Abstract
Diabetes mellitus is a complex and heterogeneous disease, which has β-cell dysfunction at its core. Glucotoxicity affects pancreatic islets, causing β-cell apoptosis. However, the role of JNK/β-catenin signaling in glucotoxic β-cell apoptosis is not well understood. Recently, we identified tetraspanin-2 (TSPAN2) protein as a proapoptotic β-cell factor induced by glucose, suggesting that TSPAN2 might contribute to pancreatic β-cell glucotoxicity. To investigate the effects of glucose concentration on TSPAN2 expression and apoptosis, we used reverted immortalized RNAKT-15 human pancreatic β cells. High TSPAN2 levels up-regulated phosphorylated (p) JNK and induced apoptosis. p-JNK enhanced the phosphorylation of β-catenin and Dickkopf-1 (Dkk1). Dkk1 knockdown by small interfering (si)RNA up-regulated nuclear β-catenin, suggesting that it is a JNK/β-catenin-dependent pathway. siRNA-mediated TSPAN2 depletion in RNAKT-15 cells increased nuclear β-catenin. This decreased BCL2-associated X protein (Bax) activation, leading to marked protection against high glucose–induced apoptosis. Bax subfamily proteins induced apoptosis through caspase-3. Thus, TSPAN2 might have induced Bax translocation and caspase-3 activation in pancreatic β cells, thereby promoting the apoptosis of RNAKT-15 cells by regulating the JNK/β-catenin pathway in response to high glucose concentrations. Targeting TSPAN2 could be a potential therapeutic strategy to treat glucose toxicity-induced β-cell failure.—Hwang, I.-H., Park, J., Kim, J. M., Kim, S. I., Choi, J.-S., Lee, K.-B., Yun, S. H., Lee, M.-G., Park, S. J., Jang, I.-S. Tetraspanin-2 promotes glucotoxic apoptosis by regulating the JNK/β-catenin signaling pathway in human pancreatic β cells.
Keywords: Bax, glucose toxicity, TSPAN2
In diabetes, glucose toxicity affects various organ systems, including pancreatic islets, where it leads to β-cell apoptosis. However, the underlying mechanisms are not fully understood. The dysfunction of β cells and impaired insulin production are the hallmarks of diabetes (1), but despite the growing diabetes epidemic, the exact molecular mechanisms by which glucose toxicity leads to β-cell apoptosis are still not fully understood (2). Tetraspanins are membrane proteins containing 4 transmembrane domains and are found in nearly all mammalian cells. Tetraspanins are involved in fundamental cellular processes, including cell growth, adhesion, and differentiation (3, 4). Members of the tetraspanin family tend to have highly conserved amino acid sequences (5). The function and expression of tetraspanin-2 (TSPAN2), however, have not yet been studied. We identified TSPAN2 as a proapoptotic β-cell factor induced by glucose, suggesting its possible role in β-cell glucose toxicity.
Apoptosis is a naturally occurring process in the body wherein a specialized intracellular signaling pathway is activated, killing the cells. It is a homeostatic mechanism to maintain cell populations in tissues. Improper apoptosis is a factor underlying various physiological conditions, including glucose toxicity, neurodegenerative diseases, metabolic stress such as ischemic damage, autoimmune disorders, and many types of cancer (6). The binding of c-Jun and β-catenin/TCF4 to the c-Jun promoter depends upon JNK activity; thus, one role for this complex is to contribute to the repression and/or activation of genes that may mediate cell maintenance, proliferation, differentiation, and death, whereas the down-regulation of these signals may contribute to carcinogenesis (7). JNK functions in the noncanonical Wnt pathway to regulate convergent extension movements in vertebrate gastrulation (8). JNK also prevents nuclear β-catenin accumulation and regulates axis formation in Xenopus embryos (9). β-Catenin enhances the survival of renal epithelial cells by inhibiting BCL2-associated X protein (Bax), which has a proapoptotic function within the cell (10).
Although the effects of glucose toxicity on β-cell dysfunction and apoptosis have been studied in detail, the precise molecular mechanisms of glucose toxicity have not been elucidated. Glucotoxicity-induced pancreatic β-cell apoptosis by the regulation of the JNK/β-catenin pathway is an aspect that remains to be understood. Therefore, we investigated the role of the TSPAN2-associated JNK/β-catenin pathway in islet β-cell apoptosis.
MATERIALS AND METHODS
Cell culture
The reverted immortalized human pancreatic β-cell line RNAKT-15 was obtained from H. S. Jun (Lee Gil Ya Cancer and Diabetes Institute, Gachon University of Medicine and Science, Inchon, Korea). Cells were grown in low-glucose (5 mM) DMEM supplemented with 10% fetal bovine serum, 1% penicillin–streptomycin, 10 mM nicotinamide, 10 μM troglitazone, and 16.7 μM zinc sulfate. Cells were cultured in a humidified incubator at 37°C and 5% CO2. Glucotoxicity was induced by treatment with high glucose (33 mM) for 48 h. The JNK inhibitor SP600125 (10 μM; Sigma-Aldrich, St. Louis, MO, USA) was administered at 48 h.
Microarray analysis
Human whole-genome microarrays (Agilent Technologies, Santa Clara, CA, USA) were used for transcription profiling analysis. For control and test RNAs, the synthesis of target cRNA probes and hybridization were performed using a Low RNA Input Linear Amplification kit (Agilent) according to the manufacturer’s instructions. In brief, 1 μg of total RNA from each sample was mixed with a T7 promoter primer mix and incubated at 65°C for 10 min. The cDNA master mix [5× first strand buffer, 0.1 M DTT, 10 mM dNTP mix, RNase-Out, and Moloney murine leukemia virus reverse transcriptase (RT)] was then prepared and added to the reaction mixture. Samples were incubated at 40°C for 2 h for RT, and dsDNA synthesis was terminated by incubation at 65°C for 15 min. Transcription of dsDNA was performed by the addition of the transcription master mix to dsDNA reaction samples, which were subsequently incubated at 40°C for 2 h. Amplified and labeled cRNA was purified using a cRNA Cleanup Module (Agilent) according to the manufacturer’s protocol. Labeled cRNA was quantified using an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). cRNA was fragmented by adding 10× blocking agent and 25× fragmentation buffer, followed by incubation at 60°C for 30 min. The fragmented cRNA was resuspended with 2× hybridization buffer and pipetted directly onto microarrays (44K). The arrays were then hybridized at 65°C for 17 h using an Agilent Hybridization Oven (Agilent). The hybridized microarrays were washed according to the manufacturer’s washing protocol. Fluorescent images were quantified and normalized as described previously. The microarray data have been submitted to the National Center for Biotechnology Information (NCBI; Bethesda, MD, USA) the Gene Expression Omnibus database (GEO accession number GSE76189; http://www.ncbi.nlm.nih.gov/geo/).
Data acquisition and analysis
Hybridized images were scanned using an Agilent DNA microarray scanner and quantified using Feature Extraction software (Agilent). Data normalization and determination of fold changes in gene expression were performed using GeneSpringGX 7.3 (Agilent). Briefly, the averages of normalized ratios were calculated by dividing the average of the normalized signal channel intensity by the average of the normalized control channel intensity. Functional annotation of genes was performed using Gene Ontology Consortium data (http://www.geneontology.org/index.shtml) with GeneSpringGX 7.3. Gene classification was based on queries applied in BioCarta (http://www.biocarta.com/), Gene Map Annotator and Pathway Profiler (GenMAPP) (http://www.genmapp.org/), Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/), and Medline databases (http://www.ncbi.nlm.nih.gov/).
Gene Ontology–related network analysis
Bioinformatics gene network analyses were performed using Ingenuity Pathway Analysis (IPA; http://www.ingenuity.com) to examine the biologic functions of the differentially regulated genes and proteins according to ontology-related interaction networks, including apoptosis signaling. IPA provides protein interaction networks based on the regularly updated Ingenuity Pathways Knowledge Base. The network is optimized to include as many proteins from the input expression profile as possible and aims to produce highly connected networks
Flow cytometric analysis using annexin V staining
To detect apoptosis, we analyzed annexin V and propidium iodide (PI)–annexin V staining with an Annexin V–FLUOS staining kit (Roche Diagnostics, Mannheim, Germany). Cells were treated with normal glucose (NG) and high glucose (HG) for 48 h and then scraped and washed twice with PBS. The cell suspension was centrifuged at 2000 g for 2 min and incubated with 0.2 mg/ml Annexin V–FLUOS or with added PI (1.4 mg/ml) for 15 min at room temperature. Analyses were performed using a MoFlo Astrios flow cytometer (Beckman Coulter, Brea, CA, USA) at a 488 nm excitation with a 530/30 nm bandpass filter to detect annexin V. Data were analyzed by Summit 6.0 software.
Acquired nuclear fraction
RNAKT-15 cells were prepared by incubation with a HG medium for 2 d. The cells were scraped and added to 2 ml of homogenization buffer A (25 mM Tris, pH 7.5, 2 mM EDTA, 0.5 mM EGTA, 1 mM DTT, protease inhibitor cocktail, 1 mM PMSF, and 0.02% Triton X-100) per culture dish; homogenized 15 times using a 15 ml Dounce homogenizer with pestle A; and centrifuged at 100,000 g for 30 min. The supernatant cytosolic fraction was transferred into a new tube, and 500 μl of homogenization buffer B (homogenization buffer A containing 1% Triton X-100) was added to the pellet. The pellet was resuspended by sonication, incubated for 30 min at 4°C by rocking, and centrifuged at 100,000 g for 30 min. The supernatant nuclear fraction was transferred into a fresh tube. The protein contents of the cytosolic and nuclear fractions were determined by using a bicinchoninic acid (BCA) assay kit (Thermo Fisher Scientific, Waltham, MA, USA) and analyzed by Western blot analysis against anti-β-catenin antibodies.
Western blot analysis
Cells were lysed in RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) containing a protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). The protein concentration of cell lysates was determined with a BCA protein assay kit. Cell lysates containing equal amounts of protein were separated by SDS-PAGE and transferred onto Immobilon NC membranes. Blots were then blocked with 5% (w/v) skim milk powder or 5% (w/v) bovine serum albumin (BSA) in Tris-buffered saline and Tween 20 (0.05% Tween 20) for 1 h, probed with antibodies against TSPAN2 (1:1000; Abnova, Taipei, Taiwan), glyceraldehyde phosphate dehydrogenase (GAPDH; 1:5000; Santa Cruz Biotechnology, Santa Cruz, CA, USA), E-cadherin (1:1000; Cell Signaling Technology, Danvers, MA, USA), caspase-3 (1:800; Cell Signaling), β-catenin (1:1000; Santa Cruz), phosphorylated (p) β-catenin (Ser33/37/Thr41, 1:200; Cell Signaling), JNK (1:1000; Santa Cruz), phosphorylated JNK (p-JNK; 1:500; Santa Cruz), truncated Bid (t-Bid; 1:1000; Santa Cruz), poly(adenosine diphosphate-ribose) polymerase (PARP; 1:2000; Santa Cruz), Bax (1:1000; Abcam, Cambridge, MA, USA), Akt (1:1000; Cell Signaling), phosphorylated Akt (p-Akt; 1:500; Cell Signaling), B-cell CLL/lymphoma 2 (Bcl-2; 1:6000; Cell Signaling), and Dickkopf-1 (Dkk1; 1:500; Cell Signaling) at 4°C overnight and incubated with peroxidase-conjugated secondary antibodies for 1 h. The membranes were rinsed 3 times with Tris-buffered saline and Tween 20 for 5 min each, and an enhanced chemiluminescence system (Thermo Fisher Scientific) was used to visualize the bands on a ChemiDoc MP system (Bio-Rad, Hercules, CA, USA). Densitometric measurements of bands were performed using ImageJ software (Image Processing and Analysis in Java; National Institutes of Health, Bethesda, MD, USA).
Plasmid construction, RNA interference, and quantitative RT-PCR
The TSPAN2 plasmid construction kit was purchased from GenScript (Piscataway, NJ, USA). pcDNA3.1 (Thermo Fisher Scientific) was used to generate expression vectors. Small interfering (si)RNAs were purchased from ST Pharm (Seoul, Korea). The nucleotide sequence for TSPAN2 siRNA was 5′-CGA UUU GGA GGU ACC AUA AAG GAU U-3′ and for Dkk1 siRNA, 5′-AAG AAC GGA AGU GUG AUA UGU-3′. The transfection of siRNA into RNAKT-15 cells was performed using Lipofectamine RNAiMax reagent (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions. RT-PCR was performed to compare the relative amounts of mRNAs in cells cultured in NG and HG medium. RT was performed in a final volume of 20 μl using Superscript III RT (Thermo Fisher Scientific) according to the manufacturer’s protocol. Subsequently, 4 μl of the final RT product mix was PCR amplified. The primer used for TSPAN2 was 5ʹ-ttg gaa ttg tcg gta ttg-3ʹ (NM_005725; 91 bp), and that for the internal standard GAPDH (NM_002046; 97 bp) was 5′-cga cca ctt tgt caa gct ca-3′.
Immunofluorescence microscopy
RNAKT-15 cells were seeded at 4 × 104 cells/well on a coverslip in a 12-well plate. After 24 h, the control cells were treated with HG (33 mM) for 48 h. TSPAN2 siRNA (25 μg)-treated cells were incubated for 24 h and then treated with HG (33 mM) for 48 h. Cells were washed twice with PBS and fixed with 4% (v/v) formaldehyde for 15 min. Cells were allowed to permeabilize with 0.1% (v/v) Triton X-100 for 15 min and were then blocked with 3% (w/v) BSA for 1 h. The cells were incubated with primary antibody (β-catenin monoclonal purified mouse IgG1; Abnova) diluted to 1:100 in 3% (w/v) BSA overnight at 4°C. After 5 washes with PBS for 5 min each, the cells were incubated in the dark for 1 h with FITC–anti-mouse secondary antibody (Thermo Fisher Scientific) diluted to 1:200 in 3% (w/v) BSA. Images of cover slipped cells were collected with a laser-scanning confocal microscope (LSM 710; Carl Zeiss GmbH, Jena, Germany) equipped with a C-Apochromat 40×/1.2 water immersion lens (488 nm argon laser/505–550 nm detection range). Data were analyzed using Zen 2009 Light Edition software (Carl Zeiss).
Statistical analysis
All results are presented as the means ± SD of at least 3 independent experiments. Statistical significance was determined using Student’s t test using Prism 5.0 software (GraphPad Software, La Jolla, CA, USA). Values of P ≤ 0.05 were considered statistically significant.
RESULTS
Differential expression of genes under NG versus HG culture conditions in RNAKT-15 cells
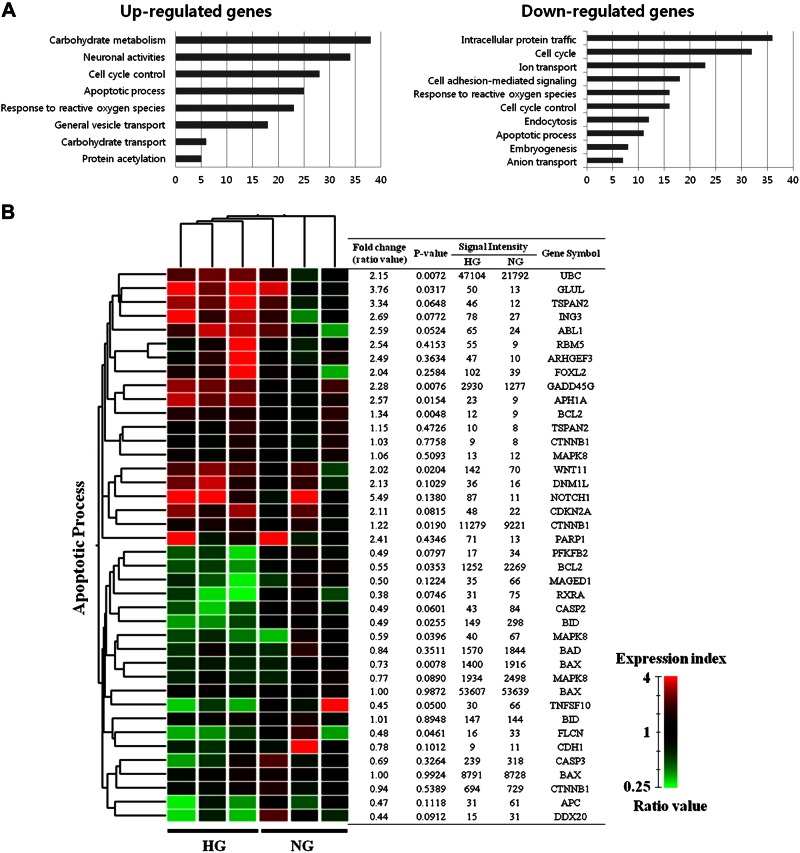
To identify the genes whose expression changed in response to HG, we performed microarray analysis in the RNAKT-15 cells. Among the 41,000 genes tested, 1394 and 741 had 2-fold up-regulation and down-regulation, respectively. In HG-treated RNAKT-15 cells, the genes that were up-regulated were in the following decreasing order: genes involved in carbohydrate metabolism > cell cycle control > apoptotic processes > response to reactive oxygen species. In contrast, the genes that were down-regulated were in the following order: genes involved in intracellular protein trafficking > cell cycle > cell adhesion-mediated signaling > cell cycle control > response to reactive oxygen species > apoptotic process (Fig. 1A). To compare our results in terms of glucose concentration and the genes potentially involved in apoptosis, we independently evaluated the association between apoptosis and genes that had a >2-fold change in HG compared with NG. The intersection obtained by hierarchical clustering is presented with the gene lists in Fig. 1B. In addition, we analyzed the signal network of the genes in response to glucose and found the expression of apoptotic genes to be modulated by glucose (Supplemental Fig. 1 and Supplemental Table 1). Microarray analysis showed that the expression of TSPAN2 increased up to 3-fold under the HG condition, and the signal network analysis suggested that TSPAN2 is involved in cellular apoptosis. Therefore, we focused on the role of TSPAN2 in HG-induced cell death.
Figure 1.
Gene expression analysis and signal network of apoptotic genes. A) Results of Gene Ontology analysis by microarray approaches in response to glucose. Gene lists corresponding to 2-fold up-regulation and down-regulation in NG- or HG-treated RNAKT-15 cells for 48 h were developed using DAVID for Gene Ontology analysis. B) Apoptotic genes and their hierarchical clustering in response to glucose. Dendrogram of hierarchical clustering revealed genes that were altered more than 2-fold as a result of apoptosis in response to HG and NG. Red and green represent more than 2-fold up-regulated and down-regulated genes, respectively. Microarray experiments were performed in triplicate. Ratio was calculated by dividing signal intensity of HG treatment by signal intensity of NG treatment.
HG induced TSPAN2 expression and apoptosis in RNAKT-15 cells
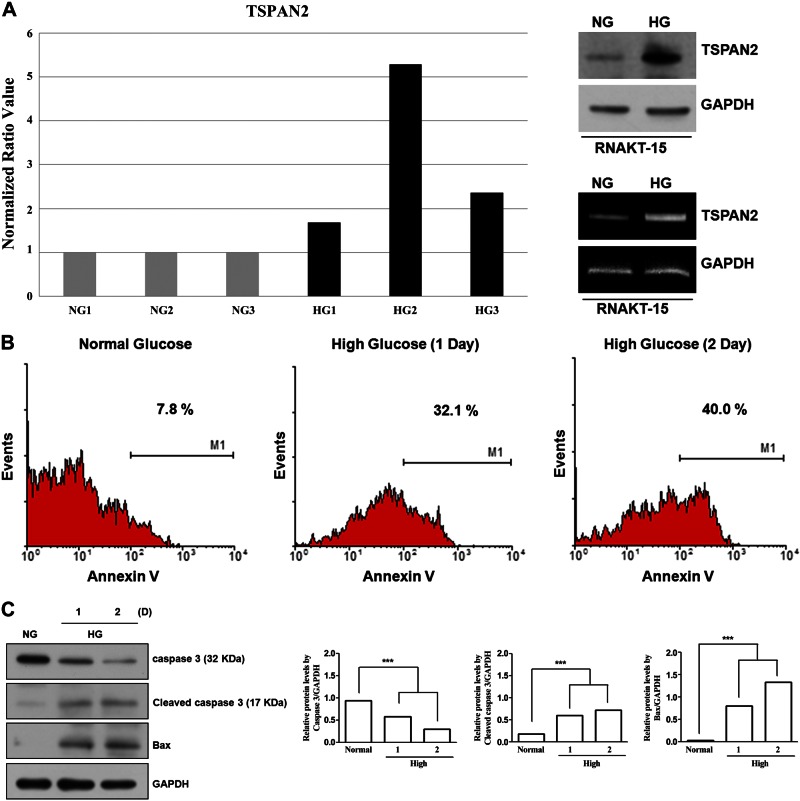
To determine the role of TSPAN2 in glucotoxicity-mediated β-cell apoptosis, we incubated RNAKT-15 islet cells under NG or HG conditions for 2 d. RNA was isolated from RNAKT-15 cells, and transcriptional profiling was performed using microarrays; RNAKT-15 cells maintained in NG were used as the control. Microarray experiments were repeated 3 times, and subsequent data analysis revealed that the expression of TSPAN2 was consistently increased in HG conditions (Fig. 2A, left). To confirm our gene expression analysis results, increased expression of TSPAN2 in HG conditions was verified by Western blot analysis (Fig. 2A, top) and RT-PCR (Fig. 2A, bottom). RNAKT-15 cells were incubated with either HG or NG medium for 2 d, after which cells were collected and equal amount of cell lysates were subjected to Western blot analysis with an anti-TSPAN2 antibody. TSPAN2 abundance was elevated up to 3-fold in HG conditions (Fig. 2A). Next, we examined mRNA levels of TSPAN2 by RT-PCR. TSPAN2 mRNA was increased up to 4-fold in HG conditions compared with control cells (Fig. 2A). Together, these results indicated that TSPAN2 was induced by HG conditions at the level of transcription. To examine the relationship between TSPAN2 and cell death, we tested whether HG conditions would induce apoptosis in RNAKT-15 cells. Cells were incubated in HG conditions for 2 d and were examined by annexin V staining. We observed that incubation at HG resulted in increased apoptosis (Fig. 2B), as measured by the increase in Bax and cleaved caspase-3 (Fig. 2C).
Figure 2.
Effect of HG on apoptosis of RNAKT-15 human pancreatic β cells. A) RNAKT-15 cells were incubated with NG (5 mM) and HG (33 mM) for 2 d. TSPAN2 levels increased in HG conditions. Elevated levels of TSPAN2 in HG medium were confirmed by Western blot (top) and RT-PCR (bottom) analyses. B) RNAKT-15 pancreatic β cells were incubated in NG or HG medium for periods indicated. Cells were then subjected to FITC–annexin V staining and analyzed by flow cytometry. C) Expression levels of Bax and cleaved caspase-3 increased in RNAKT-15 cells exposed to HG conditions. Elevated protein levels in HG medium were confirmed by Western blot analysis; NG vs. HG. ***P < 0.001.
Elevated TSPAN2 expression increased Bax levels under HG conditions
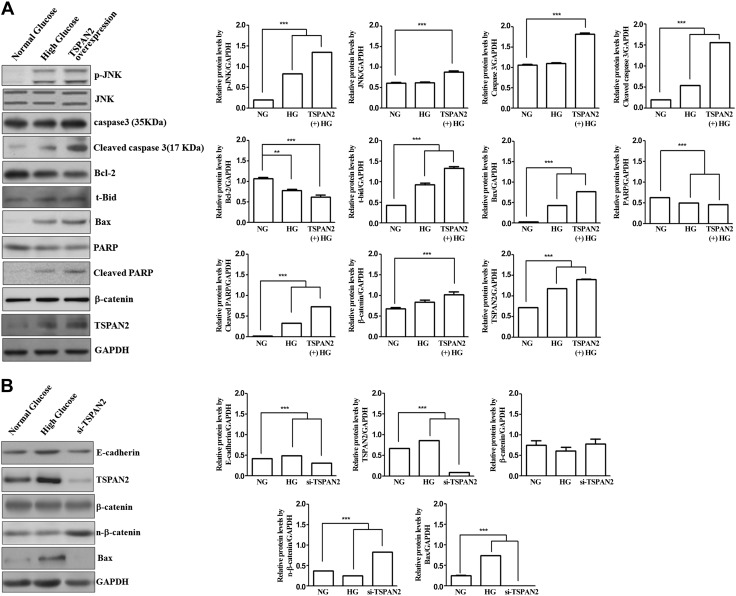
Because the expression of TSPAN2 is increased under HG conditions, we examined the cell signaling modulated by TSPAN2. RNAKT-15 cells were incubated under either NG or HG condition, and the cells transfected with plasmids expressing TSPAN2 were prepared for the comparison. The cell lysates were probed with various antibodies to examine the cell signaling. Western blot analysis showed that p-JNK, Bax, t-Bid, and cleaved PARP were increased under HG. These genes also were induced by the overexpression of TSPAN2 under NG (Fig. 3A). In addition, the expression of Bcl-2 and PARP were reduced by the HG condition and TSPAN2 overexpression. These results indicate that HG induces glucotoxicity-induced apoptosis, and the increased expression of TSPAN2 shows similar effects. To confirm that the up-regulation of TSPAN2 contributes to glucotoxicity-induced apoptosis, we silenced TSPAN2 expression by TSPAN siRNA under HG. TSPAN2 siRNA efficiently silenced the expression of TSPAN (Fig. 3B). We then examined the expression of Bax. Although HG increased the level of Bax protein, the silencing of TSPAN2 decreased the expression of Bax. These results suggest that the expression of TSPAN2 contributes to the expression Bax under HG conditions (Fig. 3B). In addition, we found that nuclear β-catenin was increased by TSPAN2 silencing.
Figure 3.
Caspase-3 activation and induction of PARP cleavage by TSPAN2 in RNAKT-15 cells exposed to glucose. A) TSPAN2 regulates expression of cleaved caspase-3 and cleaved PARP via JNK/β-catenin-Bax pathway. Western blot analysis was used to detect expression of caspase-3, Bcl-2, Bax, JNK, p-JNK, t-Bid, PARP, β-catenin, and TSPAN2 proteins in RNAKT-15 cells cultured in 5 and 33 μg/ml glucose as well as overexpression of TSPAN2 under NG. B) Effects of TSPAN2, β-catenin nuclear translocation, and Bax activation under NG, HG-up-regulated TSPAN2, and HG with si-TSPAN2 were detected by Western blot analysis. E-cadherin and GAPDH were used as protein loading control; NG vs. HG. ***P < 0.001.
JNK regulates nuclear β-catenin translocation signaling through Dkk1 expression
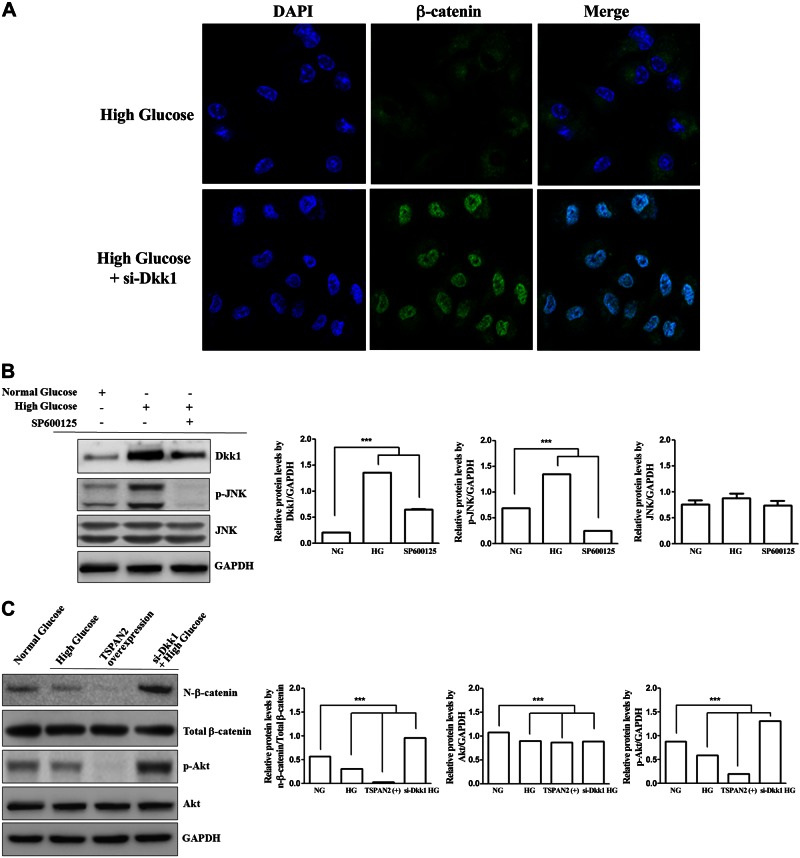
To elucidate the potential role of JNK in the regulation of β-catenin signaling, JNK inhibitor and DKK siRNA were transfected into RNAKT-15 cells. As shown in Fig. 4, Dkk1 was down-regulated in JNK inhibitor SP600125-treated RNAKT-15 cells and siRNA inhibition of Dkk1 up-regulated nuclear β-catenin protein levels and p-Akt under HG. Consistent with this finding, β-catenin nuclear translocation was inhibited under HG, whereas increases in both cytosolic and nuclear β-catenin under HG with si-Dkk1 were found by confocal microscopy (Fig. 4A). HG-induced p-JNK up-regulated p-β-catenin and Dkk1, and then, si-Dkk1 enhanced the nuclear translocation of β-catenin (Fig. 4B, C). These results show that TSPAN2 effectively attenuates nuclear β-catenin translocation by modulating the JNK-Dkk1 signaling pathway.
Figure 4.
JNK phosphorylated β-catenin and regulated nuclear β-catenin translocation. A) Effect of Dkk1 on β-catenin nuclear translocation in RNAKT-15 cells exposed to HG. Effects of β-catenin nuclear translocation under HG and HG with si-Dkk1 were observed by confocal microscopy. Representative analyses are of at least 4 separate experiments. B) JNK-Dkk1 signaling in RNAKT-15 cells in culture. Glucose-induced elevation of p-JNK up-regulated Dkk1. C) TSPAN2-mediated accumulation and nuclear translocation of active β-catenin inhibition in response to TSPAN2 overexpression in RNAKT-15 cells. si-Dkk1 up-regulated accumulation and nuclear translocation of active β-catenin and p-Akt.
TSPAN2 activates JNK to induce Bax by regulating β-catenin activation
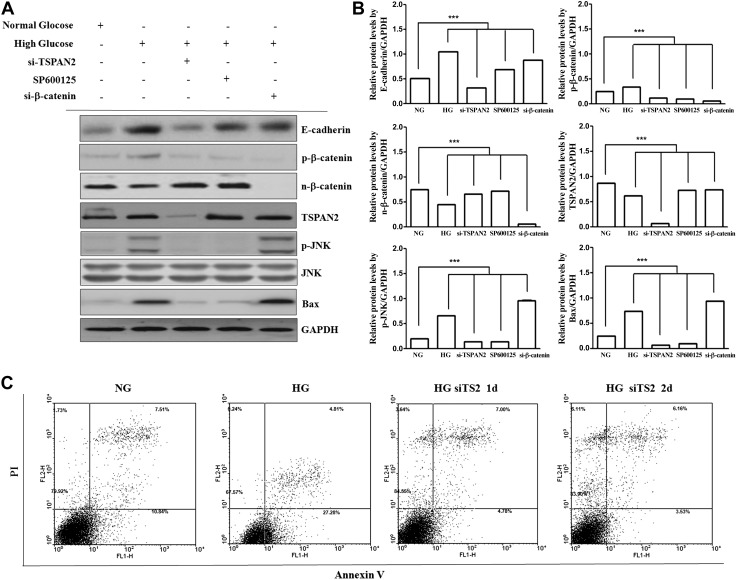
To further investigate whether TSPAN2 is linked functionally to JNK signaling, we examined the effect of HG on JNK (Fig. 4B). HG enhanced p-JNK. We next performed a loss-of-function analysis using TSPAN2 knockdown by siRNA. si-TSPAN2 suppressed p-JNK, and the selective JNK inhibitor SP600125 blocked JNK and Bax. Immunoblots confirmed the reduction in p-JNK protein in RNAKT-15 cells (Fig. 5A, B), whereas TSPAN2 overexpression enhanced the p-JNK protein-mediated increase in Bax (Fig. 3A). In addition, si-β-catenin significantly decreased nuclear β-catenin and increased Bax, whereas SP600125 significantly increased nuclear β-catenin and decreased Bax (Fig. 5A), indicating that β-catenin signaling promotes pancreatic β-cell survival by inhibiting Bax in RNAKT-15 cells. Taken together, these results indicate that HG-induced TSPAN2 overexpression up-regulated the p-JNK- and p-JNK-alleviated nuclear β-catenin translocation, leading to the up-regulation of Bax.
Figure 5.
Effect of β-catenin on Bax activation under HG. A) si-TSPAN2 attenuated JNK, whereas JNK inhibition attenuated p-β-catenin but enhanced β-catenin nuclear translocation. Moreover, si-β-catenin activated Bax. Protein levels in NG and HG medium were confirmed by Western blot analysis. B) Bar graphs indicate densitometric analysis of at least 3 separate experiments (means ± sd). NG vs. experimental treatment. ***P < 0.001. C) Analysis of apoptotic protection by si-TSPAN2 using double-labeled flow cytometry in RNAKT-15 cells treated with NG and HG. Cells were untreated (left), treated with si-TSPAN2 for 1 d (middle), or treated with si-TSPAN2 for 2 d (right). Cells were double-stained with PI and annexin V. Data are representative of 3 independent experiments.
TSPAN2 is involved in JNK and β-catenin signaling
Previous results showed that the phosphorylation of JNK is increased under HG conditions, indicating that HG activates JNK. Because TSPAN2 expression is increased under HG conditions, we examined the role of JNK on Bax expression. Cells were either mock treated or transfected with TSPAN2 siRNA or β-catenin siRNA. Later, cells were treated with mock or SP600125 (a JNK inhibitor). The cell lysates were then subjected to Western blot analysis. Both the silencing of TSPAN2 or SP600125 treatment inhibited JNK activation, and the expression of Bax decreased with similar patterns. These results suggest that JNK activation is involved in Bax expression. In addition, the level of nuclear β-catenin was increased by the silencing of TSPAN2 or JNK inhibitor. These results indicate that the inhibition of JNK-mediated β-catenin signaling activates Bax (Fig. 5A, B).
TSPAN2 expression contributes to glucotoxicity-induced apoptosis
Because HG treatment induced apoptosis in RNAKT-15 cells, we next examined the role of TSPAN2 in apoptosis. HG significantly stimulated the expression of TSPAN2, resulting in excess production of p-JNK, which down-regulated nuclear β-catenin and its transcriptional activity. JNK-mediated β-catenin signaling inhibition increases the protein level of proapoptotic Bax by regulating the JNK/β-catenin pathway, which is negatively correlated with the effect of antiapoptotic Bcl-2 (Fig. 3A). This results in the activation of caspase-3, which further cleaves PARP in nuclei, leading to RNAKT-15 cell apoptosis. Moreover, siRNA-mediated knockdown of a negative regulator of nuclear β-catenin translocation signaling, TSPAN2, efficiently protected against HG-induced apoptosis (Fig. 5C). TSPAN2, therefore, positively regulated apoptosis in RNAKT-15 cells and contributed to glucotoxicity-induced apoptosis.
DISCUSSION
To our knowledge, this is the first demonstration of the role of TSPAN2 in glucotoxicity-induced β-cell apoptosis. Although the effects of glucose toxicity on β-cell dysfunction and apoptosis have been described extensively (2, 6, 11), the exact molecular mechanisms of glucose toxicity remain unclear. Here, we used the RNAKT-15 human pancreatic β cell line to study the precise molecular mechanisms of glucose toxicity. This cell line proliferates easily in vitro and has been established from human pancreatic islet cells by the incorporation of immortalizing genes (SV40T and hTERT) into the genome and subsequent reversion into a nontumorigenic, reverted, immortalized human β cell line (12). We investigated the role of the TSPAN2-associated JNK/β-catenin pathway in islet β-cell apoptosis. The principal findings of this study are as follows: 1) TSPAN2 up-regulated p-JNK, which attenuated nuclear β-catenin translocation and induced apoptosis; 2) TSPAN2-depleted RNAKT-15 islet cells had high levels of nuclear β-catenin, which decreased Bax activation, suggesting the role of a JNK/β-catenin-dependent pathway; and 3) Bax subfamily proteins induced apoptosis through caspase-3. These findings suggest that TSPAN2 induced Bax translocation, as well as the caspase-3 activation pathway. Therefore, HG-induced apoptosis regulated the JNK/β-catenin pathway through TSPAN2 in RNAKT-15 islet cells.
In our study, a number of genes were differentially expressed upon treatment with high levels of glucose; however, we selected TSPAN2 for further analysis owing to its consistent increase in our microarray experiments. Although TSPAN2 is closely related with CD9 and CD81, targeted deletion indicates that they have diverse individual functions (5). Specifically, although CD81-null mice have impaired B cell function and enhanced T cell function, CD9-knockout mice produce sperm–egg fusions (3, 4). Hence, it is difficult to speculate about the function of TSPAN2 by sequence homology alone.
Here, we showed that HG promoted RNAKT-15 apoptosis, along with the up-regulation of cleaved caspase-3 and a higher Bax and lower Bcl2 expression (Figs. 2 and 3). These findings are consistent with those of previous studies (13, 14). We also found that TSPAN2 knockdown significantly suppressed HG-induced Bax expression in RNAKT-15 cells (Fig. 3B). Recent studies have demonstrated that JNK plays a crucial role in apoptosis, which is mediated by mitochondria in response to cellular stress (15). Although Bax has been shown to be essential for this JNK activation, the precise interplay between these 2 proteins has not been elucidated thus far. We now provide several lines of evidence that demonstrate that TSPAN2-mediated JNK phosphorylation induces the up-regulation of Bax and triggers its translocation to the mitochondria.
The inhibition of JNK by SP600125 attenuated Bax, strongly indicating that JNK regulates the activity of Bax. The up-regulation of Bax may increase mitochondrial membrane permeability and release cytochrome-c, leading to the activation of caspase-9 and, subsequently, of caspase-3 (16). Caspases are a class of cysteine proteases that play a potential role in the apoptotic signaling pathway (17). Thus, to unravel the possible downstream event of Bax, we examined the activation of caspase-3 (Fig. 3A). Treatment of RNAKT-15 cells with HG increased caspase-3 activity. Our present results are consistent with reports on HG resulting in apoptosis via the activation of caspase-3 and Bax protein (11).
Because β-catenin/Wnt signaling activates Akt, a kinase that phosphorylates and inhibits Bax (18), the effect of Akt inactivation on Bax activation was assessed in the presence of a PI3K inhibitor, LY-294002, an agent that inhibits PI3K-mediated Akt activation (19). Our results demonstrated that Dkk1, a negative regulator of Wnt signaling, affected β-catenin expression, which in turn affected Akt activation in RNAKT-15 cells under HG (Fig. 4C). Nuclear β-catenin translocation decreased under HG and TSPAN2 overexpression, whereas recovered in si-Dkk1-transfected cells under HG. siRNA mediated Dkk1 inhibition also activated Akt (Fig. 4C). The results of the present study suggest that the inhibition of p-JNK-mediated β-catenin/Wnt signaling inactivates Akt, a kinase that phosphorylates and enhances Bax, and that TSPAN2 regulated JNK/β-catenin signaling is essential for glucotoxicity-induced β-cell apoptosis.
Here, we also showed that the expression of TSPAN2 contributes to apoptosis in pancreatic β cells in response to HG; however, further research will be required to fully elucidate the function of TSPAN2. JNK binds to the E-cadherin/β-catenin complex and up-regulates the phosphorylation of β-catenin (9, 20), and it can also down-regulate canonical Wnt/β-catenin signaling (9, 21, 22). In general, JNK interacts with and suppresses β-catenin signaling in vitro and in vivo, and GSK3β, a β-catenin-phosphorylating kinase, plays a key role in this signaling (20, 21). However, some studies have shown distinct functions of JNK1 and JNK2, and have provided novel insights into the crosstalk between Wnt/β-catenin and MAPK/JNK signaling (21). In this study, to evaluate the fundamental role of JNK in the regulation of β-catenin signaling (Figs. 4B and 5A), RNAKT-15 cells were treated with the JNK inhibitor SP600125. As shown in Fig. 5A (lane 2 vs. lane 4), the protein level of nuclear β-catenin was dramatically increased in SP600125-treated RNAKT-15 cells. Dkk1 is an antagonist of the Wnt/β-catenin signaling pathway (23, 24). JNK enhanced Dkk1, and si-Dkk1 increased both cytosolic and nuclear β-catenin (Fig. 4A, C), suggesting subsequent downstream inactivation of GSK3β. These data demonstrated that JNK activation down-regulated nuclear β-catenin translocation signaling, whereas JNK inhibition up-regulated nuclear β-catenin translocation. These findings suggest that GSK3β might play a fundamental role in the crosstalk between Wnt/β-catenin and MAPK/JNK signaling.
In the present study, we have identified TSPAN2 as a critical factor mediating glucotoxicity-induced β-cell death and have elucidated the pathways by which TSPAN2 induces β-cell apoptosis. We found that HG led to the activation of JNK and Dkk1, and that JNK inhibitor SP600125 down-regulated Dkk1 (Fig. 4B) and si-TSPAN2 suppressed HG-mediated JNK activation (Fig. 5A). These findings suggest that TSPAN2 plays a key role in this signaling, which promotes the signaling pathway of Bax-associated apoptosis. Bcl-2-mediated processes of cell death were negatively correlated with Bax and TSPAN2 (Fig. 3A). These findings not only provide new evidence of the role of TSPAN2 in β-cell biology and the mechanisms involved but also identify TSPAN2 as a potential target to counter the deleterious effects of glucose toxicity by preventing pancreatic islet β-cell apoptosis (Fig. 5C). Thus, TSPAN2 silencing can potentially enhance β-cell mass. However, since we have found that TSPAN2 induces β-cell apoptosis, it is also possible that a lack of TSPAN2 may slow the natural process of β-cell apoptosis and turnover, leading to an accumulation of β cells and increased basal insulin production.
To conclude, TSPAN2 was demonstrated to be a critical mediator of glucotoxicity-induced β-cell apoptosis. Moreover, TSPAN2 protein levels dramatically increased in response to HG. These findings provide novel insights into the molecular mechanisms of β-cell glucose toxicity and apoptosis. Controlling TSPAN2 expression may therefore provide new approaches and strategies to protect pancreatic β cells from progressive destruction and to preserve a sufficient insulin-producing β-cell mass in patients with type 2 diabetes.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by Ministry of Education Grants 2015R1D1A1A01058744 and 2014R1A1A2058114, and supported by Korea Basic Science Institute Grant D36403. Author contributions: I.-H. Hwang and J. Park performed the experiments, including cell culture, Western blot analyses, and confocal microscopy, and analyzed the data; J. M. Kim and S. I. Kim performed microarray and Gene Ontology analyses; J.-S. Choi, K.-B. Lee, S. H. Yun, and M.-G. Lee supported the experiments and prepared the figures; and S. J. Park and I.-S. Jang conceived and designed the experiments, contributed reagents, sourced materials, provided analysis tools, and wrote the paper. The authors declare no conflicts of interest.
Glossary
- Bax
BCL2-associated X protein
- BCA
bicinchoninic acid
- Bcl-2
B-cell CLL/lymphoma 2
- BSA
bovine serum albumin
- Dkk1
Dickkopf-1
- GAPDH
glyceraldehyde phosphate dehydrogenase
- HG
high glucose
- NG
normal glucose
- p
phosphorylated
- PARP
poly(ADP-ribose) polymerase
- PI
propidium iodide
- RT
reverse transcriptase
- si
small interfering
- t-Bid
truncated Bid
- TSPAN2
tetraspanin-2
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1.Xu G., Chen J., Jing G., Shalev A. (2013) Thioredoxin-interacting protein regulates insulin transcription through microRNA-204. Nat. Med. 19, 1141–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J., Saxena G., Mungrue I. N., Lusis A. J., Shalev A. (2008) Thioredoxin-interacting protein: a critical link between glucose toxicity and beta-cell apoptosis. Diabetes 57, 938–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Todd S. C., Doctor V. S., Levy S. (1998) Sequences and expression of six new members of the tetraspanin/TM4SF family. Biochim. Biophys. Acta 1399, 101–104 [DOI] [PubMed] [Google Scholar]
- 4.Maecker H. T., Todd S. C., Levy S. (1997) The tetraspanin superfamily: molecular facilitators. FASEB J. 11, 428–442 [PubMed] [Google Scholar]
- 5.Hemler M. E. (2001) Specific tetraspanin functions. J. Cell Biol. 155, 1103–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elmore S. (2007) Apoptosis: a review of programmed cell death. Toxicol. Pathol. 35, 495–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saadeddin A., Babaei-Jadidi R., Spencer-Dene B., Nateri A. S. (2009) The links between transcription, beta-catenin/JNK signaling, and carcinogenesis. Mol. Cancer Res. 7, 1189–1196 [DOI] [PubMed] [Google Scholar]
- 8.Yamanaka H., Moriguchi T., Masuyama N., Kusakabe M., Hanafusa H., Takada R., Takada S., Nishida E. (2002) JNK functions in the non-canonical Wnt pathway to regulate convergent extension movements in vertebrates. EMBO Rep. 3, 69–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao G., Tao Q., Kofron M., Chen J. S., Schloemer A., Davis R. J., Hsieh J. C., Wylie C., Heasman J., Kuan C. Y. (2006) Jun NH2-terminal kinase (JNK) prevents nuclear beta-catenin accumulation and regulates axis formation in Xenopus embryos. Proc. Natl. Acad. Sci. USA 103, 16313–16318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z., Havasi A., Gall J. M., Mao H., Schwartz J. H., Borkan S. C. (2009) Beta-catenin promotes survival of renal epithelial cells by inhibiting Bax. J. Am. Soc. Nephrol. 20, 1919–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim W. H., Lee J. W., Suh Y. H., Hong S. H., Choi J. S., Lim J. H., Song J. H., Gao B., Jung M. H. (2005) Exposure to chronic high glucose induces beta-cell apoptosis through decreased interaction of glucokinase with mitochondria: downregulation of glucokinase in pancreatic beta-cells. Diabetes 54, 2602–2611 [DOI] [PubMed] [Google Scholar]
- 12.Narushima M., Kobayashi N., Okitsu T., Tanaka Y., Li S. A., Chen Y., Miki A., Tanaka K., Nakaji S., Takei K., Gutierrez A. S., Rivas-Carrillo J. D., Navarro-Alvarez N., Jun H. S., Westerman K. A., Noguchi H., Lakey J. R., Leboulch P., Tanaka N., Yoon J. W. (2005) A human beta-cell line for transplantation therapy to control type 1 diabetes. Nat. Biotechnol. 23, 1274–1282 [DOI] [PubMed] [Google Scholar]
- 13.Kang B. P., Frencher S., Reddy V., Kessler A., Malhotra A., Meggs L. G. (2003) High glucose promotes mesangial cell apoptosis by oxidant-dependent mechanism. Am. J. Physiol. Renal Physiol. 284, F455–F466 [DOI] [PubMed] [Google Scholar]
- 14.Khera T., Martin J., Riley S., Steadman R., Phillips A. O. (2006) Glucose enhances mesangial cell apoptosis. Lab. Invest. 86, 566–577 [DOI] [PubMed] [Google Scholar]
- 15.Davis R. J. (2000) Signal transduction by the JNK group of MAP kinases. Cell 103, 239–252 [DOI] [PubMed] [Google Scholar]
- 16.Adams J. M., Cory S. (2007) Bcl-2-regulated apoptosis: mechanism and therapeutic potential. Curr. Opin. Immunol. 19, 488–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z. B., Liu Y. Q., Cui Y. F. (2005) Pathways to caspase activation. Cell Biol. Int. 29, 489–496 [DOI] [PubMed] [Google Scholar]
- 18.Gardai S. J., Hildeman D. A., Frankel S. K., Whitlock B. B., Frasch S. C., Borregaard N., Marrack P., Bratton D. L., Henson P. M. (2004) Phosphorylation of Bax Ser184 by Akt regulates its activity and apoptosis in neutrophils. J. Biol. Chem. 279, 21085–21095 [DOI] [PubMed] [Google Scholar]
- 19.Sinha D., Wang Z., Ruchalski K. L., Levine J. S., Krishnan S., Lieberthal W., Schwartz J. H., Borkan S. C. (2005) Lithium activates the Wnt and phosphatidylinositol 3-kinase Akt signaling pathways to promote cell survival in the absence of soluble survival factors. Am. J. Physiol. Renal Physiol. 288, F703–F713 [DOI] [PubMed] [Google Scholar]
- 20.Lee M. H., Koria P., Qu J., Andreadis S. T. (2009) JNK phosphorylates beta-catenin and regulates adherens junctions. FASEB J. 23, 3874–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu D., Bi X., Fang W., Han A., Yang W. (2009) GSK3beta is involved in JNK2-mediated beta-catenin inhibition. PLoS One 4, e6640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu D., Fang W., Han A., Gallagher L., Davis R. J., Xiong B., Yang W. (2008) c-Jun N-terminal kinase 1 interacts with and negatively regulates Wnt/beta-catenin signaling through GSK3beta pathway. Carcinogenesis 29, 2317–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Q. G., Wang R., Khan M., Mahesh V., Brann D. W. (2008) Role of Dickkopf-1, an antagonist of the Wnt/beta-catenin signaling pathway, in estrogen-induced neuroprotection and attenuation of tau phosphorylation. J. Neurosci. 28, 8430–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niida A., Hiroko T., Kasai M., Furukawa Y., Nakamura Y., Suzuki Y., Sugano S., Akiyama T. (2004) DKK1, a negative regulator of Wnt signaling, is a target of the beta-catenin/TCF pathway. Oncogene 23, 8520–8526 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.