were analyzed with Prism 6.0 (GraphPad Software, La Jolla, CA, USA). Data are presented as the mean ± sem. Significant differences between treatment groups were determined by Student’s t test or 1-way ANOVA followed by Newman-Keuls post hoc test. Differences reaching P < 0.05 were considered statistically significant.
RESULTS
Down-regulation of DNMT3a disrupts sarcomere assembly in embryonic cardiomyocytes
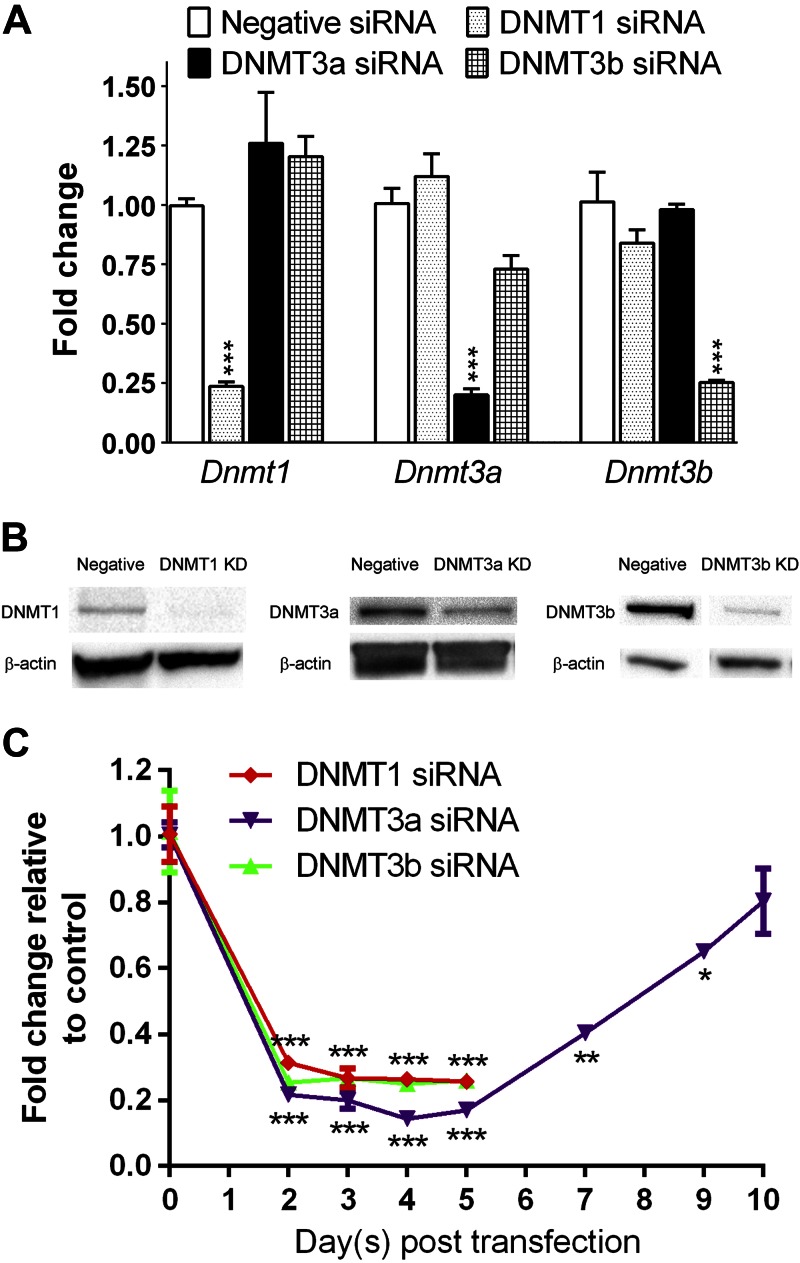
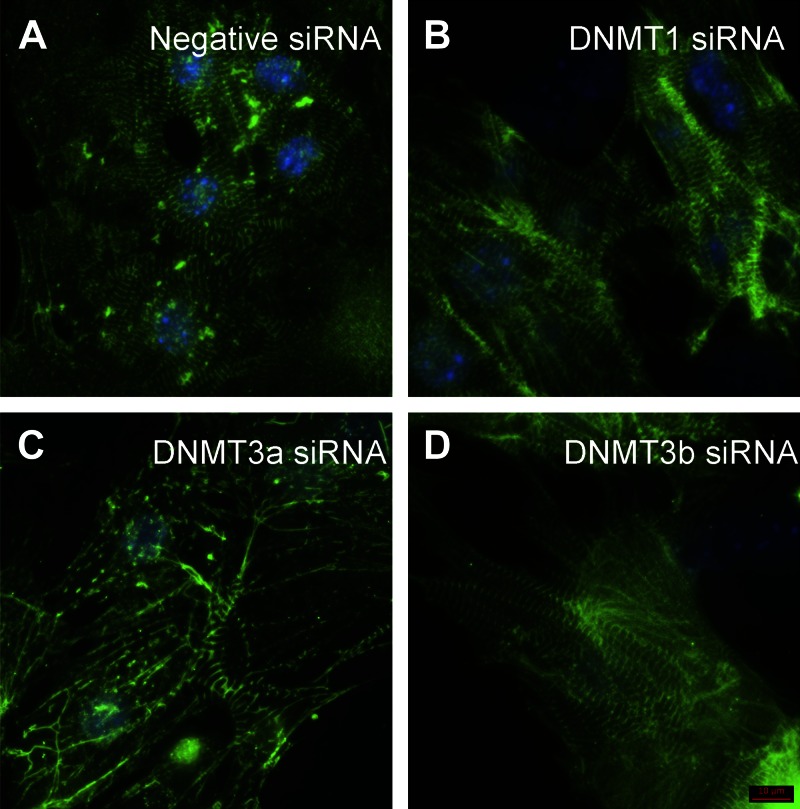
In E13.5 hearts, expression of Dnmt1 was 14.1 times higher than that of Dnmt3a and 159.8 times higher than that of Dnmt3b (n = 3; P < 0.05). To investigate the role of DNMT isoforms in regulating cardiac morphology, we reduced individual DNMT expression by using pooled siRNAs in cultured E13.5 cardiomyocytes. This method decreased target mRNA and protein expression by >70% at 72 h after siRNA transfection (Fig. 1A, B). Using the AllStars Mm/Rn Cell Death Control siRNA, we found that the transfection efficiency was near 100%. The suppression of DNMT expression by >70% lasted up to 5 d after siRNA transfection (Fig. 1C). DNMT3a expression gradually increased to the normal level at 10 d after transfection. Immunofluorescence staining analysis showed that knockdown of DNMT3a disrupted the organization of Z bands in the sarcomeres at 72 h after siRNA transfection (Fig. 2C). In contrast, abnormal morphology was not observed in cells treated with DNMT1 or -3b siRNA (Fig. 2B, D) or negative control siRNA (Fig. 2A).
Figure 1.
Validation of gene knockdown. A, B) qPCR (A) and Western blot (B) analyses showed that siRNA treatments for 72 h reduced DNMT expression in embryonic cardiomyocytes. C) Time course of Dnmt mRNA expression after siRNA transfection (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 vs. negative siRNA.
Figure 2.
Knockdown of DNMT3a expression alters sarcomere assembly in embryonic cardiomyocytes. At 72 h after negative control (A), DNMT1 (B), DNMT3a (C), or DNMT3b (D) siRNA transfection, cardiomyocytes were stained with antisarcomeric α-actinin antibody (sarcomere, green) and DAPI (nucleus, blue). Disarray of sarcomere was observed in the cardiomyocytes treated with DNMT3a siRNA (C). Scale bar, 10 μm.
Down-regulation of DNMT expression increases cytotoxicity and apoptosis in embryonic cardiomyocytes
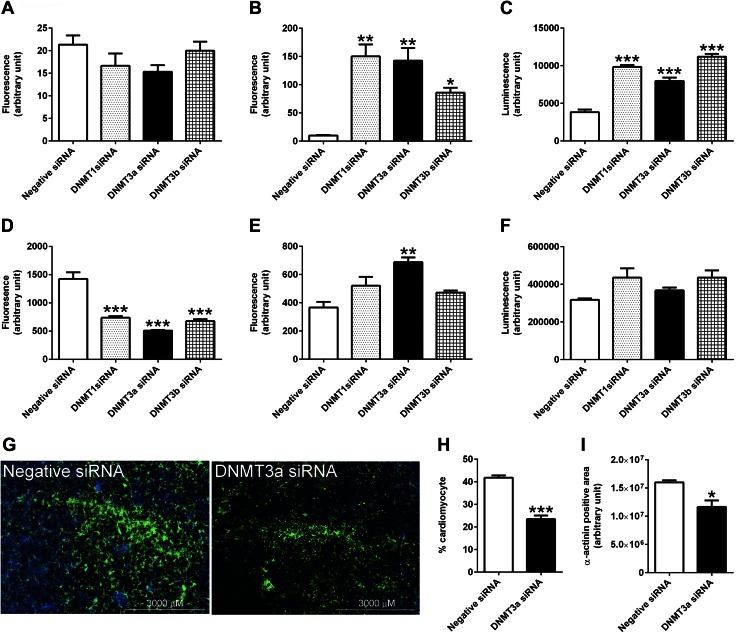
Next, cell viability, cytotoxicity, and apoptosis were assessed with an ApoTox-Glo Triplex Assay kit (Promega). This kit measures live- and dead-cell protease activities that are proportional to the number of living (cell viability) and dead (cytotoxicity) cells, respectively. The kit also measures caspase-3/7 as an indicator of apoptosis. At 24 h after siRNA transfection, cell viability was not significantly affected by knockdown of DNMTs (Fig. 3A), but cytotoxicity and apoptosis were markedly increased by treatment with DNMT1, -3a, or -3b siRNA (Fig. 3B, C).
Figure 3.
Knockdown of DNMT3a expression affects the survival of embryonic cardiomyocytes. A–F) Cell viability at 24 (A) and 72 h (D) after siRNA transfection; cell viability at 24 (B) and 72 h (E); and apoptosis at 24 (C) and 72 h (F) were measured with an ApoTox-Glo Triplex Assay. G) At 72 h, the cardiomyocytes transfected with DNMT3a siRNA or negative siRNA were stained with α-actinin (green) and DAPI (blue) and imaged by a multimode plate reader. H, I) The percentage of cardiomyocytes (α-actinin+ cells/DAPI+ cells) (H) and total area of cardiomyocytes (α-actinin+ area) (I) were decreased in DNMT3a siRNA-treated cultures. (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 vs. negative siRNA.
At 72 h, cell death ceased or slowed down, as no significant changes in cytotoxicity or apoptosis were observed to be associated with DNMT1 and -3b knockdown (Fig. 3E, F). Only DNMT3a siRNA treatment caused elevated cytotoxicity at 72 h, but this effect was significantly less than that observed at 24 h. The elevated level of cell death induced by siRNA treatment at 24 h led to a decreased number of viable cells at 72 h. Viable cells were only 51.8, 35.7, and 47.6% of control cells after knockdown of DNMT1, -3a, and -3b, respectively, for 72 h (Fig. 3D). In addition to a reduced overall number of cells, the percentage of cardiomyocytes in the culture was decreased from 41.8 to 23.5% in the DNMT3a siRNA treatment group at 72 h after transfection, as measured by staining for the cardiomyocyte marker α-actinin (Fig. 3G, H). Consistent with the decreased number and percentage of cardiomyocytes, the total cellular area of cultured cardiomyocytes was decreased by DNMT3a siRNA treatment (Fig. 3I).
Down-regulation of DNMT3a reduces contractility in embryonic cardiomyocytes
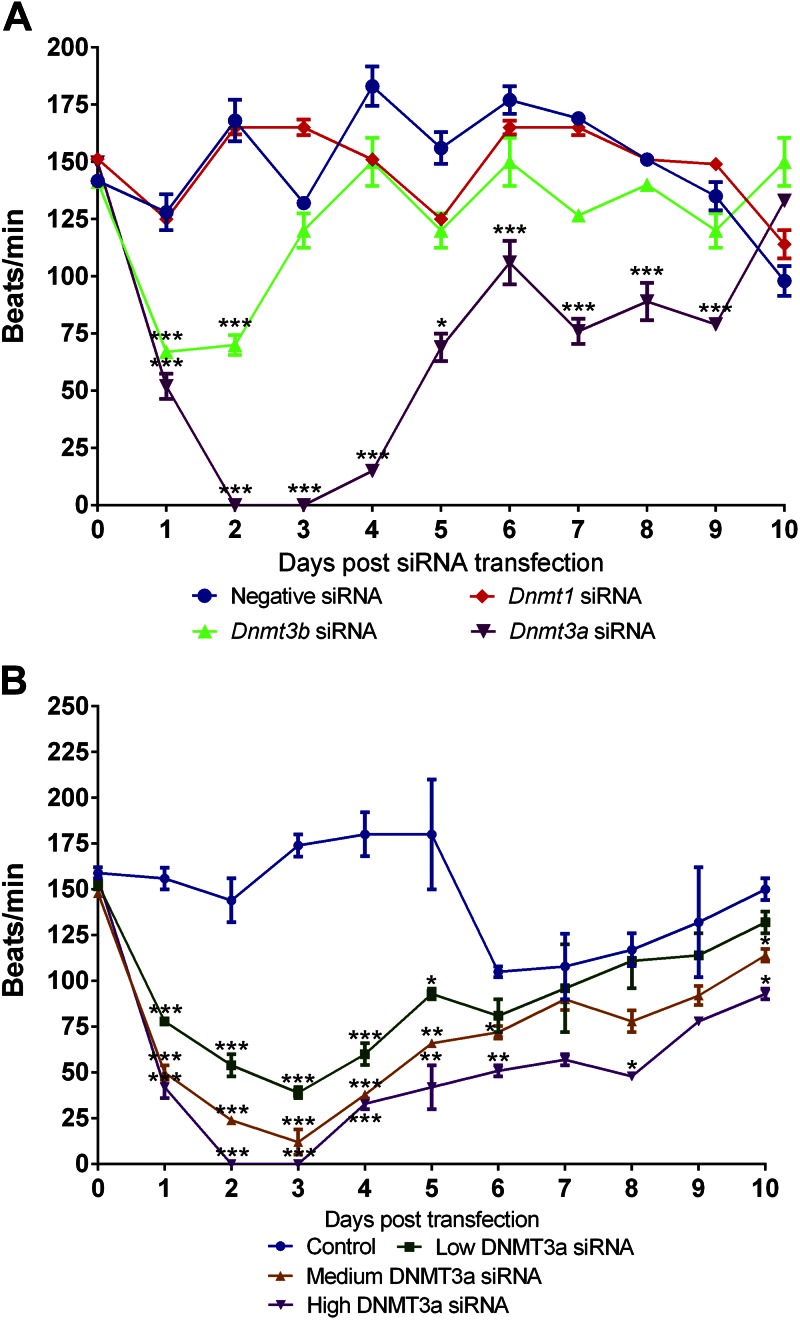
We next examined the contractile function of cardiomyocytes after knockdown of individual DNMT expression. Counting cell contraction under a phase-contrast microscope, we found that transient knockdown of DNMT3a expression led to dramatic, rapid loss of contractility, which resulted in a 66% decrease in beating frequency after 1 d and a 100% inhibition in contractility after 2 d (Fig. 4A). The beating frequency gradually returned to normal as the effect of DNMT3a siRNA subsided (Figs. 1C, 4A). The inhibition of contractility was dependent on the concentration of DNMT3a siRNA (Fig. 4B). The lowest concentration of DNMT3a siRNA (3 nM) reduced beat frequency by 74.5% at 3 d after transfection.
Figure 4.
Knockdown of DNMT3a expression decreases the beating rate of embryonic cardiomyocytes. A) Counting cell contractions under a phase-contrast microscope, we found that transient knockdown of DNMT3a expression (12 nM siRNA) led to dramatic and rapid decline of beating rate. The beating rate recovered over time as the knockdown effect subsided. B) The inhibitory effect of DNMT3a siRNA was concentration dependent (low DNMT3a siRNA, 3 nM; medium DNMT3a siRNA, 6 nM; high DNMT3a siRNA, 12 nM; n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 vs. negative siRNA.
Cell contractility was also inhibited by DNMT3b siRNA at 1–2 d after transfection, but it quickly recovered to normal levels at d 3 after transfection (Fig. 4A). Changes in beating frequency were not seen in the cardiomyocytes treated with DNMT1 siRNA.
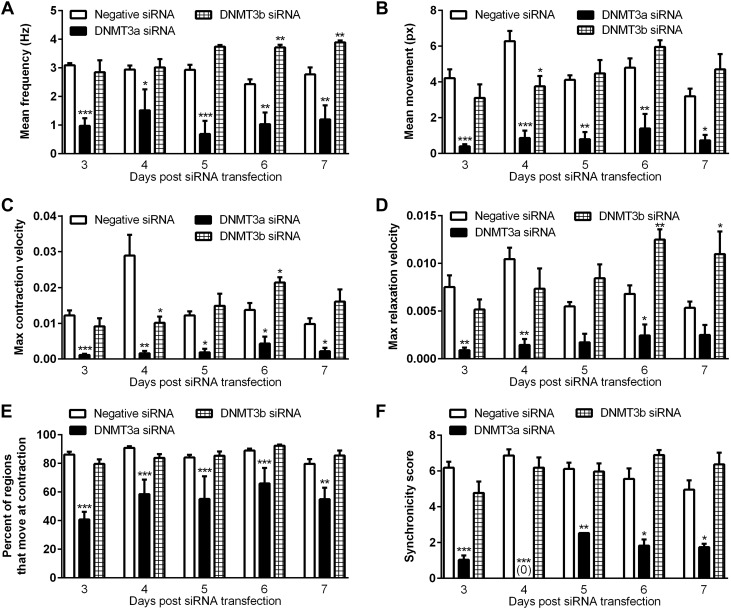
Using a video-based imaging assay, we found that transient knockdown of DNMT3a markedly decreased the beating frequency, movement (contractile force), maximum cell contraction and relaxation velocity, percentage of area of movement, and beating synchronicity (Fig. 5). At 3 d after siRNA transfection, down-regulation of DNMT3a caused a 90.4% reduction in mean movement per beat (Fig. 5B) and a 52.7% decrease in percentage of area of movement vs. negative siRNA treatment (Fig. 5E). The maximum velocity of cell movement during contraction and relaxation were also decreased by DNMT3a siRNA treatment (Fig. 5C, D). The reduced beating synchronicity score indicated that the cells were not contracting as a syncytium across the interrogated region (Fig. 5F). Representative movies from the video-based imaging assay were used to produce tracings of cell movement (Fig. 6A, C, E) and of the amount of contraction area in the region of interest (Fig. 6B, D, F).
Figure 5.
Knockdown of DNMT3a expression inhibits the contractility of embryonic cardiomyocytes. The results from a video-based imaging assay revealed that the mean beating frequency (A), mean movement (px: pixel; B), maximum cell contraction velocity (C), maximum relaxation velocity (D), percentage of regions that move at contraction (E), and synchronicity score (F) for beating in the interrogated region were decreased in the cardiomyocytes treated with DNMT3a siRNA compared to negative-siRNA–treated cardiomyocytes (n = 3 wells, 2 imaged fields per well). *P < 0.05, **P < 0.01, ***P < 0.001 vs. negative siRNA.
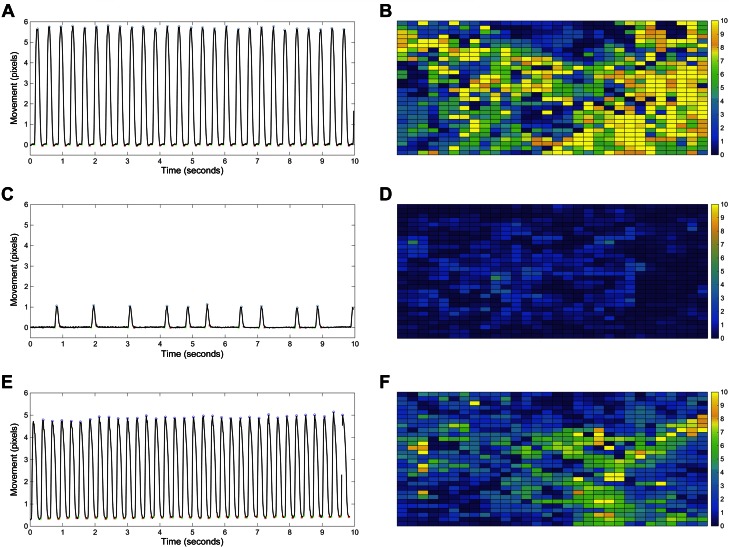
Figure 6.
Knockdown of DNMT3a expression increases beating rate variability and decreases movement in embryonic cardiomyocytes. A, C, E) The trace of cell movement from the video-based imaging assay revealed that transient knockdown of DNMT3a expression for 72 h in cardiomyocytes decreased beating frequency and cell movement and increased beating rate variability (C) compared with the cells treated with negative siRNA (A) or DNMT3b siRNA (E). B, D, F) The heatmap of cell movement across the region of interest also indicated less contractile movement in the DNMT3a siRNA–treated cardiomyocytes (D) than that in negative siRNA (B) or DNMT3b siRNA–treated cells (F). Yellow, high-movement pixels; blue, low-movement pixels.
At 72 h, the decreased movement distance per contraction was also accompanied by increased beating rate variability in the peak-to-peak intervals in DNMT3a-siRNA–treated cells (Fig. 6C). The standard deviation of the mean interpeak interval for the negative-siRNA– vs. the DNMT3a-siRNA–treated cells was 13.4 vs. 46.9% at d 3 after transfection, 24.2 vs. 42.0% at d 4, 14.5 vs. 30.5% at d 5, 22.8 vs. 37.4% at d 6, and 22.5 vs. 41.7% at d 7 (n = 3; 2 interrogated regions per biologic sample; P < 0.05).
Down-regulation of DNMT3a disrupts field action potential in embryonic cardiomyocytes
To study the electrophysiology of cardiomyocytes after the knockdown of DNMT3a expression, we examined field potential recordings with an MEA assay with 60 electrodes. At 72 h after siRNA transfection, 57 of 60 electrodes recorded strong signals from the cells treated with negative siRNA, but for the cells treated with DNMT3a siRNA, only 35 of 60 electrodes detected measurable electric signals (Table 2). The MEA data confirmed the reduced beat rate observed with the video assay. In addition, knockdown of DNMT3a expression decreased the maximum, minimum, and peak-to-peak amplitudes. In addition, the mean interspike interval was increased in the DNMT3a-siRNA–treated cells. The variable of interspike intervals, as indicated by standard deviation, was increased in the DNMT3a-siRNA–treated cells compared with the negative-siRNA–treated cells. This finding is consistent with the variable interpeak intervals of beating rate observed in the video assay.
TABLE 2.
MEA analysis of embryonic cardiomyocytes after knockdown of DNMT3a for 72 h
| Parameter | Negative siRNA | DNMT3a siRNA |
|---|---|---|
| Passed criteria/electrode [n (%)] | 57/60 (95) | 35/60 (58) |
| Beating rate (bpm) | 182.9 ± 34.3 | 125.0 ± 48.0 |
| Maximum peak amplitude (μV) | 268.8 ± 145.7 | 53.5 ± 32.6 |
| Minimum peak amplitude (μV) | −289.6 ± 174.5 | −106.5 ± 136.6 |
| Peak-peak amplitude (μV) | 558.4 ± 286.1 | 160.1 ± 133.3 |
| Interspike intervals (ms) | 336.2 ± 34.4 | 1263.3 ± 3255.9 |
Calcium signaling
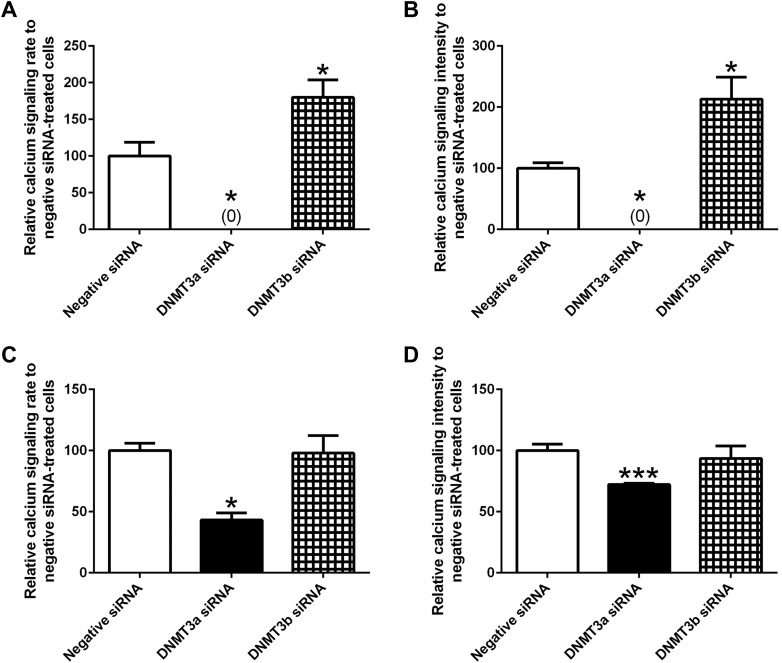
The Fluo-4 Direct Calcium assay showed that down-regulation of DNMT3a expression reduced the rate and intensity of calcium transient peak signaling at 72 and 96 h after siRNA transfection (Fig. 7). After DNMT3b siRNA treatment, the rate and intensity of calcium signaling were increased at 72 h, but remained unchanged at 96 h.
Figure 7.
Knockdown of DNMT3a expression decreases the rate and intensity of calcium signaling in embryonic cardiomyocytes. A calcium assay kit was used to examine calcium transient peak signaling in the cardiomyocytes transfected with various siRNAs for 72 (A, B) and 96 h (C, D). (n = 4–8). *P < 0.05, ***P < 0.001 vs. negative siRNA.
Differential gene expression in embryonic cardiomyocytes after knockdown of DNMT3a or -3b expression
We next performed transcriptomic mRNA sequencing on the embryonic cardiomyocytes at 72 h after transfection with either DNMT3a, DNMT3b, or negative siRNA. Before RNA isolation, cardiomyocytes were enriched with the Miltenyi MACS technology. Flow cytometry analysis confirmed that 95% of the enriched cells were cardiomyocytes. The mapping efficiency of sequencing reads was >80%, and the samples were well clustered by treatments (data not shown). DE analysis by DEseq2 and EdgeR software revealed that 524 genes were up-regulated, and 586 genes were down-regulated after knockdown of DNMT3a expression for 72 h (Supplemental Tables 1, 2). The same DE analysis showed that knockdown of DNMT3b increased the expression of 655 genes and decreased the expression of 378 genes. Expression changes of 11 genes were confirmed by qPCR (Supplemental Table 3).
Comparison of the up-regulated DE genes between DNMT3a and -3b siRNA treatments identified 292 overlapping genes, indicating that their transcriptions are regulated by both DNMT3a and -3b (Supplemental Fig. 1A). The comparative analysis of down-regulated DE genes identified another 90 common genes regulated by DNMT3a and -3b. These analyses also identified genes that are solely regulated by either DNMT3a or -3b, which may explain the distinctive phenotypes observed in cell morphology and function after the knockdown of the 2 different DNMT isoforms.
DEU genes in embryonic cardiomyocytes after DNMT knockdown
It has been reported that methylation in intragenic regions contributes to alternative splicing (7). Thus, we next studied the effects of reduced DNMT3a expression on alternative splicing, as indicated by different levels of exon usage. Analysis by DexSeq identified 209 DEU exons after knockdown of DNMT3a expression for 72 h (Supplemental Tables 1 and 2). Expression was up-regulated in 144 exons and down-regulated in 65 exons. These exons belong to 162 unique genes. DAVID analysis indicated that the DEU genes are involved in the pathways related to actin binding, cytoskeleton organization, and cell junction, among others (Supplemental Table 4). Knockdown of DNMT3b resulted in 165 up-regulated and 53 down-regulated exons (Supplemental Tables 1 and 2). These exons belong to 136 unique genes. Comparison of the DEU genes revealed that 25 genes were affected by both DNMT3a and -3b knockdown at the exon level (Supplemental Fig. 1A).
Overlap of DE and DEU genes
We next compared the genes affected by altered splicing and the genes that were differentially expressed after knockdown of DNMT3a expression. We found that 43 (26.5%) DEU genes overlapped DE genes (Supplemental Fig. 1B), indicating that these genes were affected at both the transcription and splicing levels. DAVID pathway analysis on these overlap genes identified several genes that are involved in the calcium binding and calcium channel activity pathways. These genes include Trpm7, Cacna1c, Ppp3r1, Fgf2, Frem2, Entpd6, and Itgav. The exon usage of 4 representative calcium genes is shown in Supplemental Fig. 2. Of the 136 DEU genes caused by knockdown of DNMT3b, 27 genes were also changed at the transcription level (Supplemental Fig. 1B).
The distribution of DE genes and DEU genes across chromosomes is shown in Supplemental Fig. 1C. DE genes were distributed evenly across the homologous chromosomes. But, the X chromosome had relatively fewer DE genes in proportion to its size, and no DE genes were found in the Y chromosome. A high percentage of DEU genes were found in chromosome 9 and 15. Similarly, fewer or no DEU genes were identified in chromosomes X and Y, respectively.
Pathway analysis of DE genes
Diseases and Bio Functions analysis by IPA revealed that many pathways related to cell death and survival, cell morphology, cell assembly and organization, and cell signaling were affected after DNMT3a siRNA treatment (Table 3). In addition, many cardiac function and disease pathways were significantly enriched (Table 4). In particular, the pathways that regulate heart rate, contraction of heart, function of cardiac muscle, cardiogenesis, morphology of heart, and organization of sarcomere, among others, were altered after suppression of DNMT3a expression. These affected functions are consistent with our observations in DNMT3a-siRNA–treated cardiomyocytes that manifest abnormal sarcomere morphology, reduced contractility, and increased beat rate variability. The enriched disease pathways suggest that cardiomyopathy, congenital heart disease, and heart failure occur as a consequence of altered gene expression.
TABLE 3.
Altered cellular morphology and function pathways in embryonic cardiomyocytes after DNMT3a siRNA treatment
| Pathway |
P |
Molecules (n) |
|---|---|---|
| Cell death and survival | ||
| Proliferation of cells | 1.14E-18 | 379 |
| Cell death | 8.18E-18 | 347 |
| Apoptosis | 2.12E-17 | 288 |
| Necrosis | 2.13E-17 | 282 |
| Cell death of cardiomyocytes | 4.87E-08 | 34 |
| Cell morphology, cellular assembly, and organization | ||
| Morphology of cells | 2.34E-21 | 240 |
| Differentiation of cells | 2.60E-19 | 261 |
| Organization of cytoskeleton | 1.59E-16 | 175 |
| Organization of filaments | 2.10E-11 | 39 |
| Formation of myofilaments | 9.52E-10 | 9 |
| Size of cells | 1.12E-09 | 61 |
| Organization of organelle | 8.39E-09 | 70 |
| Organization of actin cytoskeleton | 9.99E-09 | 45 |
| Organization of actin filaments | 4.62E-08 | 21 |
| Morphology of sarcomere | 6.96E-07 | 9 |
| Formation of cytoskeleton | 9.91E-07 | 48 |
| Cell signaling | ||
| Quantity of Ca2+ | 1.30E-11 | 63 |
| Homeostasis of Ca2+ | 1.54E-09 | 26 |
| Release of Ca2+ | 2.28E-07 | 32 |
| Concentration of cyclic AMP | 9.44E-07 | 29 |
TABLE 4.
Altered cardiac function and disease pathways in embryonic cardiomyocytes after DNMT3a siRNA treatment
| Pathway |
P |
Molecules (n) |
|---|---|---|
| Cardiac function | ||
| Heart rate | 6.73E-29 | 74 |
| Contraction of heart | 6.85E-29 | 58 |
| Hypertrophy of heart | 4.22E-28 | 82 |
| Function of cardiac muscle | 2.57E-22 | 51 |
| Cardiogenesis | 2.55E-17 | 82 |
| Morphology of heart | 2.58E-17 | 74 |
| Arrhythmia | 1.28E-15 | 44 |
| Fibrosis of heart | 5.14E-14 | 40 |
| Abnormal morphology of cardiomyocytes | 3.24E-13 | 25 |
| Organization of sarcomere | 7.17E-13 | 18 |
| Response of heart | 2.37E-09 | 19 |
| Function of cardiomyocytes | 1.51E-07 | 12 |
| Cardiac disease | ||
| Nonischemic cardiomyopathy | 9.48E-20 | 53 |
| Familial heart disease | 1.74E-19 | 29 |
| Dilated cardiomyopathy | 1.87E-18 | 45 |
| Familial cardiomyopathy | 1.76E-17 | 22 |
| Hypertrophic cardiomyopathy | 5.25E-14 | 26 |
| Familial dilated cardiomyopathy | 9.64E-14 | 15 |
| Failure of heart | 8.44E-10 | 41 |
| Coronary disease | 8.51E-10 | 51 |
| Myocardial infarction | 4.22E-08 | 33 |
| Congenital heart disease | 2.53E-06 | 30 |
At the molecular level, IPA analysis revealed that knockdown of DNMT3a altered many canonical pathways that are related to regulation of cardiac function (Table 5). Specifically, the calcium, PKA, renin–angiotensin, endothelin-1, cardiac β-adrenergic, NO, and phospholipase C signaling networks were predicted to be inhibited based on the gene expression patterns, indicating that the DNMT3a-deficient cardiomyocytes may be less responsive to external stimulations than the control cells. The impaired calcium signaling pathway is consistent with the reduced intensity of calcium signals that we observed in the DNMT3a-siRNA–treated cells (Fig. 7).
TABLE 5.
Top 30 most significantly altered canonical cardiac pathways in embryonic cardiomyocytes after DNMT3a siRNA treatment
| Pathway | −log(P) | Ratio (%)a | Activation z score |
|---|---|---|---|
| Calcium signaling | 9.15 | 17.4 | −0.258 |
| Actin cytoskeleton signaling | 6.53 | 13.8 | 1.800 |
| Role of NFAT in cardiac hypertrophy | 5.63 | 14.0 | −0.600 |
| PKA signaling | 5.53 | 10.6 | −0.898 |
| Cardiac hypertrophy signaling | 4.85 | 12.1 | 0.000 |
| G-protein coupled receptor signaling | 4.6 | 11.3 | - |
| Integrin signaling | 4.28 | 11.9 | 0.000 |
| Inhibition of matrix metalloproteases | 4.05 | 23.1 | — |
| Epithelial adherens junction signaling | 4.01 | 13.0 | — |
| PAK signaling | 3.98 | 15.7 | 0.277 |
| Factors promoting cardiogenesis in vertebrates | 3.82 | 15.2 | — |
| SAPK/JNK signaling | 3.72 | 14.9 | −0.535 |
| Renin-angiotensin signaling | 3.55 | 13.8 | −1.941 |
| Endothelin-1 signaling | 3.51 | 11.6 | −1.500 |
| Tight junction signaling | 3.25 | 11.4 | NaN |
| Cardiac β-adrenergic signaling | 3.09 | 12.0 | −0.905 |
| Insulin receptor signaling | 3.05 | 11.9 | −0.775 |
| Dopamine-DARPP32 feedback in cAMP signaling | 3.02 | 11.2 | −2.183 |
| NO | 2.93 | 10.0 | 1.342 |
| NO signaling in the cardiovascular system | 2.92 | 13.0 | −0.577 |
| ERK/MAPK signaling | 2.66 | 10.2 | −1.606 |
| Cardiomyocyte differentiation via BMP receptors | 2.63 | 25.0 | 2.000 |
| IGF-1 signaling | 2.56 | 12.4 | −1.414 |
| Phospholipase C signaling | 2.50 | 9.28 | −0.577 |
| α-adrenergic signaling | 2.46 | 12.6 | 0.302 |
| Sphingosine-1-phosphate signaling | 2.14 | 11.0 | 0.905 |
| eNOS signaling | 2.00 | 9.86 | −0.333 |
| TGF-β signaling | 2.00 | 11.5 | −1.414 |
| STAT3 pathway | 1.58 | 11.0 | −1.414 |
| Embryonic stem cell differentiation into cardiac lineages | 1.08 | 20.0 | — |
Percentage of genes in the pathway that were changed in expression.
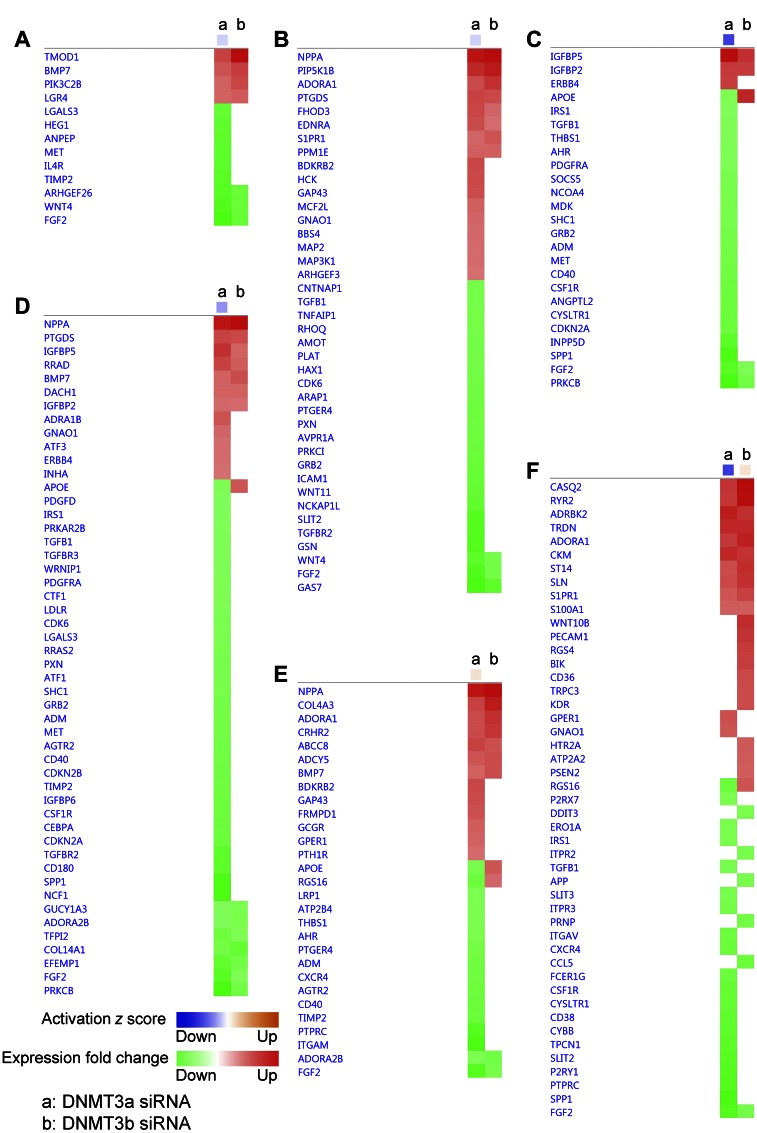
The comparison of the affected biofunction pathways after DNMT3a or -3b siRNA treatment revealed a marked difference that may be associated with the different phenotypes induced by the 2 DNMTs. Knockdown of DNMT3a expression caused more down-regulated genes in the pathways related to morphogenesis of cardiovascular tissue (Fig. 8A) and formation of actin filaments (Fig. 8B), compared with the knockdown of DNMT3b, which is consistent with the abnormal morphology seen in DNMT3a-deficient cardiomyocytes, but not in the DNMT3b deficient cells. DNMT3a siRNA also caused more down-regulated genes involved in mitogenesis (Fig. 8C) and synthesis of DNA (Fig. 8D) than that of DNMT3b siRNA. Similarly, DNMT3a siRNA suppressed more genes in the concentration of cAMP (Fig. 8E) and release of Ca2+ pathways (Fig. 8F), indicating more severely impaired cAMP and calcium signaling in the cardiomyocytes with reduced DNMT3a expression.
Figure 8.
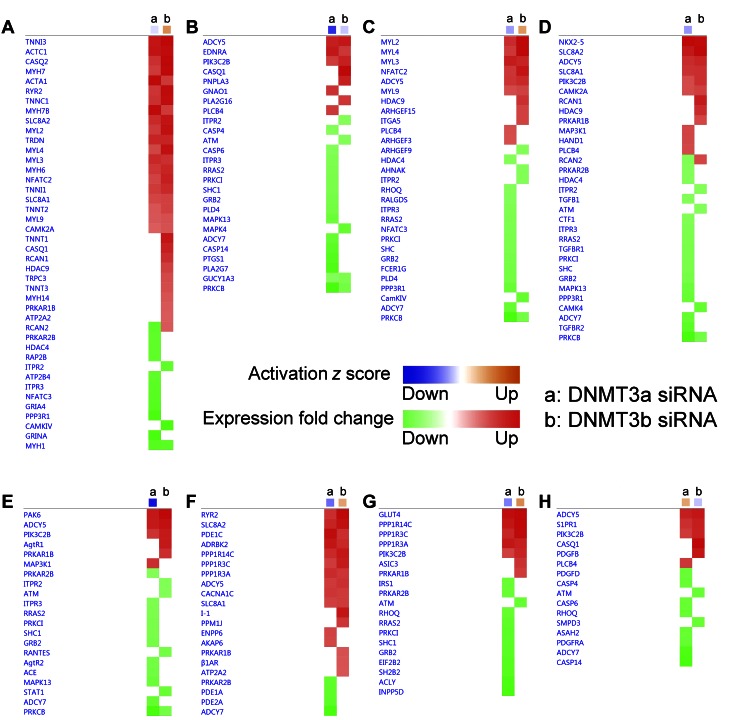
IPA identified the biologic functions that are differentially affected by knockdown of DNMT3a and -3b expression. IPA analysis of DE genes revealed that the morphogenesis of cardiovascular tissue (A), formation of actin filaments (B), mitogenesis (C), synthesis of DNA (D), concentration of cyclic AMP (E), and release of Ca2+ (F) pathways were differentially affected by knockdown of DNMT3a (a) and DNMT3b (b) expression. Orange, pathway has an activated z score; blue pathway has an inhibited z score; red, up-regulated DE gene; green, down-regulated DE gene.
The comparison of the affected canonical pathways after DNMT3a or -3b siRNA treatment also showed that compared with DNMT3b siRNA, DNMT3a siRNA suppressed more genes involved in calcium, endothelin-1, phospholipase C, nuclear factor of activated T-cells (NFAT), renin-angiotensin, cardiac β-adrenergic receptor, insulin receptor, and sphingosine-1 phosphate signaling (Fig. 9).
Figure 9.
IPA identified the canonical cardiac pathways that are differentially affected by knockdown of DNMT3a and -3b expression. IPA analysis on DE genes revealed that the calcium (A), endothelin-1 (B), phospholipase C (C), role of NFAT in cardiac hypertrophy (D), renin-angiotensin (E), cardiac β-adrenergic receptor (F), insulin receptor (G), and sphingosine-1-phosphate (H) signaling pathways were differentially affected by knockdown of DNMT3a (a) and DNMT3b (b) expression. Orange, pathway has an activated z score; blue pathway has an inhibited z score; red, up-regulated DE gene; green, down-regulated DE gene.
Gene network analysis of DE genes
One mechanism that leads to gene inactivation by DNMT is suppression of transcription factor binding to the gene promoter (6). With the DE genes identified with RNA-Seq, we used the iRegulon and IPA software to find the common transcription factors that may be activated or suppressed after DNMT knockdown. The iRegulon analysis on the up-regulated genes after DNMT3a siRNA treatment revealed that the transcription factors 2310045N01RIK, MEF2A, MEF2C, ESRRA, FOXN2, HSF4, NR3C1, and HNF4A were enriched (Table 6), suggesting enhanced activity of these factors. The transcription binding sites (motifs) and target genes of these transcription factors were also identified. For the down-regulated genes after knockdown of DNMT3a, the transcription factors ZIM1, HP1BP3, EVX2, TEAD1, FLIQ1, and TFAP2A were enriched, suggesting deactivation of these factors by DNMT3a siRNA.
TABLE 6.
iRegulon predicted activation or inhibition of transcription factors in embryonic cardiomyocytes after DNMT3a or DNMT3b siRNA treatment
| Transcription factor |
Normalized enrichment score |
Target genes (n) |
Motifs (n) |
|---|---|---|---|
| Up-regulated genes after DNMT3a siRNA treatment | |||
| 2310045N01RIK | 7.206 | 150 | 2 |
| MEF2A | 7.018 | 321 | 22 |
| MEF2C | 6.218 | 296 | 10 |
| ESRRA | 3.933 | 260 | 8 |
| FOXN2 | 3.683 | 60 | 3 |
| HSF4 | 3.287 | 34 | 1 |
| NR3C1 | 3.220 | 118 | 3 |
| HNF4A | 3.139 | 66 | 2 |
| Down-regulated genes after DNMT3a siRNA treatment | |||
| ZIM1 | 4.555 | 308 | 4 |
| HP1BP3 | 3.969 | 90 | 3 |
| EVX2 | 3.437 | 163 | 4 |
| TEAD1 | 3.401 | 126 | 2 |
| FLIQ1 | 3.356 | 276 | 7 |
| TFAP2A | 3.000 | 34 | 1 |
| Up-regulated genes after DNMT3b siRNA treatment | |||
| MEF2A | 7.906 | 368 | 33 |
| SRF | 7.803 | 281 | 6 |
| ESRRB | 3.798 | 288 | 6 |
| HNF1B | 3.329 | 260 | 1 |
| FOXN3 | 3.241 | 242 | 2 |
| Down-regulated genes after DNMT3b siRNA treatment | |||
| RUNX2 | 4.555 | 308 | 4 |
| HP1BP3 | 3.969 | 90 | 3 |
| EVX2 | 3.437 | 163 | 4 |
| TEAD1 | 3.401 | 126 | 2 |
| GATA5 | 3.356 | 276 | 7 |
| TFAP2A | 3.000 | 34 | 1 |
The DE genes induced by knockdown of DNMT3b shared some common transcription factors with the DE genes associated with DNMT3a knockdown, including MEF2A, HP1BP3, EVX2, TEAD1, and TFAP2A, but DNMT3b siRNA also influenced some other transcription factors that were not affected by DNMT3a siRNA (Table 6).
IPA analysis also predicted activation or inhibition of transcription factors by DNMT3a siRNA, even though these regulators may not be altered at the transcription level (Supplemental Table 5). These results indicate that several transcription factors known to be related to cardiac gene regulation, including GATA2, GATA4, MEF2A, MEF2C, MEF2D, HAND2, and NKX2.5, are predicted to be activated.
DNMT changes in embryonic cardiomyocytes after DNMT knockdown
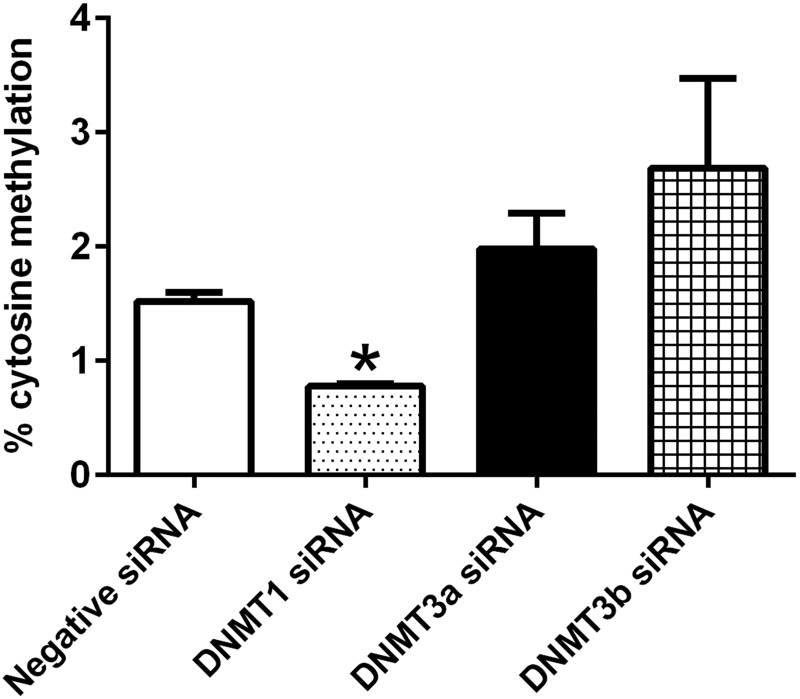
We next examined the global and gene-specific DNA methylation changes after knockdown of DNMTs. At 72 h after siRNA transfection, global cytosine methylation was significantly reduced by DNMT1 siRNA, but it remained unchanged after knockdown of DNMT3a or -3b expression (Fig. 10).
Figure 10.
Changes in global cytosine methylation after knockdown of DNMT expression. Knockdown of DNMT1 expression for 72 h significantly decreased global DNMT in embryonic cardiomyocytes. Knockdown of DNMT3a or -3b did not change global DNMT (n = 3). *P < 0.05 vs. negative siRNA.
We used multiplex targeted bisulfite sequencing to analyze promoter methylation of 15 genes that are related to cardiomyocyte morphology and proliferation. Two proximal promoter regions of each gene were interrogated. After knockdown of DNMT3a for 72 h, decreased methylation was observed in the promoter region of Myh7, Myh7b, Tnni3, and Tnnt2 (Table 7). The expression of these genes was up-regulated by knockdown of DNMT3a. Increased CpG methylation was found in the promoters of Nppa and Nppb, and their expression was significantly increased as well. Although decreased CpG methylation was identified in Mef2c, Mef2d, and Cdkn1C promoters, expressions of these genes were unchanged by DNMT3a siRNA treatment at the mRNA level.
TABLE 7.
Methylation changes in promoter CpGs after knockdown of DNMT3a expression for 72 h
| Gene | CpG location in amplicona | FDR of methylation change | Methylation difference (%) | Fold change in gene expressionb | FDR of fold change |
|---|---|---|---|---|---|
| Myh7_1 | 102 (2) | 0 | −6.20 | 1.99 | 0.015563 |
| Myh7_1 | 118 (3) | 0 | −6.64 | ||
| Myh7_1 | 127 (4) | 0 | −9.72 | ||
| Myh7_1 | 234 (6) | 0 | −5.05 | ||
| Myh7b_1 | 161 (3) | 0 | −5.79 | 3.19 | 0.00021 |
| Myh7b_2 | 352 (4) | 0 | −7.86 | ||
| Tnni3_1 | 244 (5) | 0 | −5.64 | 3.44 | 6.15E-15 |
| Tnni3_1 | 355 (6) | 0 | −5.07 | ||
| Tnnt2_1 | 94 (1) | 0 | −6.96 | 1.64 | 0.000163 |
| Tnnt2_1 | 195 (3) | 0 | −9.75 | ||
| Tnnt2_2 | 58 (1) | 0 | −5.58 | ||
| Tnnt2_2 | 209 (3) | 0 | −6.72 | ||
| Tnnt2_2 | 224 (4) | 0 | −6.00 | ||
| Tnnt2_2 | 233 (5) | 0 | −7.83 | ||
| Nppa_1 | 77 (2) | 0 | 5.978 | 4.25 | 1.39E-24 |
| Nppb_2 | 328 (4) | 0 | 11.20 | 7.64 | 6.30E-52 |
| Nppb_2 | 341 (5) | 0 | 11.71 | ||
| Nppb_2 | 381 (6) | 0 | 10.21 | ||
| Mef2c_1 | 348 (6) | 2.43E-05 | −6.30 | — | — |
| Mef2d_1 | 26 (1) | 0.000177 | −11.27 | — | — |
| Mef2d_2 | 304 (7) | 0 | −6.02 | ||
| Cdkn1C_1 | 26 (2) | 0.000329 | −6.55 | — | — |
| Cdkn1C_1 | 40 (4) | 6.85E-06 | −6.55 | ||
| Cdkn1C_1 | 66 (8) | 3.68E-06 | −5.35 | ||
| Cdkn1C_1 | 69 (9) | 5.18E-05 | −5.63 | ||
| Cdkn1C_1 | 104 (11) | 6.05E-08 | −7.27 | ||
| Cdkn1C_1 | 265 (36) | 0 | −5.93 | ||
| Cdkn1C_1 | 287 (40) | 0 | −5.56 | ||
| Cdkn1C_1 | 291 (41) | 0 | −5.83 | ||
| Cdkn1C_1 | 340 (48) | 9.23E-07 | −5.20 |
Numbers in parentheses indicate the order of CpG site within the amplicon.
Values from RNA-Seq analysis.
After knockdown of DNMT3b expression for 72 h, decreased methylation was observed in the promoter region of Myh6, Myh7b, Tnni3, Tnnt2, Nppa, Nppb, and Cdkn1C (Table 8). The expression of these genes was up-regulated by knockdown of DNMT3b. Although decreased CpG methylation was identified in Mef2d and Cdkn1A, expression of these genes was unchanged by DNMT3b siRNA treatment.
TABLE 8.
Methylation changes in promoter CpGs after knockdown of DNMT3b expression for 72 h
| Gene | CpG location in amplicona | FDR of methylation change | Methylation difference (%) | Fold change in gene expressionb | FDR of fold change |
|---|---|---|---|---|---|
| Myh6_1 | 43 (1) | 0 | −5.12 | 2.68 | 1.45E-16 |
| Myh7b_1 | 145 (2) | 0 | −41.27 | 2.01 | 0.039445 |
| Myh7b_1 | 161 (3) | 0 | −12.51 | ||
| Tnni3_1 | 66 (1) | 0 | −5.48 | 4.15 | 4.87E-25 |
| Tnni3_1 | 194 (4) | 0 | −5.32 | ||
| Tnni3_1 | 355 (6) | 0 | −6.71 | ||
| Tnnt2_1 | 195 (3) | 0 | −6.89 | 1.98 | 6.72E-11 |
| Tnnt2_1 | 201 (4) | 0 | −5.42 | ||
| Tnnt2_1 | 350 (5) | 0 | −5.35 | ||
| Tnnt2_1 | 420 (6) | 9.28E-14 | −5.52 | ||
| Tnnt2_2 | 224 (4) | 0 | −5.53 | ||
| Tnnt2_2 | 233 (5) | 0 | −5.47 | ||
| Nppa_1 | 125 (3) | 0 | −7.13 | 4.76 | 4.04E-36 |
| Nppa_1 | 133 (4) | 0 | −6.85 | ||
| Nppa_1 | 296 (6) | 0 | −7.27 | ||
| Nppa_1 | 303 (7) | 0 | −7.62 | ||
| Nppa_1 | 310 (8) | 0 | −7.40 | ||
| Nppa_1 | 352 (9) | 0 | −7.53 | ||
| Nppb_2 | 328 (4) | 0 | −8.12 | 3.62 | 4.17E-19 |
| Nppb_2 | 341 (5) | 0 | −9.99 | ||
| Nppb_2 | 381 (6) | 1.29E-08 | −5.84 | ||
| Mef2d_1 | 26 (1) | 0.033702 | −6.91 | — | — |
| Cdkn1A_1 | 374 (4) | 0 | 13.00 | — | — |
| Cdkn1C_1 | 24 (1) | 5.11E-05 | −9.39 | 1.81 | 0.000374 |
| Cdkn1C_1 | 26 (2) | 2.19E-05 | −7.52 | ||
| Cdkn1C_1 | 33 (3) | 0.000303 | −6.04 | ||
| Cdkn1C_1 | 40 (4) | 1.10E-13 | −10.41 | ||
| Cdkn1C_1 | 54 (5) | 2.27E-08 | −6.65 | ||
| Cdkn1C_1 | 60 (7) | 5.78E-10 | −8.07 | ||
| Cdkn1C_1 | 66 (8) | 9.62E-07 | −5.42 | ||
| Cdkn1C_1 | 69 (9) | 3.42E-10 | −8.45 | ||
| Cdkn1C_1 | 96 (10) | 2.52E-07 | −6.39 | ||
| Cdkn1C_1 | 104 (11) | 2.15E-07 | −6.64 | ||
| Cdkn1C_1 | 106 (12) | 7.63E-10 | −7.55 | ||
| Cdkn1C_1 | 115 (16) | 2.61E-06 | −5.66 | ||
| Cdkn1C_1 | 123 (18) | 4.22E-10 | −7.73 | ||
| Cdkn1C_1 | 131 (19) | 1.32E-06 | −5.39 | ||
| Cdkn1C_1 | 203 (27) | 0 | -5.48 | ||
| Cdkn1C_1 | 209 (28) | 0 | −6.69 | ||
| Cdkn1C_1 | 213 (29) | 0 | −7.78 | ||
| Cdkn1C_1 | 237 (31) | 0 | −5.15 | ||
| Cdkn1C_1 | 258 (34) | 0 | −5.43 | ||
| Cdkn1C_1 | 262 (35) | 0 | −5.47 | ||
| Cdkn1C_1 | 265 (36) | 0 | −10.30 | ||
| Cdkn1C_1 | 269 (37) | 0 | −10.10 | ||
| Cdkn1C_1 | 273 (38) | 0 | −8.45 | ||
| Cdkn1C_1 | 283 (39) | 0 | −7.51 | ||
| Cdkn1C_1 | 287 (40) | 0 | −11.05 | ||
| Cdkn1C_1 | 291 (41) | 0 | −11.00 | ||
| Cdkn1C_1 | 298 (42) | 0 | −6.08 | ||
| Cdkn1C_1 | 307 (44) | 0 | −8.74 | ||
| Cdkn1C_1 | 313 (45) | 0 | −9.62 | ||
| Cdkn1C_1 | 327 (47) | 0 | −6.15 |
Numbers in parentheses indicate the order of CpG site within the amplicon.
Values from RNA-Seq analysis.
Using the PROMO software, we identified the transcription factor binding sites in the PCR-amplified promoter regions of our target genes. We found several binding sites for the transcription factors predicted to be activated after knockdown of DNMT3a expression. For instance, the target region of Myh7 contains the binding sites for MEF2C, MEF2D, HNF4A, FOXN2, GATA2, and NKX2-5; Myh7b contains binding sites for MEF2C, MEF2D, HNF4A, FOXN2, and GATA2; Tnni3 contains binding sites for MEF2D, HNF4A, FOXN2, GATA2, and NKX2–5; Tnnt2 contains binding sites for HNF4A, HNF1B, GATA2, and NKX2-5; Nppa contains binding sites for MEF2C, MEF2D, HNF4A, FOXN2, GATA2, SRF, and NKX2-5; and Nppb contains binding sites for HNF1B, FOXN2, GATA2, and NKX2-5. These results indicate that methylation changes in the promoter sequences of these genes may facilitate binding of transcription factors and gene expression.
DISCUSSION
Our previous studies demonstrated that down-regulation of DNMTs in embryonic ventricles by in utero caffeine exposure correlates with altered DNMT patterns, gene expression, and cardiac function in adulthood (18, 19). To examine the role of DNMTs in cardiac development, we investigated the effects of knockdown of DNMTs on gene expression, morphology, and function in embryonic cardiomyocytes. We used E13.5 cardiomyocytes isolated from embryonic ventricles, because this time point marks the complete separation of atria and ventricles in the developing heart (35), and the cardiomyocytes are under rapid proliferation, differentiation, and dynamic DNMT changes at this stage (2, 3). In the current study, our results showed that DNMT3a plays a critical role in cardiomyocyte development.
After validation of DNMT gene knockdown in the cardiomyocytes, we examined the effects of siRNA treatment on cell viability, cytotoxicity, and apoptosis. We find that survival of embryonic cardiomyocytes is sensitive to decreased DNMT expression. These results are consistent with other reports, which show that inhibition of DNMTs promotes apoptosis in several cell types (36).
Morphologically, after DNMT3a siRNA treatment, we observed the loss of Z-band organization, which is linked to decreased function in cells, including decreased beating frequency, contractility, contraction synchronicity, field action potential, and calcium signaling. Thus, our studies indicate an essential role for DNMT3a in cardiac development.
In contrast to DNMT3a, we found that DNMT1 and -3b siRNA treatments had much less severe adverse effects on either cardiomyocyte morphology or function. To date, we are unaware of studies that investigated the relationship between DNMT1 expression and cardiac function. It has been reported that DNMT3b KO mice die between E13.5 and E16.5 of cardiac ventricular septal defects (9), and tamoxifen-induced cardiomyocyte-specific loss of DNMT3b in adult heart for 3–4 wk results in compromised systolic function, increased interstitial fibrosis, and myosarcomeric disarray (10). We observed reduced cardiomyocyte beating at 24 and 48 h after DNMT3b siRNA transfection in vitro, but it returned to normal at 72 h, and the video assay data show that the beating frequency and velocity of maximum contraction and relaxation were increased at d 6 and 7 after transfection (Fig. 5). It is possible that the effects of DNMT3b knockdown on cardiomyocytes may progress more slowly, thus requiring long-term inhibition to observe effects on function. Therefore, a stable knockdown of DNMT3b may be needed to further characterize the role of DNMT3b in cardiomyocyte function.
Because DNMT3a and -3b caused different phenotypes at 72 h after siRNA transfection, we performed RNA-Seq analysis on the isolated cardiomyocytes to detect the underlying mechanism. We identified both common and distinctive DE genes for the 2 treatments. The common DE genes included increased expression of contractile genes and fetal cardiac genes, such as Nppa, Nppb, Myh7, Myh7b, and Cacna1h, which is a hallmark of adult cardiac disease (37). The overexpression of cardiac contractile genes and fetal cardiac genes may potentiate development of pathologic hypertrophy and compromise contractility in cells (38).
The comparison of the affected biologic functions revealed that the pathways for morphogenesis of cardiovascular tissue and formation of actin filaments were inhibited by DNMT3a siRNA, but not by DNMT3b siRNA, which is consistent with the abnormal morphology of DNMT3a-deficient cardiomyocytes. DNMT3a siRNA also inhibited the mitogenesis and synthesis of DNA pathways, suggesting that DNMT3a plays a more active role than DNMT3b in cell proliferation. The altered expression of genes in the cell death and survival pathways may be responsible for the increased cell death and apoptosis observed with knockdown of DNMT3a expression. Pathway analysis identified inhibition of the genes in the calcium release pathway, which may explain the reduced calcium signaling observed in DNMT3a-siRNA–treated cells.
At the gene expression level, DNMT3a siRNA altered the expression of genes that would lead to inhibition of the pathways involved in calcium, endothelin-1, phospholipase C, NFAT, renin-angiotensin, cardiac β-adrenergic receptor, and insulin receptor signaling. Inhibition of these pathways usually associates with decreased cardiac function (39). In contrast, DNMT3b siRNA affected genes that activate calcium, phospholipase C, cardiac β-adrenergic receptor, and insulin receptor signaling networks, which may explain the different effects on function observed in the DNMT3b siRNA-treated cells compared to DNMT3a siRNA treated cells.
In a prior study, we found that DNMT3a is the DNMT isoform that is most severely affected by in utero caffeine exposure in the developing heart, with a greater than 4-fold reduction in expression (18). In addition, in utero caffeine exposure increased the expression of cardiac genes, including Myh6, Myh7, Tnni3, Nppa, and Nppb (18). In this study, we found that these genes were also upregulated by knockdown of DNMT3a and -3b expression, which provides the first indication that DNMTs may play a role in mediating in utero caffeine effects on cardiac gene expression.
Our findings show that global cytosine methylation was affected by knockdown of DNMT1 expression, but not by knockdown of either DNMT3a or -3b. Even though the overall methylation levels were not changed, DNMT3a and 3b induced methylation changes in specific gene promoters that correlated with changes in gene expression.
Overall, we have demonstrated that knockdown of DNMT3a, but not of DNMT1 or -3b, disrupts sarcomere assembly, alters cardiac gene expression and splicing, and inhibits beating frequency, contractile movement, and field action potential. Collectively, these data indicate that DNMT3a plays an important role in regulating cardiomyocyte gene expression, morphology, and function.
ACKNOWLEDGMENTS
The authors thank Dr. Jason Coleman (University of Florida) for technical assistance in analyzing the calcium signaling data. This work was supported by U.S. National Institutes of Health, National Institute of Child Health and Human Development Grant R01 HD058086 (to S.A.R. and C.C.W.), and by the Children’s Miracle Network. Author contributions: X. Fang, S. Rivkees, and C. Wendler designed the research; X. Fang, J. Wang-Hu, O. Shi, and N. Calvo performed the research; X. Fang and R. R. Poulsen analyzed the data; X. Fang and C. Wendler wrote the paper; and N. Calvo and C. Simmons developed the software necessary to perform and record the experiments.
Glossary
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- DE
differential expression
- DEU
differential exon usage
- DNMT
DNA methyltransferase
- E
embryonic day
- FDR
false-discovery rate
- fps
frames per second
- IPA
Ingenuity pathway analysis
- KO
knockout
- qPCR
quantitative PCR
- MEA
multielectrode array
- NFAT
nuclear factor of activated T-cells
- siRNA
small interfering RNA
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1.Evans S. M., Yelon D., Conlon F. L., Kirby M. L. (2010) Myocardial lineage development. Circ. Res. 107, 1428–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilsbach R., Preissl S., Grüning B. A., Schnick T., Burger L., Benes V., Würch A., Bönisch U., Günther S., Backofen R., Fleischmann B. K., Schübeler D., Hein L. (2014) Dynamic DNA methylation orchestrates cardiomyocyte development, maturation and disease. Nat. Commun. 5, 5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chamberlain A. A., Lin M., Lister R. L., Maslov A. A., Wang Y., Suzuki M., Wu B., Greally J. M., Zheng D., Zhou B. (2014) DNA methylation is developmentally regulated for genes essential for cardiogenesis. J. Am. Heart Assoc. 3, e000976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haas J., Frese K. S., Park Y. J., Keller A., Vogel B., Lindroth A. M., Weichenhan D., Franke J., Fischer S., Bauer A., Marquart S., Sedaghat-Hamedani F., Kayvanpour E., Köhler D., Wolf N. M., Hassel S., Nietsch R., Wieland T., Ehlermann P., Schultz J. H., Dösch A., Mereles D., Hardt S., Backs J., Hoheisel J. D., Plass C., Katus H. A., Meder B. (2013) Alterations in cardiac DNA methylation in human dilated cardiomyopathy. EMBO Mol. Med. 5, 413–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koczor C. A., Lee E. K., Torres R. A., Boyd A., Vega J. D., Uppal K., Yuan F., Fields E. J., Samarel A. M., Lewis W. (2013) Detection of differentially methylated gene promoters in failing and nonfailing human left ventricle myocardium using computation analysis. Physiol. Genomics 45, 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kass S. U., Pruss D., Wolffe A. P. (1997) How does DNA methylation repress transcription? Trends Genet. 13, 444–449 [DOI] [PubMed] [Google Scholar]
- 7.Flores K., Wolschin F., Corneveaux J. J., Allen A. N., Huentelman M. J., Amdam G. V. (2012) Genome-wide association between DNA methylation and alternative splicing in an invertebrate. BMC Genomics 13, 480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lei H., Oh S. P., Okano M., Jüttermann R., Goss K. A., Jaenisch R., Li E. (1996) De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122, 3195–3205 [DOI] [PubMed] [Google Scholar]
- 9.Okano M., Bell D. W., Haber D. A., Li E. (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 [DOI] [PubMed] [Google Scholar]
- 10.Vujic A., Robinson E. L., Ito M., Haider S., Ackers-Johnson M., See K., Methner C., Figg N., Brien P., Roderick H. L., Skepper J., A Ferguson-Smith, Foo R. S. (2015) Experimental heart failure modelled by the cardiomyocyte-specific loss of an epigenome modifier, DNMT3B. J. Mol. Cell. Cardiol. 82, 174–183 [DOI] [PubMed] [Google Scholar]
- 11.Childhood Overgrowth Consortium (2014) Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat. Genet. 46, 385–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheng W., Qian Y., Wang H., Ma X., Zhang P., Chen L., Ma D., Huang G. (2013) Association between mRNA levels of DNMT1, DNMT3A, DNMT3B, MBD2 and LINE-1 methylation status in infants with tetralogy of Fallot. Int. J. Mol. Med. 32, 694–702 [DOI] [PubMed] [Google Scholar]
- 13.Tao H., Yang J. J., Chen Z. W., Xu S. S., Zhou X., Zhan H. Y., Shi K. H. (2014) DNMT3A silencing RASSF1A promotes cardiac fibrosis through upregulation of ERK1/2. Toxicology 323, 42–50 [DOI] [PubMed] [Google Scholar]
- 14.Ponikowski P., Anker S. D., Chua T. P., Szelemej R., Piepoli M., Adamopoulos S., Webb-Peploe K., Harrington D., Banasiak W., Wrabec K., Coats A. J. (1997) Depressed heart rate variability as an independent predictor of death in chronic congestive heart failure secondary to ischemic or idiopathic dilated cardiomyopathy. Am. J. Cardiol. 79, 1645–1650 [DOI] [PubMed] [Google Scholar]
- 15.Lachance G., Uniacke J., Audas T. E., Holterman C. E., Franovic A., Payette J., Lee S. (2014) DNMT3a epigenetic program regulates the HIF-2α oxygen-sensing pathway and the cellular response to hypoxia. Proc. Natl. Acad. Sci. USA 111, 7783–7788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castillo P., Ibáñez F., Guajardo A., Llanos M. N., Ronco A. M. (2012) Impact of cadmium exposure during pregnancy on hepatic glucocorticoid receptor methylation and expression in rat fetus. PLoS One 7, e44139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng Y., Zhao L. Z., Hong L., Shan C., Shi W., Cai W. (2013) Alteration in methylation pattern of GATA-4 promoter region in vitamin A-deficient offspring’s heart. J. Nutr. Biochem. 24, 1373–1380 [DOI] [PubMed] [Google Scholar]
- 18.Fang X., Mei W., Barbazuk W. B., Rivkees S. A., Wendler C. C. (2014) Caffeine exposure alters cardiac gene expression in embryonic cardiomyocytes. Am. J. Physiol. Regul. Integr. Comp. Physiol. 307, R1471–R1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buscariollo D. L., Fang X., Greenwood V., Xue H., Rivkees S. A., Wendler C. C. (2014) Embryonic caffeine exposure acts via A1 adenosine receptors to alter adult cardiac function and DNA methylation in mice. (Published correction available at http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0097212). PLoS One 9, e87547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang X., Robinson J., Wang-Hu J., Jiang L., Freeman D. A., Rivkees S. A., Wendler C. C. (2015) cAMP induces hypertrophy and alters DNA methylation in HL-1 cardiomyocytes. Am. J. Physiol. Cell Physiol. 309, C425–C436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burridge P. W., Metzler S. A., Nakayama K. H., Abilez O. J., Simmons C. S., Bruce M. A., Matsuura Y., Kim P., Wu J. C., Butte M., Huang N. F., Yang P. C. (2014) Multi-cellular interactions sustain long-term contractility of human pluripotent stem cell-derived cardiomyocytes. Am. J. Transl. Res. 6, 724–735 [PMC free article] [PubMed] [Google Scholar]
- 22.Galaxy Team (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11, R86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S., Pyl P. T., Huber W. (2015) HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson M. D., McCarthy D. J., Smyth G. K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love M. I., Huber W., Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 27.Fang X., Corrales J., Thornton C., Clerk T., Scheffler B. E., Willett K. L. (2015) Transcriptomic changes in zebrafish embryos and larvae after benzo[a]pyrene exposure. Toxicol. Sci. 146, 395–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janky R., Verfaillie A., Imrichová H., Van de Sande B., Standaert L., Christiaens V., Hulselmans G., Herten K., Naval Sanchez M., Potier D., Svetlichnyy D., Kalender Atak Z., Fiers M., Marine J. C., Aerts S. (2014) iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLOS Comput. Biol. 10, e1003731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Messeguer X., Escudero R., Farré D., Núñez O., Martínez J., Albà M. M. (2002) PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18, 333–334 [DOI] [PubMed] [Google Scholar]
- 30.Anders S., Reyes A., Huber W. (2012) Detecting differential usage of exons from RNA-seq data. Genome Res. 22, 2008–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu J. L., Liu L. P., Yang S. L., Fang X., Wen L., Ren Q. G., Yu C. (2016) Hepatitis B virus induces hypoxia-inducible factor-2α expression through hepatitis B virus X protein. Oncol. Rep. 35, 1443–1448 [DOI] [PubMed] [Google Scholar]
- 32.Fang X., Thornton C., Scheffler B. E., Willett K. L. (2013) Benzo[a]pyrene decreases global and gene specific DNA methylation during zebrafish development. Environ. Toxicol. Pharmacol. 36, 40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krueger F., Andrews S. R. (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akalin A., Kormaksson M., Li S., Garrett-Bakelman F. E., Figueroa M. E., Melnick A., Mason C. E. (2012) methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13, R87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Savolainen S. M., Foley J. F., Elmore S. A. (2009) Histology atlas of the developing mouse heart with emphasis on E11.5 to E18.5. Toxicol. Pathol. 37, 395–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu S., Ren J., Chen H. B., Wang Y., Liu Q., Zhang R., Jiang S. W., Li J. (2014) Cytostatic and apoptotic effects of DNMT and HDAC inhibitors in endometrial cancer cells. Curr. Pharm. Des. 20, 1881–1887 [DOI] [PubMed] [Google Scholar]
- 37.Kuwahara K., Nishikimi T., Nakao K. (2012) Transcriptional regulation of the fetal cardiac gene program. J. Pharmacol. Sci. 119, 198–203 [DOI] [PubMed] [Google Scholar]
- 38.Akazawa H., Komuro I. (2003) Roles of cardiac transcription factors in cardiac hypertrophy. Circ. Res. 92, 1079–1088 [DOI] [PubMed] [Google Scholar]
- 39.Van Berlo J. H., Maillet M., Molkentin J. D. (2013) Signaling effectors underlying pathologic growth and remodeling of the heart. J. Clin. Invest. 123, 37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]